Abstract
Mitochondria import hundreds of cytosolically synthesized proteins via the mitochondrial protein import apparatus. Expression analysis in various organs of 19 components of the Arabidopsis mitochondrial protein import apparatus encoded by 31 genes showed that although many were present in small multigene families, often only one member was prominently expressed. This was supported by comparison of real-time reverse transcriptase-polymerase chain reaction and microarray experimental data with expressed sequence tag numbers and massive parallel signature sequence data. Mass spectrometric analysis of purified mitochondria identified 17 import components, their mitochondrial sub-compartment, and verified the presence of TIM8, TIM13, TIM17, TIM23, TIM44, TIM50, and METAXIN proteins for the first time, to our knowledge. Mass spectrometry-detected isoforms correlated with the most abundant gene transcript measured by expression data. Treatment of Arabidopsis cell culture with mitochondrial electron transport chain inhibitors rotenone and antimycin A resulted in a significant increase in transcript levels of import components, with a greater increase observed for the minor isoforms. The increase was observed 12 h after treatment, indicating that it was likely a secondary response. Microarray analysis of rotenone-treated cells indicated the up-regulation of gene sets involved in mitochondrial chaperone activity, protein degradation, respiratory chain assembly, and division. The rate of protein import into isolated mitochondria from rotenone-treated cells was halved, even though rotenone had no direct effect on protein import when added to mitochondria isolated from untreated cells. These findings suggest that transcription of import component genes is induced when mitochondrial function is limited and that minor gene isoforms display a greater response than the predominant isoforms.
The majority of the thousand or more proteins that are present in mitochondria are required to be imported from nuclear-encoded cytosolically synthesized precursors (Emanuelsson et al., 2000; Werhahn and Braun, 2002; Heazlewood et al., 2003; Taylor et al., 2003c). The import of these proteins is achieved by the mitochondrial protein import apparatus, comprising a multisubunit translocase on both the outer and inner membranes, a variety of chaperone proteins present in the cytosol and mitochondria, and a number of peptidases that remove the “transient” targeting information present on many, but not all, mitochondrial precursor proteins (Neupert, 1997; Pfanner and Geissler, 2001). Despite the fact that mitochondrial targeting signals contain little primary sequence similarity, the mitochondrial protein import apparatus specifically recognizes and imports up to 1,000 proteins (Sjoling and Glaser, 1998; Zhang et al., 2001).
The mitochondrial protein import apparatus has been studied intensively in yeast (Saccharomyces cerevisiae) using biochemical and genetic approaches. A single translocase operates on the outer mitochondrial membrane (TOM), which contains seven proteins with two primary receptors, TOM20 and TOM70. With few exceptions, these receptors recognize all precursor proteins studied so far and transfer them to the central TOM40 pore via TOM22, which can also act as a receptor for a small number of proteins. Two translocases on the inner mitochondrial membrane (TIM), called TIM17:23 and TIM22, function in the general and carrier import pathways, respectively (Pfanner and Geissler, 2001). Although not clearly understood mechanistically, TOM40 mediates the transfer of proteins to either of these TIM complexes and plays some role in sorting between the TIM complexes (Ahting et al., 2001; Gabriel et al., 2003). The TIM17:23 complex also contains a 50-kD protein, TIM50, and binds the matrix-located TIM44 (Milisav et al., 2001; Geissler et al., 2002; Yamamoto et al., 2002; Mokranjac et al., 2003). The TIM17:23 complex is responsible for the import of precursor proteins that contain N-terminal targeting signals that are generally removed after import by the mitochondrial processing peptidase (MPP; Neupert, 1997; Pfanner and Geissler, 2001). The TIM22 complex contains at least three other inner membrane proteins (TIM54, 18, and 12) that together with the small TIM proteins of the intermembrane space (TIM8, 9, 10, and 13) are responsible for the import of carrier proteins into mitochondria (Bauer et al., 1999; Kerscher et al., 2000; Koehler et al., 2000; Rehling et al., 2003a). A third less well-characterized translocase of the inner membrane, Oxa1p, consists of a homooligomeric complex that appears to be related to the YidC protein of the secretory apparatus in Escherichia coli (Luirink et al., 2001; Yen et al., 2001; Nargang et al., 2002). Sixteen of the 33 import components are essential for viability in yeast and constitute over one-third of the 40 mitochondrial proteins thought to be essential for yeast viability (Rehling et al., 2003b), whereas TIM23 also has been implicated in programmed cell death in yeast (Lohret et al., 1997).
In plants, biochemical approaches have characterized the TOM and MPP components of the plant import apparatus (Braun and Schmitz, 1995; Glaser and Dessi, 1999; Werhahn et al., 2001). Unlike yeast, the plant MPP is an integral component of the cytochrome bc1 complex, although a specific matrix activity also appears to exist (Braun and Schmitz, 1995; Szigyarto et al., 1998). The plant TOM complex differs slightly from the yeast complex because it lacks any apparent ortholog of the TOM70 receptor (Jansch et al., 1998; Werhahn et al., 2001), whereas the plant TOM22 homolog, known as TOM9, does not contain the cis-receptor domain that is present in yeast and mammalian systems (Mascasev et al., 2000). Genetic approaches indicate that an Oxa1p ortholog from Arabidopsis can complement a yeast mutant for this protein (Sakamoto et al., 2000). In addition, modifications to the TIM17 and TIM23 genes of Arabidopsis allows complementation of yeast mutants (Murcha et al., 2003). A bioinformatics approach using all the known yeast components of the mitochondrial protein import apparatus identified 27 Arabidopsis orthologs of the 33 components present in yeast (Lister et al., 2003). Overall, it was notable that many of the components of the carrier import pathway were not identifiable from the genome of this model plant, i.e. TOM70, TIM12, 18, and 54. In addition, many of the components identified were encoded in small multigene families and did not contain many of the motifs that either have been shown to be important for function in yeast or that are conserved between yeast and mammalian systems.
Protein import into mitochondria can be affected by organ, developmental, and diurnal factors (Dessi and Whelan, 1997; Dudley et al., 1997; Murcha et al., 1999). It also has been demonstrated recently that protein import into mitochondria is altered under conditions of environmental stress that also inhibited important mitochondrial functions (Taylor et al., 2003a). An analysis of the effects of oxidative stress and respiratory inhibitors on the Arabidopsis mitochondrial proteome found changes in protein abundance and also documented losses of mitochondrial function (Sweetlove et al., 2002). Using an array which contained over 11,000 genes from Arabidopsis, it was observed that changes in gene expression after antimycin A treatment were similar to those observed with diverse biotic and abiotic stresses. It was proposed that mitochondria may be an important point in mediating responses to a variety of stresses (Yu et al., 2001). If mitochondria undergo alterations to adapt to such stresses, newly synthesized proteins in the cytosol may need to be imported into the mitochondria. However, the mechanisms by which mitochondrial protein import is modulated either by development, organ type, imposed stress or mitochondrial dysfunction have not yet been elucidated.
Therefore, to gain a better understanding of the structure of the plant mitochondrial protein import apparatus and how it may change under conditions that inhibit mitochondrial function, we conducted expression analysis of the 31 genes encoding the Arabidopsis import components identified by homology to the yeast import machinery. We analyzed their expression in various organs and looked for the presence of the proteins in mitochondria and submitochondrial compartments isolated from Arabidopsis cell cultures. Arabidopsis cell cultures were treated with the mitochondrial electron transport chain inhibitors rotenone and antimycin A, and transcript abundance was measured over time by quantitative real-time PCR. Transcriptomic analysis and the rate of protein import into isolated mitochondria were investigated after rotenone treatment.
