Abstract
The Arabidopsis AtSUC3 gene encodes a sucrose (Suc) transporter that differs in size and intron number from all other Arabidopsis Suc transport proteins. Each plant species analyzed so far possesses one transporter of this special type, and several functions have been discussed for these proteins, including the catalysis of transmembrane Suc transport, and also Suc sensing and regulation of other Suc transporters. Here, we show that the AtSUC3 protein is localized in the sieve elements of the Arabidopsis phloem and is not colocalized with the companion cell-specific AtSUC2 phloem loader. Even stronger AtSUC3 expression is observed in numerous sink cells and tissues, such as guard cells, trichomes, germinating pollen, root tips, the developing seed coat, or stipules. Moreover, AtSUC3 expression is strongly induced upon wounding of Arabidopsis tissue. The physiological role of AtSUC3 in these different cells and tissues is discussed.
In most higher plants, Suc is the main type or even the exclusive form of carbohydrate that is partitioned between the different sinks after its synthesis in the mature source leaves and its subsequent loading into the sieve element-companion cell complex (SE-CCC). Since the cloning of the first higher plant Suc transporter cDNA (ps21; Riesmeier et al., 1992), genes and cDNAs encoding homologous proteins have been cloned from more than 20 different plant species (Kühn, 2003), and the Arabidopsis and the rice (Oryza sativa) genome projects (The Arabidopsis Genome Initiative, 2000; Yu et al., 2002) identified mediumsized families of Suc transporter genes in these plants (nine genes in the Arabidopsis genome and at least five genes in the rice genome).
All dicot plants seem to possess one Suc transporter gene (named SUC3 or SUT2) that differs from all other gene family members in an unusually large number of introns and in the length of its open reading frame (Davies et al., 1999; Barker et al., 2000; Meyer et al., 2000; Barth et al., 2003). The encoded protein sequences are significantly longer than those of the classical Suc transporters, and secondary structure predictions suggest that this results from enlarged cytoplasmic domains (Barker et al., 2000; Meyer et al., 2000). Interestingly, these transporters are closely related to the Suc transporters found in monocots such as rice, barley (Hordeum vulgare), oat (Avena sativa) or maize (Zea mays), where most Suc transporters seem to belong to this subcluster (Aoki et al., 2002, 2003a, 2003b; Hirose et al., 1997; Weschke et al., 2000; Barth et al., 2003; Kühn, 2003). Due to the differences observed between these special Suc transporters (from now on, called SUT2/SUC3-type transporters) and all other Suc transport proteins, it was intriguing to speculate on specific functions that these proteins might exhibit in addition to or instead of the simple catalysis of transmembrane Suc transport. Several papers have been published describing a putative sensor function for SUT2/SUC3-type transporters (Lalonde et al., 1999; Barker et al., 2000; Schulze et al., 2000) and data on the interaction between different Suc transporters seemed to support this idea (Reinders et al., 2002; Schulze et al., 2003). Using the split-ubiquitin system (Johnsson and Varshavsky, 1994), Reinders et al. (2002) showed the possible interaction of the tomato (Lycopersicon esculentum) Suc transporters LeSUT1, LeSUT2, and LeSUT4, which had all been immunolocalized in tomato SEs (Kühn et al., 1997; Barker et al., 2000; Weise et al., 2000). In similar analyses, Schulze et al. (2003) proposed an interaction between the Arabidopsis Suc transporters AtSUC2, AtSUT2 (= AtSUC3), and AtSUT4. Using β-glucuronidase (GUS)-histochemical analyses these authors showed expression of all three genes in Arabidopsis CCs. However, only the AtSUC2 protein had been immunolocalized in these cells (Stadler and Sauer, 1996).
Only recently, Barth et al. (2003) showed that in Plantago major, the situation is different from that described in tomato (all transporters in the SEs) and from that postulated for Arabidopsis (all transporters in the CCs; Schulze et al., 2003). In Plantago, the SUT2/SUC3-type transporter, PmSUC3, is not colocalized with PmSUC2, the putative ortholog of AtSUC2, which is responsible for phloem loading. PmSUC2 and its Arabidopsis ortholog AtSUC2 had been immunolocalized in CCs (Stadler et al., 1995; Stadler and Sauer, 1996), whereas PmSUC3 was found exclusively in the SEs of the Plantago phloem. This showed that physical interaction between PmSUC2 and PmSUC3 is not possible in planta and that phloem loading by PmSUC2 is unlikely to be regulated by PmSUC3. Moreover, PmSUC3 protein was immunolocalized in embryos and root tips, supporting the idea that PmSUC3 is in fact a Suc transporter rather than a Suc sensor.
In the present paper, we approached the question of whether or not AtSUC3 is a regulator of phloem loading. Our data show that inside the phloem, At-SUC3 is localized only within the SEs, which is inconsistent with its predicted interaction with the CC-specific AtSUC2 transporter and with a regulatory role of AtSUC3. Moreover, we found strong AtSUC3 expression in several nonphotosynthetic cells and tissues such as guard cells, trichomes, germinating pollen, root tips, the seed coat, and stipules, suggesting a role for AtSUC3 in the Suc import into sink tissues. This interpretation is supported by the observed induction of AtSUC3 expression upon wounding.
RESULTS
Localization of AtSUC3 in SEs
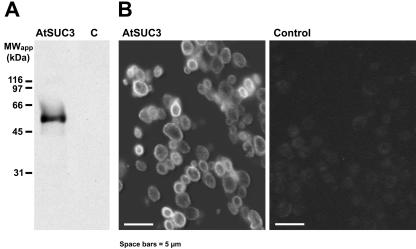
In a previous paper, AtSUC3 protein had immunolocalized in individual large cells along the phloem (Meyer et al., 2000), but signals in specific cells within the phloem had not been obtained. Due to the repeatedly described activity of the AtSUC3 promoter within the phloem (Meyer et al., 2000; Schulze et al., 2003), we assumed that this lack of antibody binding to individual phloem cells may result from a low antibody titer or from an inaccessibility of the AtSUC3 antigen within this tissue. Therefore, we raised a new antiserum against a 15-amino acid peptide from the AtSUC3 N terminus (residues 8–22 of the protein). This sequence is specific for AtSUC3, which has a longer N terminus than all other Arabidopsis Suc transporters, and a BLAST search against all available Arabidopsis protein sequences found this peptide in no other protein (not shown). Moreover, the specificity of the obtained anti-AtSUC3 antiserum-2 was tested on western blots, where plasma membrane proteins from AtSUC3-expressing yeast cells (SMY36) and from control cells harboring the empty vector (SMYET1) had been blotted after separation on polyacrylamide gels (Fig. 1A). The new serum recognized a single band of about 55 kD, which was absent in extracts from controls, suggesting that the serum is specific for AtSUC3 denatured with SDS. In western analyses with SDS-solubilized proteins from Arabidopsis source leaves, no protein band was labeled with this antiserum (not shown). This suggests that only small amounts of AtSUC3 protein are present in leaves and that no crossreaction with other proteins occurs. To test whether this serum will also identify AtSUC3 protein that has been fixed and denatured for immunohistochemical analyses, AtSUC3-expressing SMY36 cells and SMYET1 control cells were harvested, fixed, and embedded in methacrylate as described (Meyer et al., 2000). Sections of these cells were incubated with anti-AtSUC3 antiserum-2 and with an anti-rabbit immunoglobulin (Ig) G-FITC-isomer 1-conjugate (Fig. 1B), yielding signals only with the AtSUC3-expressing yeast cells. Moreover, the obtained fluorescence was seen almost exclusively on the surface of these cells, suggesting a localization of the AtSUC3 protein in the plasma membranes of these cells. No signals were detected when an excess of AtSUC3 peptide (see above) was added during incubation with the second antibody (data not shown).
