Abstract
Tobacco smoking and reduced methylation of long interspersed element-1 (LINE-1) are crucial in oral carcinogenesis. 5′UTR of human LINE-1 sequence contains several CpG dinucleotides which are methylated in various proportions (0–100%). Methylation levels of many LINE-1s in cancer were reduced, hypomethylated. The hypomethylation of each LINE-1 locus can promote instability of genome and repress expression of a gene located on that same chromosome. This study investigated if cigarette smoking influences LINE-1 methylation of oral mucosal cells. The methylation of human LINE-1 in clinically normal oral mucosa of current smokers was compared to non-smokers. By using the combined bisulphite restriction analysis, each LINE-1 sequence was categorised into 4 patterns depending on the methylation status and location of the two 18-bp successive CpG from 5′ to 3′ including mCmC, uCuC, mCuC and uCmC. Of these, mC and uC represent methylated and unmethylated CpG, respectively. The DNA bisulphite sequence demonstrated that most CpGs of mCmC and uCuC were methylated and unmethylated, respectively. Nevertheless, some CpGs of each mCuC or uCmC allele were methylated. Imaging of the digestion products was used to generate %methylation value. No significant difference in the overall LINE-1 methylation level but the differences in percentages of some methylation patterns were discovered. The %mCmC and %uCuC increased, while the %mCuC decreased in current smokers (p = 0.002, 0.015, and <0.0001, respectively). Additionally, the lower %mCuC still persisted in persons who had stopped smoking for over 1 year (p = 0.001). The %mCuC also decreased in the higher pack-year smokers (p = 0.028). Smoking possibly altered mCuC to mCmC and uCuC forms, and changes uCmC to uCuC forms. In conclusion, smoking changes methylation levels of partial methylated LINE-1s and increased the number of hypo- and hypermethylated loci. These hypomethylated LINE-1s may possess carcinogenesis potential. Moreover, LINE-1 methylation patterns may be useful for monitoring oral carcinogenesis in smokers.
Introduction
Tobacco smoking is a predisposing factor of many malignancies [1]–[4]. The risk of upper aerodigestive cancers increases with the higher pack-years cigarette smoking [3], [5], [6]. However, this risk decreases after discontinuation of smoking and reverts to the non-smoker risk level if smoking is ceased for more than 15 years [3], [6]. Additionally, smoking increases the number of keratinised cells in the epithelium of the tongue and hard palate [7]. This effect varied in different regions, depending on the extent of direct exposure to smoke [8]. Interestingly, oral mucosal lesions resolved after cessation of smoking for a period of time [9], [10].
Mutations, promoter methylation and global hypomethylation are three crucial DNA modification events that lead to cancer development [11]–[13]. Smoking promotes mutations and alterations of gene promoter methylation [11], [14], [15]. Moreover, the evidence suggesting the association between the degree of global hypomethylation and smoking history of head and neck squamous cell carcinoma (HNSCC) patients was shown [16]. Long interspersed element-1s (LINE-1s) are repetitive transposable elements which are widely distributed in the genome [17]. There are 500,000 copies of LINE-1 in the human genome [18]. More than 10,000 LINE-1s contain a 5′UTR [19]. Most LINE-1 methylation studies, including this one, evaluated methylation at 5′UTR. The reduction of methylation levels of LINE-1, which reflects global hypomethylation, in various types of cancers had been extensively studied [20]. In most cancers, LINE-1 methylation levels diminish early and progressively which correlate significantly with tumour phenotype, including tumour progression and prognosis [12], [16], [21]–[26]. The hypomethylation of LINE-1 significantly increases the risk for HNSCC [27]. On the other hand, events associating LINE-1 hypermethylation with carcinogenesis have also been found in malignant peripheral nerve sheath tumor, myelodysplastic syndrome and partial hydatidiform moles [28]–[30].
In blood samples of HNSCC patients, LINE-1 methylation levels slightly increased with higher pack-years of smoking [27]. The effects of smoking on LINE-1 methylation levels in non-cancerous cells have also been reported. No changes were observed in blood cells or in the colonic epithelium of smokers in vivo [31]–[33]. However, an in vitro study revealed minimal reduction of LINE-1 methylation levels in the respiratory epithelium under high dosage cigarette smoke condensate treatment [34]. The oral mucosa is directly exposed to tobacco smoke and its chemical agents. Therefore, it is interesting to clarify whether this epigenetic change occurs before malignant transformation.
Currently, most LINE-1 methylation studies have measured the genome-wide methylation levels of LINE-1s. However, methylation of LINE-1s can be influenced by multiple mechanisms. The measurement of the methylation level alone may not be able to detect LINE-1 methylation changes in certain events, even if such changes can promote cancer development. In normal cells, some functions of LINE-1 methylation are to maintain genomic integrity and regulate gene expression in cis [20], [35]–[37]. Consequently, genomic instability and repression of gene expression can be observed on chromosomes in which LINE-1s are hypomethylated. Therefore, in theory, certain conditions that stochastically alter LINE-1 methylation levels will promote carcinogenesis on chromosomes with LINE-1 hypomethylation, but these hypomethylated LINE-1s may be undetectable because the methylation levels are counterbalanced by other hypermethylated LINE-1 loci. Locus-specific mechanisms causing variations in methylation levels among LINE-1s in different loci has also been reported [38].
Recently, a wide range of approaches to obtain quantitative information of genomic DNA methylation have been developed [39]. Most standard techniques measure several CpGs in each LINE-1. Pyrosequencing often measures 4 CpG dinucleotides [40], whereas combined bisulphite restriction analysis (COBRA) polymerase chain reaction often measures 2 CpGs [12]. Compared to previously reported LINE-1 sequences [38], the methylation state of 2 of the CpG dinucleotides detected by COBRALINE-1 correlated directly with other CpG dinucleotides on 5′LINE-1s. COBRA classifies LINE-1alleles into four groups depending on the methylation status of 2 CpG dinucleotides on each strand from 5′ to 3′ detected by COBRALINE-1. The first class contains 2 unmethylated CpGs (uCuC) and the second class contains 2 methylated CpGs (mCmC), representing hypomethylated and hypermethylated LINE-1 loci, respectively. The third and fourth classes are partially methylated LINE-1s including 5′methylated with 3′unmethylated CpGs (mCuC) and 5′ unmethylated with 3′methylated CpGs (uCmC) (Figure 1A and 1B). Recently, our group found that %uCuC is more effective in determining cancer risk than overall methylation levels [41]–[43].