RESULTS
Experimental Definition of the Mitochondrial Protein Import Apparatus
Gene Expression of Components of the Mitochondrial Protein Import Apparatus
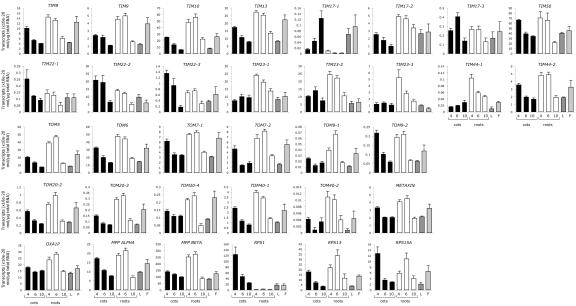
Bioinformatic approaches have identified 19 putative components of the Arabidopsis mitochondrial protein import machinery encoded by 31 genes (Lister et al., 2002). To investigate the expression patterns of the putative protein import machinery genes, quantitative real-time reverse transcriptase (RT)-PCR analysis was used to measure transcript abundance in Arabidopsis cotyledons, roots, leaves, and flowers (Fig. 1). Initially, we measured the transcript abundance of ribosomal proteins that are required in the three cellular compartments where translation takes place. Transcripts of RPS13 and RPS15A, nuclear-encoded ribosomal proteins that reside in the mitochondria and cytosol, respectively, were found in all Arabidopsis organs examined. In cotyledons, the transcript abundance of these two genes was highest at 4 d after germination and decreased as the plant aged. In roots, the transcript abundance increased to peak at 6 d, then decreased. Message levels of the nuclear-encoded chloroplast ribosomal protein, RPS1, were similar to the cytosolic and mitochondrial RPS genes in cotyledons, leaves, and flowers, whereas the transcript abundance in roots was very low, as expected.
Figure 1.
Transcript abundance of the cDNAs encoding the protein import machinery. Quantitative real-time RT-PCR analysis of the cDNAs encoding the Arabidopsis mitochondrial protein import machinery was performed on Arabidopsis cotyledons (C; 4, 6, and 10 d after germination), roots (R; 4, 6, and 10 d after germination), leaves (L; 10 d after germination), and flowers (F).
Message levels for the 31 genes encoding the plant mitochondrial protein import machinery were measured. With only one exception, transcripts for all genes were found in all samples examined, indicating that all were expressed. The exception was TOM20-1, which displayed expression below reliable detection levels. In roots, transcript abundance of the TIM and TOM components predominantly peaked at 4 and 6 d after germination, then decreased significantly by 10 d. Interestingly, like the chloroplast RPS1, TIM17-1 message was present at very low levels in roots compared with other organs, possibly indicating that it was not utilized in nongreen organs. As in roots, message levels in cotyledons for most components were generally highest at 4 to 6 d but significantly decreased by 10 d. However, TIM23-1, TIM23-3, TIM44-1, and TOM20-4 displayed a more constant transcript abundance over cotyledon development. TIM17-1 again showed an expression profile differing from the general pattern in cotyledons, increasing over development. All genes were expressed in leaves and flowers. Included in this analysis was the Arabidopsis homolog of METAXIN (At2g19080), the outer membrane receptor for animal mitochondrial protein import (Armstrong et al., 1997). The Arabidopsis homolog displays 23% amino acid identity to human (Homo sapiens) and fruitfly (Drosophila melanogaster) METAXIN proteins. Previously uncharacterized in plants, the similarity of the Arabidopsis METAXIN expression pattern to that of other import components provides preliminary evidence of it being the plant homolog of the METAXIN protein import receptor in animals.
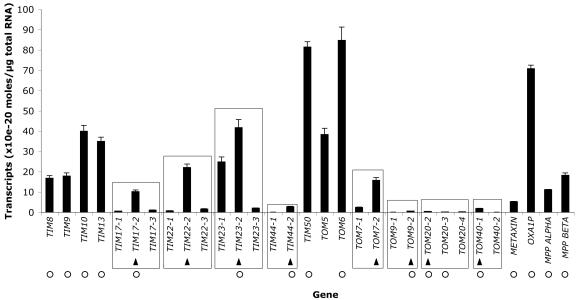
Quantitative real-time RT-PCR was also used to compare the absolute transcript abundance for each import component in Arabidopsis cell culture (Fig. 2). Most import components displayed similar transcript levels, except for the low quantity of TIM44, TOM9, TOM20, TOM40, and METAXIN. In yeast, it has been calculated that TOM complexes are approximately 4 times more abundant than TIM complexes (Dekker et al., 1998). This stoichiometry was not observed in the transcript abundance of TOM and TIM import components in Arabidopsis cell culture (Fig. 2). Therefore, the low message abundance of these components may indicate other means of regulation, such as RNA stability, translation efficiency, and/or protein turnover.
Figure 2.
Absolute transcript abundance of the cDNAs encoding the protein import machinery. Quantitative real-time RT-PCR analysis of Arabidopsis cell culture was used to measure the absolute transcript abundance for cDNAs encoding the Arabidopsis mitochondrial protein import machinery. Multiple gene families are encompassed by a rectangle. An arrow indicates the isoform with the highest transcript abundance in each gene family. A white circle indicates the protein isoform found by mass spectrometry.
Many import components are encoded in multiple gene families (Fig. 2, rectangles) that generally display high sequence similarity (Lister et al., 2003). The gene families each possess a predominant isoform that had the highest transcript abundance in cell culture (Fig. 2, arrows). The predominance of specific isoforms was also evident in whole plants as supported by a comparison of the message abundance in cell culture with the levels measured in Arabidopsis cotyledons, roots, leaves, and flowers (data not shown). With the exception of TOM7, the majority of these organs displayed the same predominant isoforms as observed in the cell culture.
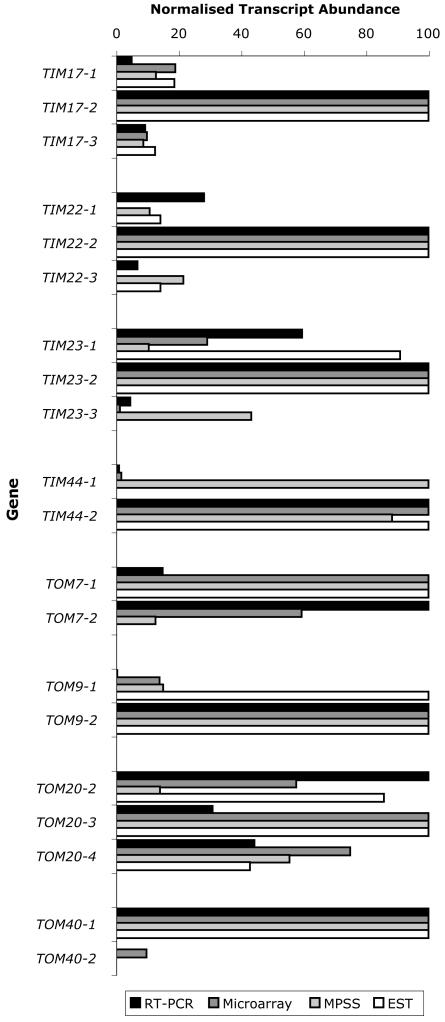
Additional support for the presence of predominant isoforms was obtained by analysis of the massively parallel signature sequence (MPSS) project data (Brenner et al., 2000), expressed sequence tag (EST) numbers, and our microarray analysis of Arabidopsis cell culture, all of which generally showed similar expression patterns to the real-time RT-PCR data (Fig. 3). Expression data were present for several different organs for both real-time RT-PCR (cell culture, cotyledon, root, leaf, and floral) and MPSS (callus, shoot, root, silique, and floral) experiments. However, because growth conditions and sample particulars varied between the two different approaches, the data were adjusted as indicated in “Materials and Methods.” As seen in the TOM20 transcript abundance in Arabidopsis cell culture (Fig. 2), the comparison of real-time RT-PCR, microarray, MPSS, and EST data did not reveal a distinct predominant isoform but indicated that they were present at similar levels. The predominant isoform within the gene families of TIM17, TIM22, TIM23, and TOM40 was the same regardless of the technique used, whereas for TIM44, TOM7, and TOM9, there was a significant discrepancy in the isoform that predominated using one technique. For TOM9, real-time RT-PCR, microarray, and MPSS data were similar, but EST numbers did not agree. For TOM7, real-time RT-PCR data did not agree with the other approaches. Real-time RT-PCR and microarray data for TIM44 indicated that TIM44-1 message levels were very low, whereas TIM44-2 transcript abundance was 100-fold higher. This was not in agreement with MPSS and EST data, which indicated that both TIM44 genes had similar transcript levels. TOM40-2 was not represented by ESTs or MPSS data, perhaps because its transcript appears to be present at very low levels. Unfortunately, probe pairs to detect TIM22-1 and TIM22-3 were not present on the Affymetrix ATH1 GeneChip (Affymetrix, Santa Clara, CA).
