Abstract
Nodulated legumes receive their nitrogen via nitrogen-fixing rhizobia, which exist in a symbiotic relationship with the root system. In tropical legumes like French bean (Phaseolus vulgaris) or soybean (Glycine max), most of the fixed nitrogen is used for synthesis of the ureides allantoin and allantoic acid, the major long-distance transport forms of organic nitrogen in these species. The purpose of this investigation was to identify a ureide transporter that would allow us to further characterize the mechanisms regulating ureide partitioning in legume roots. A putative allantoin transporter (PvUPS1) was isolated from nodulated roots of French bean and was functionally characterized in an allantoin transport-deficient yeast mutant showing that PvUPS1 transports allantoin but also binds its precursors xanthine and uric acid. In beans, PvUPS1 was expressed throughout the plant body, with strongest expression in nodulated roots, source leaves, pods, and seed coats. In roots, PvUPS1 expression was dependent on the status of nodulation, with highest expression in nodules and roots of nodulated plants compared with non-nodulated roots supplied with ammonium nitrate or allantoin. In situ RNA hybridization localized PvUPS1 to the nodule endodermis and the endodermis and phloem of the nodule vasculature. These results strengthen our prediction that in bean nodules, PvUPS1 is involved in delivery of allantoin to the vascular bundle and loading into the nodule phloem.
Availability of reduced nitrogen is an important determinant in the growth and development of plants. Although in most vascular plant species the major transport form of reduced/organic nitrogen is as amino acids (including amides), tropical and subtropical legumes like cowpea (Vigna unguiculata), soybean (Glycine max), and French bean (Phaseolus vulgaris) transport large amounts of the nitrogenous compounds called ureides. The dominant forms of ureides in these species are allantoin and allantoic acid (Pate et al., 1980). In legumes that are adapted to temperate climates (e.g. pea [Pisum sativum] and faba bean [Vicia faba]), the amides Gln and Asn take on the major transport function (Herridge et al., 1978; Schubert, 1986). Ureides can comprise up to 90% of the total nitrogen transported in the xylem of nitrogen-fixing tropical legumes (Herridge et al., 1978; Pate et al., 1980) and can be stored in high amounts in the different plant organs (Matsumoto et al., 1977a; Streeter, 1979; Layzell and LaRue, 1982). In soybean, ureides have been found to be in concentrations of 20 or 10 mm in the stem tissue or xylem (Layzell and LaRue, 1982; Rainbird et al., 1984) and 94 mm in nodule exudate (Streeter, 1979). In leaves, total ureide concentrations varied from approximately 1 to 3 mm, but analyses of the various leaf cells have shown that ureides can reach concentration levels up to 59 mm in the paraveinal mesophyll (Matsumoto et al., 1977b; Thomas and Schrader, 1981; Costigan et al., 1987). Due to the high concentrations in the vascular system and in certain plant tissues, ureides are interpreted to have an important function in nitrogen transport and nitrogen storage in tropical legumes.
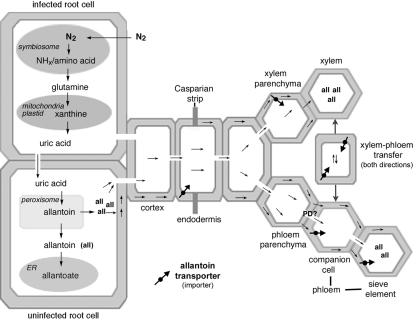
The synthesis of ureides occurs mainly in root nodules through the coordination of the plant-bacteria association (Atkins and Smith, 2000; see also Fig. 6). After nitrogen fixation in bacteroids of infected root cells, ammonia (NH3), ammonium (NH4+), or amino acids are released or transported from the symbiosome into the cytosol, where they are utilized for Gln synthesis (Smith and Emerich, 1993; Day et al., 2000; Lodwig et al., 2003; see Fig. 6). After formation of Gln, the nitrogen goes through the de novo purine synthesis pathway in the plastids (Shelp et al., 1983) or mitochondria (Atkins et al., 1997), followed by purine degradation via xanthine in the plastids (Schubert, 1986) or cytosol of infected or uninfected root cells (Matsumoto et al., 1977c; Atkins et al., 1980; Shelp et al., 1983). The ureide allantoin is finally synthesized in the peroxisomes of noninfected root cells from the purine degradation product uric acid (Hanks et al., 1981). Allantoic acid is probably produced in the smooth endoplasmic reticulum of the noninfected cells after import of allantoin into this compartment (Hanks et al., 1981). From the place of synthesis, allantoin and allantoic acid are transferred into the cytosol and are then transported to the root xylem and phloem for long-distance transport (McClure and Israel, 1979; Streeter, 1979; Atkins et al., 1982). The underlying mechanisms of ureide transport in root nodules have not been resolved.
Figure 6.
Predicted model of allantoin transport in French bean nodules. PvUPS1 is strongly expressed in the vascular endodermis and phloem of the nodule vasculature and is suggested to function in (a) loading of allantoin into the symplast for passage of the Casparian strip of the endodermis surrounding the vascular bundles, (b) in import of allantoin into the xylem parenchyma cells, (c) in phloem loading of allantoin, in (d) phloem retrieval of allantoin leaked into the apoplast, and in (e) in transfer of allantoin from xylem to phloem. PvUPS1 is also expressed in the nodule endodermis (not shown in model) where it is predicted to function in uptake of allantoin from the apoplast to redirect allantoin to the vascular system.
Molecular studies on organic nitrogen transport have mainly concentrated on amino acid transport in Arabidopsis. A minimum of 53 amino acid transporters are predicted to be present (Wipf et al., 2003). Another type of transporter of nitrogenous compounds that has been isolated is the Arabidopsis AtPUP1 nucleobase transporter specific for adenine, guanine, hypoxanthine, and probably purine derivates, and Lpe1 from maize (Zea mays) that is specific for uric acid, xanthine, and also recognizes ascorbate (Gillissen et al., 2000; Argyrou et al., 2001). Recently, an Arabidopsis transporter (AtUPS1) was identified that mediates transport of derivates of heterocylic nitrogen compounds including allantoin, uric acid, and xanthine (Desimone et al., 2002), but Arabidopsis does not have significant pools of these compounds in its tissues. The only organic nitrogen transporters characterized in legumes so far are members of the AAP family that are probably involved in translocation of neutral amino acids across the plasma membrane, and two peptide transporters (VfPTRs, VfAAPs, and PsAAPs from faba bean and pea; Montamat et al., 1999; Tegeder et al., 2000; Miranda et al., 2001, 2003). Molecular studies on ureide transport in legumes have not been reported at all. In this publication, we describe the characterization of an allantoin transporter from French bean (PvUPS1) as well as the regulation and localization of the transporter.