Figure 1. Methylation patterns of COBRALINE-1.
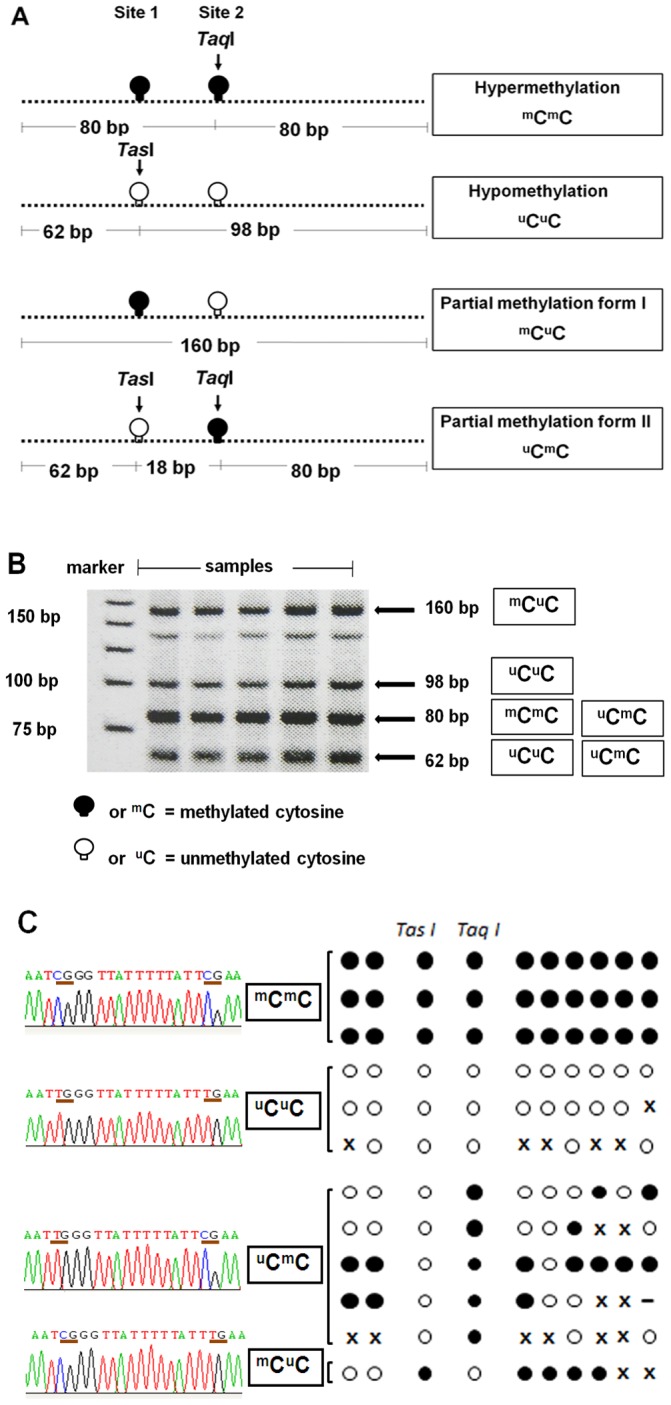
(A) The LINE-1 amplicons were 160 bp and had 2 CpG dinucleotides. Four patterns of methylated CpGs were detected, including hypermethylation (mCmC), hypomethylation (uCuC), and two forms of partial methylation (mCuC and uCmC). The TasI enzyme targets unmethylated cytosine site 1, and TaqI targets methylated cytosine site 2. (B) After restriction digestion with TasI and TaqI, four sizes of products (160, 98, 80, and 62 bp) were identified, depending on the methylation status of both CpG loci. (C) Examples of bisulfite sequencing. Left side represents sequences of the two CpG dinucleotides at TasI and TaqI cut site whereas the right side represents the CpG dinucleotides in the amplified PCR products. Each circle exemplifies the methylation status of each selected clone. Black and white circles are methylated and unmethylated CpG dinucleotides, respectively. x is mutated sequence and – is deleted sequence.
However, studies that evaluated the association between smoking and repetitive sequence methylation changes in vivo have not yet been conclusive. Herein, we evaluated the possibility that smoking may promote cancer development via genomic hypomethylation by evaluating the human LINE-1 methylation pattern found in the oral mucosa of smokers.
Results
Methylation Status of LINE-1 Methylation Patterns
Twenty CpG dinucleotides of COBRALINE-1 clones were analysed and compared with mCmC, uCuC, mCuC and uCmC patterns. Examples of bisulphite DNA sequencing were demonstrated (Figure 1C). Most of the CpG dinucleotides of mCmC were methylated. On the contrary, uCuC were enriched in unmethylated CpGs. Finally, all alleles of mCuC and uCmC were partially methylated (Figure 1C).
The Percentage of Each LINE-1 Methylation Pattern by Gender, Age and Alcohol Consumption
Oral rinses were collected from 60 non-smoker (NS) volunteers (35 males and 25 females) and 96 current smoker (CS) volunteers (80 males and 16 females) (Table 1). The imaging of the digestion products was used to generate %methylation value of each pattern. We evaluated if sex and age influenced LINE-1 methylation patterns. No significant difference in the percentage of LINE-1 products was detected between the males and females (Table S1). Furthermore, no correlation between each methylation pattern and age was found except a borderline increase in LINE-1 hypomethylation by age of CS group (p = 0.047, Table S2).
