Abstract
Besides the systemic acquired resistance (SAR) induced in response to microbial stimulation, host plants may also acquire resistance to pathogens in response to endogenous stimuli associated with their own development. In tobacco (Nicotiana tabacum), the vegetative-to-flowering transition comes along with a susceptibility-to-resistance transition to the causal agent of black shank disease, the oomycete Phytophthora parasitica. This resistance affects infection effectiveness and hyphal expansion and is associated with extracellular accumulation of a cytotoxic activity that provokes in vitro cell death of P. parasitica zoospores. As a strategy to determine the extracellular events important for restriction of pathogen growth, we screened the tobacco genome for genes encoding secreted or membrane-bound proteins expressed in leaves of flowering plants. Using a signal sequence trap approach in yeast (Saccharomyces cerevisiae), 298 clones were selected that appear to encode for apoplastic, cell wall, or membrane-bound proteins involved in stress response, in plant defense, or in cell wall modifications. Microarray and northern-blot analyses revealed that, at late developmental stages, leaves were characterized by the coordinate up-regulation of genes involved in SAR and in peroxidative cross-linking of structural proteins to cell wall. This suggests the potential involvement of these genes in extracellular events that govern the expression of developmental resistance. The analysis of the influence of salicylic acid on mRNA accumulation also indicates a more complex network for regulation of gene expression at a later stage of tobacco development than during SAR. Further characterization of these genes will permit the formulation of hypotheses to explain resistance and to establish the connection with development.
Establishment of acquired resistance to pathogens may be induced by physiological and/or developmental changes taking place in growing plants. The occurrence of a transition from susceptibility to resistance during development is a widely reported phenomenon in monocotyledons and dicotyledons in the case of viruses (Leisner et al., 1992, 1993), bacteria (Century et al., 1999; Kus et al., 2002), fungi (Moose and Sisco, 1994; Abedon and Tracy, 1996), and oomycetes (Reuveni et al., 1986; Wyatt et al., 1991: Hugot et al., 1999). Although this phenomenon, age-related resistance (ARR), is well documented from a pathological point of view, few studies have dealt with the genetic and molecular bases of disease control during plant development.
On the contrary, numerous studies have investigated defense mechanisms activated in response to pathogen infection and associated with plant disease resistance (Hammond-Kosack and Parker, 2003). These studies have underlined the key role of the host extracellular space as sorting compartment of plant-pathogen interactions. At the early step of infection, the host plant recognizes some pathogen-secreted molecules that elicit the coordinate activation of defense reactions. At late stages, a number of host-secreted molecules accumulate in the extracellular space and contribute to the control of invasion of pathogens. Synthesis and secretion of defense-related proteins are a critical part of the establishment of resistance. A defensive role of some proteins encoded by pathogenesis-related (PR) genes is strongly suggested by their antimicrobial activity in vitro and in vivo. It is the case for certain PR proteins (Van Loon and Strien, 1999) of classes 1 (Alexander et al., 1993; Niderman et al., 1995), 2 (Mauch et al., 1988; Sela-Buurlage et al., 1993), and 5 (Abad et al., 1996).
Physiological and/or developmental modifications leading to the expression of developmental acquired resistance involve extracellular events and probably also require accumulation of defense compounds. Reuveni et al. (1986) have proposed that the activation of defense mechanisms associated with developmental resistance leads to the accumulation of antimicrobial substances inhibiting the pathogen development. Different observations are consistent with this hypothesis. Some PR genes are known to be up-regulated by both plant/pathogen interactions and developmental signals. PR-1 and PR-2 proteins are expressed in floral tissue (Lotan et al., 1989; Côté et al., 1991) and in leaves (Fraser, 1981; Uknes et al., 1993) during floral development of healthy tobacco (Nicotiana tabacum). The establishment of tobacco resistance to Phytophthora parasitica is associated with the expression of an extracellular cytotoxic activity effective against P. parasitica zoospores and with accumulation of PR-1 proteins in the apoplasm (Hugot et al., 1999). In Arabidopsis, ARR to Pseudomonas syringae is also associated with the expression of an extracellular antibacterial activity (Kus et al., 2002). These changes are initiated either independently of interactions with pathogens (Hugot et al., 1999) or in response to infection (Kus et al., 2002).
Systemic acquired resistance (SAR) is one of the best characterized plant defense responses activated by pathogen infection. It occurs when pathogens induce localized plant necrosis during initial infection. Plant defense responses are activated locally and also may occur in distal uninfected tissues leading to SAR, which is long lasting and effective against a broad spectrum of virulent and avirulent pathogens (Ross, 1961; Kuc, 1982). During the last decade, extensive studies have greatly improved our knowledge of plant signaling networks involved in the repression or activation of SAR (Ryals et al., 1996; Dong, 2001). Salicylic acid (SA) has been identified first as a positive component playing an essential role in the SAR transduction pathway (Malamy et al., 1990; Métraux et al., 1990; Rasmussen et al., 1991; Ward et al., 1991; Gaffney et al., 1993; Delaney et al., 1994). Genetic studies on Arabidopsis notably have led to the identification of MPK4 and NPR1/NIM genes that are required for SAR repression and activation, respectively. MPK4 encodes a peptide with a kinase activity (Petersen et al., 2000), and NPR1 encodes an ankyrin repeat protein involved in SA perception and downstream SAR responses (Cao et al., 1997; Ryals et al., 1997). Nuclear localization of NPR1 is essential for SA-induced gene expression (Kinkema et al., 2000). This protein interacts with members of the TGA family of basic Leu zipper transcription factors (Després et al., 2000) that bind to PR1 promoter elements, suggesting that NPR1-mediated DNA binding of TGA factors is critical for activation of defense genes (Qin et al., 1994; Jupin and Chua, 1996; Lebel et al., 1998; Fan and Dong, 2002). Positive and negative regulatory elements in promoter sequences of SAR genes also have been characterized based on transcriptome analysis during SAR (Maleck et al., 2000; Petersen et al., 2000). Downstream of the activation of a transduction pathway leading to SAR, a number of extracellular events regulate the plant/pathogen interactions. Different host-secreted molecules accumulate in the extracellular space and contribute to the control of invasion by the pathogen, among which are some PR proteins of classes 1, 2, and 5 (Uknes et al., 1992).
Preliminary studies on the mechanisms involved in acquired resistance occurring during plant development have underlined similarities and differences with SAR. Thus, as in the case of SAR, SA is required for activation of the signaling transduction pathway leading to resistance against P. parasitica and P. syringae in tobacco and in Arabidopsis, respectively (Hugot et al., 1999; Kus et al., 2002). However, unlike SAR, Arabidopsis ARR to P. syringae does not depend on a functional NPR1 gene and is not caused by constitutive PR-1 gene expression in mature leaves (Kus et al., 2002). In tobacco, resistance to P. parasitica is associated with SA-dependent accumulation of PR-1 proteins in the apoplasm and also with SA-independent extracellular cytotoxic activity, which could not be detected from leaves induced to SAR (Hugot et al., 1999).
To have an overview of the extracellular modifications associated with transition from susceptibility to developmental resistance, we screened the tobacco genome for genes encoding secreted or membrane-bound proteins. Membrane proteins and proteins intended for secretion are targeted to the appropriate cellular localization by their signal peptides (Blobel and Dobberstein, 1975). We used a yeast (Saccharomyces cerevisiae) signal peptide selection system to isolate 5′-cDNA fragments that encode signal peptides (Klein et al., 1996; Jacobs et al., 1997, 1999) from leaf tissue acquiring resistance to P. parasitica and to identify cell surface and secreted proteins. By microarray and northern-blot analyses, we examined the changes that occur in the abundance of transcripts corresponding to genes coding for secreted proteins after induction of developmental resistance and compared them with changes occurring after SAR induction. Taken together, the results show that at late stages of tobacco development, resistance to oomycete is associated with activation of a different signaling network than during SAR. This network controls gene activation for extracellular defenses and for cell wall modifications without previous pathogen attack.
RESULTS
Selection of Tobacco cDNAs with Functional Signal Peptides
During vegetative growth, tobacco is highly susceptible to P. parasitica, the causal agent of black shank disease. Throughout flowering growth, leaves and stem develop resistance against this pathogen. At this stage, inoculated zones do not develop symptoms or develop symptoms with an area reduced by up to 80% when compared with those observed during vegetative growth. Resistance phenotype is associated with SA-independent extracellular cytotoxic activity and with SA-dependent accumulation of PR1 proteins in the apoplasm (Hugot et al., 1999). To undertake an analysis of extracellular events important for resistance, a cDNA library was constructed in pSUC2T7M130RI vector by using mRNA from 15-week-old and P. parasitica-resistant leaves. Synthesis of first strand cDNA used random hexamers, which increases the likelihood of obtaining a 5′ end signal peptide sequence and decreased the likelihood of producing stop codons in the pSUC2T7M130RI vector. pSUC2T7M130RI vector contained a modified invertase gene and lacked the region encoding an initiator Met and signal peptide (Klein et al., 1996). cDNA cloning efficiency (45%) and insert diversity were estimated by PCR analysis and sequence determination on 96 clones.