RESULTS
Identification of a Putative Allantoin Transporter from French Bean
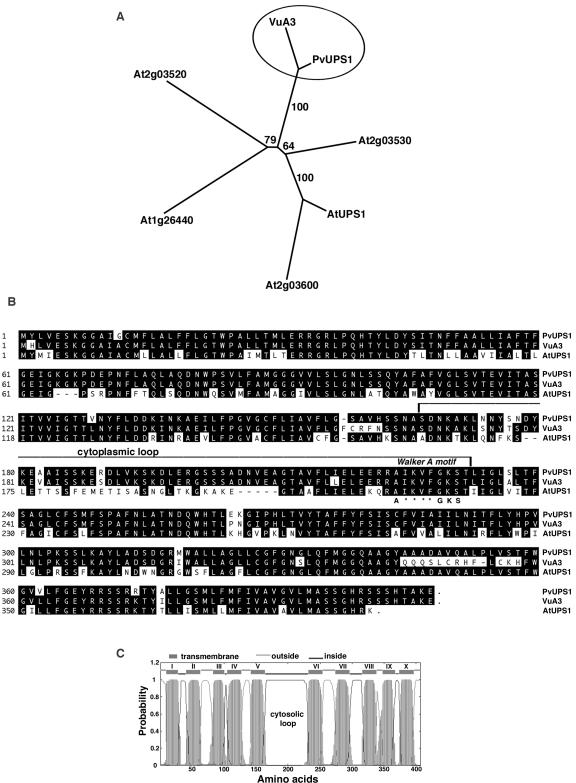
A putative allantoin transporter (PvUPS1) was isolated using RNA from nodulated roots of French bean and an reverse transcriptase (RT)-PCR approach with degenerate primers based on AtUPS1 (Desimone et al., 2002) and legume expressed sequence tags (ESTs). PvUPS1 (P. vulgaris ureide permease 1) has an open reading frame of 1,224 bp and encodes a protein of 407 amino acids with a predicted molecular mass of 44 kD (accession no. AY461734). A search through the databases shows 60% to 75% similarity of the amino acid sequence of PvUPS1 to members of the UPS family from Arabidopsis (At2g03590, At2g03530, At2g03600, At2g03520, and At1g26440) from which AtUPS1 (At2g03590) has been characterized and interpreted to be a transporter of derivates of heterocylic nitrogen compounds including allantoin (Desimone et al., 2002). PvUPS1 was 93% similar to VuA3 (X90487) from cowpea, an ATP-/GTP-binding protein with unknown function. The close relatedness between PvUPS1 and VuA3 was confirmed by phylogenetic analysis using the UPS proteins. The legume proteins group together, whereas the UPS proteins from Arabidopsis form a separate group (Fig. 1A). A number of additional legume genes related to this family of proteins were identified in the database of ESTs that show high similarity on the nucleotide level to PvUPS1 (e.g. ESTs from soybean [AW707051, AW311266, BM095010, AW311266, and CA801058] and Medicago truncatula [BG448192, BF647275, and BF646038]). Alignment of the derived full-length proteins PvUPS1, VuA3, and AtUPS1 (Fig. 1B) shows homology of the C and N terminus between all sequences, whereas the legume sequences are very different from the Arabidopsis AtUPS1 in the mid part of the proteins from about amino acid 175 to 210. All sequences contain the conserved Walker A motif (= P-loop; Saraste et al., 1990) that is thought to be responsible for ATP/GTP binding. Hydropathy analysis indicates that the predicted PvUPS1 protein is highly hydrophobic and contains 10 putative membrane-spanning regions with the C and N terminals protruding outside of the cell in the apoplast (Fig. 1C). Five sequence stretches form cell inward-directed loops and the third inward loop (amino acids 165–231) is predicted to be 67 amino acids long. This cytoplasmic loop represents the above discussed sequence that is highly conserved between the two legume proteins, but is very different between the legume and Arabidopsis proteins. The Walker A motif is located at the end region of the cytoplasmic loop, as also shown for AtUPS1 from Arabidopsis (Desimone et al., 2002).
Figure 1.
Analysis of PvUPS1 and related proteins. A, Phylogenetic analysis. The tree is based on maximum parsimony analysis (PAUP 4.0b5, Rogers and Swofford, 1998) of aligned protein sequences from French bean (PvUPS1), cowpea (VuA3 protein/X90487), and Arabidopsis (AtUPS1/At2g03590, At2g03530, At2g03600, At2g03520, and At1g26440). The numbers indicate the occurrence of a given branch in 10,000 bootstrap replicates of the given data set. B, Amino acid sequence of PvUPS1 and alignment with two related members of the UPS family. Using PvUPS1 from French bean, AtUPS1 from Arabidopsis (At2g03590) and VuA3 from cowpea (X90487), a sequence alignment was generated by Clustal algorithm (LASERGENE software; DNASTAR, Madison, WI). Black background shows identical amino acids. Dashes indicate gaps in the sequences to allow maximal alignment. The third cytoplasmic loop is highlighted as well as the “Walker A” motif (A-x(4)-G-K-S), in which asterisks refer to variable amino acids. C, Topology of PvUPS1 using a transmembrane prediction program (TMHMM version 2.0; http://www.cbs.dtu.dk/services). PvUPS1 contains a large central loop probably located in the cytosol. The C and N terminus are predicted to be outside.
PvUPS1 Transports Allantoin
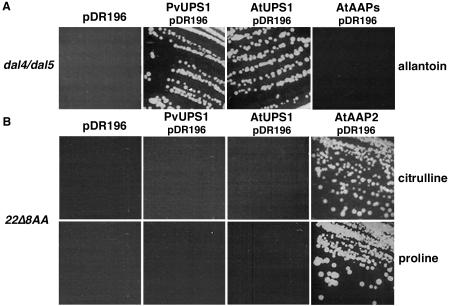
To analyze its substrate specificity, PvUPS1 was expressed in two yeast mutants lacking different amino acid or ureide uptake systems (Fig. 2). Amino acids were tested to analyze if PvUPS1 mediates growth of yeast cells on these different substrates as has been shown for the amino acid permeases from Arabidopsis (AtAAPs). AtAAPs transport amino acids including citrulline (Wipf et al., 2003). The yeast strain 22Δ8AA is unable to grow on γ-amino butyric acid (GABA), Pro, Asp, Arg, and citrulline as sole nitrogen source (Tegeder et al., 2000). The mutant dal4/dal5 cannot grow on minimal media supplemented only with allantoin. The yeast strains were transformed with the empty expression vector pDR196, PvUPS1 in pDR196, or the positive controls (AtUPS1 and Arabidopsis amino acid permease 2 [AtAAP2] in pDR196) followed by complementation on the different amino acids and ureides, respectively. PvUPS1 was not able to restore growth on selective amino acids, whereas the positive controls were able to mediate yeast growth (data only shown for citrulline and Pro complementation, Fig. 2B). The complementation experiments using ureides demonstrated that PvUPS1 does mediate growth of cells on allantoin as nitrogen source (Fig. 2A).
Figure 2.
Yeast complementation of PvUPS1. Growth assays were performed with the yeast strains dal4/dal5 (A) and 22Δ8AA (B). A, dal4/dal5 that is deficient in allantoin transport was transformed with PvUPS1/pDR196, AtUPS1, and AtAAP2 in pDR196 (Fischer et al., 1995; positive controls) or pDR196 only (negative control) and were grown on media containing allantoin as sole nitrogen source. B, To test if PvUPS1 mediates growth of yeast cells on amino acids, 22Δ8AA (deficient in citrulline, Pro, Arg, Asp, and GABA transport) expressing PvUPS1/pDR196, AtUPS1/pDR196, AtAAP2/pDR196, or the empty pDR196 vector were grown on media supplemented with citrulline, Pro, Arg, Asp, and GABA as sole nitrogen source (here shown only yeast complementation on citrulline and Pro).