Figure 3.
Comparison of quantitative real-time RT-PCR, microarray, MPSS, and EST data. For each experimental approach, the transcript abundance of every gene was averaged over the different tissue types, then within each gene family, these average message levels of the different isoforms were normalized, with the most abundant isoform given the arbitrary value of 100. EST numbers and microarray signal values (from control arrays) within each gene family were normalized against the isoform with the highest transcript abundance, allowing comparison of EST and microarray values in each gene family with real-time RT-PCR and MPSS data.
Detection of Proteins for Components of the Mitochondrial Protein Import Apparatus
Highly enriched mitochondrial preparations were isolated from Arabidopsis cell culture and fractionated into four sub-compartments: inner membrane, outer membrane, matrix, and intermembrane space. Although the purity of the mitochondrial preparation is approximately 98% (Millar et al., 2001), the subfractionation procedure is an enrichment process rather than a high-purity separation; hence, the intermembrane space is nearly 20% contaminated by matrix, the outer membrane contains 20% to 50% inner membrane, and the inner membrane contains a significant amount of still-attached outer membrane portions. Protein aliquots from whole mitochondria and the sub-fractions were trypsin digested to identify peptides from import components by an liquid chromatography tandem mass spectrometry (MS/MS) strategy of shotgun sequencing of peptides (Heazlewood et al., 2003). Peptides from 17 different import components were identified in the mitochondria and sub-fraction aliquots (Werhahn and Braun, 2002). In all cases of import components encoded in multiple gene families, the peptides identified were from the predominant isoform apparent from the cell culture transcript analysis (Fig. 2, peptide presence indicated by white circle). Notably, with TIM44, the proteomic identification of TIM44-2 matches the expression data we obtained by real-time PCR and microarray analysis. This reveals a close correlation between the highly expressed members of the gene families and the presence of the encoded protein in the mitochondria. Subfractionation of the mitochondria assisted in the identification of six import components that were clearly too low in abundance to be readily detected in whole mitochondrial isolates. Enrichment of intermembrane space proteins, which normally constitute approximately 5% of the total mitochondrial protein content (Sweetlove et al., 2001), enabled identification of TIM8, TIM9, and TIM10, known in yeast to function in the intermembrane space. Inner membrane enrichment allowed identification of TIM17-2 and TIM44-2, whereas TOM20-2 was only identified after enrichment of outer membranes.
The identification of import components from the mitochondrial sub-compartments in which they are found in yeast reinforces the hypothesis that they are the authentic plant mitochondrial protein import components. The small TIM proteins (8, 9, 10, and 13) were readily identified in the intermembrane space but were largely absent from other compartments. The outer membrane-enriched fractions contained the three key TOM complex subunits, but we did not identify the small TIMs or the TIM translocase subunits in this fraction. The inner membrane TIM subunits (TIM17-2, TIM23-1, TIM44-2, and TIM50) were found in the enriched inner membrane and whole mitochondrial samples but were not identified in the soluble fractions investigated. The MPP subunits were identified in all fractions, which, in our opinion, does not suggest localization outside the inner membrane but rather their high abundance in mitochondrial samples.
The import components directly identified in mitochondrial samples encompass nearly the entire plant mitochondrial protein import apparatus as characterized by comparison with yeast and mammalian systems. Specific peptides from both TOM20-2 and TOM20-3 were sequenced in our analysis, which correlates with the similar transcript abundance of both isoforms of this family observed in the real-time RT-PCR, microarray, MPSS, and EST data. This also correlates with the dominant isoforms of TOM20 detected by Werhahn et al. (2003) in the purified TOM complex from Arabidopsis. The absence of two of the small TOM components (TOM5 and TOM7) may be explained by the relative paucity of suitable peptides generated by trypsin digestion of small proteins (data not shown). However, TOM7-1 and TOM5 have been identified in purified TOM complex from Arabidopsis mitochondria (Werhahn et al., 2003). The data presented in Table I constitute the first experimental identification of the Arabidopsis TIM8, TIM13, TIM17, TIM23, TIM44, TIM50, and METAXIN proteins.
Table I.
Import component peptides identified by mass spectrometry of trypsin-digested mitochondria
| Component
|
Locus
|
Peptides Found
|
Mowse Score
|
Found in
|
||||
|---|---|---|---|---|---|---|---|---|
| Mitochondria | Outer Membrane | Intermembrane Space | Inner Membrane | Matrix | ||||
| TIM8 | At5g50810 | 1 | 35 | x | ||||
| TIM9 | At3g46560 | 1 | 22 | x | ||||
| TIM10 | At2g29530 | 2 | 70 | x | ||||
| TIM13 | At1g61570 | 2 | 129 | x | x | |||
| TIM17-2 | At2g37410 | 1 | 13 | x | ||||
| TIM23-2 | At1g72750 | 2 | 87 | x | x | |||
| TIM44-2 | At2g36070 | 1 | 50 | x | ||||
| TIM50 | At1g55900 | 2 | 70 | x | x | |||
| TOM6 | At1g49410 | 1 | 60 | x | ||||
| TOM9-2 | At5g43970 | 1 | 50 | x | ||||
| TOM20-2 | At1g27390 | 1 | 70 | x | x | |||
| TOM20-3 | At3g27080 | 2 | 61 | x | x | x | ||
| TOM40-1 | At3g20000 | 3 | 99 | x | x | x | ||
| METAXIN | At2g19080 | 2 | 132 | x | x | |||
| Oxa1p | At5g62050 | 1 | 32 | x | ||||
| MPPα | At3g16480 | >10 | naa | x | x | x | x | x |
| MPPβ | At3g02090 | >10 | na | x | x | x | x | x |
na, Not available.
The Gene Expression of Components of the Mitochondrial Protein Import Apparatus Responds to Mitochondrial Dysfunction
We investigated if gene expression of components of the mitochondrial protein import apparatus responded to changes in mitochondrial activity. To assess this, we added inhibitors to the mitochondrial electron transport chain and assessed changes in gene expression at 1, 3, and 12 h after addition. Gene expression was measured in this period to assess if changes in expression occurred immediately, i.e. 1 to 3 h, or were a secondary affect, i.e. 12 h. Mitochondria were chemically stressed with 40 μm rotenone or 5 μm antimycin A, compounds that inhibit the action of complexes I and III of the mitochondrial electron transport chain, respectively. Both chemicals were added at concentrations intended to perturb mitochondrial function and did not result in cell death, as determined by vital stains and oxygen consumption assays (data not shown). Cell samples were taken 0, 1, 3, and 12 h after treatment for quantitative real-time PCR analysis. In addition, cells treated with rotenone were collected after 12 h for analysis using full-genome Affymetrix GeneChip ATH1 microarrays.