The Trp- and invertase-deficient yeast YTK12 strain was transformed with the tobacco cDNA library constructed in pSUC2T7M130RI. The yeast strain was able to grow on a Suc medium only when a cDNA encoding an initiator Met and a signal peptide were cloned in-frame with the invertase sequence of the vector (Klein et al., 1996). To estimate the rate of yeast transformants with trapped signal sequence, YTK12 cells were selected either on a medium without Trp and with Glc or on a medium without Trp and with raffinose. Approximately 2% of yeast colonies were able to grow in the second medium compared with those growing in the first medium. At the same time, we applied the Sequence Signal Trap (SST) strategy and isolated 298 clones able to synthesize and to secrete invertase.
After DNA sequencing, the 298 clones were found to represent 131 distinct genes. Determination BLAST search programs (Altschul et al., 1990; National Center for Biotechnology Information) were used to search for sequence homologies with known genes. Forty-three represented known tobacco genes (20) or orthologous genes from other plant species (23), 40 clones presented significant similarities with previously reported sequences encoding unknown proteins or resulting from expressed sequence tag (EST) programs, and 48 sequences were unrelated to any known proteins (Table I; see supplemental data Table II). All 131 sequences were analyzed with PSORT program (Nakai and Kanehisa, 1992; http://psort.nibb.ac.jp/) to predict the subcellular localization of the corresponding proteins and the presence of a signal peptide using both McGeoch (1985) and Nielsen et al. (1997) prediction methods. The cloning and signal peptide selection were very efficient: 81% of peptides corresponded to the known or were related to known cDNAs containing signal peptides and were addressed to the secretory pathway, the cell surface, or the extracellular environment, and 19% were predicted to be intracellular among which proteins targeted to mitochondrial inner membrane or to endoplasmic reticulum membrane. Among the 20 sequences representing tobacco genes, 17 corresponded to proteins containing a cleavable signal peptide (Table II). Deduced amino acid sequences from SST clones with no similarities with any known proteins were also analyzed for the presence of a putative signal peptide: 66% of proteins appeared to have a cleavable N-terminal signal sequence, 31% appeared to have none, and 9% of sequences could not be classified as having or not a signal peptide because they were too short or because of the discrepancy between the results obtained with McGeoch and Nielsen methods. Thus, the SST strategy clearly leads to select sequences enriched in tobacco genes encoding membrane-bound or secreted proteins: 76% of the 131 sequences analyzed contained a signal peptide.
Table I.
Sequence similarities to Signal Sequence Trap (SST) clones
| Clone Number | Previously Reported Proteins Identical or Related to SST Clones | GenBank Accession No. |
|---|---|---|
| Tobacco | ||
| 1 | Beta 1-3 glucanase | M20619 |
| 2 | PR-5d | D76437 |
| 3 | Elicitor-inducible protein | AB040407 |
| 4 | Lipid transfer protein | AF151214 |
| 5 | PAR-1a | X83853 |
| 6 | PAR-1b | X83851 |
| 7 | Putative prepro-Cys proteinase CPR2 | AJ242994 |
| 8 | Cys protease NTCP-23 | AB032168 |
| 9 | Cathepsin B-like Cys proteinase | X81995 |
| 0 | Lignin-forming peroxidase | J02979 |
| 11 | Pectin methylesterase | AJ401158 |
| 12 | Cell wall protein TLRP | Y19032 |
| 13 | Cys-rich protein | AF213464 |
| 14 | Nectarin I precursor | AF132671 |
| 15 | Arabinogalactan protein precursor | U13066 |
| 16 | Receptor-like kinase CHRK1 | AF088885 |
| 17 | Cell-type guard Gly-rich protein GRP1 | AF151215 |
| 18 | Invertase inhibitor homolog | Y12806. |
| 19 | Glutathione transferase PAR C | D85911 |
| 20 | Ribosomal protein L11 like | AJ295006 |
| Arabidopsis | ||
| 21 | Xylose isomerase | AB011482 |
| 22 | Putative receptor-like protein kinase | AB026644 |
| 23 | Similar to Ser/Thr kinases | AC009894 |
| 24 | Protein kinase homolog | F17M5.190 |
| 25 | Protein kinase-like protein | AL132960 |
| 26 | Pectinesterase | AB013388 |
| 27 | Ser carboxypeptidase II | AB023032 |
| 28 | Similar to Gly metalloendoproteinase | AC002062 |
| 29 | Ser carboxypeptidase precursor | AC011560 |
| 30 | Similar to protein disulfide isomerase | U63815 |
| 31 | Dehydration-responsive protein | D10703 |
| 32 | Cytosolic monodehydroascorbate reductase | AP000371 |
| Tomato | ||
| 33 | Osmotin-like protein | L76632 |
| 34 | Aspartic protease (LeAspP) | L46681 |
| 35 | Cystatin LTC | AF198388 |
| Others | ||
| 36 | Ser caboxypeptidase | AF141384 |
| 37 | Leu-rich receptor-like protein kinase | AF053127 |
| 38 | Leu-rich repeat protein kinase | AF172282 |
| 39 | Pectin acetylesterase precursor | S68805 |
| 40 | Plastidic ATP/ADP transporter | Y10821 |
| 41 | Fractalkine/neurotactin | AF071549 |
| 42 | Alpha amylase | X53049 |
| 43 | SINA1 p | Y18471 |
| Arabidopsis | ||
| 44 | Membrane protein | AC010795 |
| 45 | Similar to unknown protein | AB017067 |
| 46 | Similar to unknown protein | AB025622 |
| 47 | Similar to unknown protein | AC016795 |
| 48 | Similar to unknown protein | AB006703 |
| 49 | Unknown protein | AC006264 |
| 50 | Unknown protein | U76300 |
| 52 | Hypothetical protein | T22A6.180 |
| 53 | Hypothetical protein | F8F16.160 |
| 54 | Hypothetical protein | AC079730 |
| 55 | Hypothetical protein | T23J7.140 |
| 56 | Hypothetical protein | F13C5.140 |
| EST from tomato | ||
| 57 | Mixed elicitor | AW443014 |
| 58 | Mixed elicitor | AW092334 |
| 59 | Mixed elicitor | AW038593 |
| 60 | Mixed elicitor | AW041820 |
| 61 | Mixed elicitor | AW039505 |
| 62 | Mixed elicitor | AW094011 |
| 63 | Mixed elicitor | AW038448 |
| 64 | Flowers, buds | AW622724 |
| 65 | Flowers, buds | AW160161 |
| 66 | Flowers, buds | AW929481 |
| 67 | Flowers, buds | AW738469 |
| 68 | Fruit mature green | AW930018 |
| 69 | Fruit mature green | AW933059 |
| 70 | Immature green fruit | BE458435 |
| 71 | Ovary | A1485184 |
| 72 | Ovary | A1483697 |
| 73 | Radicle, postimbibition | AW625982 |
| 74 | Radicle, postimbibition | AW625778 |
| 75 | Root, plants pre-anthesis | BE344515 |
| 76 | Germinating seedlings | AW651276 |
| 77 | Callus | AI894912 |
| 78 | Callus | AI896368 |
| Others | ||
| 79 | cDNA (pepper [Capsicum annuum]) | AA840643 |
| 80 | EST (potato [Solanum tuberosum]) | BE923903 |
| 81 | EST (potato) | BE471805 |
| 82 | EST (potato) | BE922431 |
| 83 | EST (rice [Oryza sativa]) | OC02B07 |
Table II.