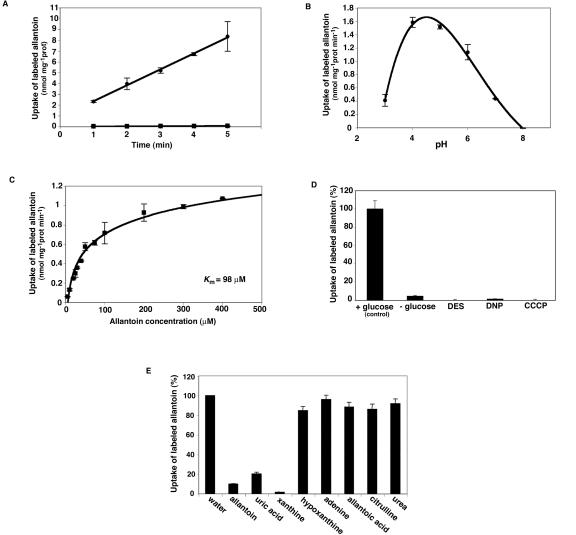
Transport studies in yeast cells with [14C]-labeled allantoin showed that PvUPS1 mediates uptake of allantoin (Fig. 3A) and that uptake activity strongly depends on the pH (Fig. 3B). The optimal pH for allantoin uptake ranged between pH 4 and 5. The transport activity determined at pH 4.5 was dependent on the substrate concentration and showed a Michaelis-Menten constant (Km) for allantoin of 98 μm (Fig. 3C). PvUPS1 transport activity was reliant on the presence of Glc (metabolic energy) and was sensitive to protonophores (2,4-dinitrophenol and carbonyl cyanide m-chlorphenyl-hydrazone) and a plasma membrane H+-ATPase inhibitor (diethylstilbestrol), suggesting that allantoin uptake is energy dependent (Fig. 3D). This data, together with the pH dependence of transport, suggest an active transport mechanism. The transport could be energized by a proton motive force, as has been shown for a number of transport systems for nitrogenous organic compounds (Desimone et al., 2002; Wipf et al., 2002; see Fig. 3, B and D). On the other hand, the Walker A consensus site for ATP binding might imply a direct role of ATP in the transport. Additional experiments need to be performed to determine if the transport mechanism involves a proton motive force or direct ATP hydrolysis.
Figure 3.
Biochemical properties of PvUPS1: analysis of allantoin transport. A, Time-dependent uptake of [14C]allantoin in yeast cells. dal4/dal5 mutant was transformed with PvUPS1/pDR196 (circles) or the vector pDR196 alone (squares) and was incubated with 200 μm [14C]allantoin in 100 mm potassium phosphate buffer (pH 4.5) and 100 mm Glc at 30°C for uptake studies. B, pH-dependent uptake of [14C]allantoin in PvUPS1-expressing yeast cells. C, Michaelis-Menten kinetics of [14C]allantoin uptake. D, Inhibition of [14C]allantoin uptake in yeast cells. [14C]allantoin uptake in dal4/dal5 yeast cells expressing PvUPS1 was measured after preincubation of yeast cells for 5 min with Glc (100 mm, control), without Glc, with Glc and 0.1 mm 2,4-dinitrophenol, 0.1 mm diethylstilbestrol, or 0.1 mm carbonyl cyanide m-chlorphenyl-hydrazone. Data are expressed as percentage of control values. E, Substrate specificity of PvUPS1 from French bean. Competition of [14C]allantoin (200 μm) uptake into yeast cells (dal4/dal5) expressing PvUPS1 in the presence of a 9-fold molar excess of purines or purine degradation products. The noncompeted uptake rate was taken as 100% corresponding to 1.2 nmol allantoin min–1 mg–1 protein. A through E, Results represent means of three independent experiments (±sd).
To study the substrate specificity of PvUPS1 further, [14C]allantoin uptake in yeast cells was determined in the presence of a 9-fold molar excess of allantoin, citrulline, purines, or purine derivatives (up- and downstream products of allantoin synthesis, Fig. 3E; see also Fig. 6). Competition with an excess of unlabeled allantoin led to a reduction of the uptake rate by 90%, confirming the notion that PvUPS1 mediates transport of allantoin. Xanthine and uric acid were also found to be effective competitors for [14C]allantoin uptake when applied in excess. Adenine, urea, citrulline, and the ureide allantoic acid showed very weak or no inhibition of allantoin uptake. These results suggest that besides allantoin, PvUPS1 binds the precursors of allantoin synthesis xanthine and uric acid, but not allantoic acid, citrulline, purines, or other purine degradation products like urea.
PvUPS1 Expression and Its Regulation
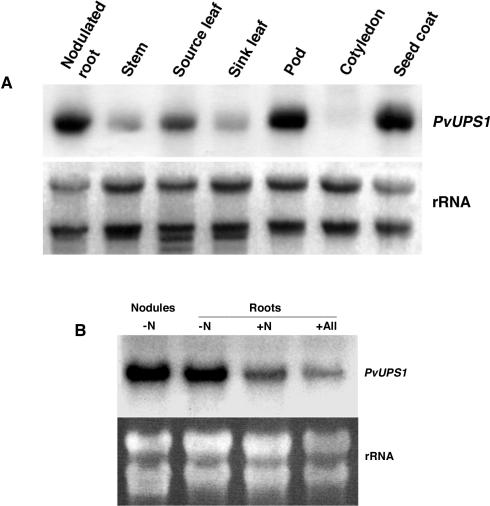
The expression of PvUPS1 throughout the plant body and its regulation were examined in French bean plants in relation to nodulation status and under different nitrogen conditions using RNA gel-blot analysis. Full-length 32P-labeled cDNAs of PvUPS1 were used as a probe. Because further members of the UPS family might exist in French bean, cross-hybridization cannot be excluded, although the experiments were performed under high stringency conditions. In nodulated, soil-grown plants, PvUPS1 was found to be expressed to varying amounts throughout the plant body with highest expression in roots, source leaves, pods, and seed coats (Fig. 4A).
Figure 4.
Northern-blot analysis of PvUPS1. A, Organ-specific expression of PvUPS1 in nodulated bean plants. Expression was analyzed by RNA gel-blot hybridization with 32P-labeled PvUPS1 full-length cDNAs as probe. At the bottom, ethidium bromide-stained rRNA is shown. B, PvUPS1 gene expression in nodules and roots under different nitrogen regimes. French bean plants were fertilized with Hoagland solution without nitrogen (-N), with ammonium nitrate (+N), or with allantoin (+All) as sole nitrogen source. Plants without nitrogen fertilization developed nodules, whereas +N and +All plants showed no nodulation. Expression was analyzed by RNA gel-blot hybridization with 32P-labeled PvUPS1 full-length cDNAs as probe. Ethidium bromide-stained rRNA is shown below the RNA blots.
Further studies were conducted with roots of plants grown in perlite with and without nitrogen supply. The non-nitrogen-fed plants were nodulated, whereas the plants supplied with allantoin or ammonium nitrate showed no nodulation. PvUPS1 was expressed in non-nodulated roots supplied with ammonium nitrate or allantoin (Fig. 4B). The differences in the expression level in roots of ammonium nitrate- or allantoin-fed plants shown in Figure 4B are due to differences in loading of total RNA. This was confirmed by densitometric measurements showing that the divergence in rRNA loading or PvUPS1 expression between ammonium nitrate- or allantoin-fed plants was about 35% for both (data not shown). PvUPS1 is most highly expressed in roots of nodulated plants and in nodules, indicating that in roots, the expression level of PvUPS1 depends on the status of nodulation, which is consistent with very high levels of allantoin present in nodules/nodulated roots (Streeter, 1979).