Table II lists groups of genes identified in the microarray analysis with altered transcript abundance after rotenone treatment, fold induction, and microarray signal value listed for each. These genes encode proteins involved in diverse mitochondrial activities: mitochondrial protein import components, molecular chaperones, proteins involved in respiratory chain complex assembly, protein degradation, and mitochondrial division. Twelve import component genes were up-regulated more than 1.5-fold after rotenone treatment, including those encoding pore-forming subunits (TIM17, TIM23, and TOM40) and outer membrane receptor components (TOM20 and METAXIN). The concurrent increase in transcript abundance of eight of 10 genes encoding mitochondrial molecular chaperone proteins suggests a requirement for the folding and assembly of proteins in the mitochondria. Also up-regulated were genes encoding Arabidopsis proteins similar to yeast proteins involved in the correct assembly of mitochondrial respiratory chain complexes: BCS required for the assembly of a functional cytochrome bc1 complex (Nobrega et al., 1992); COX15, 17, 19, SCO1, and SURFEIT 1 required for the assembly of the cytochrome oxidase complex (Schulze and Rodel, 1988; Nobrega et al., 1992; Glerum et al., 1996, 1997; Nijtmans et al., 2001; Balandin and Castresana, 2002); and an m-AAA protease required for the assembly of ATP synthase (Arlt et al., 1996; Kolodziejczak et al., 2002). A zinc metalloprotease (At3g19170) involved in the rapid degradation of mitochondrial presequences after they have been removed by MPP was up-regulated 3.4-fold (Stahl et al., 2002). In addition, genes possibly involved in mitochondrial division (ADL2a and FIS1) were up-regulated (Mozdy et al., 2000; Arimura and Tsutsumi, 2002).
Table II.
Microarray measurements of changes in transcript abundance after rotenone treatment
| Gene
|
Locus
|
Fold Change
|
Microarray Signal
|
|
|---|---|---|---|---|
| Control | Rotenone | |||
| Import components | ||||
| TIM10 | At2g29530 | 1.6 | 1,244 | 1,990 |
| TIM17-1 | At1g20350 | 3.9 | 583 | 2,259 |
| TIM17-2 | At2g37410 | 1.6 | 3,081 | 5,049 |
| TIM17-3 | At5g11690 | -1.6 | 309 | 193 |
| TIM23-1 | At1g17530 | 2.4 | 556 | 1,323 |
| TIM23-3 | At3g04800 | 5.6 | 24 | 137 |
| TIM44-2 | At2g36070 | 1.5 | 415 | 625 |
| TIM50 | At1g55900 | 1.7 | 894 | 1,562 |
| TOM20-4 | At5g40930 | 1.7 | 1,046 | 1,768 |
| TOM40-2 | At1g50400 | 5.3 | 160 | 850 |
| TOM7-2 | At1g64220 | 5.8 | 161 | 933 |
| METAXIN | At2g19080 | 1.9 | 1,247 | 2,348 |
| Oxa1p | At5g62050 | 1.6 | 1,013 | 1,591 |
| Mitochondrial molecular chaperones | ||||
| CPN10 | At1g23100 | 4.6 | 128 | 588 |
| HSP60-2 | At2g33210 | 2.0 | 2,187 | 4,322 |
| HSP60-3a | At3g13860 | 1.6 | 1,281 | 2,050 |
| HSP60-3b | At3g23990 | 2.1 | 1,225 | 2,608 |
| HSP60-3c | At3g13470 | -1.8 | 1,159 | 644 |
| HSP70-4 | At4g37910 | 2.2 | 3,659 | 8,026 |
| HSP70-5 | At5g09590 | 1.9 | 1,795 | 3,451 |
| HSP90-3 | At4g24190 | -2.5 | 4,753 | 1,901 |
| GRPE | At5g55200 | 4.2 | 916 | 3,846 |
| MGE | At5g17710 | 1.9 | 847 | 1,573 |
| Respiratory chain assembly | ||||
| BCS1a | At3g50930 | 12.2 | 124 | 1,516 |
| BCS1b | At3g50940 | 2.5 | 90 | 231 |
| BCS1c | At5g17760 | 1.5 | 960 | 1,469 |
| COX15 | At5g56090 | 3.2 | 706 | 2,228 |
| COX17 | At1g53030 | 1.9 | 352 | 668 |
| COX17 | At3g15352 | 2.2 | 245 | 532 |
| COX19 | At1g69750 | 1.8 | 1,379 | 2,483 |
| SCO1 | At3g08950 | 2.2 | 648 | 1,409 |
| SURFEIT1 | At3g17910 | 1.8 | 730 | 1,291 |
| m-AAA protease | At1g07510 | 1.5 | 1,150 | 1,712 |
| Mitochondrial division | ||||
| ADL2a | At4g33650 | 1.8 | 380 | 684 |
| Fis1 homolog | At3g57090 | 2.8 | 925 | 2,590 |
| Fis1 homolog | At5g12390 | 2.0 | 116 | 232 |
| Mitochondrial proteases | ||||
| Lon protease | At5g26860 | 2.3 | 301 | 693 |
| Lon protease | At3g05780 | 3.7 | 13 | 49 |
| ClpC1 | At5g50920 | 1.9 | 1,769 | 3,362 |
| ClpX2 | At5g49840 | 1.7 | 912 | 1,550 |
| DegP10 | At5g36950 | 2.1 | 221 | 465 |
| FtsH protease | At1g06430 | 3.7 | 420 | 1,554 |
| PreP | At3g19170 | 3.4 | 894 | 3,041 |
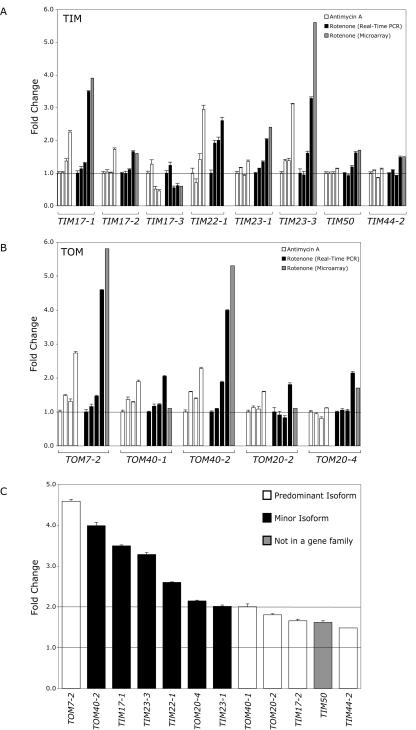
Quantitative real-time RT-PCR analysis of the transcript abundance of import components in cells treated with antimycin A or rotenone at 0, 1, 3, and 12 h was performed to confirm the microarray data and determine whether the changes observed in the microarray analysis were primary or secondary responses to the chemical treatments (Fig. 4). Shown are the import components that displayed significant changes in transcript abundance in comparison with the control (fold change > 1.5). In addition, the microarray fold change is indicated for comparison with the fold change measured by real-time RT-PCR. Probe pairs to detect TIM22-1 were not present on the Affymetrix ATH1 GeneChip, so real-time RT-PCR was used to quantify the changes in transcript abundance. The fold changes in transcript abundance as measured by real-time RT-PCR were very similar in most cases to the fold changes observed in the microarray analysis (Dekker et al., 1998). The transcript abundance changes of TIM10, METAXIN, and Oxa1p observed by microarray analysis were not supported by the quantitative real-time RT-PCR analysis. Of the TIM components, seven genes showed significant up-regulation after rotenone treatment, whereas only TIM17-3 showed a decrease in transcript abundance (Fig. 4A). Moreover, except for TIM22-1, the highest fold change was observed after 12 h, indicating that this was likely to be a secondary response. TIM22-1 transcript abundance doubled only 1 h after rotenone treatment, possibly indicating an early response of the carrier import pathway. The message levels of the small TIM components (8, 9, 10, and 13) did not change after inhibitor treatment, but they may not be rate limiting in the carrier import pathway. The five TOM components changed primarily 12 h after treatment (Fig. 4B). Comparison of the fold changes of the import components 12 h after rotenone treatment revealed an interesting trend (Fig. 4C). The fold change in up-regulated minor import component isoforms was greater than that of the induced predominant isoforms (white), perhaps indicating a specialized role for the minor import component isoforms as stress-responsive genes. In addition, the highest inductions of the minor isoforms involved the pore-forming import components TOM40, TIM17, and TIM23.
Figure 4.