Subcellular localization
Selected clones homologous or similar to tobacco sequences with the corresponding deduced amino acid sequence. Subcellular localization based on protein sorting signals and associated score were determined from complete protein sequence with PSORT program (http://psort.nibb.ac.jp/). Signal peptide is underlined.
| Name | Deduced Amino Acid Sequence | Subcellular Localization | |
|---|---|---|---|
| PR2 | MSTSDKHNTPQMAAITLLGLLLVASTIEIAGAQSIGVCYGMLGNNLPNHWEVIQLYKSRN | Outside | 0.820 |
| PR-5d | MRSLITCLVFFLLPCVTYTHASGVFEVHNNCPYTVWAAATPVGGGRRLE | Outside | 0.738 |
| LTP1 | MVKVALLVVMCIAAATSVMLTPHADAAISCGQVVTRLSPCANY | Outside | 0.820 |
| Elicitor-inducible protein | MFNHILGXLXHIIIFFSFSISSISGSDDDCVYTAYVRTSSIIKGGTDSIISLTLYDADGYGIRIKN | Outside | 0.814 |
| PAR-1a | MVASFHSFTLAIVACALVFCVQVTLGSITCENLNKDSCTFAISSTGKRCVLEKHLRRSGEEVYACKTLEIEADKLKNWIETDQC | Outside | 0.820 |
| PAR-1b | MVASFHNFTLAIVACALVFCVQVTLGSITCENLNKDSCAFAISSTGKRCVLEKRLQRSGEEV | Outside | 0.820 |
| Cys proteinase CPR2 | MERLFLLSLLAFVLFSSAIAFSDEDPLIR QVVSETDDSHLLNAEHHFSLFKSKFG KIYASEEEHDHRFKVFKANLRRARRHQ LLDPSAEHGITKFSDLTPSEFRRTYLGLHKPK | Outside | 0.820 |
| Cys protease NTCP -23 | MSSRFTLLLALVVAGGLAAALAGPATFAVENPIRQVVSDGLHELENGILQVVGQSRHALSFVRFAHRYGKRYESVEEIEQRLEV | Outside | 0.820 |
| Cathepsin B-like Cys proteinase | MALXSEVFSQLLLLLGAFFILVLQVVAEKPISEAKVESAILKESIIKEVNENAKAGWKAAFNPQFSNFTVSQFKRLLGVKPAREGDLEG | Outside | 0.820 |
| Lignin-forming peroxidase | MSFLRFVGTILFLVAIFAASNAQLSATFYDSTCPNVTSIVRGVMDQRQRTD | Plasma membrane | 0.460 |
| Pectin methylesterase | MTRVKDFFAGILDSGKNVNFSKGKKKFLA VVASVLLVAAVIGVVAGVKSSRSNNSNDH ADIQAITSAAHAIVKSACENTLHPELCYSTI ASVSDFSKKVTSQKDVIELSLNITCXAVQHNFFTVEKLIKTRKGLTPREKVAL | Outside | 0.820 |
| Cell wall protein TLRP | MGSKAFLFLGLFLAIFFLISFEVVAAELAETSNSMKSDNENEVHFDGRSG | Outside | 0.820 |
| Cys-rich protein | MATMKMLLTLLVAAILFCSHQQVATAREVVVADDGNELQLWPWKIPCYL | Plasma membrane | 0.460 |
| CHRK1 | MNIIILFFFHTIFQSQLHNSIAAETWIKAGY YQANDISYSPNIQSINSTLYTHLIYGFANI DFNTSTDQMIVSISDDEQLKNFNNIVKK KNPSLKTLLSIGGKDQSSYDSG | Plasma membrane | 0.460 |
| Arabinogalactan protein | MAYSRMMFAFIFALVAGSAFAQAPGASPAASPKASPVAPVASPPT | Plasma membrane | 0.919 |
| GRP1 | MGYKAFLFLGLFLAIFLMISSEVLARELSETSTTSAEDSKKSTSKNEVHEAQYGGYP | Outside | 0.820 |
| Nectarin I oxalate oxidase-like protein | MAAFGINKIFESMVTTMFFLLAISIDRYCFAADEDMLQDVCVADLNSMVKVNGFPCKKNFTAADFSSLAISKPGATNNTFGSVVTA | Lysosome (lumen) | 0.865 |
| Outside | 0.786 | ||
| Invertase inhibitor | MRNLFPIFMLITNLAFNDNNNSNNIINTTCRATTNYP | Endoplasmic reticulum (membrane) | 0,550 |
| Glutathione S-Transferase Par-C | GRVDHNGKPICESIIAVEYIEEVWKDKAPSLLPSDPYDRAQARFWADYIDKKLYDFGRKLWATK | Cytoplasm | 0.450 |
| L11-like | MDFYVVLERPGYRVARRRRCK | Nucleus | 0.300 |
Functional Classification of SST-Selected Genes
Screening of the tobacco cDNA library resulted in the isolation of sequences that were identical or similar to genes reported in databases (BLAST, National Center for Biotechnology Information file servers) and involved in defense or stress reactions (33%), in cell wall modification (17%), or in plant signaling (16%; Table I). Among genes involved in defense responses are those encoding tobacco PR-2, PR-5d, and tomato (Lycopersicon esculentum) osmotin-like proteins exhibiting antifungal activities (Mauch et al., 1988; Sela-Buurlage et al., 1993; Abad et al., 1996). Additional related defense genes were also selected such as PAR-1a and PAR-1b, previously reported to be induced by infection with potato virus Y (Herbers et al., 1995) and genes corresponding to an elicitor-inducible protein (Takemoto et al., 2001). One protein is also similar to a tomato systemic response aspartic protease expressed during defense response to herbivore attacks (Schaller and Ryan, 1996). Seven sequences are identical or related to tobacco and Arabidopsis proteins involved in synthesis and modification of the cell wall. These genes encode defense-related lignin biosynthetic peroxidase (LFP, Lagrimini et al., 1987), a Tyr- and Lys-rich protein (NtTLRP) able to cross-link structural proteins to cell wall and with putative specialized function in the formation of xylem tissue (Domingo et al., 1999), and pectin acetyl- or methylesterases which, through their interactions with the TMV-encoded movement protein, mediate cell-to-cell movement through plasmodesmata (Chen et al., 2000). Several selected genes encode kinase-like proteins and are putatively involved in signal recognition and transduction at late stages of plant development. Among them, CHRK1 encodes a tobacco membrane-bound chitinase-related receptor-like kinase and is strongly stimulated by infection with fungal pathogen and tobacco mosaic virus (Kim et al., 2000). Finally, when the SST strategy was applied to resistant leaf tissues that were healthy and unchallenged with pathogens, 12 of 43 selected genes encoding known proteins were found to be involved or suspected to be involved at different steps of signal transduction pathways in cellular defense reactions.
Transcriptional Fluctuations
To profile secretion gene expression in developmental acquired resistance we constructed a microarray slide containing the 131 tobacco SST sequences, each representing a unique gene. Using targeted mRNA from Xanthi nc line, we determined the mRNA abundance in acquired resistance situations (cryptogein-treated and flowering plants) and compared that with the susceptible situations (water-treated and vegetative plants, respectively).
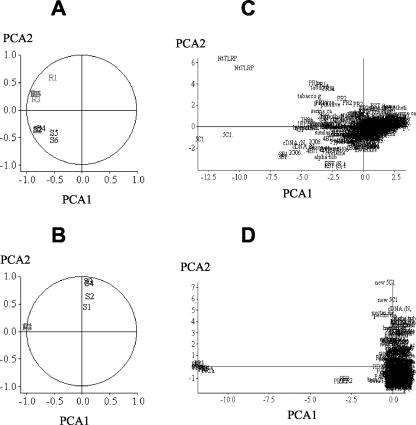
To establish correlations between the treatments and the genes analyzed, a principal component factor analysis (PCA) was performed. Figure 1A shows the results of PCA for each of six replicates from vegetative (S) and flowering (R) plants. Variables from flowering (R) plants are close together and correlated. In addition, they are poorly correlated with variables from water-treated plants (S). Thus, analysis of transcriptional variations with this subset of genes may distinguish vegetative growth and flowering growth. Figure 1B shows a similar distribution after analysis of replicates from cryptogein-treated (R) and water-treated (S) plants. As expected, variables from cryptogein-treated (R) plants are highly correlated together and orthogonal (poorly correlated) with variables from water-treated plants (S). Whatever the conditions, most genes are near the origin of the biplot (Fig. 1, C and D). These genes are either not expressed differentially between susceptible and resistance situations or are explained poorly by the PCA. Few genes are close to the head of a vector as NtTLRP (Fig. 1C) and PR-1 and PR-2 (Fig. 1D).
Figure 1.
PCA of microarray data. Variables (normalized intensity values or individuals [genes] were plotted separately on graph by report of the first two principal components (PC1 and PC2). A, Distribution of normalized intensity values; C, distribution of genes from water (S) and cryptogein (R)-treated plants. B, Distribution of normalized intensity values; D, distribution of genes from vegetative (S) and flowering plants (R). PCA1 and PCA2 represent 78% and 69% of the total variability in A and C and B and D, respectively.