In Nodules, PvUPS1 Is Localized to the Endodermis and Phloem
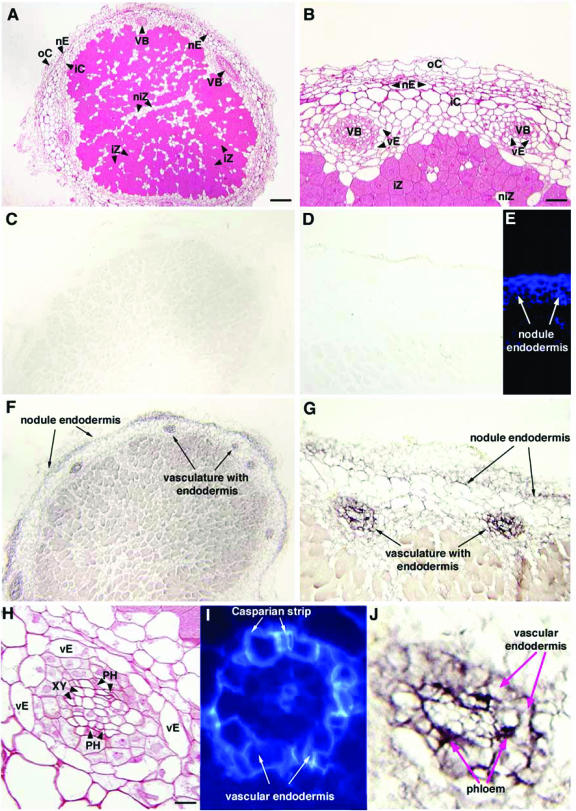
To further define the functional role of PvUPS1 in nodules, in situ RNA localization was performed (Fig. 5). French bean nodules are determinate in structure, and have an endodermis layer that separates the inner cortex from outer cortex of the nodules. Within the nodule inner cortex, vascular bundles are present (Fig. 5A, B, and H). Using DIG-labeled PvUPS1 antisense riboprobes, PvUPS1 expression was located in two positions: the nodule endodermis and the vasculature of the nodules (Fig. 5, F, G, and J). To confirm PvUPS1 expression in the nodule endodermis, the suberin of this cell layer was visualized by autofluorescence under UV light and its location was consistent with the color detection of DIG-labeled PvUPS1 antisense probes (Fig. 5, E–G).
Figure 5.
In situ RNA localization of PvUPS1 in French bean nodules. Nodule paraffin sections were treated with digoxygenin (DIG)-labeled PvUPS1 sense (C and D) and antisense (F, G, and J) riboprobes. Suberin of nodule endodermis (E) and vascular endodermis (I) is visualized by autofluorescence under UV light. F, G, and J, PvUPS1 is strongly expressed in the nodule endodermis, vascular endodermis, and in the phloem of the vascular bundles. A, B, and H, Resin sections stained with safranin O reveal structure of nodules. nE, Nodule endodermis; vE, vascular endodermis; VB, vascular bundles; iZ, infected zone; iC, inner cortex; oC, outer cortex; PH, phloem; XY, xylem. Bar in A = 60 μm, bar in B = 24.5 μm, and bar in H = 9.5 μm.
Structural analysis of the vasculature and examining the vascular system of the nodule under UV light demonstrated that each vascular bundle is surrounded by an endodermis (Fig. 5, H and I) similar to what was shown in indeterminate nodules of broad bean (Vicia faba; Hartmann et al., 2002). PvUPS1 expression was found in the vascular endodermis and in the phloem of the vascular nodule tissue (Fig. 5J), suggesting a role of PvUPS1 in importing allantoin into the symplast to pass the endodermis and in loading of allantoin into the sieve element/companion cell complex of the phloem for long distance transport.
DISCUSSION
Nitrogen partitioning in plants requires long-distance translocation of reduced nitrogen via the phloem and xylem. Although most vascular plants (including temperate legumes) transport primarily amino acids/amides, major transport forms of organic nitrogen in tropical legumes are the ureides allantoin and allantoic acid. Yet, little is known about the mechanisms of ureide transport, and this is the first study describing the characterization of PvUPS1, an allantoin transporter from French bean. PvUPS1 shows similarity of approximately 93% to the cowpea homolog VuA3, encoding a protein with unknown function (Chaya et al., 1996), indicating a close relationship of these legume proteins. This close relationship was confirmed by phylogenetic analysis where the peptide sequences of the two “ureide-transporting” legumes group together, whereas the UPS proteins of the “non-ureide transporting” species Arabidopsis form separate groups (see Fig. 1A). Equally important with respect to the phylogenetic study, molecular analysis of the UPS amino acid sequences shows variation of the large third cytoplasmic loop between the Arabidopsis and the legume proteins (see Fig. 1, B and C). The role of this protein segment is currently not known, however, it may possibly function in regulation of transport activity, especially because a putative binding site for the phosphate residue of ATP or GTP (P-loop motif) exists in the end region of the loop. Although PvUPS1 and AtUPS1 recognize similar substrates (this study; Desimone et al., 2002), the high similarity of the third loop between legume sequences and their divergence from the Arabidopsis sequences may be a reflection of differences in regulation of transporter function in these species.
Even though protein analyses are predictive, we cannot exclude that the actual protein structure differs from the model, and only further studies such as x-ray structure analysis (Dencher et al., 2000) will resolve the final configuration of the transporter protein. However, all these data indicate the existence of different UPS gene families and are consistent with probable differences in function of the transporters between the ureide-transporting legumes and the non-ureide transporting Arabidopsis. Further sequence data of UPS proteins from various ureide versus non-ureide plant species are needed to verify our prediction that different UPS gene families with differences in their function or regulation of function exist.
Homology is a powerful tool for initial identification of protein candidates, but it does not prove function. Therefore, the putative function of PvUPS1 was explored by yeast complementation and uptake studies into yeast cells. The results clearly demonstrate that PvUPS1 transports allantoin but not citrulline or allantoic acid and urea (downstream products of the purine pathway; see Figs. 2 and 3). However, products of the purine pathway upstream of allantoin synthesis, uric acid and xanthine, are competitors for allantoin uptake when applied in 9-fold excess. This indicates that PvUPS1 binds or even transports these compounds in addition to allantoin. Similar results were obtained from competition experiments with AtUPS1 and were further confirmed by uric acid transport studies (Desimone et al., 2002). The recognition of allantoin and its precursors by PvUPS1 could be dependent on the oxoderivation of the purine/imidazole ring, present in all three compounds, but not in adenine, hypoxanthine, and downstream products of the purine pathway (see also Desimone et al., 2002).
Although uric acid and xanthine are recognized, these compounds might not represent important substrates for PvUPS1 in beans. PvUPS1 might be swamped by an excess of allantoin present in the nodule apoplast and symplast. In nodules of tropical legumes, ureides are present in extremely high amounts (Herridge et al., 1978; Streeter, 1979; Atkins et al., 1982). For example, the allantoin concentration in soybean nodule exudates were shown to be 94 mm (Streeter, 1979). Our hypothesis is supported by PvUPS1 mRNA in situ hybridization studies where PvUPS1 was localized to the nodule and vascular endodermis as well as to the nodule phloem. This suggests a role in uptake from an apoplast that is enriched with allantoin. Thus, PvUPS1 would be expected to predominantly import allantoin into the nodule cells and the sieve element-companion cell complex of the nodule phloem. Loading of allantoin into the nodule symplast for passage of the Casparian strip of the vascular endodermis and subsequent ureide flux into the xylem parenchyma and then out into the xylem vessel is perhaps the most important transport pathway in nodules (see Fig. 6) as indicated by the high allantoin concentrations present in the xylem sap of tropical legumes (Herridge et al., 1978; Layzell and LaRue, 1982; Ceccatto et al., 1988). However, to determine if xanthine and uric acid as well as allantoin are transported by PvUPS1 under physiological conditions, future studies need to be performed in bean plants repressing or overexpressing PvUPS1.