Changes in import component transcript abundance over time after treatment with mitochondrial electron transport chain inhibitors. Transcript abundance of import components were measured by quantitative real-time PCR at 0, 1, 3, and 12 h after treatment of Arabidopsis cell culture with 5 μm antimycin A or 40 μm rotenone. The fold change in transcripts of TIM components (A) and TOM components (B), which changed significantly in comparison with the transcript abundance in control cells over the same period. The fold change measured by microarray analysis after 12 h of rotenone treatment is shown (M). C, Comparison of the increase in transcript abundance of minor (indicated in black) and predominant (indicated in white) import component isoforms after 12 h of rotenone treatment as measured by quantitative real-time PCR.
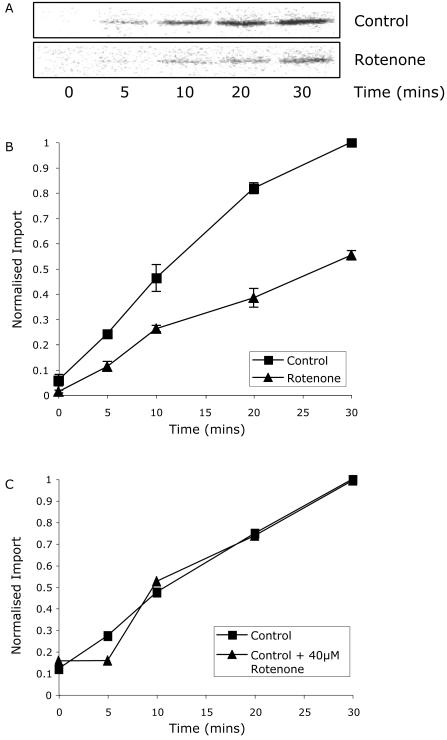
The microarray gene expression profiles after rotenone treatment of cell culture indicates an increase in the expression of genes encoding mitochondrial protein import components and mitochondrial biogenesis in general. The increase of several mitochondrial proteases may be required to replace real damaged components of mitochondria because of the rotenone treatment or the perception of damage because of the partial inactivation of electron transport capacity. To test if rotenone treatment of cells affects protein import in vitro, protein import assays were performed with mitochondria isolated from control and rotenone-treated cells (Fig. 5). Import of the Arabidopsis nuclear-encoded mitochondrial protein RPS10 was lower in mitochondria isolated from Arabidopsis cell culture 12 h after rotenone treatment (Fig. 5A). Time course experiments indicated that the rate of import into mitochondria isolated from rotenone-treated cells was approximately 50% of the control rate (Fig. 5B). Addition of rotenone to mitochondria isolated from untreated cells with 40 μm rotenone immediately before import did not result in a significant change in the rate of protein import (Fig. 5C), indicating import per se was not being directly inhibited by rotenone.
Figure 5.
The rate of mitochondrial protein import is decreased in rotenone-treated mitochondria. A, In vitro import of RPS10 into mitochondria isolated from control or rotenone-treated cell culture. B, Rate of import into mitochondria isolated from control or rotenone-treated cell culture. C, Rate of import in the presence or absence of 40 μm rotenone of mitochondria isolated from cell culture.
DISCUSSION
Molecular or genetic approaches are required to characterize the majority of components of the mitochondrial protein import apparatus because of their scarcity. This is especially true for the translocases of the inner membrane, where the abundant respiratory chain complexes mean that even in yeast, the identification of these components was achieved largely by genetic means (Rehling et al., 2001). Subsequent tagging and knockout and mutational approaches have allowed the determination of biochemical mechanisms. The identification of these components in yeast also has greatly facilitated efforts in plant and mammalian systems in the identification and study of components of the mitochondrial protein import apparatus. This in silico approach shows that although there are many similarities, some major differences also exist, and even when orthologs can be identified in Arabidopsis for components, close analysis indicates that they either differ structurally to their yeast counterpart or lack motifs or residues shown to be essential for function in yeast (Rapaport et al., 2001; Gabriel et al., 2003; Taylor et al., 2003b). Therefore, to identify which genes are expressed in Arabidopsis whose products comprise the mitochondrial protein import apparatus and how either expression changes with mitochondrial function, we carried out detailed analysis of the expression patterns of all genes identified by homology to the yeast import components. Transcriptomic analysis was used to investigate additional changes in components required for mitochondrial biogenesis, and a proteomic approach to identify many of the import apparatus proteins actually present in mitochondria.
A comprehensive expression analysis of all the genes identified in the Arabidopsis genome involved in the mitochondrial protein import apparatus indicates that they are all expressed in all organs examined, except for TOM20-1. Notable differences to the general pattern observed was evident for low TIM17-1 expression in roots and the high level of TIM23-3 and TIM44-1 message levels in roots compared with other organs. This indicates, at least for some members of the small gene families, that specialization in expression is evident. Examination of the actual transcript levels indicates that a 10-fold difference in the level of message for some components is observed despite the fact that protein levels would be expected to be the same in a functioning complex (Dekker et al., 1998; Moro et al., 1999). The transcript abundance of TIM44 was almost 10-fold lower than TIM50 and TIM23, whereas the levels of TOM9, TOM20, and TOM40 were almost 10-fold lower than TOM6. However, within the gene families themselves, there was a good correlation between the member of each gene family with the highest transcript level and the protein isoforms detected in purified mitochondria. Therefore, although many import components are encoded by small multigene families, it appears that largely one member functions to form the mitochondrial protein import apparatus.
The general question of why some components are encoded by small gene families is pertinent given that expression of some isoforms is extremely low. To investigate this, we treated cells with the mitochondrial poisons rotenone and antimycin A and analyzed the transcript abundance of the import components. It was evident that on treatment with both compounds, the expression of many import components was up-regulated significantly. The minor isoforms displayed the greater induction by these treatments, most notably for the pore-forming subunits TOM40, TIM17, TIM23, and TIM22. Rotenone appeared to have a slightly greater effect than antimycin A, with higher stimulation observed for TIM17-1, TIM23-1, TIM50, TIM44-2, TOM7-2, TOM40-2, and TOM20-4. We observed that isolated mitochondria from rotenone-treated cells imported proteins at 50% of the rate of mitochondria isolated from control cells. Thus, rotenone treatment may be causing mitochondrial damage and protein turnover that require replacement through mitochondrial biogenesis. Thus, the signal driving the up-regulation of expression of genes for components of the mitochondrial import apparatus is the decreased rate of import, which is still evident after 12 h. We propose that the up-regulation of gene expression observed is an attempt to overcome this decreased rate of import.
Diverse changes upon inhibition of complex I have been reported previously (Dutilleul et al., 2003). It has been demonstrated that cytoplasmic male-sterile mutant plants impaired in complex I function lost diurnal patterns of alternative oxidase activity, had altered antioxidant enzyme expression and activity, and showed higher tolerance to ozone levels and tobacco mosaic virus. Recently, the association of the terminal enzyme in ascorbate biosynthesis, l-galactono-1,4-lactone dehydrogenase, with complex I has been discovered (Millar et al., 2003). Treatment with rotenone impeded ascorbate biosynthesis, demonstrating a link between complex I activity and synthesis of a key cellular antioxidant. The reduction of ascorbate levels may result in damage to proteins by reactive oxygen species, necessitating replacement of the damaged components. Such damage to the protein import machinery may be the mechanism by which the rate of protein import into rotenone-treated mitochondria is reduced. With the results presented here, this indicates that perturbation of complex I activity may have widespread effects on gene expression and, thus, could be an important point for retrograde signaling between the mitochondrion and the nucleus.
The presence of multigene families is not unique to nuclear-encoded proteins destined for mitochondria several of the chloroplast import components are encoded in small gene families (Jackson-Constan and Keegstra, 2001; Davila-Aponte et al., 2003). In the case of the plant mitochondrial protein import components, it is likely that some components encoded by gene families will have overlapping but distinct roles. It is possible that the multiple forms of TOM20 compensate for the lack of a receptor domain of TOM9 and the apparent lack of TOM70 in plants (Werhahn et al., 2001; Werhahn and Braun, 2002). We have shown previously that the different isoforms of TIM17 and TIM23 differ in their ability to complement knockout mutants for the orthologous subunits in yeast, which further suggests some specialization of function between different isoforms (Murcha et al., 2003). In addition, we have demonstrated here that the various isoforms display differential induction upon inhibitor treatment that also may account for common changes in the mitochondrial proteome observed upon oxidative stress and inhibitor treatments (Sweetlove et al., 2002) and changes in import of various precursor proteins observed under environmental stress (Taylor et al., 2003a).