Figure 1.
Analysis of the specificity of the anti-AtSUC3 antiserum-2. A, Western-blot analysis of proteins extracted with SDS from plasma membrane preparations of the AtSUC3-expressing yeast strain SMY36 (AtSUC3) or of untransformed control cells (C). Three micrograms of protein were separated per lane on SDS-polyacrylamide gels, transferred to nitro-cellulose filters, and incubated with anti-AtSUC3 antiserum-2. B, Sections from the same yeast strains as in A were fixed and embedded in methacrylate. Semi-thin sections were treated with anti-AtSUC3 antiserum-2, and binding of antibodies to AtSUC3 was visualized fluorescein isothiocyanate (FITC)-conjugated second antibody under appropriate excitation light.
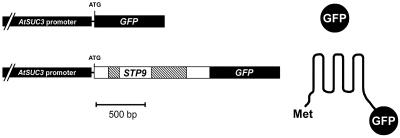
For a parallel analysis of the AtSUC3 expression pattern, we used AtSUC3-promoter::GUS plants (Meyer et al., 2000) and generated Arabidopsis plants expressing the green fluorescent protein (GFP) reporter gene or a GFP variant encoding a GFP fusion targeted to the plasma membrane (TM-GFP; Fig. 2). This GFP variant was obtained by fusing the GFP cDNA to the 3′ end of a genomic fragment encoding the N-terminal one-half of the AtSTP9 monosaccharide transporter gene (Schneidereit et al., 2003) including the first two introns and exon sequences for 232 amino acids (= six transmembrane helices). Transgenic Arabidopsis plants express GFP or TM-GFP in the very same cells, but in cells with large plasmodesmata, e.g. in cells of the vascular tissue, free GFP might traffic cell-to-cell (Imlau et al., 1999) thereby blurring the correct expression pattern of AtSUC3. In contrast, TM-GFP should not traffic cell-to-cell, and, therefore, should indicate the precise site of promoter activity. Thus, comparison of TM-GFP and GFP fluorescence will not only provide data on the precise expression pattern of the promoter in question, but will also information on the quality of possible symplastic connections of fluorescent cells.
Figure 2.
AtSUC3-promoter::GFP constructs and resulting proteins. Schematic presentation of the two constructs used for analysis of AtSUC3 expression via GFP fluorescence. The top construct represents an in-frame fusion of GFP to the start ATG of the AtSUC3 gene yielding the free, hydrophilic form of GFP (right). The bottom construct represents an in-frame fusion of GFP to a genomic fragment of the AtSTP9 gene that codes for the N-terminal six-transmembrane helices of the AtSTP9 protein (232 amino acids and two introns) and that has the N-terminal Met of AtSTP9 and the C-terminal GFP fusion exposed to the cytoplasmic side of the plasma membrane. The transmembrane helices of this GFP variant (TM-GFP) make this a lipophilic fusion that is targeted to the membrane system and not freely mobile in the cytoplasm.
Figure 3A shows the fluorescence of TM-GFP in an Arabidopsis leaf. Weak fluorescence is detected in the vasculature, but stronger fluorescence is seen in individual, larger cells along the veinal network. These larger cells seem to represent the cells previously described by Meyer et al. (2000). Figure 3B shows the fluorescence of TM-GFP in an Arabidopsis root, where two files of fluorescent cells represent the two vascular strands. In AtSUC3-promoter::GFP plants, this vascular bundle-specific fluorescence is much weaker (data not shown), possibly because the mobile form of GFP is transported away together with the assimilates.
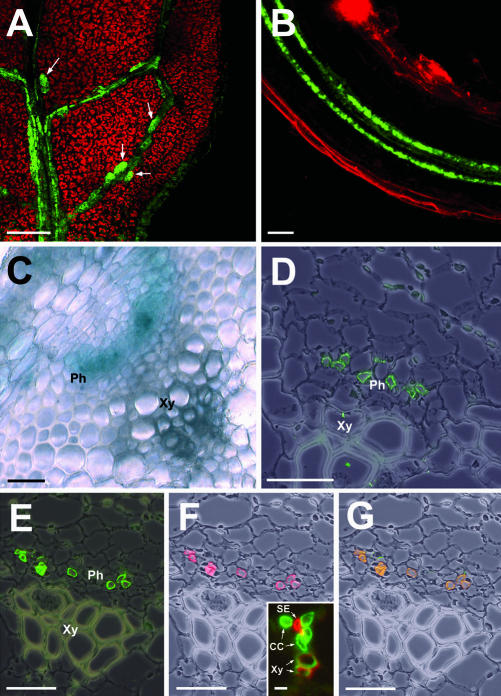
Figure 3.
Localization of AtSUC3 in the SEs of the Arabidopsis phloem. A, AtSUC3 promoter-dependent GFP fluorescence in a leaf of an Arabidopsis plant expressing TM-GFP. Fluorescence is seen in the vasculature and in several larger cells (CLSM) along the veins. B, GFP fluorescence in a root of an Arabidopsis plant expressing TM-GFP. Fluorescence is restricted to the two vascular bundles in the center of the root (CLSM). C, GUS-histochemical staining in a cross section through a stem of an AtSUC3-promoter::GUS plant. The blue GUS staining is seen only in the phloem part of the vascular bundle. D, Immunolocalization of AtSUC3 protein in individual cells of the Arabidopsis phloem. The stem section was treated with anti-AtSUC3-antiserum-2 and with FITC-conjugated second antibody. E through G, Cross sections through the stems of Arabidopsis wild-type plants. Sections were incubated with anti-AtSUC3 antiserum-2/FITC-conjugated second antibody (green fluorescence in E) and with the SE-specific monoclonal antibody RS6/Cy3-conjugated second antibody (red fluorescence in F). The insert in F represents a cross section through an Arabidopsis minor vein and is shown to confirm the SE specificity of the RS6 antiserum (details in the text). RS6/tetramethylrhodamine isothiocyanate (TRITC)-conjugated second antibody (red fluorescence) labels the small SEs in the center of the Arabidopsis minor vein. G, Merging E and F results in a yellow label showing that AtSUC3 is localized in the SEs of the phloem. Ph, Phloem; Xy, Xylem. Scale bars in A = 40 μm, = 20 μm in B, and = 25 μm in C through G. The scale bar in the insert of F = 5 μm.