Table 1. Demographic and behavioral characteristics of the subjects.
| Non-smokers | Current smokers | Former smokers | ||
| Total Subjects | 60 | 96 | 17 | |
| Gender | Male | 35 (58.33%) | 80 (83.33%) | 15 (88.24%) |
| Female | 25 (41.67%) | 16 (16.67%) | 2 (11.76%) | |
| Age | Mean ± SD | 44.63±14.19 | 41.60±4.60 | 46.59±16.39 |
| History of alcohol drinking | Currently drink | 36 (60%) | 86 (89.58%) | 14 (82.35%) |
| Previously drink | - | - | 3 (17.65%) | |
| Never drink | 24 (40%) | 10 (10.42%) | - | |
| History of betel chewing | Currently chew | - | - | - |
| Previously chew | - | - | - | |
| Never chew | - | - | - |
Most of the CS subjects in this study were also alcohol drinkers (Table 1). We found that in NS group, only %mCuC was significantly lower in the current drinker than in the never drink (p = 0.006). For CS group, only overall methylation level was higher in current drinker (p = 0.005, Table 2). However, the association between smoking and alcohol consumption and its contribution to malignant potency has not been completely elucidated. Therefore, before analysing the possible impact of smoking, we assessed the interaction between alcohol and smoking on every LINE-1 methylation patterns by using two-way ANOVA. No interactions between alcohol and smoking consumption were found for any of the patterns (p>0.05). Accordingly, the smoking factor could be independently studied, while controlling for alcohol consumption. The percentages of all the patterns are presented in Table S1.
Table 2. The percentages of LINE-1 products between never drinks and current drinkers.
| Non-smokers | Current smokers | |||||
| Never drink | Currently drink | p-valuea | Never drink | Currently drink | p-valueb | |
| Number of subjects | 24 | 36 | 10 | 86 | ||
| % mC (mean ± SD) | 41.82±1.99 | 41.56±2.99 | 0.71 | 40.66±1.00 | 42.23±2.65 | <0.01 |
| % mCmC (mean ± SD) | 15.89±2.87 | 14.81±5.83 | 0.52 | 15.61±4.64 | 17.87±4.74 | 0.23 |
| %uCuC (mean ± SD) | 32.24±2.91 | 31.70±2.78 | 0.54 | 34.29±5.98 | 33.41±3.49 | 0.71 |
| % mCuC (mean ± SD) | 28.41±2.69 | 25.82±3.72 | <0.01 | 24.28±4.89 | 24.11±3.56 | 0.91 |
| % uCmC (mean ± SD) | 23.47±4.34 | 27.67±8.81 | 0.11 | 25.81±9.20 | 24.61±7.71 | 0.70 |
| % mCuC+uCmC (mean ± SD) | 51.88±4.19 | 53.49±6.91 | 0.29 | 50.10±10.51 | 48.72±6.42 | 0.61 |
t-test was used to compare the percentage of LINE-1 products between never drink and currently drink of non-smokers.
t-test was used to compare the percentage of LINE-1 products between never drink and currently drink of current smokers.
The Percentages of Loci of Each LINE-1 Methylation Pattern in NS and CS were Different
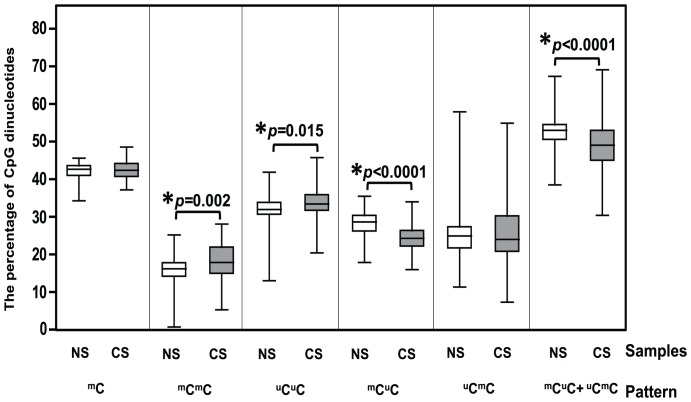
Here we compared LINE-1 methylation between CS and NS. The CS had significantly higher %mCmC and %uCuC and lower %mCuC and %mCuC+uCmC than the NS (p = 0.002, 0.015, <0.0001, and <0.0001, respectively). However, no significant difference was found in overall methylation (%mC) and %uCmC (p = 0.33 and 0.84, respectively, Figure 2 and Table S1).
Figure 2. Percentage of each methylated CpG pattern in NS and CS.
mC represents the overall methylation level of the amplified LINE-1s. mCmC and uCuC represent hypermethylated and hypomethylated CpG of the amplified region, respectively. mCuC and uCmC represent two forms of partial methylation. mCuC+uCmC is the sum of partially methylated loci of both forms. The horizontal line within each box indicates the mean of the percentage. Stars indicate statistical significance at p<0.05. The results demonstrated that the CS had a significantly higher %mCmC and %uCuC and a lower %mCuC and %mCuC+%uCmC than the NS.
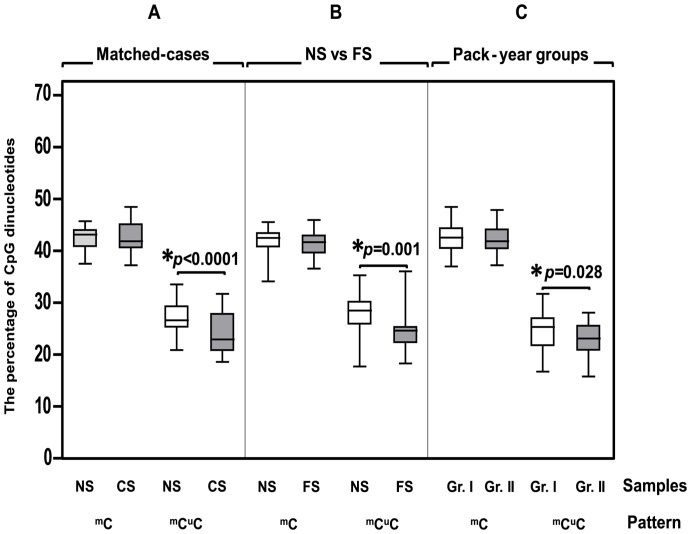
To account for potential confounding by age, gender and alcohol consumption, the NS subjects were matched to the CS subjects based on age, gender and alcohol drinking behaviour, which produced 29 pairs. The same tendency of differences in the LINE-1 methylation patterns as those found in the total sample was found. However, only %mCuC resulted in a significant difference at p<0.0001 (Figure 3A and Table S1).
Figure 3. %mCuC in the matched cases, FS, and pack-year groups.
mCuC represented %mCuC. The percentage of overall LINE-1 methylation level was depicted as mC. (A) To account for potential confounding factors, we matched the NS to the CS based on age, gender and alcohol drinking. The CS showed a significantly lower %mCuC. (B) The FS also had a significantly lower %mCuC. (C) %mCuC was significantly lower in the higher pack-year smoking (group II) than in lower pack-year group (group I).