We know from studies of other transporter genes that their expression in plants can be organ specific and/or regulated by a number of factors, including nutritional status, metabolism, or developmental stage of the plant (for review, see Delrot et al., 2000; Smith et al., 2000). Although physiological studies in French bean on allantoin accumulation and partitioning are rare (Thomas et al., 1979; 1980), a number of experiments have been done in soybean and cowpea demonstrating the connection between ureide concentration and nodulation, nitrogen supply or exogenous factors like water stress (Matsumoto et al., 1977a; McClure and Israel, 1979; Kaur et al., 1985; for review, see Atkins and Beevers, 1990; Serraj et al., 1999; Lima et al., 2000). These studies clearly suggest a relationship between ureide synthesis/amount and transporter activity, which is regulated developmentally and through nutrient availability. In this study, we examined levels and location of gene expression of PvUPS1 in response to varying nodulation status and nitrogen supply (no nitrogen, ammonium nitrate, or allantoin, see Fig. 4). Although expression studies were performed under highly stringent conditions, we cannot exclude that cross-hybridization with other uncharacterized members of the PvUPS family occurred. It was found that PvUPS1 is expressed at some level throughout the plant body, but by far the strongest expression was seen in nodulated roots, source leaves, pods, and seed coats. This expression pattern is consistent with sites of ureide synthesis and utilization (for review, see Atkins and Beevers, 1990; Smith and Atkins, 2002). Of relevance to this, the level of expression of PvUPS1 in roots and nodules was influenced by different nitrogen regimes or nodulation status. Plants that grow without nitrogen fertilization developed large numbers of nodules and showed strong transporter expression in the nodules and the roots. The expression level was reduced in ammonium nitrate-fertilized plants and bean plants fed allantoin at the root zone. These molecular data are strongly supported by physiological studies done in some leguminous species showing that the ureide concentration in the xylem sap relative to all transported nitrogen compounds increases with nodulation (Matsumoto et al., 1977a; Pate et al., 1980). It is also well known that nitrogen fertilization has a significant inhibitory effect on nodule formation and activity, and should have an effect on transporter expression as well. In nodulated soybean plants, the number of nodules and the N2 fixation rate increase with decreasing nitrate concentrations in the culture media (Herridge, 1982).
The synthesis of ureides can also take place in plants without symbiosis, although much smaller amounts are produced. Data from McClure and Israel (1979) show that the xylem ureide content in nodulated soybean plants receiving their nitrogen exclusively via N2 fixation was about 78%, whereas the sap of nitrate fertilized, non-nodulated legume plants still consisted of 6% ureides. However, most of the soil nitrogen (NO3– or NH4+) seems to be assimilated to amides (Gln and asparagines) that are loaded into the xylem and phloem (Atkins and Smith, 2000). The reduced synthesis of ureides presumably results in reduction in the need for allantoin transport. This is precisely what was found in roots of nitrogen-fertilized plants in comparison with nodulated bean plants where a reduction of PvUPS1 expression occurred in response to nitrogen supply. PvUPS1 expression was also detected in roots of plants grown on allantoin as sole nitrogen source, indicating a role of PvUPS1 in allantoin uptake and translocation to root or shoot. However, there seems to be no difference in expression levels between allantoin and nitrate/ammonia-fed plants. It could be that not all nitrogen reached the roots in the form of allantoin because the plants were not grown axenically, and this may have mitigated any allantoin inductive effect. However, we tried to reduce the conversion of allantoin into other nitrogen forms by fertilizing the plants every 2 d with a freshly prepared nutrient solution. On the other hand, this expression pattern might be due to metabolism of some allantoin in the root cortex after being taken up by the plant root, such the allantoin amounts reaching the endodermis of the root were low.
Differences in PvUPS1 expression in allantoin-fed and nodulated plants are presumably due to differences in allantoin concentrations in the roots of the differently treated plants. Although a role in allantoin uptake has also been suggested for AtUPS1, Arabidopsis seedlings grown on allantoin as sole nitrogen source showed a very repressed growth compared with plants cultured on ammonium nitrate as sole nitrogen source (Desimone et al., 2002). In contrast, in our feeding experiments with French bean differences in plant growth and habit could not be observed, indicating that allantoin is taken up by roots, metabolized in the root cells, and/or transported throughout the plant and used by the plant for growth and development. It is also worth noting that PvUPS1 expression in French bean plants was high compared with AtUPS1 transcript levels in the non-ureide-transporting Arabidopsis. AtUPS1 was only detectable by RT-PCR (Desimone et al., 2002). This supports the hypothesis that PvUPS1 is particularly important to the general nitrogen physiology of French bean plants, even in the absence of nodulation. However, as clearly demonstrated by various means here, in French bean roots, PvUPS1 expression is regulated by the nodulation status and the different nitrogen sources, and is directly related to allantoin transport.
The identification of the cellular localization of PvUPS1 is important in understanding how its function is integrated into tissue physiology. The assimilation of bacterial fixed N2 into allantoin takes place in the peroxisomes of nodules (Hanks et al., 1981; Shelp et al., 1983; Fig. 6). After synthesis, allantoin is transported to the xylem for long-distance translocation to the shoot. The transport of allantoin to the xylem parenchyma can be symplasmic (Shelp et al., 1983), although loading into the apoplast might occur in addition to leakage of the ureides into the apoplastic space (Fig. 6). Loading of allantoin into the symplast has to take place in any case where the Casparian strip in the endodermis surrounding the nodule vascular tissue makes apoplastic passage of allantoin impossible. The high expression of PvUPS1 in nodules and roots as well as the localization studies are consistent with the transport function demonstrated here and makes PvUPS1 a candidate for involvement in transfer of high amounts of ureides produced in nodulated French bean roots to the vascular system for export. Upon reaching the xylem parenchyma of the vasculature, allantoin is loaded into the xylem apoplast and is distributed to the shoot via the transpiration stream (Smith and Atkins, 2002). Allantoin levels in the phloem can also be high, therefore active loading of the ureide into the sieve element/companion cell complex via PvUPS1 as well as xylemphloem transfer in both directions, as postulated for the shoot, cannot be excluded (Pate et al., 1975; Atkins et al., 1982; Van Bel, 1984). The localization of PvUPS1 mRNA in the endodermis surrounding the vasculature as well as in the phloem (see Fig. 5) clearly support our predicted model of allantoin transport in the nodules and the involvement of PvUPS1 in this process. In addition, the endodermis in the outer nodule presents a barrier for large amounts of ureides synthesized in the nodule cells and released into the apoplast that might otherwise leak out of the nodule. The expression of PvUPS1 in this nodule endodermis points to its role in recovery of allantoin from the apoplast for redistribution to the vascular system.
In conclusion, in tropical legumes, the transport of allantoin is important for ultimately supplying nitrogen to the developing sinks for growth and development. To understand the underlying mechanisms of ureide transport and its regulation, an allantoin transporter (PvUPS1) was isolated from nodulated roots of French bean and was functionally characterized in an allantoin transport-deficient yeast mutant showing that PvUPS1 transports allantoin but also binds the allantoin precursors xanthine and uric acid. However, our expression and localization studies suggest that allantoin is an important substrate for PvUPS1. In beans, PvUPS1 expression levels are consistent with sites of synthesis and utilization of allantoin. Strong expression of PvUPS1 in root nodules, where allantoin is synthesized and translocated to the vascular system for long-distance transport, indicates the importance of PvUPS1 function in these tissues. The localization of PvUPS1 mRNA to the nodule and vascular endodermis and nodule phloem supports the proposed role of PvUPS1 in allantoin transport.