The cross section through the stem of an AtSUC3-promoter::GUS plant shown in Figure 3C clearly demonstrates that the site of AtSUC3 promoter activity within the vascular bundle is restricted to the phloem. Additional immunohistochemical analysis with anti-AtSUC3 antiserum-2-labeled individual cells within this tissue (Fig. 3D). These cells could be identified as SEs by double labeling with anti-AtSUC3 antiserum-2 (Fig. 3E) and the SE-specific monoclonal antibody RS6 (Fig. 3F). The RS6 antibody has previously been shown to be specific for the SEs of several Brassicaceae (R.D. Sjolund, University of Iowa, unpublished data) and its SE specificity was reconfirmed by the analysis presented in the insert of Figure 3F. This insert shows the double labeling of a minor vein from a transgenic Arabidopsis plant expressing the Plantago PmSUC1 Suc transporter cDNA (Gahrtz et al., 1996) in its CCs with an anti-PmSUC1 antiserum (green fluorescence in the large CCs) and with RS6 (red fluorescence in the small SEs in the center). Merging Figure 3E (green AtSUC3-derived fluorescence) and Figure 3F (red RS6-derived fluorescence) yields a yellow staining in all labeled cells, confirming that AtSUC3 protein is located only in the SEs of the phloem (Fig. 3G).
AtSUC3 Is Also Expressed in Sink Cells and Sink Tissues of Arabidopsis
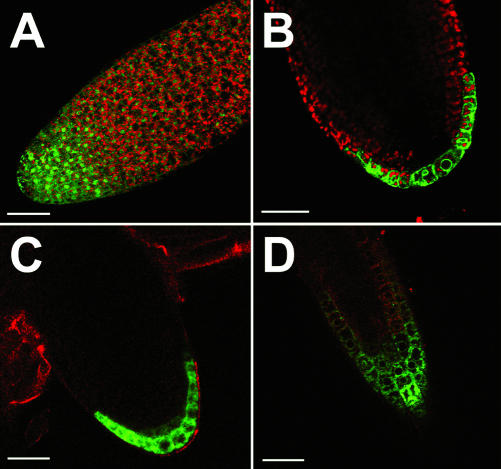
Analyses of our different AtSUC3-promoter:: reporter gene plants (GUS, GFP, and TM-GFP) revealed that AtSUC3 is also expressed in numerous cells and tissues outside the phloem (Fig. 4). AtSUC3-promoter-driven GUS activity was found in pollen grains (Fig. 4, A and B) and tubes (Fig. 4C), and Figure 4D confirms this localization using anti-AtSUC3 antiserum-2. Furthermore, expression was also seen in guard cells (Fig. 4, E and F), trichomes (Fig. 4, G and H), root tips of embryos (Fig. 4, I and J) and lateral roots (Fig. 4, K and L), and in stipules (Fig. 4, M and N). Guard cell-specific and trichome-specific expression were only observed in very young leaves and were absent in the larger leaves of rosettes. Further analyses with the confocal laser scanning microscope (CLSM) revealed AtSUC3-promoter-driven fluorescence also in the suspensor of the young embryo (Fig. 4O) and in the seed coat (Fig. 4P).
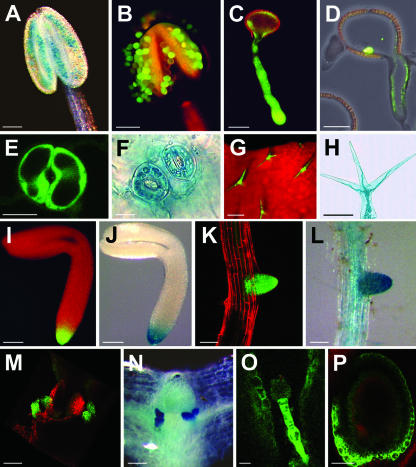
Figure 4.
Identification of AtSUC3 promoter activity or AtSUC3 protein in various sink cells and tissues of Arabidopsis. A, GUS-histochemical staining of an anther from an AtSUC3-promoter::GUS plant showing blue GUS staining in the pollen grains of a still closed anther. B, GFP fluorescence in pollen grains still attached to an open anther from an AtSUC3-promoter::GFP plant (EF, Epifluorescence). The yellow fluorescence results from cell wall phenolics deposited during anther maturation. C, GFP fluorescence in the tube of a germinating pollen from an AtSUC3-promoter::GFP plant (CLSM). D, Immunolocalization of AtSUC3 protein in the pollen tube of an Arabidopsis wild-type plant. E, GFP fluorescence in a pair of guard cells from an AtSUC3-promoter::GFP plant (CLSM). F, GUS-histochemical staining of two pairs of guard cells from an AtSUC3-promoter::GUS plant. G, GFP fluorescence in the trichomes from an AtSUC3-promoter::GFP plant (EF). H, GUS-histochemical staining in the detached trichome of an AtSUC3-promoter::GUS plant. I, GFP fluorescence in the root tip of an embryo from an AtSUC3-promoter::GFP plant (EF). J, GUS-histochemical staining in the root tip of an embryo from an AtSUC3-promoter::GUS plant. K, GFP fluorescence in the tip of an emerging lateral root from an AtSUC3-promoter::GFP plant (CLSM). L, GUS-histochemical staining in the tip of an emerging lateral root from an AtSUC3-promoter::GUS plant. M, GFP fluorescence in the stipules of an AtSUC3-promoter::GFP plant (CLSM). N, GUS-histochemical staining in the stipules of an AtSUC3-promoter::GUS plant. O, GFP fluorescence in the suspensor of an ovule from an AtSUC3-promoter::TM-GFP plant (CLSM). P, GFP fluorescence in the seed coat of an developing ovule from an AtSUC3-promoter::TM-GFP plant (CLSM). Scale bars = 100 μm in A, B, H, I, and J, = 20 μm in C and O, = 10 μm in D, = 8 μm in E and F, = 200 μm in G, and = 80 μm in K, L, M, N, and P.
All AtSUC3-expressing cells and tissues shown in Figure 4 represent strong sinks that depend on the import of organic carbon from the source leaves, suggesting a role for AtSUC3 in the delivery of Suc to these cells and tissues from the apoplast.
Cells of the Root Tip Have Plasmodesmata, Allowing the Cell-to-Cell Trafficking of GFP
The epifluorescence analysis of intact root tips from AtSUC3-promoter::GFP embryos (Fig. 4I), as well as the GUS-histochemical staining of intact root tips from corresponding AtSUC3-promoter::GUS embryos (Fig. 4J), showed a strong AtSUC3 expression in these tips. Optical sections of AtSUC3-promoter::GFP root tips (Fig. 5A) obtained with the CLSM confirmed the gradient of GFP fluorescence seen in Figure 4I. In contrast, a slightly different expression pattern was obtained in CLSM analyses of embryonic root tips from AtSUC3-promoter::TM-GFP plants. In the tips from AtSUC3-promoter::TM-GFP embryos, only the outermost cell layer is labeled (Fig. 5B). This suggests that free GFP synthesized in the outermost cell layer of the root tip in AtSUC3-promoter::GFP plants can move out of these cells, thereby forming the observed gradient of decreasing GFP fluorescence (Figs. 4I and 5A).