Overall, our investigations showed that with few exceptions, there is one prominent expressed isoform for each import component in Arabidopsis, which is confirmed by the protein detected in mitochondria. This suggests a similar import apparatus in the majority of organs. The expression of the additional isoforms was induced upon respiratory inhibitor treatment. The induction of genes encoding components of the mitochondrial protein import apparatus may allow recovery from such treatments. It also may contribute to differences observed in mitochondrial proteomes and to protein import upon chemical oxidant and environmental stress treatments. Thus, various members of the gene families may produce an import apparatus with different characteristics to that observed under normal conditions.
MATERIALS AND METHODS
Plant Growth
Arabidopsis plants were grown in media (Gamborg et al., 1968) at 22°C with a 16-h-light (100 μE m–2 s–1) and 8-h-dark photoperiod. Arabidopsis suspension cell culture was maintained as described (Sweetlove et al., 2002).
Chemical Treatment of Arabidopsis Cell Culture
Arabidopsis cell cultures were treated by the addition of 12 μL of 50 mm antimycin A (final concentration 5 μm) or 480 μL of 10 mm rotenone (final concentration 40 μm) to 120 mL of suspension cell culture 4 d after subculturing.
Cloning and Expression Analysis of Components of the Mitochondrial Protein Import Apparatus
Cloning and transcript abundance for components of the mitochondrial protein import apparatus were carried out as previously described (Murcha et al., 2003). Primers for amplification of the cDNAs and determination of transcript abundance are listed in the supplemental material, available in the online version of this article at http://www.plantphysiol.org.
To compare the expression patterns of the various genes obtained from quantitative RT-PCR, MPSS, and microarrays, the data were normalized as follows. For each experimental approach, the transcript abundance of every gene was averaged over the different organ types, then within each gene family, these average message levels of the different isoforms were normalized, with the most abundant isoform given the arbitrary value of 100. This enabled comparison of the average transcript abundance of different isoforms within a gene family in all organs analyzed and between real-time RT-PCR and MPSS data. Within each gene family, microarray signal values and EST numbers were normalized against the isoform with the highest transcript abundance, allowing comparison of microarray data and EST numbers in each gene family with real-time RT-PCR and MPSS data.
Microarray Analysis
Microarray analysis of the changes in transcript abundance in Arabidopsis cell culture was performed using Affymetrix GeneChip Arabidopsis ATH1 Genome Arrays (catalog no. 510690, Affymetrix). Total RNA was isolated from Arabidopsis cell culture 12 h after treatment with 40 μm rotenone using the RNeasy Plant mini protocol (Qiagen, Clifton Hill, Victoria, Australia). The high quality of the total RNA was verified by using both an Agilent Bioanalyzer (Agilent Technologies, Palo Alto, CA) and spectrophotometric analysis of the A260 to A280 ratio. Six flasks of Arabidopsis cell culture were treated with rotenone and combined into two separate pools. Total RNA was isolated from each of the two pools; after cRNA sample preparation, each was analyzed on separate GeneChips, resulting in two biological replicates of rotenone-treated cells. Six untreated control cell culture flasks were prepared similarly, resulting in two biological replicates of control treated cells. Double-stranded cDNA synthesis, biotin-labeled cRNA target synthesis, target hybridization, washing, staining, and scanning were performed exactly as described in the Affymetrix GeneChip Expression Analysis Technical Manual, using the kits, chemicals, and reagents precisely as outlined. Control Oligo B2 and Biotinylated Hybridization Controls (Affymetrix) were included in the hybridization. Before hybridization to an ATH1 GeneChip, the cRNA target quality was assessed by hybridization of an aliquot of the prepared cRNA to a Test3 array (catalog no. 510599, Affymetrix). Hybridization was performed in an Affymetrix GeneChip hybridization Oven 640. Washing and staining were performed using an Affymetrix Fluidics Station 400. Scanning was performed with an Agilent GeneArray Scanner G2500A. GeneChip scans were initially analyzed using the Affymetrix Microarray Suite 5.1 software, from which PivotData tables were exported. Raw data from the PivotData Tables were analyzed in GeneSpring software version 6 (Silicon Genetics, Redwood City, CA), using the parameters suggested by Silicon Genetics for analysis of Affymetrix Microarrays. All quoted changes in transcript abundance between control and rotenone-treated cells were significant, with Student's t test P values < 0.05.
Isolation and Fractionation of Arabidopsis Mitochondria
For protein identification, Arabidopsis cell culture mitochondria were isolated from cultures 7 d after subculturing as described (Sweetlove et al., 2002). Sub-fractionation of mitochondria into inner membrane, matrix, outer membrane, and intermembrane space was performed as described previously (Lister et al., 2002) based on the methods outlined by Sweetlove et al. (2001). For in vitro import experiments, Arabidopsis cell culture mitochondria were isolated after 12 h of chemical treatment.
Trypsin Digestion and Liquid Chromatography-MS/MS Analysis of Whole Mitochondrial Protein Extracts
Mitochondrial protein extracts were acetone precipitated at –20°C overnight. Fifty micrograms of protein was digested with 5 μg of trypsin (Roche, Sydney) overnight at 37°C in 100 mm Tris (pH 8.5). Resulting tryptic peptides were differentially separated over 10 h using a 0.3- × 150-mm Zorbax C18 column (Agilent, Sydney) and injected directly into Q-Star Pulsar I MS/MS (Applied Biosystems, Sydney) via an electrospray source. Peptides were automatically selected by Analyst QS (Applied Biosystems) for MS/MS analysis and fragmented with N2. Mass spectra and collision MS/MS data were analyzed and matched to predicted gene products with BioAnalyst and ProID software (Applied Biosystems), using mass accuracy cutoffs of peptide mass ± 0.15 and MS/MS ± 0.05. Collison-induced dissociations were also analyzed by Mascot (Matrix Science, London) for independent matching.
[35S]-Met-Labeled Precursor Proteins
Arabidopsis ribosomal protein S10 (RPS10) was amplified using primers designed to sequences obtained from cDNA to the RPS gene (At3g22300; Adams et al., 2002). [35S]-Met labeled precursor proteins were synthesized using the rabbit reticulocyte TNT in vitro transcription/translation kit (Promega, Madison, WI) as described previously (Whelan et al., 1995).
In Vitro Import into Mitochondria
Import of precursor proteins was carried out in import master mix using intact mitochondria isolated from control or rotenone-treated Arabidopsis cell culture as described previously (Whelan et al., 1995). Rotenone treatment of mitochondria was performed by addition of 10 mm rotenone to a final concentration of 40 μm in the final import reaction. Protein import, protease treatment of mitochondria, separation, and detection of imported proteins using phosphor imaging was performed as previously outlined (Murcha et al., 1999). The use of equivalent amounts of control and rotenone-treated mitochondria was ensured by measuring protein concentration with the Coomassie Protein Assay Reagent as per the manufacturer's instructions (Pierce, Rockford, IL). Three independent cell culture treatments, mitochondrial isolations, and in vitro import reactions were performed.
Supplementary Material
Acknowledgments
Use of the facilities of the Lotterywest State Microarray Facility and assistance from Violet Peeva and William Kenworthy in Affymetrix Gene-Chip Microarray use and analysis are gratefully acknowledged.
Article, publication date, and citation information can be found at http://www.plantphysiol.org/cgi/doi/10.1104/pp.103.033910.
This work was supported by the Australian Research Council (funds to A.H.M. and J.W. and Australian Postdoctoral Fellowship to A.H.M.), by Australian Postgraduate Awards (to R.L. and R.C.), by University Postgraduate Awards (to O.C. and M.N.L.), and by University of Western Australia (Small Grants Scheme to A.H.M. and J.W.).
The online version of this article contains Web-only data.