Smoking Influenced the Methylation of LINE-1s
To study the persisting effect of smoking, an additional investigation was performed on 17 former smokers (FS),15 males and 2 females, who had quit smoking for no less than 1 year and who had no mucosal lesions. We found that the FS had a significant lower level of %mCuC than the NS (p = 0.001) while other patterns did not reveal any significant differences (Figure 3B and Table S1). This result implied the prolong effect of smoking on oral mucosa.
Based on the intensity of smoking, all of the smoking subjects were categorised into 2 groups based on the average pack-year (group I≤13.23and group II>13.23 pack-years). Only %mCuC was significantly different between the groups; %mCuC was significantly lower in group II, p = 0.028 (Figure 3C and Table S3). This result demonstrated the influence of smoking intensity on oral mucosa.
The Possible Direction of LINE-1 Methylation Pattern Changes
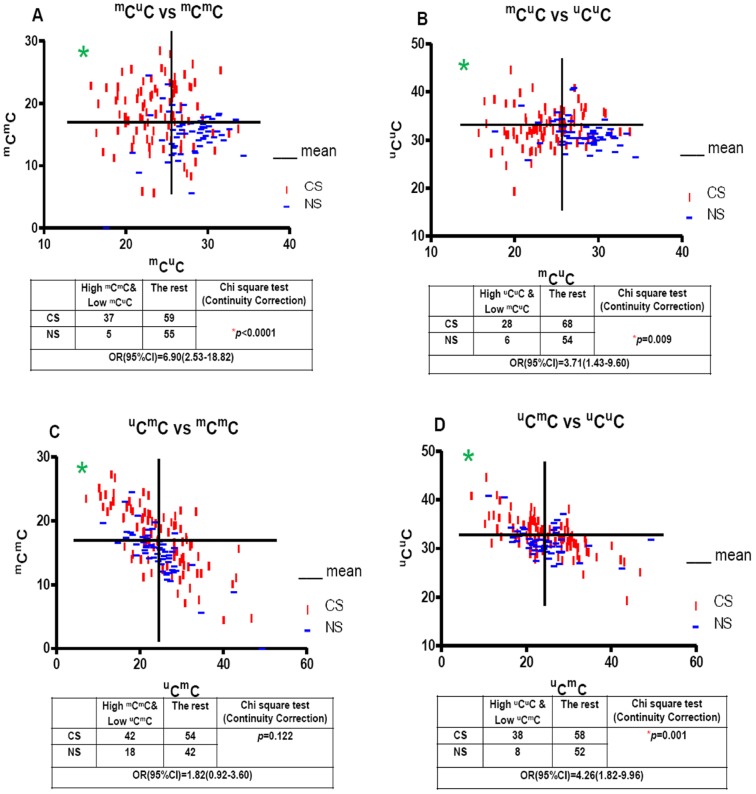
Encouraged by the information that the change in mCuC was opposite that of mCmC and uCuC, we checked whether the bivariate distributions of hetero-methylated/homo-methylated allelic proportions differed by smoking status. The number of CS who had a lower %mCuC concomitantly a higher %mCmC than the group means were counted and compared with the remainder of the group, also those of NS using the chi-squared test. We found that the odds ratio (OR) was 6.90 and the 95% confidential interval (CI) was 2.53–18.82 with p<0.0001 (Figure 4A). We performed the same test for the proportion of low mCuC/high uCuC, the OR and 95% CI were 3.71 and 1.43–9.60, respectively with p = 0.009 (Figure 4B). These results implied that a reduction of mCuC in the CS was associated with an increase in mCmC or uCuC. Moreover, to clarify the possibility that the reduction of uCmC is related to the increase in mCmC or uCuC in CS, the same analysis was performed in the group with low uCmC/high either mCmC or uCuC. Though elevated in CS, the proportion of low uCmC/high mCmC subjects was not statistically different between CS and NS individuals (OR = 1.82, 95% CI = 0.92–3.60, p = 0.122, Figure 4C), while CS had a higher proportion of low uCmC/high uCuC relative to NS (OR = 4.26, 95% CI = 1.82–9.96, p = 0.001, Figure 4D). This result suggested that the reduction of uCmC might associate with the increase in uCuC but not mCmC.
Figure 4. The possible direction of LINE-1 methylation pattern changes.
mCuC, uCmC, mCmC and uCuC, represented %mCuC, %uCmC, %mCmC and %uCuC, respectively. The graphs were plotted for the percentages of either mCuC or uCmC on the X-axis and either mCmC or uCuC on the Y-axis. The vertical and horizontal lines indicate the mean percentages of each axis. The graph is divided into 4 quadrants. The numbers of NS and CS who fell in the upper left quadrant were counted and compared to the remainder of the group using the chi-squared test. The results are shown in the tables below the graphs.
Discussion
This study confirms that COBRALINE-1 does not only analyse the overall methylation level, but it is also able to show the methylation pattern of LINE-1s.We found that uCuC and mCmC represent the hypomethylation and hypermethylation of LINE-1, respectively. Moreover, mCuC and uCmC also represent partial methylation.
Although we did not find differences in LINE-1 methylation level by age and gender, the influences by these two factors in blood cells were demonstrated [44]–[46]. However, the degrees of differences by ages and gender were minimal when comparing with differences by environmental insults or diseases [12], [47], [48]. Therefore, the differences by age and gender could be found only when large cohorts were studied. Here, although the relationship of age and LINE-1 methylation pattern of oral mucosa was not found in this study, further investigation should be performed in larger sample size and in a variety of age.
Similar to blood cells and colonic epithelium [31]–[33], cigarette smoke does not change overall LINE-1 methylation level of oral mucosa. However, there are alterations in patterns of LINE-1 methylation that we found both uCuC and mCmC loci increased. The unchanged overall methylation level can be explained by the fact that the LINE-1 methylation level is a sum of the methylation from all the LINE-1s. Therefore, the increases in both the uCuC and mCmC loci counterbalance each other, neutralising their effect on the overall LINE-1 methylation levels. This evidence supported by our previous study, which revealed that the overall LINE-1 methylation level measurement was not sufficiently sensitive or accurate to determine the LINE-1 methylation changes in pathological conditions [20]. We recommend re-evaluating LINE-1 methylation pattern to most of the previous studies reported unaltered overall methylation level.