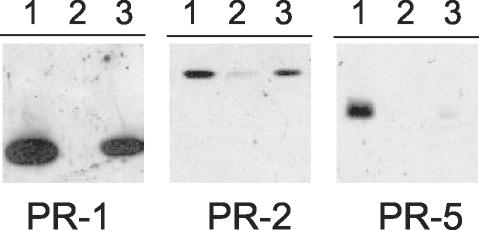
Determination of the relative mRNA abundance between vegetative (susceptible) and flowering (developmental resistance) Xanthi plants also revealed the extent to which development regulates the expression of genes for secreted and membrane-bound proteins (see supplemental data Table IV). Twelve sequences from the DNA array displayed differential expression of at least 2-fold or more, and all of them exhibited flowering-inducible expression. Among these, six had mRNA abundance significantly different between flowering-resistant plants and vegetative susceptible plants based on a Student's t test (P < 0.001). This set of genes included PR-1a and PR-2, with relative mRNA abundance for up-regulation of 3.17 and 2.34, respectively. Genes coding for NtTLRP, for LFP (a lignin forming peroxidase), for CPR2 (a putative prepro-Cys proteinase), and for an unknown protein were also found up-regulated during developmental resistance: 3.89-fold for NtTLRP, 2.65-fold for LFP, 2.53-fold for CPR2, and 2.22-fold for the unreported gene (clone no. 2H01). Concordant results were obtained by northern-blot analysis for PR-1a, PR-2, NtTLRP, and LFP, with ratios for up-regulation of 2.39, 2.19, 2.23, and 1.95, respectively. The specific extracellular accumulation of peptides in flowering plants was confirmed by western-blot analysis for PR-1a and PR-2 (Fig. 2). Based on these data, we considered these six genes as markers for developmental resistance. The encoded proteins may have functions in developmental resistance.
Figure 2.
Protein gel-blot analysis of extracellular PR proteins of tobacco leaf. Intercellular fluids were prepared from leaves of tobacco plants: lane 1 corresponds to proteins (1 μg) from Xanthi leaves 48 h after stem treatment with cryptogein and SAR induction; lanes 2 and 3 correspond to proteins (25 μg) from leaves of Xanthi plants 50 or 110 d after seeding, respectively.
Microarray analyses were also performed between water-treated (susceptible) and cryptogein-treated (SAR) tobacco plants (see supplemental data Table V). As expected, a high relative mRNA abundance was found for SAR genes PR-1a, PR-2, and PR-5 (34, 14, and 35, respectively). In these conditions, no other gene presented an increase in expression of at least 2-fold or more, excepted Par-1a (2.86). Up-regulated genes during developmental resistance (NtTLRP, LFP, CPR2, and the unreported one) were found only fairly induced during SAR with relative mRNA abundance ranging between 1.3 and 1.8.
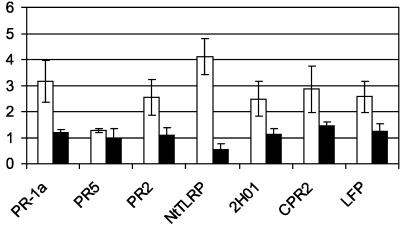
Similar analyses performed with the transgenic line NahG-8 revealed that at least two signaling pathways regulate the activation of the marker genes during developmental resistance. After inoculation with tobacco mosaic virus, the transgenic line (NahG-8) does not accumulate SA nor develop SAR to subsequent inoculations (Gaffney et al., 1993). The influence of SA on the accumulation of mRNA during establishment of developmental resistance and SAR was determined by comparison with the results obtained with the Xanthi line. For most tested genes for secreted and membrane-bound proteins, the inability to accumulate SA did not modify relative mRNA abundance. However, transcriptional activities were disturbed for few genes in each resistance conditions. Concerning transcriptional fluctuations through development, among the six genes that were found to be induced at flowering, all were moderately affected in the transgenic line. Nevertheless, after statistical treatment, we found significant reduction of relative mRNA abundance only for PR-1a and NtTLRP (Fig. 3). This suggests that an additional SA-independent pathway controlled activation of the four remaining genes during expression of developmental resistance. As expected, comparison between cryptogein- and water-treated plants showed a strong reduction of relative mRNA accumulation of PR-1a, PR2, and PR5. In the transgenic line, relative mRNA abundance for each of the three PR genes was 2.03, 2.04, and 7.51, whereas they were 34, 14, and 35, respectively, in the Xanthi line.
Figure 3.
Influence of SA accumulation on gene expression during developmental resistance. Each bar represents the values of relative abundance for the indicated gene, between vegetative and flowering Xanthi (white bars) or NAHG-8 (black bars) plants. Values are the means ± sd of two replicates of three different experiments. Differences between Xanthi NAHG-8 plants are statistically significant (P < 0,01, n = 3) for PR-1a and NtTLRP genes.
DISCUSSION
The goal of the present study was to identify genes encoding secreted proteins and to analyze transcriptional fluctuations to characterize extracellular modifications associated with establishment of developmental acquired resistance in tobacco. For this purpose, we first used a yeast genetic system (Klein et al., 1996; Jacobs et al., 1997) in which secretion of invertase depends on reconstitution of extracellular invertase activity via cDNA cloning. This was accomplished by fusion of tobacco coding sequences for functional signal peptides in frame with a truncated yeast invertase coding sequence missing information for an initiator Met and signal peptide. In the experiments reported here, we cloned 131 different tobacco cDNAs; among these, 76% appeared to encode proteins including known functional or predicted signal peptides.
Based on the numbers of colonies selected on medium without Trp and then on medium with raffinose, we estimated that approximately 2% of cDNAs from the library encode proteins with signal peptides. This rate is rather high compared with the previously reported of 0.1% using the SST technology with rat and nematode tissues (Klein et al., 1996; Wang et al., 2001). It could correspond to the important role of the apoplast and cell wall in plants. It could also indicate a specialization of cellular activities at late stages of leaf development that required the synthesis and the secretion of a number of proteins.
Clearly, the signal sequence trap system is very effective and gives the opportunity to identify most of extracellular structural or signaling molecules. Nevertheless, in this work, it did not lead to the identification of all secretion genes expressed in tobacco leaves at flowering. For example, PR-1a gene was not identified, whereas northern-blot and microarray analysis confirmed its expression at late developmental stages. The possibility of a complete overview could be increased by the analysis of a bigger number of yeast-selected clones. The absence of PR-1a from this screen could also indicate that some plant signal peptides may not be functional in yeast. This observation is inconsistent with the idea that signal sequences can function across kingdoms. Several studies have demonstrated that signal sequences from one species can function with the translocation machinery of other species (Wiedmann et al., 1984; Garcia and Walter, 1988). However, Al-Qahtani et al. (1998) have shown that signal sequences from Trypanosoma brucei and Leishmania chagasi glycoproteins are not able to mediate translocation into canine pancreas microsomes, indicating that in some cases signal peptides are not exchangeable.
The gene expression profiling approach allows delineating transduction pathways and transcriptional events that could determine tobacco resistance to P. parasitica controlled by development. This analysis is not exhaustive. Other genes are doubtless activated, and their characterization will be useful in allowing understanding of other important aspects of the resistance. Moreover, confirmation of the role of all activated genes during developmental resistance requires further functional characterizations. However, our results reveal that among genes whose expression is coordinated at late phases of development (PR-1a, PR-2, NtTLRP, LFP, CPR2, and an unreported one), four can be grouped in at least two functional groups, one that would include PR defense-related genes and the other that would bring together genes involved in cell wall modifications (NtTLRP and LFP). CPR2 could be also involved in cell wall modifications. It encodes a putative proteinase showing significant similarity to CYP15a, a protein located in the cell wall of stem cortical cells in peas (Pisum sativum; Jones and Mullet, 1995). The last one shows no significant similarity to any known sequence. However, it encodes a secreted or a membrane-bound protein and shows a specific expression profile at late developmental stages. Moreover, hierarchical clustering for expression patterns in the different tested conditions (establishment of developmental resistance or SAR in Xanthi or NahG-8 lines) indicated that this gene is in a group of 10 genes that include all genes defined as markers of developmental resistance (data not shown). Taken together, these results suggest a role for this unreported protein in a function such as defense or in elaboration of cell wall architecture. In broad outline, the resistance could be associated with two simultaneous events: activation of the process leading to cell wall modifications and induction of defense mechanisms also activated during SAR.
The strong transcriptional activation of PR-1a and PR-2 genes is a characteristic of SAR in tobacco (Ward et al., 1991) as the activation of PR-1 and BGL2 (PR-2; orthologous genes in Arabidopsis; Uknes et al., 1992). Our results are in agreement with previous studies describing the regulation of individual defense genes both by plant/pathogen interaction and by developmental signals (Fraser, 1981; Reuveni et al., 1986; Lotan et al., 1989; Côté et al., 1991; Uknes et al., 1993; Yapalni et al., 1993). They demonstrate the ability of host plants to stimulate SAR-associated defense responses without microbial perception. Quantification of transcriptional fluctuations of SAR genes had revealed a clear induction in leaves of flowering tobacco. However, these genes are less activated during developmental resistance than during SAR. In flowering plants, relative mRNA abundance of PR-1a and PR-2 were 12 and 7 times less plentiful, respectively, than in cryptogein-treated plants. This difference could indicate that plants may overcome some pathogenic infections with moderate activation of defense mechanisms. It confirms that during flowering growth, mechanisms other than those involved in SAR are activated and probably participate in the expression of the developmental resistance (Hugot et al., 1999).