Figure 5.
Comparison of GFP and TM-GFP fluorescence in the tip of an Arabidopsis root (all CLSM). A, Optical section and detection of GFP fluorescence in the root tip an AtSUC3-promoter::GFP embryo. A continuous gradient of GFP fluorescence is seen in the root tip. The soluble, hydrophilic GFP used in this analysis accumulates in the nuclei of all cells and is seen up to 100 μm behind the tip. B, Optical section and detection of TM-GFP fluorescence in the root tip of an AtSUC3-promoter::TM-GFP embryo. The fluorescence is restricted to the outermost cell layer, and TM-GFP labels primarily the cellular membrane systems. C, Optical section and detection of TM-GFP fluorescence in the tip of a very young lateral root of an AtSUC3-promoter::TM-GFP plant. As in the embryo shown in B, TM-GFP fluorescence is restricted to the outermost cell layer. D, Optical section and detection of TM-GFP fluorescence in the tip of a lateral root (slightly older than that shown in C) from an AtSUC3-promoter::TM-GFP plant. At this later stage of lateral root development, TM-GFP fluorescence is seen in the outermost cell layer, but in addition, TM-GFP fluorescence is also detected in the columella cells in the center of the root tip. Scale bars = 40 μm in A, and = 20 μm in B through D.
A similar difference was observed in the tips of lateral roots emerging from the roots of mature Arabidopsis plants. Epifluorescence analyses and longitudinal sections obtained with the CLSM (not shown) show GFP fluorescence in all cells of the root tips in AtSUC3-promoter::GFP plants (Fig. 4K). However, CLSM analyses of AtSUC3-promoter::TM-GFP plants revealed that during the early stages of lateral root formation, AtSUC3 expression is also restricted to the outermost cell layer (Fig. 5C). During root elongation, this expression pattern is modified and GFP fluorescence is seen in the epidermis and in several subepidermal cells, most likely the columella. In the tips of fully developed roots, AtSUC3 expression is no longer detectable.
AtSUC3 Expression Is Induced upon Wounding
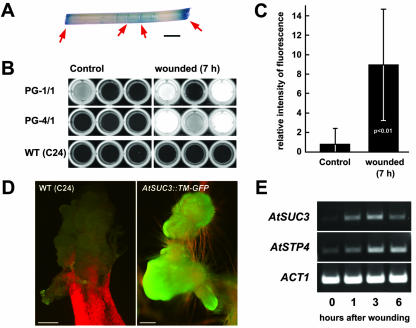
Analyses of AtSUC3-promoter::GUS plants reproducibly gave very strong GUS staining at all sites where leaves or stem sections were cut before the incubation with the staining solution. This result suggested that the activity of the AtSUC3 promoter driving this GUS expression might be enhanced upon wounding. Therefore, we tested the induction of GUS activity in leaves and stems of AtSUC3-promoter::GUS plants. Figure 6A shows an example of wound-induced GUS staining in a stem section that had been cut on both ends and injured at several positions with a scalpel. Similar results were obtained with wounded leaves (data not shown).
Figure 6.
Induction of AtSUC3 expression upon wounding of Arabidopsis tissue. A, GUS-histochemical staining of a stem section showing strong staining at both ends and at several wounding sites along the stem (red arrows). B, Detection of methylumbelliferone fluorescence in cell-free extracts from nine unwounded and nine wounded plants (wild-type plants and two different AtSUC3-promoter::GUS lines). Fluorescence was detected under UV light. C, Quantification of the fluorescence data obtained with the extracts from the six unwounded and the six wounded transgenic plants shown in B (mean ± sd; Student's p test < 0.01). D, Strong GFP fluorescence in a callus that formed at a wounded root of an AtSUC3-promoter::TM-GFP plant. A callus that had formed on an injured wild-type plant is shown for comparison. E, Reverse transcriptase (RT)-PCR analysis on total RNA preparations isolated from Arabidopsis stem sections that were wounded and incubated for the indicated periods. Scale bars = 2 mm in A, and = 500 μm (wild type) or 200 μm (AtSUC3::TM-GFP) in D.
To exclude the possibility that this increased GUS activity at the sites of wounding may simply result from a better diffusion of substrate into the plant tissue, cell-free extracts were prepared from untreated control tissues and from tissues that had been wounded and incubated for 7 h. GUS activities were determined in these extracts using the GUS substrate 4-methylumbelliferyl-β-d-glucuronide (MUG), which yields a fluorescent product after hydrolysis by GUS. Figure 6B shows these analyses that were performed with six independent plants from each of two different AtSUC3-promoter::GUS lines (PG-1/1 and PG-1/4) and with six wild-type plants. No GUS-derived fluorescence could be observed in the extracts from wild-type plants, from unwounded or wounded tissues. One of the extracts from the six unwounded transgenic plants showed some background fluorescence, and the fluorescence in all other extracts was comparable with wild-type levels. In contrast, strongly enhanced GUS activities were observed in five of six extracts from transgenic lines. A quantification of these fluorescence data (Fig. 6C) showed that the increase in GUS activity is significant.
Finally, we tried to confirm these data by analyzing potential changes in AtSUC3 mRNA levels in tissue of Arabidopsis wild-type plants at different times after wounding. To this end, Arabidopsis inflorescence stems were incision wounded with a scalpel and were incubated in 50 mm Na+-phosphate buffer at 21°C. Figure 6E shows one out of several RT-PCR analysis that were performed on independent RNA preparations isolated from Arabidopsis inflorescence stems after different times of incubation. Controls were performed with primers specific for the mRNAs of the previously described, wound-induced AtSTP4 monosaccharide transporter (Truernit et al., 1996) or for the Arabidopsis ACT1 actin gene. The mRNA levels for AtSUC3 and AtSTP4 increased already during the first 3 h after wounding, and, in all analyses, induction was already detected after 60 min. This result confirms that the activity of the AtSUC3 promoter is enhanced upon wounding of Arabidopsis tissues.
Strong AtSUC3 expression was also observed in callus tissue forming after incubation of wounded Arabidopsis tissues on petri plates (Fig. 6D). This tissue is not present during normal development and the observed GFP fluorescence in this tissue is a further example of AtSUC3 expression in sinks.
DISCUSSION
AtSUC3 Cannot Regulate AtSUC2 in Vivo
The aim of the present paper was to gain insight into the function and the physiological role of the protein encoded by the Arabidopsis AtSUC3 gene. Although phylogenetic analyses and sequence comparisons clearly characterized the AtSUC3 protein as a member of the Arabidopsis Suc transporter family, several characteristics, such as the size of the protein, the number of introns in the gene, and the codon bias of the mRNA, made AtSUC3 unique within this transporter family (Barker et al., 2000; Meyer et al., 2000). Together with the observation that the cDNA of a homologous gene from tomato (LeSUT2) could not complement the defect of a yeast mutant to grow on Suc as sole carbon source, these differences led to the model that this special type of Suc transporter-like proteins might be sensors of extracellular Suc concentrations (Barker et al., 2000) or flux sensors (Schulze et al., 2000), modulating the activity of other Suc transporters during phloem loading. The observation that different Arabidopsis Suc transporters, including AtSUC2 and AtSUC3, have the potential to interact, when their cDNAs are coexpressed in bakers' yeast, was used as an additional argument for this sensor theory (Schulze et al., 2003).