Contrary to the reduction of genome-wide methylation levels caused by some chemical agents [49], [50], smoking could promote both an increase and decrease methylation in certain LINE-1s. Interestingly, while smoking-induced uCuC, hypomethylated LINE-1 loci, possibly originated from both forms of partially methylated LINE-1(mCuC and uCmC), the mCmC might derive from only one partially methylated form, mCuC. These observations suggest that the mechanisms which increase or decrease methylation are different. The methylation loss mechanism seems to be a generalised process that affects many LINE-1s regardless of the original methylation patterns and is similar to the global hypomethylation found in cancer [20], [24], [38]. Accordingly, cancer and smoking may reduce genome-wide methylation by the same mechanism.
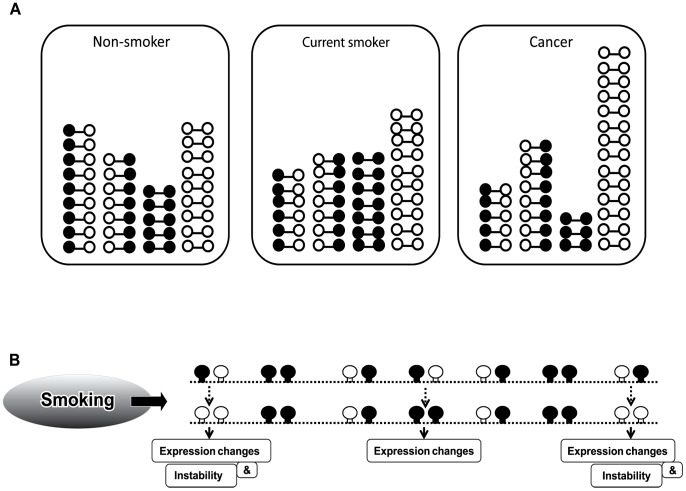
Even though the methylation differences between smokers and non-smokers are just a few percentage points difference, the alteration may play a role in carcinogenesis. Global hypomethylation can cause cancer by promoting genomic instability and by altering gene expression in cis, on that same chromosome [20]. There are evidences suggesting that DNA methylation maintains genomic integrity in cis. First, a close correlation between the site of the chromosome translocation and the loss of the methylation of satellite DNA has been reported [51], [52]. Recently, we found that the repair of the replication of independent DNA double-strand breaks occurring within hypomethylated regions was more error prone [36]. For gene expression, we reported the repression of mRNA production by hypomethylated intragenic LINE-1s [37]. Therefore, the increasing number of hypomethylated LINE-1 loci, which implies the hypomethylation, induced by smoking should promote cancer at certain loci in cis (Figure 5).
Figure 5. The influence of smoking on the epigenetic progression of multistep carcinogenesis.
(A) Models of LINE-1 methylation patterns in oral mucosal cells of a NS, the oral mucosal cells of a CS and in cancer cells (HeLa) are shown. Although the overall methylation level did not change in the CS, some alterations in the methylation patterns were detected. While the numbers of mCmC and uCuC were increased, only one form of partial methylation, mCuC, was decreased. Moreover, the addition of mCmC and uCuC correlated with the depletion of mCuC. In contrast, a reduction in the overall methylation level was found in cancer cells. The numbers of mCmC and mCuC were significantly decreased, while the numbers of uCuC were significantly increased. The numbers of uCmC were slightly increased. (B) The smoking-induced hypomethylated loci could be derived from both classes of the partial methylation patterns and could result in genome instability and gene expression changes. However, the smoking-induced hypermethylated loci were from mCuC only and, consequently, effected gene expression.
Although no interaction between alcohol consumption and cigarette smoking was found in LINE-1 methylation, similar to a report in colon [53], the result revealed some significant change in alcohol drinkers. When there is no effect of smoking, the oral mucosa of drinkers has lower %mCuC than never drink persons. For the persons who drink and smoke, the overall methylation level of LINE-1s increases while the patterns are not affected. This information also needs further investigation of the result of chemical exposure to oral mucosa.
In conclusion, smoking alters LINE-1 methylation by increasing or decreasing methylation of certain loci. The mechanisms causing LINE-1 loss or gain methylation are different. The alteration can persist after stop smoking. Moreover, the higher intensity of smoking results in the higher alteration degree. Further exploration of methylation pattern changes of other intersperse repetitive sequences and gene promoters whether they are related to other smoking-associated malignancies, as well as other carcinogens, is necessary. Finally, a better understanding of the causes and mechanisms of genome-wide methylation changes will be crucial for cancer prevention.
Materials and Methods
Ethics Statement
The Ethics Committee of the Faculty of Dentistry, Chulalongkorn University approved this study (approval number: 7/2010). Written informed consent was obtained from every participant to the study.
Oral Mucosal Cell Collection
The patients who received dental treatment in the Faculty of Dentistry during December 2010 to May 2011 were voluntarily enrolled. After oral examination and history taking, the patients who had no oral mucosal lesion and no history of malignancy of any organs were recruited in this study. The number and duration of cigarette smoking were recorded if they had the smoking history. The alcohol consumption was also recorded as never or ever drinks. CS included the patients who had still smoked at the time of interview. NS included the patients who had never smoked. FS, the patients who quit smoking longer than 1 year were also included in this study. Oral epithelia were collected from oral rinses. Ten millilitres of sterile 0.9% normal saline solution was gargled for 15 seconds. This solution was kept in a sterile tube and stored at 4°C until the DNA extraction process.
Genomic DNA Extraction
After oral rinses were centrifuged at 4°C, 2500 g for 15 minutes, the supernatant was discarded. The cell pellets were washed twice in sterile PBS. One millilitre of the DNA extraction buffer with 10% SDS and proteinase K (0.5 mg/ml) was added to the cell pellets. The mixtures were then incubated at 50°C for two nights. A phenol-chloroform extraction was used to purify and desalt the digested cell pellets. After centrifuging at 4°C, 14000 g for 15 minutes, 10 M ammonium acetate and cold absolute ethanol were added to the upper aqueous phase for DNA precipitation. The precipitated DNA was washed with 70% ethanol. The air-dried DNA was then resuspended in Tris-EDTA-treated water.