The activation of LFP and NtTLRP genes indicates that peroxidative cross-linking of cell wall takes place at late stages of development and can be important for resistance. It is known that activation of defense mechanisms is associated with modifications of cell wall, which is the first barrier against pathogens. A rapid oxidative cross-linking mechanism for cell wall insolubilization of preexisting structural proteins is mediated by the action of extracellular peroxidases and is also under developmental control (Bradley et al., 1992). This mechanism would act as a rapid defense response against pathogenic attack by reducing cell wall digestibility (Brisson et al., 1992). NtTLRP that mediates cross-linking of proteins to the cell wall (Domingo et al., 1999) and the extracellular lignin-forming peroxidase LFP (Lagrimini et al., 1987) could be involved in this process of peroxidative cross-linking of cell wall proteins at late developmental stages. The deposition of the structural cell wall proteins mainly occurs once cellular growth has ceased and elongation is complete (Cassab and Varner, 1987; Carpita and Gibeaut, 1993) and may be a developmentally regulated process associated with the establishment of resistance. These modifications resulting in alterations of the functional properties of the cell wall could perturb the oomycete cycle in plants by at least two ways. A first one is that cross-linking of structural proteins to the cell wall protects plants from infections by leading to an increase in the tensile strength of the cell wall and to a reduction of its degradation by the microorganism. A second one is that the oomycete cell wall is also a target for these modifications. Thus, during the interaction between tobacco and P. parasitica at late developmental stages, an altered pathogen cell wall could become much less extensible and then lock the growing hyphal tip. This would result in altered mycelium elongation.
The distribution of up-regulated genes during flowering growth in two functional groups does not mean that gene expression for each group results from parallel activation of two different signaling pathways: one ending in the expression of the PR genes also expressed during SAR and the other one in the expression of genes for cell wall modifications. Clearly, the resistance does not result from a simple superimposition of two responses, one associated with SAR and the other associated with elaboration of cell wall architecture. The set of genes activated during developmental resistance does not include all genes that are induced during SAR. Only PR-1a and PR-2 but not PR-5 are expressed during developmental resistance. Furthermore, although PR-1a is regulated via an SA-dependent pathway, PR-2 expression does not depend mainly on SA accumulation at late developmental stages. From these results, it appears that: (a) PR genes could be regulated through different pathways in SAR and in late developmental stages, (b) and gene expression associated with developmental resistance results from complex networks of regulation. The interlacing of networks regulating resistance and development mechanisms was highlighted in other studies describing up-regulation of defense genes both by plant/pathogen interactions and by developmental signals (Fraser, 1981; Lotan et al., 1989; Côté et al., 1991; Uknes et al., 1993). Other studies have led to characterization of genes that positively regulate a developmental process and negatively regulate disease resistance. For example, in Arabidopsis, the gene AtTIP49a is essential for both sporophyte and female gametophyte viability and also acts as negative regulator of resistance functions during cultivar-specific resistance to strains of P. syringae mediated via RPM1 (Holt et al., 2002). The Arabidopsis MPK4 gene is involved in development and in negative regulation of SAR, and its inactivation leads to a dwarf phenotype and constitutive expression of SAR (Petersen et al., 2000). Silencing of NPK1 required for cytokinesis in tobacco (Nishihama et al., 2001), results in the attenuation of the N-mediated and Bs2-mediated disease resistance pathways, suggesting an additional role in plant defense responses (Jin et al., 2002).
What is the functional meaning of the activation of a plant defense response either in a pathological or in a developmental context? Our results show that there is a coordinated expression of genes for secreted proteins involved in defense and cell wall modifications through development without microbial stimulation. This subset of secreted and membrane-bound proteins plays a role both in plant defense and plant development. The only function that studies in physiopathology have revealed for defense proteins (the antimicrobial effect for some of them) could only be one of the roles of these proteins. It is possible that they are also involved in plant physiology or plant development.
MATERIALS AND METHODS
Plant Material
Experiments were performed with tobacco (Nicotiana tabacum) cv Xanthi nc. tobacco plants or with the transgenic line NahG-8 expressing the nahG gene (Gaffney et al., 1993). Plants were grown in a growth chamber at 24°C with a 16-h photoperiod at a light intensity of 100 mE–2 s–1. Experiments were performed on 7- to 8- or 14- to 15-week-old tobacco plants that were susceptible or resistant to Phytophthora parasitica, respectively (Hugot et al., 1999). Induction of SAR was performed by elicitin application: four 7- to 8-week-old plants were decapitated and stem treated with 20 μL of water or a 5 μm aqueous solution of cryptogein (Bonnet et al., 1996). For each point, leaf tissues were taken from 10 leaves taken from four different plants. The positions from the base of the leaves ranged between five and 10 when the plants were 7 to 8 weeks old or 25 and 30 when the plants were 14 to 15 weeks old.
The pSUC2T7M130RI Vector
This vector has been designed to identify sequences in cDNA clones that mediate synthesis and transport of invertase into the endoplasmic reticulum and, hence, into the secretion pathway (Jacobs et al., 1997; Genetics Institut). It carries the β-lactamase and Trp (trp1)-selectable markers for Escherichia coli and yeast (Saccharomyces cerevisiae), respectively. The invertase gene is transcribed from the alcohol dehydrogenase promoter. The protein, however, is not expressed and secreted because it lacks a translation start codon and a signal sequence, respectively. These elements must be provided by inserted cDNA fragments.
SST Library Construction
For cDNA synthesis, total RNA and mRNA were purified from tobacco leaves (15-week-old tobacco plants) using the RNeasy Maxi kit and the oligotex kit (Qiagen USA, Valencia, CA), respectively. A random-primed directional cDNA library was synthesized from 4 μg of polyadenylated mRNA by using the SuperScript Choice System for cDNA synthesis (Life Technologies/Gibco-BRL, Cleveland) as previously described by Jacobs et al. (1997, 1999). The primer used for the first strand synthesis corresponds to an equimolecular mixture of the oligonucleotides 5′CGATTGAATTCTAGACCTGCCTCGAGNNNNNNNNNX3′ including an XhoI site (underlined), a random ninemer, and a degenerated 3′ end (X= A, C, G, or T). After synthesis of the second strand, ligation to an EcoRI -NotI adaptator, 5′ phosphorylation with T4 polynucleotide kinase, and digestion with the restriction endonuclease XhoI, cDNAs were size fractionated by column chromatography to allow for 5′-untranslated regions of mRNA and translation termination codons at the end of the protein-coding region.
The cDNAs were ligated to the EcoRI- and XhoI-digested pSUC2T7M130RI vector (Jacobs et al., 1997). The E. coli ElextroMAX DH10B strain (Life Technologies/Gibco-BRL) was transformed with the ligation mixture by electroporation (E. coli Apparatus, Bio-Rad Laboratories, Hercules, CA), and the size of the library was estimated to 4.106 plaque-forming units. Fifty clones from the E. coli library were checked for the presence, the size, and the sequence of the cDNA insert. The rest of clones were used for plasmid DNA isolation and yeast transformation.
Plasmid DNA was recovered from bacterial colonies and introduced into the yeast YTK12 strain (Jacobs et al., 1999) using Frozen-EZ Yeast Transformation II kit (Zymo Research, Orange, CA). The pSUC2T7M130RI vector provides selection for Trp+ transformants and requires a cDNA insert with secretion signal sequence to be cloned with the vector invertase gene for functional selection in yeast on Suc media. To select complementation for SUC2 deletion, Trp+ yeast transformants were first selected after growth at 30°C over 3 d on solid medium in the absence of Trp (0.67% [w/v] yeast nitrogen base without amino acids, 0.075% [w/v] –Trp dropout supplement [CLONTECH Laboratories, Palo Alto, CA], 0.1% [w/v] Glc, 2% [w/v] saccharose, and 2% [w/v] agar). The Trp+ transformants were replicated on solid raffinose medium (1% [w/v] yeast extract, 2% [w/v] peptone, 2% [w/v] raffinose, 2% [w/v] agar, and 2.10%–4% [w/v] antimycin) and grown at 30°C for 3 d (Jacobs et al., 1999).
For each yeast colony that survived to selection on Trp– and raffinose media, the sequence of the cDNA clone was determined after PCR amplification. An aliquot of the colonies was incubated for 10 min at 100°C in 20 μL of lysis buffer (1% [w/v] Triton X-100, 2 mm EDTA, and 20 mm Tris-HCl [pH 8.9]). Amplification was performed after addition of 38 μL of PCR mixture including the oligonucleotide primers 5′-GGTGTGAAGTGGACCAAAGGTCTA-3′ and 5′-CCTCGTCATTGTTCTCGTTCCCTT-3 derived from the pSUC2T7M130RI vector. The PCR consisted of 30 cycles of 94°C for 1 min, 55°C for 1 min, and 68°C for 5 min. The PCR products were purified on a SOPE resin (Quick Step PCR purification, Edge BioSystems, Gaithersburg, MD). DNA sequencing was performed using the same primers that were used in PCR reaction with an automated DNA sequencer (Beckman CQE2000, Beckman Instruments, Fullerton, CA).