MATERIALS AND METHODS
Plant Materials
French bean (Phaseolus vulgaris cv Redland) plants were grown in the greenhouse at 26°C to 28°C with light conditions between 300 and 400 μmol photons m–2 s–1. The plants were cultured in potting soil, fertilized once a week (Peters Fertilizer 20–20-20; J.R. Peters, Allentown, PA), and used for organ-specific RNA expression and localization studies. For studies on regulation of transporter expression, plants were grown in perlite and were fertilized with a Hoagland solution (Hoagland and Arnon, 1938) containing no nitrogen, 6 mm KNO3–, and 6 mm NH4+NO3– or 4.5 mm allantoin. The total amount of molecular nitrogen was 18 mm in both nitrogen treatments. Plants were fertilized every 2 d with 250 mL of the different Hoagland solutions. In addition, plants without nitrogen supply were inoculated 5 d after germination with Rhizobium leguminosarum phaseoli (American Type Culture Collection no. TCC 8002, Rockville, MD) to guarantee nodulation.
Isolation of a Putative Allantoin Transporter
Total RNA was isolated from nodulated roots of French bean plants and RT-PCR was performed with degenerate primers according to the manufacturer's protocol (RETROscript; Ambion, Austin, TX). The primers were designed using ESTs from legumes identified in the National Center for Biotechnology Information database after “blasting” AtUPS1, a putative transporter of oxo-derivatives of nitrogen heterocyclic compounds including allantoin in Arabidopsis (At2g03590; Desimone et al., 2002). The alignment of the legume sequences revealed homology downstream of the start codon and upstream of the stop codon, resulting in the following primer sequences: forward primer Uri 1 (5′-ATGTATDTGRTRGAGAGCAARGGAGG-3′), based on the alignment of sequences from cowpea (Vigna unguiculata; VuA3, accession no. X90487), soybean (Glycine max; AW707051), and Medicago truncatula (BF646038); and reverse primer Uri 3 (5′-CAATTATTCCTTGGCAGTG-3′), based on the alignment of sequences from cowpea (X90487) and soybean (AW311266). The amplified cDNAs from French bean were cloned into the pGEM-T-Easy vector (Promega, Madison, WI) and sequenced. The newly isolated putative transporter (PvUPS1) was analyzed by sequence alignment (LASERGENE software; DNASTAR), hydrophobicity plots (TMHMM program, Sonnhammer et al., 1998; TMPred program, Hofmann and Stoffel, 1993; and the DAS program, Cserzö et al., 1997), and phylogenetic analysis (PAUP program, Rogers and Swofford, 1998).
Yeast Transformation and Complementation
Saccharomyces cerevisiae strains dal4/dal5 (Desimone et al., 2002) and 22Δ8AA (Tegeder et al., 2000) were used to investigate the substrate specificity of PvUPS1 in yeast. The yeast mutant dal4/dal5 is unable to grow on media containing allantoin as sole nitrogen source. The yeast strain 22Δ8AA (MATα, ura3-1, gap1-1, put4-1, uga4-1, can1::HisG, lyp/alp::HisG, hip1::HisG, and dip5:: HisG; Tegeder et al., 2000) can be used for analysis of cell growth on citrulline, Pro, Arg, Asp, and GABA. PvUPS1 was cloned into the yeast expression vector pDR196 (kindly provided by Doris Rentsch, unpublished data), and the yeast mutants dal4/dal5 and 22Δ8AA were transformed according to the protocol of Dohmen et al. (1991). AtUPS1 (Desimone et al., 2002) and AtAAP2 (Fischer et al., 1995) in pDR196 as well as the empty vector pDR196 were used as controls. dal4/dal5 transformants were grown on nitrogen-free minimal media supplemented with 0.5 g L–1 allantoin. Medium for 22Δ8AA transformants contained 0.5 g L–1 citrulline, Pro, Arg, Asp, or GABA as sole nitrogen source.
Synthesis and Purification of [14C]Allantoin for Transport Studies
[14C]allantoin was produced from 8-[14C]uric acid in a catalytic reaction with uricase using a modified protocol of Rainbird et al. (1984). Thirty μCi of 8-[14C]uric acid (Moravek Biochemicals, Brea, CA; specific activity 55 mCi mmol–1) were incubated for 2 h at 25°C in 1.5 mL of 0.1 m NaP04 buffer containing 1 unit of uricase (catalog no. U–0880; Sigma, St. Louis). The decrease of uric acid in the reaction mixture was monitored spectrophotometrically at 293 nm. The mixture was added to columns filled with Dowex H+ and Dowex formate resin (AG 50W-X8, catalog no. 142–1451; and AG1-X8, catalog no. 731–6221; Bio-Rad, Hercules, CA) to remove the uricase as well as remaining uric acid and allantoic acid. Allantoin was eluted with 50 mL of water, brought to dryness with a rotary evaporator (Büchi, Flawil, Switzerland), and resuspended in water. Purity of the synthesized allantoin was determined by HPLC analysis, and the measurement showed that [14C]allantoin represented 99% of the total labeled compounds. For HPLC, nonlabeled allantoin (catalog no. A–7878; Sigma) and uric acid (catalog no. U–2625; Sigma) were used as standards. The newly synthesized [14C]allantoin was used for transport studies in yeast.
Yeast Transport Measurements
For [14C]allantoin uptake studies, yeast cells were harvested at OD600 of 0.8 by centrifugation for 5 min at 4,500g, washed, and resuspended in water to a final OD600 of 4. The cells (100 μL) were then mixed with 20 μL of 1 m potassium phosphate buffer, pH 4.5, 20 μL of 1 m Glc, and 60 μL of water, and they were then preincubated for 5 min at 30°C. To start the uptake reaction, 20 μL of 2 mm [14C]allantoin were added to the yeast cells and incubated. Fifty-microliter samples were removed after 1, 2, 3, and 4 min, transferred to 4 mL of ice-cold water on fiberglass filters in a vacuum filtration unit, filtered, and washed with 8 mL of water.
Radioactivity of [14C]allantoin taken up in yeast cells was determined using liquid scintillation spectrometry (Beckman Instruments, Fullerton, CA). Endogenous uptake activity of yeast transformed with empty vector pDR196 was subtracted as background activity. Transport measurements were repeated independently and represent a mean of at least three experiments. Competition experiments were performed with a concentration of 200 μm [14C]allantoin and a 9-fold excess of respective amino acids according to Fischer et al. (1995).
RNA Gel-Blot Analysis
Total RNA was extracted according to the method described in Frommer et al. (1994) from plant organs of 3-month-old, soil-grown and nodulated French bean plants. Developing cotyledons, seed coats, and pods were harvested over a period of 2 weeks at seed water content of 75% to 85%. Extracted RNA from each organ (20 μg) was electrophoresed and transferred to Hybond-N+ membranes (Amersham Pharmacia Biotech, Uppsala) by standard procedure. Membranes were hybridized with 32P-labeled full-length PvUPS1 cDNA probes according to Tegeder et al. (2000). Membranes were washed at 50°C with 2× SSC and 0.2% (w/v) SDS (for 30 min), followed by a wash in 1× SSC and 0.1% (w/v) SDS at 56°C (30 min). Membranes were exposed to x-ray film (Eastman Kodak, Rochester, NY) for 3 to 7 d. Total RNA of nodules and roots was additionally extracted and blotted from nodulated and non-nodulated French bean plants grown in perlite and under controlled nitrogen conditions. The non-nodulated plants were supplied with ammonium nitrate as sole nitrogen source or allantoin. The nodulated plants were not supplied with nitrogen and relied on nitrogen fixation only (see also “Plant Materials”).