COBRALINE-1
COBRA for LINE-1 was performed as previously described, the 5′UTR of LINE-1.2 (L1Hs) sequence from NCBI Accession Number M80343 was used [12]. In brief, the DNA samples were converted by a bisulphite reaction such that unmethylated cytosine (uC) would be converted to uracil (U), whereas methylated cytosine (mC) would remain as cytosine. One microlitre of bisulphite DNA was then subjected to 35 cycles of PCR, at a 50°C annealing temperature using the following primer sets: LINE-1-F (5′-CCGTAAGGGGTTAGGGAGTTTTT-3′) and LINE-1-R (5′-RTAAAACCCTCCRAACCAAATATAAA-3′). The LINE-1 amplicons (160 bp) were digested with 2 U of TaqI and 2 U of TasI in NEB3 buffer (New England Biolabs, Ontario, Canada) at 65°C overnight. The products were identified by polyacrylamide gel electrophoresis (8% non-denaturing) and stained with SYBR green nucleic acid stain (Sigma-Aldrich, St. Louis, Missouri). Distilled water was used as a negative control. The same preparation of DNA from 3 cell lines, HeLa (cervical cancer), Daudi (Human Burkitt’s lymphoma), and Jurkat (acute T cell leukemia) (ATCC, Manassas, VA, USA) were used as positive controls in all experiments and for inter-assay variation adjustment [41].
COBRALINE-1 Product Analysis and Bisulphite DNA Sequencing
Here, we classified LINE-1s into four groups depending on the methylation status of 2 CpG dinucleotides on each strand from 5′ to 3′ detected by COBRALINE-1 as described previously [42]. These COBRA-detected LINE-1s were categorised into the following four classes: 2 unmethylatedCpGs (uCuC), 2 methylated CpGs (mCmC), 5′methylated and 3′unmethylated CpGs (mCuC), or 5′unmethylated and 3′methylated CpGs (uCmC) (Figure 1A). LINE-1 methylation levels and the percentage of loci of each class were calculated from COBRALINE-1 digested products. Intensities of COBRALINE-1 bands were measured by a phosphoimager using ImageQuant Software (Molecular Dynamics, GE Healthcare, Slough, UK). After enzymatic digestion, the COBRALINE-1 amplicons were separated into 5 DNA strands depending on their length, 160, 98, 80, 62, and 18 bp (Figure 1B). The 18 bp band was not used in the following calculation. The 160 bp band contains 2 CpGs, in which the 5′CpG is methylated and the other 3′CpG is unmethylated. The 98 bp band contains 2 unmethylated CpGs. The 80 bp and 62 bp bands each contain 1 methylated and 1 unmethylated CpG. The CpGs of the 160 bp and 98 bp bands were derived from mCuC and uCuC, respectively. The CpGs of the 80 bp band were derived from 3′methylated CpGs of mCmC and uCmC, respectively, while the CpGs of the 62 bp band were derived from 5′unmethylated CpGs of uCuC and uCmC (Figure 1). To normalise each band to represent the total number of CpG dinucleotides present, the intensity of each band was divided by the number of base pairs of double stranded DNA as follows: %160/160 = A, %98/94 = B, %80/78 = C, and %62/62 = D. Then, the LINE-1 methylation levels were computed with the following formula: percentage of overall LINE-1 methylation level (%mC) = 100×(C+A)/(C+A+A+B+D), percentage number of mCuC loci (%mCuC) = 100×(A)/(((C-D+B)/2)+A+D), %uCmC = 100×(D-B)/((C-D+B)/2)+A+D, %uCuC = 100×B/(((C-D+B)/2)+A+D), and %mCmC = 100×((C-D+B)/2)/(((C-D+B)/2)+A+D).
To analyse methylation status of each LINE-1 methylation pattern, COBRALINE-1 PCR products were cloned into the pGEM-T easy vector (Promega, Santhan, UK) and sequenced.
Statistical Analysis
Statistical analyses were performed using SPSS version 17.0 (SPSS Inc., Chicago, IL). All p-values less than 0.05 were considered significant. All the variables were normally distributed (Kolmogorov-Smirnov Test). We used a two-way analysis of variance (ANOVA) to determine the effects of two factors, alcohol and smoking, on the methylation levels of LINE-1. The independent sample t-test was performed to compare each LINE-1 methylation pattern between males and females, NS and CS, NS and FS, the 2 pack-year groups also never drinks and current drinker. In addition, the paired t-test was used for a matched-case analysis. The relationship of age and LINE-1 methylation was investigated with Pearson’s correlation. The chi-squared test and OR were used to test the association among LINE-1 methylation variables in CS and NS.
Supporting Information
The percentage of LINE-1 products in smokers and non-smokers.
(DOC)
Relationship of the percentage of LINE-1 products with age status.
(DOC)
Demographic characteristics and the percentage of LINE-1 products in pack-year smoking groups.