Immunoblot Analysis
For the detection of PR proteins, intercellular fluid from leaves was prepared in 50 mm Tris-HCl buffer (pH 7) as described by Hammond-Kosak (1992). Protein extracts were run on 15% (w/v) SDS-PAGE gels and transferred electrophoretically onto nictrocellulose membranes (Amersham, Buckinghamshire, UK). The membranes were first incubated with rabbit purified IgG antibodies against PR1 proteins (Abad et al., 1989) and PR2 or PR5 proteins (Kauffmann et al., 1990). They were then incubated with goat peroxidase-conjugated IgG against rabbit immunoglobulins (Amersham). The bound antibodies were revealed using the ECL western detection system (Amersham) according to the manufacturer's instructions.
Northern-Blot Hybridization
Total RNA was purified as described by Logemann et al. (1987) and then treated with 7 units of soluble RNase-free DNase I (Amersham-Pharmacia Biotech, Uppsala). RNA (10 μg) was subjected to electrophoresis in a 1.4% (w/v) agarose-formaldehyde gel, transferred and cross-linked onto Hybond N+ filters (Amersham-Pharmacia Biotech). Hybridizations were performed under stringent conditions according to the manufacturer's instructions with the random-primed probes corresponding to the PCR products of different SST clones that were obtained as described above. Blots were analyzed on a PhosphorImager (Fuji Photo Film, Tokyo). rRNA 28S was chosen as loading control. The northern probes and RNA were identical to the cDNAs spotted onto the array and cDNA targets used for the hybridization step.
Microarray Analysis of Genes for Secreted Proteins
Microarrays were produced on the INRA/CNRS Genomics Platform (Sophia-Antipolis, France; R. Feyereisen and P. Barbry teams). Each glass slide contained two copies of all SST probes and included control cDNAs corresponding to either SAR tobacco genes for PR-1a, PR2, and PR5 proteins (Ward et al., 1991) or genes for alpha- and gamma-tubulins (AB052822 and AB051679). They also included 36 copies of FaNAC (Lingueglia et al., 1995), DgNac (Darboux et al., 1998), and DmdNac (Darboux et al., 1998) cDNAs corresponding to sodium channels from Helix aspersa (FaNac) or fruitfly (Drosophila melanogaster; DgNac and DmdNac), which were used as external controls for hybridization.
SST probes were obtained by PCR amplification from yeast transformants. After growth in liquid medium (1% [w/v] yeast extract, 2% [w/v] peptone, and 2% [w/v] Suc) at 30°C for 2 d, an extract containing plasmid was prepared as described by Hoffman and Winston (1987). For each transformant, an amplification by PCR was performed in a 100-μL reaction using 5 μL of extract as template and the 5′ amino-modified C6 oligonucleotides 5′-GGTGTGAAGTGGACCAAAGGTCTA-3′ and 5′-CCTCGTCATTGTTCTCGTTCCCTT-3′ as primers. The PCR consisted of 30 cycles of 94°C for 1 min, 55°C for 1 min, and 68°C for 5 min. PCR products were analyzed by gel electrophoresis before purification using the Millipore MAFNOB 96-well PCR Cleanup kit (Millipore, Bedford, MA) according to the manufacturer's instructions. PCR products were analyzed again to determine the final quantities by using PicoGreen reagent (Molecular Probes, Eugene, OR), lyophilized, and resuspended in 8 μL of 3× SSC (1× SSC is 0.15 m NaCl and 0.015 m sodium citrate) in a 96-well microtiter plate at a mean final concentration of 0.2 μg μL–1.
Microarray printing was performed using a high-speed printer (SDDC2, Virtek, Bochholt, Belgium) with pins supplied by Telechem (Sunnyvale, CA). Each pin withdraws a volume of about 250 nL and deposits a spot volume of about 0.6 nL, with a diameter of approximately 90 to 130 μm. Printing was performed on aldehyde coating slides. We printed cDNAs clones on the slides with an element center-center spacing of 180 μm. After printing, slides were allowed to dry and then used immediately, or were stored desiccated at room temperature in darkness. Just before hybridization, slides were sequentially post-processed: (a) Unbound DNA was removed by washing with 0.2% (w/v) SDS and double-distilled water, (b) covalently bound DNA was denatured for 2 min in boiling water, (c) free aldehydes were reduced by soaking slides for 5 min in 68 mm sodium borohydride (dissolved in PBS containing 25% [v/v] ethanol), and (d) free glasses sites were blocked by an incubation of 30 min at 60°C in the presence of 0.2% (w/v) casein. Several washing steps were performed with 0.2% (w/v) SDS and double-distilled water; then, slides were dried by centrifugation at 500g for 5 min.
For each hybridization, cDNA targets were prepared from RNA (10 μg) isolated using the method of Logemann et al. (1987) and treated with RNase-free DNase I (Amersham-Pharmacia Biotech). At least two independent RNA preparations for each biological sample were made and used for probe synthesis. Using the CyScribe First Strand cDNA Labeling Kit (Amersham-Pharmacia Biotech), mRNA targets were converted into fluorescent cDNA by incorporation of either Cy3-dCTP or Cy5-dCTP in the presence of FaNac and DgNac mRNA (0.5 ng) or FaNac and DmdNac (0.5 ng), respectively. Single-stranded cDNA was purified using QIaqick Nucleotide Removal Kit (Qiagen USA, Valencia, CA), dried under vacuum, and finally dissolved in 4 μL of water.
For hybridization to the SST array, the hybridization mix containing 4 μL of each labeled cDNA and 12 μL of DigEasy Hyb buffer (Boehringer Mannheim/Roche, Basel) was added to the microarray surface and covered with a standard 22- × 32-mm coverslip. Slides were placed in hybridization chambers (Corning, Corning, NY), and 20 μL of 3× SSC was placed inside each chamber before sealing. Slides were incubated for 14 to 16 h in a water bath at 42°C and then were sequentially washed in the following solutions: 1× SSC, 0.03% (w/v) SDS for 5 min, 0.2× SSC for 5 min, and 0.05× SSC for 5 min. Slides were dried by centrifugation at 600g for 2 min before they were scanned. For each experiment, three arrays were probed with cDNAs labeled with Cy-3 or Cy-5 dCTP and corresponding to both situations to be compared. Three others were probed with cDNAs swapped for Cy-3 and Cy-5 dCTP.
Microarrays were scanned with a scanning laser microscope (Pacard 4000, ScanArray3000, GSI Lumonics, Watertown, MA). Separate images were acquired for fluorochromes cyanine-3 and cyanine-5 at a resolution of 10 μm per pixel. Intensity values were quantified from the resultant pairs of TIFF files using Quant Array 2.0.0.0a (GSI Lumonics). The signal intensity for each fluorochrome was determined by subtracting the local background from signal intensity values. Genes showing a signal value < 500 in both Cy3 and Cy5 channels were not considered for the analyses. The data analysis was carried out using Winstat 2 (CIRAD, Montpelier, France). To calculate the correlation between treatments, normalized intensity values for each gene were ordered into a table (matrix) in which the six replicates for each condition were represented as columns, and the genes were represented as rows. The data was mean centered for each variable before analysis, and the table was used for the calculation of correlations between the individual experiments and PCA. The association between genes and normalized intensity values was visualized in a Euclidean space by a low-dimension graphic representation.
The relative abundance that corresponds to the resistance signal intensity/susceptibility signal intensity ratio was determined for each mRNA target after three different types of normalization that gave similar results. (a) The first one consisted of dividing the ratio obtained for each gene by the average of the ratios of all examined genes. (b) The second mode of normalization was based on use of an external control. During the reverse transcription procedure, the FaNAC cDNA was labeled with Cy3 or Cy5 and mixed for hybridization. Thus, this target indifferently hybridized with the FaNAc cDNA spotted on the slides leading to theoric equal signal intensity values in both Cy3 and Cy5 channels. A correction factor corresponding to the average of the Cy5/Cy3 ratio for the 36 copies of FaNAC was determined to centralize on 1 for effects of global variations between Cy3 and Cy5 for incorporation and hybridization. (c) To normalize the effects of global variations between Cy3 and Cy5 fluorescence intensities, relative abundance was also corrected by determination of the root square of the R-Cy3 × R-Cy5/S-Cy3 × S-Cy5 ratio. R-Cy3 and R-Cy5 corresponded to normalized intensity values from microarrays hybridized with cDNAs from resistant plants and labeled with Cy-3 or Cy-5 dCTP. S-Cy3 and S-Cy5 corresponded to normalized intensity values obtained with cDNAs from susceptible plants. In the three normalizations made, we defined induction or repression of a gene as a minimum 2-fold change in its relative abundance. The data presented here result form the first type of normalization.