In Situ mRNA Hybridization
French bean nodules were fixed in formaldehyde-acetic acid solution (10% formaldehyde, 50% ethanol, and 5% glacial acetic acid, all v/v), and subsequently embedded in paraffin. DIG-labeled PvUPS1 sense and antisense riboprobes were synthesized by in vitro transcription according to the manufacturer's instructions (Roche Diagnostics, Mannheim, Germany). Tissue sections (8 μm) were probed with DIG-labeled PvUPS1 RNAs at 37°C for 16 h, as described in Harrington et al. (1997), and were then washed in 4× SSC, 2× SSC, and 1× SSC (at room temperature for each 30 min), and in 0.1× SSC (at 37°C for 30 min). For mRNA localization, tissue was preincubated in Tris-buffered saline-Tween (TBST; 10 mm Tris-HCl, 500 mm NaCl, and 0.05% [v/v] Tween 20, pH 7.5 at room temperature for 1 h) containing 1.25% (w/v) bovine serum albumin, followed by incubation in TBST-bovine serum albumin with anti-DIG-antibodies conjugated to alkaline phosphatase (1:500 dilution of secondary antibody at room temperature for 1 h). After four washes for 15 min each in TBST, PvUPS1 was visualized by color development using 5-bromo-4-chloro-3-indolyl phosphate and nitroblue tetrazolium as substrates (Roche Diagnostics). Micrographs were taken using a microscope (Leitz; Wetzlar, Germany), equipped with a camera (DKC5000; Sony, Tokyo).
For structural studies, nodules were embedded in London Resin White acrylic resin according to Harrington et al. (1997), and cut sections (1 μm) were stained for 1 min in safranin O. To visualize the endodermis in nodules, paraffin sections (8 μm) in VECTASHIELD mounting medium (Vector Laboratories Inc., Burlingame, CA) were observed under UV light.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Acknowledgments
M.T. gratefully acknowledges the productive environment provided by Dr. Wolf Frommer (University of Tuebingen) during the initiation of this project. We also thank Carole Bidal (Washington State University) for technical support, and we strongly acknowledge the use of the Electron Microscopy Center at Washington State University.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.033365.
This work was supported by the National Research Initiative Competitive Grants Program-U.S. Department of Agriculture (grant no. 2001–35318–10990 to M.T.).
References
- Argyrou E, Sophianopoulou V, Schultes N, Diallinas G (2001) Functional characterization of a maize purine transporter by expression in Aspergillus nidulans. Plant Cell 13: 953–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins CA, Beevers L (1990) Synthesis, transport and utilization of translocated solutes of nitrogen. In YP Abrol, ed, Nitrogen in Higher Plants. Research Studies Press, Somerset, UK, pp 233–295
- Atkins CA, Pate JS, Ritchie A, Peoples MB (1982) Metabolism and translocation of allantoin in ureide-producing grain legumes. Plant Physiol 70: 476–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins CA, Rainbird R, Pate JS (1980) Evidence for a purine pathway of ureide synthesis in N2-fixing nodules of cowpea (Vigna unguiculata L. Walp). Z Pflanzenphysiol 97: 249–260 [Google Scholar]
- Atkins CA, Smith PMC (2000) Ureide synthesis in legume nodules. In EJ Triplett, ed, Prokaryotic Nitrogen Fixation: A Model System for the Analysis of a Biological Process. Horizon Scientific Press, Wymondham, Norfolk, UK, pp 559–587
- Atkins CA, Smith PMC, Storer PJ (1997) Reexamination of the intracellular localization of de novo purine synthesis in cowpea nodules. Plant Physiol 113: 127–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccatto VM, Gomes JE, Sarriés GA, Moon DH, Tsai SM (1988) Effects of host plant origin on nodulin activities and nitrogen in Phaseolus vulgaris L. Plant Soil 204: 79–87 [Google Scholar]
- Chaya MS, Broughton WJ, Krause A (1996) VuA3 (accession no. X90487), a gene of Vigna unguiculata encoding a protein with unknown function (PGR96–072). Plant Physiol 112: 8618883395 [Google Scholar]
- Costigan SA, Franceschi VR, Ku MSB (1987) Allantoinase activity and ureide content of mesophyll and paraveinal mesophyll of soybean leaves. Plant Sci 50: 179–187 [Google Scholar]
- Cserzö M, Wallin E, Simon I, von Heijne G, Elofsson A (1997) Prediction of transmembrane α-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng 10: 673–676 [DOI] [PubMed] [Google Scholar]
- Day DA, Poole PS, Hendriks JHM, Tyerman SD, Rosendahl L (2000) Ammonia and amino acid transport across the membranes in nitrogen-fixing nodules. Cell Mol Life Sci 58: 61–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delrot S, Atanassova R, Maurousset L (2000) Regulation of sugar, amino acid and peptide plant membrane transporters. Biochim Biophys Acta 1465: 281–306 [DOI] [PubMed] [Google Scholar]
- Dencher NA, Sass HJ, Büldt G (2000) Water and bacteriorhodopsin: structure, dynamics, and function. Biochim Biophys Acta 1460: 192–203 [DOI] [PubMed] [Google Scholar]
- Desimone M, Catoni E, Ludewig U, Hilpert M, Schneider A, Kunze R, Tegeder M, Frommer WB, Schumacher K (2002) A novel superfamily of transporters for allantoin and other oxo derivatives of nitrogen heterocyclic compounds in Arabidopsis. Plant Cell 14: 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmen RJ, Strasser AWM, Honer CB, Hollenberg CP (1991) An efficient transformation procedure enabling long-term storage of competent cells of various yeast genera. Yeast 7: 691–692 [DOI] [PubMed] [Google Scholar]
- Fischer WN, Kwart M, Hummel S, Frommer WB (1995) Substrate specificity and expression profile of amino acid transporter (AAPs) in Arabidopsis. J Biol Chem 270: 16315–16320 [DOI] [PubMed] [Google Scholar]
- Frommer WB, Hummel S, Rentsch D (1994) Cloning of an Arabidopsis histidine transporter protein related to nitrate and peptide transporters. FEBS Lett 347: 185–189 [DOI] [PubMed] [Google Scholar]
- Gillissen B, Burkle L, Andre B, Kuhn C, Rentsch D, Brandl B, Frommer WB (2000) A new family of high-affinity transporters for adenine, cytosine, and purine derivatives in Arabidopsis. Plant Cell 12: 291–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks JF, Tolbert NE, Schubert KR (1981) Localization of enzymes of ureide biosynthesis in peroxisomes and microsomes of nodules. Plant Physiol 68: 65–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington GN, Nussbaumer Y, Wang XD, Tegeder M, Franceschi VR, Frommer WB, Patrick JW, Offler CE (1997) Spatial and temporal expression of sucrose transport-related genes in developing cotyledons of Vicia faba L. Protoplasma 200: 35–50 [Google Scholar]
- Hartmann K, Peiter E, Koch K, Schubert S, Schreiber L (2002) Chemical composition and ultrastructure of broad bean (Vicia faba L.) nodule endodermis in comparison to the root endodermis. Planta 215: 14–25 [DOI] [PubMed] [Google Scholar]