(DOC)
Funding Statement
This study was financially supported by the 90th Anniversary of Chulalongkorn University Fund (Ratchadaphiseksomphot Endowment Fund). KS is supported by the Grant for Development of New Faculty Staff (Ratchadapiseksomphot Endowment Fund) and the Thailand Research Fund (Young Scientific Researcher Grant No. MRG5380176). Apiwat Mutirangura is supported in part by the Research Chair Grant 2011, the National Science and Technology Development Agency, Thailand, the Four Seasons Hotel Bangkok's 4th Cancer Care charity fun run in coordination with the Thai Red Cross Society and Chulalongkorn University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Olshan AF, Weissler MC, Watson MA, Bell DA (2000) GSTM1, GSTT1, GSTP1, CYP1A1, and NAT1 polymorphisms, tobacco use, and the risk of head and neck cancer. Cancer Epidemiol Biomarkers Prev 9: 185–191. [PubMed] [Google Scholar]
- 2. DeMarini DM (2004) Genotoxicity of tobacco smoke and tobacco smoke condensate: a review. Mutat Res 567: 447–474. [DOI] [PubMed] [Google Scholar]
- 3. Schlecht NF, Franco EL, Pintos J, Kowalski LP (1999) Effect of smoking cessation and tobacco type on the risk of cancers of the upper aero-digestive tract in Brazil. Epidemiology 10: 412–418. [DOI] [PubMed] [Google Scholar]
- 4. Spitz MR, Wei Q, Li G, Wu X (1999) Genetic susceptibility to tobacco carcinogenesis. Cancer Invest 17: 645–659. [DOI] [PubMed] [Google Scholar]
- 5. Macfarlane GJ, Zheng T, Marshall JR, Boffetta P, Niu S, et al. (1995) Alcohol, tobacco, diet and the risk of oral cancer: a pooled analysis of three case-control studies. Eur J Cancer B Oral Oncol 31B: 181–187. [DOI] [PubMed] [Google Scholar]
- 6. Spitz MR, Fueger JJ, Goepfert H, Hong WK, Newell GR (1988) Squamous cell carcinoma of the upper aerodigestive tract. A case comparison analysis. Cancer 61: 203–208. [DOI] [PubMed] [Google Scholar]
- 7. Banoczy J (1962) Observations sur l'alteration de la cornification de la muqueuse buccale sous l'influence du tabac. Bull Group Int Rech Sci Stomatol 5: 543–553. [Google Scholar]
- 8. Meyer J, Rubinstein AS, Medak H (1970) Early effects of smoking on surface cytology of the oral mucosa: II. Cell changes in smokers. Oral Surgery, Oral Medicine, Oral Pathology 30: 700–710. [DOI] [PubMed] [Google Scholar]
- 9. Martin GC, Brown JP, Eifler CW, Houston GD (1999) Oral leukoplakia status six weeks after cessation of smokeless tobacco use. J Am Dent Assoc 130: 945–954. [DOI] [PubMed] [Google Scholar]
- 10. Shibly O, Cummings KM, Zambon JJ (2008) Resolution of oral lesions after tobacco cessation. J Periodontol 79: 1797–1801. [DOI] [PubMed] [Google Scholar]
- 11. Greenblatt MS, Bennett WP, Hollstein M, Harris CC (1994) Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res 54: 4855–4878. [PubMed] [Google Scholar]
- 12. Chalitchagorn K, Shuangshoti S, Hourpai N, Kongruttanachok N, Tangkijvanich P, et al. (2004) Distinctive pattern of LINE-1 methylation level in normal tissues and the association with carcinogenesis. Oncogene 23: 8841–8846. [DOI] [PubMed] [Google Scholar]
- 13. Dammann R, Strunnikova M, Schagdarsurengin U, Rastetter M, Papritz M, et al. (2005) CpG island methylation and expression of tumour-associated genes in lung carcinoma. Eur J Cancer 41: 1223–1236. [DOI] [PubMed] [Google Scholar]
- 14. Toyooka S, Tokumo M, Shigematsu H, Matsuo K, Asano H, et al. (2006) Mutational and epigenetic evidence for independent pathways for lung adenocarcinomas arising in smokers and never smokers. Cancer Res 66: 1371–1375. [DOI] [PubMed] [Google Scholar]
- 15.Salskov A, Hawes SE, Stern JE, Feng Q, Jordan CD, et al.. (2011) Hypermethylation of CCND2 May Reflect a Smoking-Induced Precancerous Change in the Lung. J Oncol: doi: 10.1155/2011/950140. [DOI] [PMC free article] [PubMed]
- 16. Smith IM, Mydlarz WK, Mithani SK, Califano JA (2007) DNA global hypomethylation in squamous cell head and neck cancer associated with smoking, alcohol consumption and stage. Int J Cancer 121: 1724–1728. [DOI] [PubMed] [Google Scholar]
- 17. Graham T, Boissinot S (2006) The genomic distribution of L1 elements: the role of insertion bias and natural selection. J Biomed Biotechnol 2006: 75327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, et al. (2001) Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- 19. Penzkofer T, Dandekar T, Zemojtel T (2005) L1Base: from functional annotation to prediction of active LINE-1 elements. Nucleic Acids Res 33: D498–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kitkumthorn N, Mutirangura A (2011) Long interspersed nuclear element-1 hypomethylation in cancer: biology and clinical applications. Clin Epigenet 2: 315–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shuangshoti S, Hourpai N, Pumsuk U, Mutirangura A (2007) Line-1 hypomethylation in multistage carcinogenesis of the uterine cervix. Asian Pac J Cancer Prev 8: 307–309. [PubMed] [Google Scholar]
- 22. Tangkijvanich P, Hourpai N, Rattanatanyong P, Wisedopas N, Mahachai V, et al. (2007) Serum LINE-1 hypomethylation as a potential prognostic marker for hepatocellular carcinoma. Clin Chim Acta 379: 127–133. [DOI] [PubMed] [Google Scholar]
- 23. Iacopetta B, Grieu F, Phillips M, Ruszkiewicz A, Moore J, et al. (2007) Methylation levels of LINE-1 repeats and CpG island loci are inversely related in normal colonic mucosa. Cancer Sci 98: 1454–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Iramaneerat K, Rattanatunyong P, Khemapech N, Triratanachat S, Mutirangura A (2011) HERV-K hypomethylation in ovarian clear cell carcinoma is associated with a poor prognosis and platinum resistance. Int J Gynecol Cancer 21: 51–57. [DOI] [PubMed] [Google Scholar]
- 25. Florl AR, Lower R, Schmitz-Drager BJ, Schulz WA (1999) DNA methylation and expression of LINE-1 and HERV-K provirus sequences in urothelial and renal cell carcinomas. Br J Cancer 80: 1312–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Subbalekha K, Pimkhaokham A, Pavasant P, Chindavijak S, Phokaew C, et al. (2009) Detection of LINE-1s hypomethylation in oral rinses of oral squamous cell carcinoma patients. Oral Oncol 45: 184–191. [DOI] [PubMed] [Google Scholar]
- 27. Hsiung DT, Marsit CJ, Houseman EA, Eddy K, Furniss CS, et al. (2007) Global DNA methylation level in whole blood as a biomarker in head and neck squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev 16: 108–114. [DOI] [PubMed] [Google Scholar]