Supplementary Material
Acknowledgments
We are grateful to René Feyereisen (INRA, Antibes, France) and to Genichi Kakefuda (BASF, Triangle Research Park, NC) for their support. We also thank Sébastien Duplessis (INRA, Nancy, France) for helpful discussion, Catherine Etienne (INRA, Antibes, France) for plant care, and Annik Lacombe (Université Laval, Canada) for revising the manuscript.
Article, publication date, and citation information can be found at http://www.plantphysiol.org/cgi/doi/10.1104/pp.103.034173.
The CNRS/INRA Microarray facility was supported by grants from Groupement d'Intérêt Public Aventis, region Provence Alpes Côte d'Azur, CNRS, and INRA. This work was supported by INRA (fellowship to M.-P.R.) and by the Association pour la Recherche sur les Nicotianées (to M.-P.R.).
The online version of this article contains Web-only data.
References
- Abad A, Cardin L, Poupet A, Ponchet M (1989) Comparison of pathogenesis related “b1” protein determination obtained by Elisa and HPLC techniques. J Phytopathol 124: 175–188 [Google Scholar]
- Abad LR, D'Urzo MP, Liu D, Narasimhan ML, Reuveni M, Zhu J, Niu X, Singh NK, Hasegawa PM, Bressan RA (1996) Antifungal activity of tobacco osmotin has specificity and involves plasma membrane permeabilization. Plant Sci 118: 11–23 [Google Scholar]
- Abedon BG, Tracy WF (1996) Congrass1 of maize (Zea mays L.) delays development of adult plant resistance to common rust (Puccinia sorgi Schw) and European corn borer (Ostrinia nubilalis Hubner). J Hered 87: 219–223 [Google Scholar]
- Alexander D, Goodman RM, Gut-Rella M, Glascock C, Weymann K, Friedrich L, Maddox D, Ahl-Goy P, Luntz T, Ward E et al. (1993) Increased tolerance to two oomycete pathogens in transgenic tobacco expressing pathogenesis-related protein 1a. Proc Natl Acad Sci USA 90: 7327–7331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Qahtani A, Teilhet M, Mensa-Wilmot K (1998) Species-specificity in endoplasmic reticulum signal peptide utilization revealed by proteins from Trypanosoma brucei and Leishmania. Biochem J 331: 521–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G, Dobberstein B (1975) Transfer of proteins across membranes: I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J Cell Biol 67: 835–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet P, Bourdon E, Ponchet M, Blein JP, Ricci P (1996) Acquired resistance triggered by elicitins in tobacco and other plants. Eur J Plant Pathol 102: 181–192 [Google Scholar]
- Bradley DJ, Kjellbom P, Lamb CJ (1992) Elicitor- and wound-induced oxidative cross-linking of a proline-rich plant cell wall protein: a novel, rapid defence response. Cell 70: 21–30 [DOI] [PubMed] [Google Scholar]
- Brisson LF, Tenhaken R, Lamb C (1992) Function of oxidative cross-linking of the cell wall structural proteins in plant disease resistance. Plant Cell 6: 1703–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong X (1997) The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88: 57–63 [DOI] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM (1993) Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J 3: 1–30 [DOI] [PubMed] [Google Scholar]
- Cassab GI, Varner JE (1987) Immunocytolocalization of extensin in developing soybean seed coats by immunogold-silver staining and by tissue printing on nitrocellulose paper. J Cell Biol 105: 2581–2588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Century KS, Lagman RA, Adkisson M, Morlan J, Tobias R, Schwartz K, Smith A, Love J, Ronald PC, Whalen MC (1999) Developmental control of Xa21-mediated disease resistance in rice. Plant J 20: 231–236 [DOI] [PubMed] [Google Scholar]
- Chen MH, Sheng J, Hind G, Handa AK, Citovsky V (2000) Interaction between the tobacco mosaic virus movement protein and host cell pectin methylesterases is required for viral cell-to-cell movement. EMBO J 19: 913–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côté F, Cutt JR, Asselin A, Klessig DF (1991) Pathogenesis-related acidic β-1,3-glucanase genes of tobacco are regulated by both stress and developmental signals. Mol Plant-Microbe Interact 4: 173–181 [DOI] [PubMed] [Google Scholar]
- Darboux I, Lingueglia E, Champigny G, Coscoy S, Barbry P, Lazdunski M (1998) dGNaC1, a gonad-specific amiloride-sensitive Na+ Channel. J Biol Chem 273: 9424–9429 [DOI] [PubMed] [Google Scholar]
- Delaney TP, Uknes S, Vernooij B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gut Rella M, Kessmann H et al. (1994) A central role of salicylic acid in plant disease resistance. Science 266: 1247–1250 [DOI] [PubMed] [Google Scholar]
- Després C, DeLong C, Glaze S, Liu E, Fobert PR (2000) The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 12: 279–290 [PMC free article] [PubMed] [Google Scholar]
- Domingo C, Sauri A, Mansilla E, Conejero V, Vera P (1999) Identification of a novel peptide motif that mediates cross-linking of proteins to cell walls. Plant J 20: 563–570 [DOI] [PubMed] [Google Scholar]
- Dong X (2001) Genetic dissection of systemic acquired resistance. Curr Opin Plant Biol 4: 309–314 [DOI] [PubMed] [Google Scholar]
- Fan W, Dong X (2002) In vivo interaction between NPR1 and transcription factor TGA2 leads to salicylic acid-mediated gene activation in Arabidopsis. Plant Cell 14: 1377–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser RSS (1981) Evidence for the occurrence of the “pathogenesis-related” proteins in the leaves of healthy tobacco during flowering. Physiol Plant Pathol 19: 69–76 [Google Scholar]
- Gaffney T, Friedrich L, Vernooij B, Negrotto D, Nye G, Uknes S, Ward E, Kessmann H, Ryals J (1993) Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261: 754–766 [DOI] [PubMed] [Google Scholar]
- Garcia PD, Walter P (1988) Full-length prepro-alpha-factor can be translocated across the mammalian microsomal membrane only if translation has not terminated. J Cell Biol 106: 1043–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond-Kosak KE (1992) Preparation and analysis of intercellular fluid. In SJ Gurr, MJ McPherson, DJ Bowles, eds, Molecular Plant Pathology: A Practical Approach, Vol II. IRL Press at Oxford University Press, UK, pp 15–22 [Google Scholar]
- Hammond-Kosack K, Parker J (2003) Deciphering plant-pathogen communication: fresh perspectives for molecular resistance breeding. Curr Opin Biotechnol 14: 177–193 [DOI] [PubMed] [Google Scholar]
- Herbers K, Monke G, Badur R, Sonnewald U (1995) A simplified procedure for the subtractive cDNA cloning of photoassimilate-responding genes: isolation of cDNAs encoding a new class of pathogenesis-related proteins. Plant Mol Biol 29: 1027–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman CS, Winston F (1987) A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57: 267–272 [DOI] [PubMed] [Google Scholar]
- Holt BF, Boyes DC, Ellerstrom M, Siefers N, Wiig A, Kauffman S, Grant MR, Dangl JL (2002) An evolutionarily conserved mediator of plant disease resistance gene function is required for normal Arabidopsis development. Dev Cell 2: 807–817 [DOI] [PubMed] [Google Scholar]
- Hugot K, Aime S, Conrod S, Poupet A, Galiana E (1999) Developmental regulated mechanisms affect the ability of a fungal pathogen to infect and colonize tobacco leaves. Plant J 20: 163–170 [DOI] [PubMed] [Google Scholar]
- Jacobs KA, Collins-Racie LA, Colbert M, Duckett M, Golden-Fleet M, Kelleher K, Kriz R, LaVallie ER, Merberg D, Spaulding V et al. (1997) A genetic selection for isolating cDNAs encoding secreted proteins. Gene 198: 289–296 [DOI] [PubMed] [Google Scholar]
- Jacobs KA, Collins-Racie LA, Colbert M, Duckett M, Evans C, Golden-Fleet M, Kelleher K, Kriz R, La Vallie ER, Merberg D et al. (1999) A genetic selection for isolating cDNA clones that encode signal peptides. Methods Enzymol 303: 468–479 [DOI] [PubMed] [Google Scholar]
- Jin H, Axtell MJ, Dahlbeck D, Ekwenna O, Zhang S, Staskawicz B, Baker B (2002) NPK1, an MEKK1-like mitogen-activated protein kinase kinase kinase, regulates innate immunity and development in plants. Dev Cell 2: 291–297 [DOI] [PubMed] [Google Scholar]
- Jones JT, Mullet JE (1995) A salt- and dehydration-inducible pea gene, Cyp15a, encodes a cell-wall protein with sequence similarity to cysteine proteases. Plant Mol Biol 28: 1055–1065 [DOI] [PubMed] [Google Scholar]