The immunohistochemical data presented in this paper demonstrate that AtSUC3 and the Arabidopsis phloem loader AtSUC2 (Stadler and Sauer, 1996; Gottwald et al., 2000) are not colocalized in the same cell type within the SE-CCC. The data in Figure 3 show that within the Arabidopsis phloem, the AtSUC3 protein is localized exclusively in the SEs, which were unequivocally identified using the SE-specific monoclonal antibody RS6. In contrast, AtSUC2 has previously been located specifically and exclusively in the CCs of the Arabidopsis phloem (Stadler and Sauer, 1996). These results are in perfect agreement with previous analyses by Schulze and coworkers (2003), who showed that the genes for AtSUC2 and AtSUC3 are expressed in the CCs. These authors determined only the sites of gene expression within the phloem by GUS histochemical staining, which was in fact confined to the CCs of the SE-CCC. However, it is well known from previous analyses of solanaceous plants (Kühn et al., 1997) and from a recent paper on the PmSUC3 Suc transporter from Plantago (Barth et al., 2003) that transporters encoded by genes expressed in the CCs can be targeted to the SEs. This is obviously also the case for the AtSUC3 protein, the first Arabidopsis Suc transporter identified in SEs. This clearly demonstrates that the physical interaction of AtSUC2 and AtSUC3 predicted from the split ubiquitin system (Schulze et al., 2003) cannot occur in vivo, and that the proposed regulation of AtSUC2-dependent phloem loading by AtSUC3 is unlikely. Thus, AtSUC3 might be responsible for the retrieval of Suc into the SE along the phloem path.
AtSUC3 Is Strongly Expressed in Several Sink Tissues
The continuous discussion on the different predicted functions (sensor, flux sensor, or transporter) of AtSUC3 and related proteins from other plants concentrated exclusively on the role of these proteins in the phloem. As a result of this discussion, other important sites of SUT2/SUC3 gene expression were completely overlooked. First evidence that these proteins might not only be found in the phloem came from analyses of the PmSUC3 localization in Plantago (Barth et al., 2003). Using a highly specific polyclonal antiserum, this protein was identified in different sink tissues of Plantago, such as embryos or root tips.
Our AtSUC3-promoter::GUS and AtSUC3-promoter::GFP analyses, as well as the immunohistochemical data, show that AtSUC3 is also expressed at several sites outside the phloem. One of these, most likely the glucosinolate-containing S cells (Koroleva et al., 2000), was mentioned in a previous paper that concentrated mainly on the functional characterization of the AtSUC3 transport activity (Meyer et al., 2000). The detailed analyses presented in the present paper (Fig. 4) show that AtSUC3 is expressed in guard cells, root tips, trichomes, stipules, pollen grains and pollen tubes, and in the suspensor and coat of the developing seed. All of these tissues are nongreen, symplastically isolated, rapidly growing, or metabolically extremely active properties describing these cells and tissues as sinks, depending on the import of organic carbon. In some of these cells or tissues, genes encoding other transporters, mainly transporters for monosaccharides, were previously shown to be expressed (AtSTP2 in pollen grains: Truernit et al., 1999; AtSUC1, AtSTP6, and AtSTP9 in pollen tubes: Stadler et al., 1999; Scholz-Starke et al., 2003; Schneidereit et al., 2003; AtSTP1 in guard cells: Stadler et al., 2003; and AtSTP4 in root tips: Truernit et al., 1996). For guard cells, an additional Suc transport activity has been postulated (Ritte et al., 1999), but thus far, no Suc transporter has been identified in these cells. The AtSUC3 expression observed in root tips and embryos is in agreement with immunohistochemical data obtained for the Plantago PmSUC3 transporter (Barth et al., 2003).
The data presented in this paper suggest that the Arabidopsis AtSUC3 transporter is most likely responsible for the import of Suc into several sink tissues, and, in addition, it seems to be involved in the retrieval of Suc along the transport phloem. Additional analyses will be necessary to understand the role of the cytoplasmic extensions present on all SUT2/SUC3 proteins that can be removed with no detectable effect on the transport properties of the recombinant protein (Meyer et al., 2000) and that may be used for the regulation of AtSUC3 transport properties in planta.
Expression of AtSUC3 Is Strongly Enhanced in Injured Tissue
Thus far, only a single carbohydrate transporter gene from Arabidopsis has been shown to be involved in the response to wounding or infection. Truernit et al. (1996) had shown that the expression of the otherwise sink-specific AtSTP4 monosaccharide transporter gene was enhanced in plants that had been injured or treated with fungal elicitors. This observation was extended only recently, when coinduction of AtSTP4 and of a cell wall invertase gene (Atβfruct1; Schwebel-Dugue et al., 1994) was found after infection of Arabidopsis plants with the fungal biotroph Erysiphe cichoracearum (Fotopoulos et al., 2003). These results suggest that carbohydrate import from the apoplast after wounding or infection might be important for the enhanced metabolic needs of the adjacent cells or may reflect a mechanism for the efficient reduction of extracellular carbohydrates, which otherwise might be used by pathogens.
The finding that the reporter protein activity was reproducibly increased at the cutting sites of AtSUC3-promoter::GUS plants (Fig. 6A) suggested that AtSUC3 expression might be up-regulated under similar conditions as shown for AtSTP4. The MUG analyses (Fig. 6B) and the increase in AtSUC3 mRNA levels (Fig. 6E) confirm this assumption and show for the first time that a plant disaccharide transporter may be involved in stress response. As for AtSTP4, the function of the stress-induced AtSUC3 may be a better supply of carbohydrates to the cells involved in the wound response or the efficient removal of extracellular carbohydrates to reduce the risk of pathogen infection.
Finally, strong AtSUC3 promoter-driven GFP fluorescence was observed in callus (Fig. 6D), representing a sink typically formed at the sites of wounding. This activity of the AtSUC3 promoter is not unexpected for a gene that is expressed in developmental and in environmentally induced sinks. Furthermore, this result explains the high level of AtSUC3 mRNA detected in suspension-cultured Arabidopsis cells in expression analyses performed with Affymetrix whole genome chips (John Ward, University of Minnesota, St. Paul; http://www.cbs.umn.edu/Arabidopsis/). In these analyses, comparable AtSUC3 expression levels were described for all Arabidopsis tissues. However, the levels found in suspension-cultured cells exceeded these tissue-specific levels more than 10-fold.