References
- Adams KL, Daley DO, Whelan J, Palmer JD (2002) Genes for two mitochondrial ribosomal proteins in flowering plants are derived from their chloroplast or cytosolic counterparts. Plant Cell 14: 931–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahting U, Thieffry M, Engelhardt H, Hegerl R, Neupert W, Nussberger S (2001) Tom40, the pore-forming component of the protein-conducting Tom channel in the outer membrane of mitochondria. J Cell Biol 153: 1151–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimura S, Tsutsumi N (2002) A dynamin-like protein (ADL2b), rather than FtsZ, is involved in Arabidopsis mitochondrial division. Proc Natl Acad Sci USA 99: 5727–5731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlt H, Tauer R, Feldmann H, Neupert W, Langer T (1996) The YTA10-12 complex, an AAA protease with chaperone-like activity in the inner membrane of mitochondria. Cell 85: 875–885 [DOI] [PubMed] [Google Scholar]
- Armstrong LC, Komiya T, Bergman BE, Mihara K, Bornstein P (1997) Metaxin is a component of a preprotein import complex in the outer membrane of the mammalian mitochondrion. J Biol Chem 272: 6510–6518 [DOI] [PubMed] [Google Scholar]
- Balandin T, Castresana C (2002) AtCOX17, an Arabidopsis homolog of the yeast copper chaperone COX17. Plant Physiol 129: 1852–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer MF, Rothbauer U, Muhlenbein N, Smith RJ, Gerbitz K, Neupert W, Brunner M, Hofmann S (1999) The mitochondrial TIM22 preprotein translocase is highly conserved throughout the eukaryotic kingdom. FEBS Lett 464: 41–47 [DOI] [PubMed] [Google Scholar]
- Braun HP, Schmitz UK (1995) Are the “core” proteins of the mitochondrial bc1 complex evolutionary relics of a processing protease? Trends Biochem Sci 20: 171–175 [DOI] [PubMed] [Google Scholar]
- Brenner S, Johnson M, Bridgham J, Golda G, Lloyd DH, Johnson D, Luo S, McCurdy S, Foy M, Ewan M et al. (2000) Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat Biotechnol 18: 630–634 [DOI] [PubMed] [Google Scholar]
- Davila-Aponte JA, Inoue K, Keegstra K (2003) Two chloroplastic protein translocation components, Tic110 and Toc75, are conserved in different plastid types from multiple plant species. Plant Mol Biol 51: 175–181 [DOI] [PubMed] [Google Scholar]
- Dessi P, Whelan J (1997) Temporal regulation of in vitro import of precursor proteins into mitochondria. FEBS Lett 415: 173–178 [DOI] [PubMed] [Google Scholar]
- Dekker PJ, Ryan MT, Brix J, Muller H, Honlinger A, Pfanner N (1998) Preprotein translocase of the outer mitochondrial membrane: molecular dissection and assembly of the general import pore complex. Mol Cell Biol 18: 6515–6524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley P, Wood CK, Pratt JR, Moore AL (1997) Developmental regulation of the plant mitochondrial matrix located HSP70 chaperone and its role in protein import. FEBS Lett 417: 321–324 [DOI] [PubMed] [Google Scholar]
- Dutilleul C, Garmier M, Noctor G, Mathieu C, Chetrit P, Foyer CH, de Paepe R (2003) Leaf mitochondria modulate whole cell redox homeostasis, set antioxidant capacity, and determine stress resistance through altered signaling and diurnal regulation. Plant Cell 15: 1212–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localisation of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Gabriel K, Egan B, Lithgow T (2003) Tom40, the import channel of the mitochondrial outer membrane, plays an active role in sorting imported proteins. EMBO J 22: 2380–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamborg OL, Miller RA, Ojima K (1968) Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res 50: 151–158 [DOI] [PubMed] [Google Scholar]
- Geissler A, Chacinska A, Truscott KN, Wiedemann N, Brandner K, Sickmann A, Meyer HE, Meisinger C, Pfanner N, Rehling P (2002) The mitochondrial presequence translocase: an essential role of Tim50 in directing preproteins to the import channel. Cell 111: 507–518 [DOI] [PubMed] [Google Scholar]
- Glaser E, Dessi P (1999) Integration of the mitochondrial-processing peptidase into the cytochrome bc1 complex in plants. J Bioenerg Biomembr 31: 259–274 [DOI] [PubMed] [Google Scholar]
- Glerum DM, Muroff I, Jin C, Tzagoloff A (1997) COX15 codes for a mitochondrial protein essential for the assembly of yeast cytochrome oxidase. J Biol Chem 272: 19088–19094 [DOI] [PubMed] [Google Scholar]
- Glerum DM, Shtanko A, Tzagoloff A (1996) Characterization of COX17, a yeast gene involved in copper metabolism and assembly of cytochrome oxidase. J Biol Chem 271: 14504–14509 [DOI] [PubMed] [Google Scholar]
- Heazlewood JL, Millar AH, Day DA, Whelan J (2003) What makes a mitochondrion? Genome Biol 4: 218–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson-Constan D, Keegstra K (2001) Arabidopsis genes encoding components of the chloroplastic protein import apparatus. Plant Physiol 125: 1567–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansch L, Kruft V, Schmitz UK, Braun HP (1998) Unique composition of the preprotein translocase of the outer mitochondrial membrane from plants. J Biol Chem 273: 17251–17257 [DOI] [PubMed] [Google Scholar]
- Kerscher O, Sepuri NB, Jensen RE (2000) Tim18p is a new component of the Tim54p-Tim22p translocon in the mitochondrial inner membrane. Mol Biol Cell 11: 103–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler CM, Murphy MP, Bally NA, Leuenberger D, Oppliger W, Dolfini L, Junne T, Schatz G, Or E (2000) Tim18p, a new subunit of the Tim22 complex that mediates insertion of imported proteins into the yeast mitochondrial inner membrane. Mol Cell Biol 20: 1187–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodziejczak M, Kolaczkowska A, Szczesny B, Urantowka A, Knorpp C, Kieleczawa J, Janska H (2002) A higher plant mitochondrial homologue of the yeast m-AAA protease: molecular cloning, localization, and putative function. J Biol Chem 277: 43792–43798 [DOI] [PubMed] [Google Scholar]
- Lister R, Mowday B, Whelan J, Millar AH (2002) Zinc-dependent inter-membrane space proteins stimulate import of carrier proteins into plant mitochondria. Plant J 30: 555–566 [DOI] [PubMed] [Google Scholar]
- Lister R, Murcha MW, Whelan J (2003) The Mitochondrial Protein Import Machinery of Plants (MPIMP) database. Nucleic Acids Res 31: 325–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohret TA, Jensen RE, Kinnally KW (1997) Tim23, a protein import component of the mitochondrial inner membrane, is required for normal activity of the multiple conductance channel, MCC. J Cell Biol 137: 377–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luirink J, Samuelsson T, de Gier JW (2001) YidC/Oxa1p/Alb3: evolutionarily conserved mediators of membrane protein assembly. FEBS Lett 501: 1–5 [DOI] [PubMed] [Google Scholar]
- Mascasev D, Newbigin E, Whelan J, Lithgow T (2000) How do plant mitochondria avoid importing chloroplast proteins? Components of the import apparatus Tom20 and Tom22 from Arabidopsis differ from their fungal counterparts. Plant Physiol 123: 811–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milisav I, Moro F, W. N., Brunner M (2001) Modular structure of the Tim23 preprotein translocase of mitochondria. J Biol Chem 276: 25856–25861 [DOI] [PubMed] [Google Scholar]