- Herridge DF (1982) Relative abundance of ureides and nitrate in plant tissues of soybean as a quantitative assay of nitrogen fixation. Plant Physiol 70: 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herridge DF, Atkins CA, Pate JS, Rainbird RM (1978) Allantoin and allantoic acid in the nitrogen economy of the cowpea (Vigna unguiculata L. Walp.). Plant Physiol 62: 495–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoagland DR, Arnon DI (1938) The water culture method for growing plants without soil. US imprint, Tallahassee, FL
- Hofmann K, Stoffel W (1993) Tmbase: a database of membrane spanning proteins segments. Biol Chem 374: 166 [Google Scholar]
- Kaur A, Sheoran IS, Singh R (1985) Effect of water stress on the enzymes of nitrogen metabolism in mung bean (Vigna radiata Wilcseck) nodules. Plant Cell Environ 8: 195–200 [Google Scholar]
- Layzell DB, LaRue TA (1982) Modeling C and N transport to developing soybean fruits. Plant Physiol 70: 1290–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima JD, Da Matta FM, Mosquim PR (2000) Growth attributes, xylem sap composition, and photosynthesis in common bean as affected by nitrogen and phosphorous deficiency. J Plant Nutr 23: 937–947 [Google Scholar]
- Lodwig EM, Hosle AH, Bourdes A, Findlay K, Allaway D, Karunakaran, Downie JA, Poole PS (2003) Amino acid cycling drives nitrogen fixation in the legume-Rhizobium symbiosis. Nature 422: 722–726 [DOI] [PubMed] [Google Scholar]
- Matsumoto T, Yatazawa M, Yamamoto Y (1977a) Distribution and change in the contents of allantoin and allantoic acid in developing nodulating and non-nodulating soybean plants. Plant Cell Physiol 18: 353–359 [Google Scholar]
- Matsumoto T, Yatazawa M, Yamamoto Y (1977b) Effects of exogenous nitrogenous-compounds on the concentrations of allantoin and various constituents in several organs of soybean plants. Plant Cell Physiol 18: 613–624 [Google Scholar]
- Matsumoto T, Yatazawa M, Yamamoto Y (1977c) Incorporation of 15N into allantoin in nodulated soybean plants supplied with 15N. Plant Cell Physiol 18: 459–462 [Google Scholar]
- McClure PR, Israel DW (1979) Transport of nitrogen in the xylem of soybean plants. Plant Physiol 64: 411–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda M, Borisjuk L, Tewes A, Dietrich D, Rentsch D, Weber H, Wobus H (2003) Peptide and amino acid transporters are differentially regulated during seed development and germination in faba bean. Plant Physiol 132: 1950–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda M, Borisjuk L, Tewes A, Heim U, Sauer N, Wobus U, Weber H (2001) Amino acid permeases in developing seeds of Vicia faba L.: expression precedes storage protein synthesis and is regulated by amino acid supply. Plant J 28: 61–71 [DOI] [PubMed] [Google Scholar]
- Montamat F, Maurousset L, Tegeder M, Frommer W, Delrot S (1999) Cloning and expression of amino acid transporters from broad bean. Plant Mol Biol 41: 259–268 [DOI] [PubMed] [Google Scholar]
- Pate JS, Atkins CA, White ST, Rainbird RM, Woo KC (1980) Nitrogen nutrition and xylem transport of nitrogen in ureide-producing grain legumes. Plant Physiol 65: 961–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pate JS, Sharkey PJ, Lewis OAM (1975) Xylem to phloem transfer of solutes in fruiting shoots of legumes, studied by a phloem bleeding technique. Planta 122: 11–26 [DOI] [PubMed] [Google Scholar]
- Rainbird RM, Thorne JH, Hardy RWF (1984) Role of amides, amino acids, and ureides in the nutrition of developing soybean seeds. Plant Physiol 74: 329–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers JS, Swofford DL (1998) A fast method for approximating maximum likelihoods of phylogenetic trees from nucleotide sequences. Syst Biol 47: 77–89 [DOI] [PubMed] [Google Scholar]
- Saraste M, Sibbald PR, Wittinghofer A (1990) The P-loop: a common motif in ATP- and GTP-binding proteins. Trends Biochem Sci 15: 430–434 [DOI] [PubMed] [Google Scholar]
- Schubert KR (1986) Products of biological nitrogen fixation in higher plants: synthesis, transport and metabolism. Annu Rev Plant Physiol 37: 539–574 [Google Scholar]
- Serraj R, Vadez V, Denison RF, Sinclair TR (1999) Involvement of ureides in nitrogen fixation inhibition in soybean. Plant Physiol 119: 289–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelp BJ, Atkins CA, Storer PJ, Canvin DT (1983) Cellular and subcellular organization of pathways of ammonia assimilation and ureide synthesis in nodules of cowpea (Vigna unguiculata L. Walp.). Arch Biochem Biophys 224: 429–441 [DOI] [PubMed] [Google Scholar]
- Smith FW, Rae AL, Hawkesford MJ (2000) Molecular mechanisms of phosphate and sulphate transport in plants. Biochim Biophys Acta 1465: 236–245 [DOI] [PubMed] [Google Scholar]
- Smith PMC, Atkins CA (2002) Purine biosynthesis: big in cell division, even bigger in nitrogen assimilation. Plant Physiol 128: 793–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MT, Emerich DW (1993) Alanine dehydrogenase from soybean bacteroids-kinetic mechanism and pH studies. J Biol Chem 268: 10746–10753 [PubMed] [Google Scholar]
- Sonnhammer ELL, von Heijne G, Krogh A (1998) A hidden Markov model for predicting transmembrane helices in protein sequences. In J Glasgow, T Littlejohn, F Major, R Lathrop, D Sankoff, C Sensen, eds, Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology. American Association for Artificial Intelligence Press, Menlo Park, CA, pp 175–182 [PubMed]
- Streeter JG (1979) Allantoin and allantoic acid in tissues and stem exudate from field-grown soybean plants. Plant Physiol 63: 478–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegeder M, Offler CE, Frommer WB, Patrick JW (2000) Amino acid transporters are localized to transfer cells of developing pea seeds. Plant Physiol 122: 319–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas RJ, Feller U, Erismann KH (1979) The effect of different inorganic nitrogen sources and plant age on the composition of bleeding sap of Phaseolus vulgaris. New Phytol 82: 657–669 [DOI] [PubMed] [Google Scholar]
- Thomas RJ, Feller U, Erismann KH (1980) Ureide metabolism in non-nodulated Phaseolus vulgaris L. J Exp Bot 31: 409–417 [Google Scholar]
- Thomas RJ, Schrader LE (1981) Ureide metabolism in higher plants. Phytochemistry 20: 361–371 [Google Scholar]
- Van Bel AJE (1984) Quantification of the xylem-to-phloem transfer of amino acids by use of inulin (14C) [carbon isotope] carboxylic acid as xylem transport marker (Lycopersicon esculentum, tomatoes). Plant Sci L 35: 81–85 [Google Scholar]
- Wipf D, Ludewig U, Tegeder M, Rentsch D, Koch W, Frommer W (2003) Conservation of amino acid transporters in fungi, plants and animals. Trends Biochem Sci 27: 139–147 [DOI] [PubMed] [Google Scholar]