As already mentioned in the paper of Barth et al. (2003), two different mutant lines (line 9.43 and line 22.6) with T-DNA insertions in the AtSUC3 gene have been isolated, and no differences between the growth of these lines and the isogenic wild type were observed under normal growth conditions. Of course, these plants will now need to be reevaluated under certain stress conditions, e.g. the infection with and the resistance to different pathogens.
Expression of AtSUC3 Promoter::GFP and AtSUC3-Promoter::TM-GFP Constructs Yields Different Expression Patterns
It has previously been shown that GFP can move cell-to-cell, if expressed in cells having plasmodesmata with a large size-exclusion limit (SEL; Imlau et al., 1999). Based on this observation, Oparka and coworkers (1999) used GFP and GFP variants to show that plasmodesmata of mesophyll cells develop from simple plasmodesmata with large SEL (up to 50 kD) to complex, branched plasmodesmata with small SEL (about 1 kD). To exclude incorrect localization data resulting from this possible cell-to-cell movement of free GFP between cells with large SEL plasmodesmata, we used plants expressing free GFP or the membrane-anchored TM-GFP.
In fact, AtSUC3-promoter::TM-GFP plants showed differences when compared with AtSUC3-promoter:: GFP plants. First, the fluorescence detected in the vascular tissue was much stronger in AtSUC3-promoter::TM-GFP plants (not shown), most likely because the membrane-bound version of the reporter is not transported away together with the stream of assimilates. Second, a clear difference was observed in the root tips of the different lines (Fig. 5). In contrast to GFP plants, fluorescence in TM-GFP plants was seen exclusively in the outermost cell layer(s) of the root tips. This suggests that these cells and the underlying cell layers are connected by plasmodesmata with SELs of at least 30 kD, because free GFP can move cell-to-cell. Together with previous data from Oparka et al. (1994) and Imlau et al. (1999), who showed symplastic connections between the root protophloem and the surrounding cell layers, these data demonstrate symplastic continuity between the phloem and the outermost cell layers of the growing root tip. Expression of AtSUC3 in the final cells of this symplastic continuum, in the root tip epidermis, might be a mechanism to minimize the loss of Suc by diffusion and to optimize carbon supply to these rapidly growing cells.
MATERIALS AND METHODS
Strains and Growth Conditions
Arabidopsis plants (Columbia wild type or C24) were grown at 21°C under short-day conditions (8 h of light/16 h of dark) and were transferred to long-day conditions (16 h of light/8 h of dark) after bolting. For cloning in Escherichia coli, we used strain DH5α (Hanahan, 1983). Transformation of Arabidopsis was performed using Agrobacterium tumefaciens strain GV3101 (Holsters et al., 1980).
Molecular Cloning of AtSUC3-Promoter::GFP and AtSUC3-Promoter::TM-GFP
For construction of the AtSUC3-promoter::GFP construct, a first AtSUC3-promoter::GFP fusion was generated in a pUC19-based construct that has previously been described (in pUC-GUS-2a; Meyer et al., 2000) by replacing the GUS fragment with a GFP fragment containing appropriate cloning sites, yielding the plasmid pCL23. From this plasmid, the AtSUC3-promoter::GFP fragment was excised by HindIII and SacI and was inserted into the corresponding sites of the plant transformation vector pGPTV-bar (Becker et al., 1992).
For construction of the AtSUC3-promoter::TM-GFP construct, a genomic AtSTP9 fragment (encoding the 232 N-terminal amino acids of AtSTP9 and harboring the first two introns of At1g50310) was PCR amplified and NcoI cloning sites were introduced on both ends. Using these NcoI sites, the genomic AtSTP9 fragment was cloned into the unique NcoI site of plasmid pCL23, yielding the plasmid pCL26. This plasmid represents a pUC19-based construct that harbors the AtSUC3 promoter and the GFP open reading frame, which are separated by an in-frame fusion of the genomic AtSTP9 fragment to the N terminus of GFP. From pCL26, the AtSUC3-promoter::AtSTP9-GFP fragment (named AtSUC3-promoter::TM-GFP) was excised by HindIII and SacI and was inserted into the corresponding sites of the plant transformation vector pGPTV-bar (Becker et al., 1992).
Analysis of GUS Activity
The GUS plants used in these analyses were previously described by Meyer et al. (2000). For analyses of GUS activity in cell free extracts, the petioles of the five largest leaves of 4-week-old Arabidopsis plants were used (about 20 mg). Sections of 1 mm were incubated for 7 h in 50 mm Na-phosphate buffer, pH 7.0, 10 mm EDTA, and 0.1% (w/v) Triton X-100 at room temperature. After that, the sections were withdrawn and homogenized in a 1.5-mL reaction tube with a driller. The homogenate was mixed with 5 μL of GUS reaction buffer per milligram of plant material used (50 mm Na-phosphate buffer, pH 7.0, 10 mm EDTA, 0.1% [w/v] Triton X-100, 0.1% [w/v] SDS, and 10 mm β-mercaptoethanol). For control extracts, petioles were treated the same way with no extra incubation. Samples were vortexed, and, after 2 min of centrifugation, 70 μL of supernatant was mixed with 7 μL of MUG solution (10 mm MUG in water) on a microwell plate and was incubated at room temperature for 45 min. The fluorescence of methylumbelliferone was detected by UV excitation and was documented with a photo documentation system (IP-910.SD; Vilber Lourmat, Torcy, France). Quantitative analysis of the fluorescence intensity was performed using the software analySIS Doku 3.2 (Soft Imaging System, Münster, Germany).
Expression in Bakers' Yeast and Isolation of Yeast Plasma Membranes
The AtSUC3-expressing yeast cells (SMY36) and the control cells (SMYET1) used for qualitative characterization were described previously by Meyer et al. (2000). Plasma membranes were isolated according to the protocol of Stolz et al. (1994). Membrane proteins were solubilized with SDS and were separated on SDS-polyacrylamide gels as published by Laemmli (1970). Protein transfers to nitrocellulose and western-blot analyses were performed according to the protocol of Dunn (1986). The nitrocellulose was incubated overnight in affinity-purified anti-AtSUC3 antiserum-2 (diluted 1:500 in blocking buffer). Binding of antibodies to AtSUC3 protein was detected by treatment of the filter with anti-rabbit IgG antiserum-peroxidase conjugate (diluted 1:4,000 in blocking buffer) followed by incubation with Lumi-Light western Blotting Substrate (Roche Diagnostics, Mannheim, Germany).
Immunohistochemical Techniques
The anti-AtSUC3 antiserum-2 used in this paper was raised in rabbits (Pineda-Antikörper-Service, Berlin) against a synthetic peptide (SVPYRNLRKEIELET; corresponding to amino acids 8–22 from the AtSUC3 protein sequence) that had been coupled to a carrier protein.