- Millar AH, Mittova V, Kiddle G, Heazlewood JL, Bartoli CG, Theodoulou FL, Foyer CH (2003) Control of ascorbate synthesis by respiration and its implications for stress responses. Plant Physiol 133: 443–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AH, Sweetlove LJ, Giege P, Leaver CJ (2001) Analysis of the Arabidopsis mitochondrial proteome. Plant Physiol 127: 1711–1727 [PMC free article] [PubMed] [Google Scholar]
- Mokranjac D, Paschen SA, Kozany C, Prokisch H, Hoppins SC, Nargang FE, Neupert W, Hell K (2003) Tim50, a novel component of the TIM23 preprotein translocase of mitochondria. EMBO J 22: 816–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moro F, Sirrenberg C, Schneider HC, Neupert W, Brunner M (1999) The TIM17.23 preprotein translocase of mitochondria: composition and function in protein transport into the matrix. EMBO J 18: 3667–3675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozdy AD, McCaffery JM, Shaw JM (2000) Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J Cell Biol 151: 367–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murcha MW, Huang T, Whelan J (1999) Import of precursor proteins into mitochondria from soybean tissues during development. FEBS Lett 464: 53–59 [DOI] [PubMed] [Google Scholar]
- Murcha MW, Lister R, Ho AY, Whelan J (2003) Identification, expression, and import of components 17 and 23 of the inner mitochondrial membrane translocase from Arabidopsis. Plant Physiol 131: 1737–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargang FE, Preuss M, Neupert W, Herrmann JM (2002) The Oxa1 protein forms a homooligomeric complex and is an essential part of the mitochondrial export translocase in Neurospora crassa. J Biol Chem 277: 12846–12853 [DOI] [PubMed] [Google Scholar]
- Neupert W (1997) Protein import into mitochondria. Annu Rev Biochem 66: 863–917 [DOI] [PubMed] [Google Scholar]
- Nijtmans LG, Artal Sanz M, Bucko M, Farhoud MH, Feenstra M, Hakkaart GA, Zeviani M, Grivell LA (2001) Shy1p occurs in a high molecular weight complex and is required for efficient assembly of cytochrome c oxidase in yeast. FEBS Lett 498: 46–51 [DOI] [PubMed] [Google Scholar]
- Nobrega FG, Nobrega MP, Tzagoloff A (1992) BCS1, a novel gene required for the expression of functional Rieske iron-sulfur protein in Saccharomyces cerevisiae. EMBO J 11: 3821–3829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfanner N, Geissler A (2001) Versatility of the mitochondrial protein import machinery. Nat Rev Mol Cell Biol 2: 339–349 [DOI] [PubMed] [Google Scholar]
- Rapaport D, Taylor RD, Kaser M, Langer T, Neupert W, Narang FE (2001) Structural requirements of Tom40 for assembly into preexisting Tom complexes of mitochondria. Mol Biol Cell 12: 1189–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehling P, Model K, Brandner K, Kovermann P, Sickmann A, Meyer HE, Kuhlbrandt W, Wagner R, Truscott KN, Pfanner N (2003a) Protein insertion into the mitochondrial inner membrane by a twin-pore translocase. Science 299: 1747–1751 [DOI] [PubMed] [Google Scholar]
- Rehling P, Pfanner N, Meisinger C (2003b) Insertion of hydrophobic membrane proteins into the inner mitochondrial membrane: a guided tour. J Mol Biol 326: 639–657 [DOI] [PubMed] [Google Scholar]
- Rehling P, Wiedemann N, Pfanner N, Truscott KN (2001) The mitochondrial import machinery for preproteins. Crit Rev Biochem Mol Biol 36: 291–336 [DOI] [PubMed] [Google Scholar]
- Sakamoto W, Spielewoy N, Bonnard G, Murata M, Wintz H (2000) Mitochondrial localisation of AtOXA1, an Arabidopsis homologue of yeast Oxa1p involved in the insertion and assembly of protein complexes in mitochondrial inner membrane. Plant Cell Physiol 41: 1157–1163 [DOI] [PubMed] [Google Scholar]
- Schulze M, Rodel G (1988) SCO1, a yeast nuclear gene essential for accumulation of mitochondrial cytochrome c oxidase subunit II. Mol Gen Genet 211: 492–498 [DOI] [PubMed] [Google Scholar]
- Sjoling S, Glaser E (1998) Mitochondrial targeting peptides in plants. Trends Plant Sci 3: 136–140 [Google Scholar]
- Stahl A, Moberg P, Ytterberg J, Panfilov O, Brockenhuus Von Lowenhielm H, Nilsson F, Glaser E (2002) Isolation and identification of a novel mitochondrial metalloprotease (PreP) that degrades targeting presequences in plants. J Biol Chem 277: 41931–41939 [DOI] [PubMed] [Google Scholar]
- Sweetlove LJ, Heazlewood JL, Herald V, Holtzapffel R, Day DA, Leaver CJ, Millar AH (2002) The impact of oxidative stress on Arabidopsis mitochondria. Plant J 32: 891–904 [DOI] [PubMed] [Google Scholar]
- Sweetlove LJ, Mowday B, Hebestreit HF, Leaver CJ, Millar AH (2001) Nucleoside diphosphate kinase III is localised to the inter-membrane space in plant mitochondria. FEBS Lett 254: 1–5 [DOI] [PubMed] [Google Scholar]
- Szigyarto C, Dessi P, Smith MK, Knorpp C, Harmey MA, Day DA, Glaser E, Whelan J (1998) A matrix-located processing peptidase of plant mitochondria. Plant Mol Biol 36: 171–181 [DOI] [PubMed] [Google Scholar]
- Taylor NL, Rudhe C, Hulett JM, Lithgow T, Glaser E, Day DA, Millar AH, Whelan J (2003a) Environmental stresses inhibit and stimulate different protein import pathways in plant mitochondria. FEBS Lett 547: 125–130 [DOI] [PubMed] [Google Scholar]
- Taylor RD, McHale BJ, Nargang FE (2003b) Characterization of Neurospora crassa Tom40-deficient mutants and effect of specific mutations on Tom40 assembly. J Biol Chem 278: 765–775 [DOI] [PubMed] [Google Scholar]
- Taylor SW, Fahy E, Zhang B, Glenn GM, Warnock DE, Wiley S, Murphy AN, Gaucher SP, Capaldi RA, Gibson BW et al. (2003c) Characterization of the human heart mitochondrial proteome. Nat Biotechnol 21: 281–286 [DOI] [PubMed] [Google Scholar]
- Werhahn W, Braun HP (2002) Biochemical dissection of the mitochondrial proteome from Arabidopsis thaliana by three-dimensional gel electrophoresis. Electrophoresis 23: 640–646 [DOI] [PubMed] [Google Scholar]
- Werhahn W, Jansch L, Braun H-P (2003) Identification of novel subunits of the TOM complex from Arabidopsis thaliana. Plant Physiol Biochem 41: 407–416 [Google Scholar]
- Werhahn W, Niemeyer A, Jansch L, Kruft V, Schmitz UK, Braun H-P (2001) Purification and characterisation of the preprotein translocase of the outer mitochondrial membrane from Arabidopsis: identification of multiple forms of Tom 20. Plant Physiol 125: 943–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan J, Hugosson M, Glaser E, Day DA (1995) Studies on the import and processing of the alternative oxidase precursor by isolated soybean mitochondria. Plant Mol Biol 27: 769–778 [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Esaki M, Kanamori T, Tamura Y, Nishikawa S, Endo T (2002) Tim50 is a subunit of the TIM23 complex that links protein translocation across the outer and inner mitochondrial membranes. Cell 111: 519–528 [DOI] [PubMed] [Google Scholar]
- Yen MR, Harley KT, Tseng YH, Saier MH Jr (2001) Phylogenetic and structural analyses of the oxa1 family of protein translocases. FEMS Microbiol Lett 204: 223–231 [DOI] [PubMed] [Google Scholar]
- Yu J, Nickels R, McIntosh L (2001) A genome approach to mitochondrial-nuclear communication in Arabidopsis. Plant Physiol Biochem 39: 345–353 [Google Scholar]
- Zhang X-P, Sjoling S, Tanudji M, Somogyi L, Andreu D, Eriksson LEG, Graslund A, Whelan J, Glaser E (2001) Mutagenesis and computer modelling approach to study determinants for recognition of signal peptides by the mitochondrial processing peptidase. Plant J 27: 427–438 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.