- 28. Feber A, Wilson GA, Zhang L, Presneau N, Idowu B, et al. (2011) Comparative methylome analysis of benign and malignant peripheral nerve sheath tumors. Genome Res 21: 515–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Romermann D, Hasemeier B, Metzig K, Schlegelberger B, Langer F, et al. (2007) Methylation status of LINE-1 sequences in patients with MDS or secondary AML. Verh Dtsch Ges Pathol 91: 338–342. [PubMed] [Google Scholar]
- 30. Perrin D, Ballestar E, Fraga MF, Frappart L, Esteller M, et al. (2007) Specific hypermethylation of LINE-1 elements during abnormal overgrowth and differentiation of human placenta. Oncogene 26: 2518–2524. [DOI] [PubMed] [Google Scholar]
- 31. Figueiredo JC, Grau MV, Wallace K, Levine AJ, Shen L, et al. (2009) Global DNA hypomethylation (LINE-1) in the normal colon and lifestyle characteristics and dietary and genetic factors. Cancer Epidemiol Biomarkers Prev 18: 1041–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hillemacher T, Frieling H, Moskau S, Muschler MA, Semmler A, et al. (2008) Global DNA methylation is influenced by smoking behaviour. Eur Neuropsychopharmacol 18: 295–298. [DOI] [PubMed] [Google Scholar]
- 33. Madrigano J, Baccarelli A, Mittleman MA, Wright RO, Sparrow D, et al. (2011) Prolonged exposure to particulate pollution, genes associated with glutathione pathways, and DNA methylation in a cohort of older men. Environ Health Perspect 119: 977–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu F, Killian JK, Yang M, Walker RL, Hong JA, et al. (2010) Epigenomic alterations and gene expression profiles in respiratory epithelia exposed to cigarette smoke condensate. Oncogene 29: 3650–3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kongruttanachok N, Phuangphairoj C, Thongnak A, Ponyeam W, Rattanatanyong P, et al.. (2010) Replication independent DNA double-strand break retention may prevent genomic instability. Mol Cancer 9: doi: 10.1186/1476-4598-1189-1170. [DOI] [PMC free article] [PubMed]
- 36. Pornthanakasem W, Kongruttanachok N, Phuangphairoj C, Suyarnsestakorn C, Sanghangthum T, et al. (2008) LINE-1 methylation status of endogenous DNA double-strand breaks. Nucleic Acids Res 36: 3667–3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Aporntewan C, Phokaew C, Piriyapongsa J, Ngamphiw C, Ittiwut C, et al. (2011) Hypomethylation of intragenic LINE-1 represses transcription in cancer cells through AGO2. PLoS One 6: e17934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Phokaew C, Kowudtitham S, Subbalekha K, Shuangshoti S, Mutirangura A (2008) LINE-1 methylation patterns of different loci in normal and cancerous cells. Nucleic Acids Res 36: 5704–5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ho SM, Tang WY (2007) Techniques used in studies of epigenome dysregulation due to aberrant DNA methylation: an emphasis on fetal-based adult diseases. Reprod Toxicol 23: 267–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Baba Y, Huttenhower C, Nosho K, Tanaka N, Shima K, et al. (2010) Epigenomic diversity of colorectal cancer indicated by LINE-1 methylation in a database of 869 tumors. Mol Cancer 9: 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pobsook T, Subbalekha K, Sannikorn P, Mutirangura A (2010) Improved measurement of LINE-1 sequence methylation for cancer detection. Clin Chim Acta 412: 314–321. [DOI] [PubMed] [Google Scholar]
- 42. Kitkumthorn N, Tuangsintanakul T, Rattanatanyong P, Tiwawech D, Mutirangura A (2012) LINE-1 methylation in the peripheral blood mononuclear cells of cancer patients. Clin Chim Acta 413: 869–874. [DOI] [PubMed] [Google Scholar]
- 43. Patchsung M, Boonla C, Amnattrakul P, Dissayabutra T, Mutirangura A, et al. (2012) Long interspersed nuclear element-1 hypomethylation and oxidative stress: correlation and bladder cancer diagnostic potential. PLoS One 7: e37009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bollati V, Schwartz J, Wright R, Litonjua A, Tarantini L, et al. (2009) Decline in genomic DNA methylation through aging in a cohort of elderly subjects. Mech Ageing Dev 130: 234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jintaridth P, Mutirangura A (2010) Distinctive patterns of age-dependent hypomethylation in interspersed repetitive sequences. Physiol Genomics 41: 194–200. [DOI] [PubMed] [Google Scholar]
- 46. El-Maarri O, Walier M, Behne F, van Uum J, Singer H, et al. (2011) Methylation at global LINE-1 repeats in human blood are affected by gender but not by age or natural hormone cycles. PLoS One 6: e16252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bollati V, Baccarelli A, Hou L, Bonzini M, Fustinoni S, et al. (2007) Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res 67: 876–880. [DOI] [PubMed] [Google Scholar]
- 48. Nakkuntod J, Avihingsanon Y, Mutirangura A, Hirankarn N (2011) Hypomethylation of LINE-1 but not Alu in lymphocyte subsets of systemic lupus erythematosus patients. Clin Chim Acta 412: 1457–1461. [DOI] [PubMed] [Google Scholar]
- 49. Teneng I, Montoya-Durango DE, Quertermous JL, Lacy ME, Ramos KS (2011) Reactivation of L1 retrotransposon by benzo(a)pyrene involves complex genetic and epigenetic regulation. Epigenetics 6: 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bollati V, Baccarelli A, Hou L, Bonzini M, Fustinoni S, et al. (2007) Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res 67: 876–880. [DOI] [PubMed] [Google Scholar]
- 51. Maraschio P, Zuffardi O, Dalla Fior T, Tiepolo L (1988) Immunodeficiency, centromeric heterochromatin instability of chromosomes 1, 9, and 16, and facial anomalies: the ICF syndrome. J Med Genet 25: 173–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ji W, Hernandez R, Zhang XY, Qu GZ, Frady A, et al. (1997) DNA demethylation and pericentromeric rearrangements of chromosome 1. Mutat Res 379: 33–41. [DOI] [PubMed] [Google Scholar]
- 53. Schernhammer ES, Giovannucci E, Kawasaki T, Rosner B, Fuchs CS, et al. (2010) Dietary folate, alcohol and B vitamins in relation to LINE-1 hypomethylation in colon cancer. Gut 59: 794–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The percentage of LINE-1 products in smokers and non-smokers.
(DOC)
Relationship of the percentage of LINE-1 products with age status.
(DOC)
Demographic characteristics and the percentage of LINE-1 products in pack-year smoking groups.
(DOC)