Arabidopsis tissue was prepared, fixed in methacrylate, sectioned, and transferred to adhesion microscope slides (Linaris, Wertheim, Germany) as previously described by Stadler and Sauer (1996). Methacrylate was removed by incubation of the slides for 3 min in 100% (w/v) acetone. Sections were rehydrated by sequential incubation in ethanol of decreasing concentrations (100%, 95%, 80%, 60%, and 30%, all w/v), blocked for 1 h (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, and 1% [w/v] skim milk powder), and incubated overnight with affinity-purified anti-AtSUC3 antiserum-2 (dilutions 1:10 or 1:50 in blocking buffer), anti-PmSUC1 antiserum (diluted 1:50 in blocking buffer), monoclonal RS6 antibody (diluted 1:5 in blocking buffer), or combinations of these. After five washes with blocking buffer, sections were incubated for 1 h with anti-rabbit IgG-FITC-isomer 1-conjugate (diluted 1:300), with Cy3 goat anti-mouse IgG (diluted 1:200), with anti-mouse IgG-TRITC-isomer conjugate (diluted 1:100), or with combinations of these (all second antibodies from Sigma-Aldrich, Deisenhofen, Germany). After five final washes with blocking buffer, the slides were rinsed with water and were mounted in antifading medium (for FITC and TRITC: ProLong Antifade kit; Molecular Probes, Leiden, The Netherlands; and for Cy3: Vectashield-Mounting Medium; Vector Laboratories, Burlingame, CA).
For antibody-peptide competition experiments, a conjugate of the specific peptide with ovalbumin was used. Before immunolocalization, the affinity-purified antiserum was incubated for 2 to 3 h at room temperature with 200 μg mL–1 conjugate or pure ovalbumin, respectively.
RS6 is a monoclonal antibody that was made by injecting BALB/c mice with phloem SEs from Streptanthus tortuosus (Brassicacea). The SEs were raised in plant tissue cultures, and enriched fractions of SEs were obtained by digesting the callus parenchyma cell walls, and then collecting the undigested SEs from the callus cultures by filtration and centrifugation. The SE fractions were used to immunize BALB/c mice, and the spleen was removed from the mouse to prepare hybridomas by fusion with NS-1 myeloma cells. The resulting population of hybridomas was screened to identify those secreting phloem-specific monoclonal antibodies by using stem sections of S. tortuosus and light-level immunofluorescence microscopy. Hybridoma colonies that secreted SE-specific (as determined by fluorescent light microscopy) antibodies were further subcultured by limiting dilution to obtain monocellular hybridoma colonies. RS6 was determined to be specific for the SE plasma membrane of several species in the family Brassicacea (including Arabidopsis).
Epifluorescence and Confocal Laser Scanning Microscopy
Images of GFP fluorescence were made with a CLSM (Leica TCS SP II; Leica Microsystems, Bensheim, Germany) using a 100-mW argon laser (488 nm) and detection windows from 497 to 526 nm (for the detection of GFP), from 571 to 623 nm (for visualization of cell wall autofluorescence), and from 682 to 730 nm (for visualization of chlorophyll). Where indicated, an epifluorescence microscope (Zeiss Axioskop; Carl Zeiss, Jena, Germany) or stereomicroscopes (Zeiss SV11; Carl Zeiss, or Leica MZFLIII; Leica Microsystems) was used with an excitation wavelength of 460 to 500 nm. Emitted fluorescence was monitored at detection wavelengths longer than 510 nm.
Wounding of Plant Material, RNA Isolation, and RT-PCR Analyses
Arabidopsis inflorescence stems were cut into 2- to 3-mm sections that were incubated in 50 mm Na+-phosphate buffer (pH 5.0) on a shaker at 21°C. At indicated time points, material was withdrawn and used for total RNA preparations with the Trizol reagent from Invitrogen (Carlsbad, CA). RT-PCRs were performed using the following pairs of primers: AtSUC3:SUC3–3: 5′-GCGCGAATTCACATGCTCTACTAGTTAAAAAAGG-3′; SUC3–5 m: 5′-CTGTGAATTCAGGACGATGAGTGACTCGG-3′; AtSTP4: STP4t5: 5′-GTTCGGAACTACAATTACAAACTGACACC-3′; STP4t3: 5′-GTTGTTGAAAGCTCCTCTTAGATTCGG-3′; AtACT1: AtAct1–5′: 5′-GCGATGAAGCTCAATCCAAACGAG-3′; and AtAct1–3′: 5′-GGTCACGACCAGCAAGATCAAGAC-3′.
Distribution of Materials
Upon request, all novel material described in this publication will be made available in a timely manner for noncommercial research purposes.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.033399.
This work was supported by the Deutsche Forschungsgemeinschaft (grant no. SPP 1108–Sa 382/12–1).
References
- Aoki N, Hirose T, Scofield GN, Whitfeld PR, Furbank RT (2003a) The sucrose transporter gene family in rice. Plant Cell Physiol 44: 223–232 [DOI] [PubMed] [Google Scholar]
- Aoki N, Hirose T, Takahashi S, Ono K, Ishimura K, Ohsugi R (2002) Molecular cloning and expression analysis of a gene for a sucrose transporter in maize (Zea mays L.). Plant Cell Physiol 40: 1072–1078 [DOI] [PubMed] [Google Scholar]
- Aoki N, Whitfeld P, Hoeren F, Scofield G, Newell K, Patrick JW, Offler CE, Clarke B, Rahman S, Furbank RT (2003b). Three sucrose transporter genes are expressed in the developing grain of hexaploid wheat. Plant Mol Biol 50: 453–462 [DOI] [PubMed] [Google Scholar]
- Barker L, Kühn C, Weise A, Schulz A, Gebhardt C, Hirner B, Hellmann H, Schulze W, Ward JM, Frommer WB (2000) SUT2, a putative sucrose sensor in sieve elements. Plant Cell 12: 1153–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth I, Meyer S, Sauer N (2003) PmSUC3: characterization of a SUT2/SUC3-type sucrose transporter from Plantago major. Plant Cell 15: 1375–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker D, Kemper E, Schell J, Masterson R (1992) New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol Biol 20: 1195–1197 [DOI] [PubMed] [Google Scholar]
- Davies C, Wolf T, Robinson SP (1999) Three putative sucrose transporters are differentially expressed in grapevine tissues. Plant Sci 147: 93–100 [Google Scholar]
- Dunn SD (1986) Effects of the modification of transfer buffer composition on the renaturation of proteins in gels on the recognition of proteins on Western blots by monoclonal antibodies. Anal Biochem 157: 144–153 [DOI] [PubMed] [Google Scholar]
- Fotopoulos V, Gilbert MJ, Pittman JK, Marvier AC, Buchanan AJ, Sauer N, Hall JL, Williams LE (2003) The monosaccharide transporter gene, AtSTP4, and the cell-wall invertase, Atβfruct1, are induced in Arabidopsis during infection with the fungal biotroph Erysiphe cichoracearum. Plant Physiol 132: 821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahrtz M, Schmelzer E, Stolz J, Sauer N (1996) Expression of the PmSUC1 sucrose carrier gene from Plantago major L. is induced during seed development. Plant J 9: 93–100 [DOI] [PubMed] [Google Scholar]
- Gottwald JR, Krysan PJ, Young JC, Evert RF, Sussman MR (2000) Genetic evidence for the in planta role of phloem-specific plasma membrane sucrose transporters. Proc Natl Acad Sci USA 97: 13979–13948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D (1983) Studies on transformation of E. coli with plasmids. J Mol Biol 166: 557–580 [DOI] [PubMed] [Google Scholar]
- Hirose T, Imaizumi N, Scofield GN, Furbank RT, Ohsugi R (1997) cDNA cloning and tissue specific expression of a gene for a sucrose transporter from rice (Oryza sativa L.). Plant Cell Physiol 38: 1389–1396 [DOI] [PubMed] [Google Scholar]
- Holsters M, Silva B, Van Vliet F, Genetello C, De Block M, Dhaese P, Depicker A, Inze D, Engler G, Villarroel R et al. (1980) The functional organization of the nopaline Agrobacterium tumefaciens plasmid pTiC58. Plasmid 3: 212–230 [DOI] [PubMed] [Google Scholar]
- Imlau A, Truernit E, Sauer N (1999) Cell-to-cell and long distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink tissues. Plant Cell 11: 309–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsson N, Varshavsky A (1994) Split-ubiquitin as a sensor of protein interactions in vivo. Proc Natl Acad Sci USA 91: 10340–10344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koroleva OA, Davies A, Deeken R, Thorpe MR, Tomos AD, Hedrich R (2000) Identification of a new glucosinolate-rich cell type in Arabidopsis flower stalk. Plant Physiol 124: 599–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn C (2003) A comparison of the sucrose transporter systems of different plant species. Plant Biol 5: 215–232 [Google Scholar]
- Kühn C, Franceschi VR, Schulz A, Lemoine R, Frommer WB (1997) Macromolecular trafficking indicated by localization and turnover of sucrose transporters in enucleate sieve elements. Science 275: 1298–1300 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lalonde S, Boles E, Hellmann H, Barker L, Patrick JW, Frommer WB, Ward JM (1999) The dual function of plant sugar carriers: transport and sugar sensing. Plant Cell 11: 707–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S, Truernit E, Hümmer C, Besenbeck R, Stadler R, Sauer N (2000) AtSUC3, a gene encoding a new Arabidopsis sucrose transporter, is expressed in cells adjacent to the vascular tissue and in a carpel cell layer. Plant J 24: 869–882 [DOI] [PubMed] [Google Scholar]
- Oparka KJ, Duckett CM, Prior DAM, Fisher DB (1994) Real-time imaging of phloem unloading in the root tip of Arabidopsis. Plant J 6: 759–766 [Google Scholar]
- Oparka KJ, Roberts AG, Boewink P, Santa Cruz S, Roberst I, Pradel K, Imlau A, Kotlizky G, Sauer N, Epel B (1999) Simple, but not branched, plasmodesmata allow the nonspecific trafficking of proteins in developing tobacco leaves. Cell 97: 1–20 [DOI] [PubMed] [Google Scholar]
- Reinders A, Schulze W, Kühn C, Barker L, Schulz A, Ward JM, Frommer WB (2002) Protein-protein interactions between sucrose transporters of different affinities colocalized in the same enucleate sieve element. Plant Cell 14: 1567–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesmeier JW, Willmitzer L, Frommer WB (1992) Isolation and characterization of a sucrose carrier cDNA from spinach by functional expression in yeast. EMBO J 11: 4705–4713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritte G, Rosenfeld J, Rohrig K, Raschke K (1999) Rates of sugar uptake by guard cell protoplasts of Pisum sativum L. related to the solute requirement for stomatal opening. Plant Physiol 121: 647–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidereit A, Scholz-Starke J, Büttner M (2003) Functional characterization and expression analyses of the glucose-specific AtSTP9 monosaccharide transporter in pollen of Arabidopsis. Plant Physiol 133: 182–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz-Starke J, Büttner M, Sauer N (2003) AtSTP6, a new pollen-specific H+-monosaccharide symporter from Arabidopsis. Plant Physiol 131: 70–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze W, Weise A, Frommer WB, Ward JM (2000) Function of the cytosolic N-terminus of sucrose transporter AtSUC2 in substrate affinity. FEBS Lett 485: 189–194 [DOI] [PubMed] [Google Scholar]
- Schulze WX, Reinders A, Ward J, Lalonde S, Frommer WB (2003) Interactions between co-expressed Arabidopsis sucrose transporters in the splitubiquitin system. BMC Biochem 18: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwebel-Dugue N, el Mtili NE, Krivitzky M, Jean-Jacques I, Williams JH, Thomas M, Kreis M, Lecharny A (1994) Arabidopsis gene and cDNA encoding cell wall invertase. Plant Physiol 104: 809–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler R, Brandner J, Schulz A, Gahrtz M, Sauer N (1995) Phloem loading by the PmSUC2 sucrose carrier from Plantago major occurs into companion cells. Plant Cell 7: 1545–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler R, Büttner M, Ache P, Hedrich R, Ivashikina N, Melzer M, Shearson SM, Smith SM, Sauer N (2003) Diurnal and light-regulated expression of AtSTP1 in guard cells of Arabidopsis. Plant Physiol 133: 525–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler R, Sauer N (1996) The Arabidopsis thaliana AtSUC2 gene is specifically expressed in companion cells. Bot Acta 109: 299–306 [Google Scholar]
- Stadler R, Truernit E, Gahrtz M, Sauer N (1999) The AtSUC1 sucrose carrier may represent the osmotic driving force for anther dehiscence and pollen tube growth in Arabidopsis. Plant J 19: 269–278 [DOI] [PubMed] [Google Scholar]
- Stolz J, Stadler R, Opekarová M, Sauer N (1994) Functional reconstitution of the solubilized Arabidopsis thaliana STP1 monosaccharide-H+ symporter in lipid vesicles and purification of the histidine tagged protein from transgenic Saccharomyces cerevisiae. Plant J 6: 225–233 [DOI] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Truernit E, Schmid J, Epple P, Illig J, Sauer N (1996) The sink-specific and stress-regulated Arabidopsis STP4 gene: enhanced expression of a gene encoding a monosaccharide transporter by wounding, elicitors, and pathogen challenge. Plant Cell 8: 2169–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E, Stadler R, Baier K, Sauer N (1999) A male gametophyte-specific monosaccharide transporter in Arabidopsis. Plant J 17: 191–201 [DOI] [PubMed] [Google Scholar]
- Weise A, Barker L, Kühn C, Lalonde S, Buschmann H, Frommer WB, Ward JM (2000) A new subfamily of sucrose transporters, SUT4, with low affinity/high capacity is localized in enucleate sieve elements of plants. Plant Cell 12: 1345–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weschke W, Panitz R, Sauer N, Wang Q, Neubohn B, Weber H, Wobus U (2000) Sucrose transport into barley seeds: molecular characterization of two transporters and implications for seed development and starch accumulation. Plant J 21: 455–467 [DOI] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X et al. (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296: 79–92 [DOI] [PubMed] [Google Scholar]