- Jupin I, Chua NH (1996) Activation of the CaMV as-1 cis-element by salicylic acid: differential DNA-binding of a factor related to TGA1a. EMBO J 15: 5679–5689 [PMC free article] [PubMed] [Google Scholar]
- Kauffmann S, Legrand M, Fritig B (1990) “Isolation and characterization of six pathogenesis-related (PR) proteins of Samsun NN tobacco.” Plant Mol Biol 14: 381–390 [DOI] [PubMed] [Google Scholar]
- Kim YS, Lee JH, Yoon GM, Cho HS, Park SW, Suh MC, Choi D, Ha HJ, Liu JR, Pai HS (2000) CHRK1, a chitinase-related receptor-like kinase in tobacco. Plant Physiol 123: 905–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinkema M, Fan W, Dong X (2000) Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 12: 2339–2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RD, Gu Q, Goddard A, Rosenthal A (1996) Selection for genes encoding secreted proteins and receptors. Proc Natl Acad Sci USA 93: 7108–7113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuc J (1982) Induced immunity to plant disease. Bioscience 32: 854–860 [Google Scholar]
- Kus JV, Zaton K, Sarkar R, Cameron RK (2002) Age-related resistance in Arabidopsis is a developmentally regulated defense response to Pseudomonas syringae. Plant Cell 14: 479–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagrimini LM, Burkhart W, Moyer M, Rothstein S (1987) Molecular cloning of complementary DNA encoding the lignin-forming peroxidase from tobacco: molecular analysis and tissue-specific expression. Proc Natl Acad Sci USA 84: 7542–7546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebel E, Heifetz P, Thorne L, Uknes S, Ryals J, Ward E (1998) Functional analysis of regulatory sequences controlling PR-1 gene expression in Arabidopsis. Plant J 16: 223–233 [DOI] [PubMed] [Google Scholar]
- Leisner SM, Turgeon R, Howell SH (1992) Long distance movement of cauliflower mosaic virus in infected plants. Mol Plant-Microbe Interact 5: 41–47 [Google Scholar]
- Leisner SM, Turgeon R, Howell SH (1993) Effects of host plant development and genetics determinants on the long-distance movement of cauliflower mosaic virus in Arabidopsis. Plant Cell 5: 191–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingueglia E, Champigny G, Lazdunski M, Barbry P (1995) Cloning of the amiloride-sensitive FMRFamide peptide-gated sodium channel. Nature 378: 730–733 [DOI] [PubMed] [Google Scholar]
- Logemann J, Schell J, Willmitzer L (1987) Improved method for the isolation of RNA from plant tissues. Anal Biochem 163: 16–20 [DOI] [PubMed] [Google Scholar]
- Lotan T, Oni N, Fluhr R (1989) Pathogenesis-related proteins are developmentally regulated in tobacco flowers. Plant Cell 9: 881–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy J, Carr JP, Klessig DF, Raskin I (1990) Salicylic acid: a likely endogenous signal in the resistance response of tobacco to viral infection. Science 250: 1002–1004 [DOI] [PubMed] [Google Scholar]
- Maleck K, Levine A, Eulgem T, Morgan A, Schmid J, Lawton KA, Dangl JL, Dietrich RA (2000) The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat Genet 26: 403–410 [DOI] [PubMed] [Google Scholar]
- Mauch F, Mauch-Mani B, Boller T (1988) Antifungal hydrolases in pea tissue: II. Inhibition of fungal growth by combinations of chitinase and beta-1,3-glucanase Plant Physiol 88: 936–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch DJ (1985) On the predictive recognition of signal peptide sequences. Virus Res 3: 271–286 [DOI] [PubMed] [Google Scholar]
- Métraux J-P, Signer H, Ryals J, Ward E, Wyss-Benz M, Gaudin J, Raschdorf K, Schmid E, Blum W, Inverardi B (1990) Increase in salicylic acid at the onset of systemic acquired resistance in cucumber. Science 250: 1004–1006 [DOI] [PubMed] [Google Scholar]
- Moose SP, Sisco PH (1994) Glossy15 controls the epidermal juvenile-to-adult phase transition in maize. Plant Cell 8: 1343–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Kanehisa M (1992) A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics 4: 897–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niderman T, Genetet I, Bruyere T, Gees R, Stintzi A, Legrand M, Fritig B, Mosinger E (1995) Pathogenesis-related PR-1 proteins are antifungal: isolation and characterization of three 14-kilodalton proteins of tomato and of a basic PR-1 of tobacco with inhibitory activity against Phytophthora infestans. Plant Physiol 108: 17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng 10: 1–6 [DOI] [PubMed] [Google Scholar]
- Nishihama R, Ishikawa M, Araki S, Soyano T, Asada T, Machida Y (2001) The NPK1 mitogen-activated protein kinase kinase kinase is a regulator of cell-plate formation in plant cytokinesis. Genes Dev 15: 352–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen M, Brodersen P, Naested H, Reasson E, Lindhart U, Johansen B, Nielsen HB, Lacy M, Austin MJ, Parker JE et al. (2000) Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103: 1111–1120 [DOI] [PubMed] [Google Scholar]
- Qin XF, Holuigue L, Horvath DM, Chua NH (1994) Immediate early transcription activation by salicylic acid via the cauliflower mosaic virus as-1 element. Plant Cell 6: 863–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen JB, Hammerschmidt R, Zook MN (1991) Systemic induction of salicylic acid accumulation in cucumber after inoculation with Pseudomonas syringae pv syringae. Plant Physiol 97: 1342–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross FA (1961) Systemic acquired resistance induced by localized virus infections in plants. Virology 14: 340–358 [DOI] [PubMed] [Google Scholar]
- Reuveni M, Tuzun S, Cole JS, Siegel MR, Kuc J (1986) The effects of plant age and leaf position on the susceptibility of tobacco to blue mould caused by Peronospora tabacina. Phytopathology 76: 455–458 [Google Scholar]
- Ryals JA, Neuenschwander UH, Willits MG, Molina A, Steiner HY, Hunt MD (1996) Systemic acquired resistance. Plant Cell 8: 1809–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals JA, Weymann K, Lawton K, Friedrich L, Ellis D, Steiner HY, Johnson J, Delaney TP, Jesse T, Vos P et al. (1997) The Arabidopsis NIM1 protein shows homology to the mammalian transcription factor inhibitor IκB. Plant Cell 9: 425–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller A, Ryan CA (1996) Molecular cloning of a tomato leaf cDNA encoding an aspartic protease, a systemic wound response protein. Plant Mol Biol 31: 1073–1077 [DOI] [PubMed] [Google Scholar]
- Sela-Buurlage MB, Ponstein AS, Bres-Vloemans SA, Melchers LS, Van den Elsen PJM, Cornelissen BJC (1993) Only specific tobacco (Nicotiana tabacum) chitinases and beta-1,3-glucanases exhibit antifungal activity. Plant Physiol 101: 857–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto D, Hayashi M, Doke N, Nishimura M, Kawakita K (2001) Characterization of elicitor-inducible tobacco genes isolated by differential hybridization. J Gen Plant Pathol 67: 89–96 [Google Scholar]
- Uknes S, Dincher S, Friedrich L, Negrotto D, Williams S, Thompson-Taylor H, Potter S, Ward E, Ryals J (1993) Regulation of pathogenesis-related protein-1a gene expression in tobacco. Plant Cell 5: 159–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J (1992) Acquired resistance in Arabidopsis. Plant Cell 4: 645–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Loon LC, Strien EAv (1999) The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol Mol Plant Pathol 55: 85–97 [Google Scholar]
- Wang X, Allen R, Ding X, Goellner M, Maier T, de Boer JM, Baum TJ, Hussey RS, Davis EL (2001) Signal peptide-selection of cDNA cloned directly from the esophageal gland cells of the soybean cyst nematode Heterodera glycines. Mol Plant-Microbe Interact 14: 536–544 [DOI] [PubMed] [Google Scholar]
- Ward ER, Uknes SJ, Williams SC, Dincher SS, Wiederhold DL, Alexander DC, Ahl-Goy P, Metraux J-P, Ryals JA (1991) Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell 3: 1085–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedmann M, Huth A, Rapoport TA (1984) Xenopus oocytes can secrete bacterial beta-lactamase. Nature 309: 637–639 [DOI] [PubMed] [Google Scholar]
- Wyatt SE, Pan SQ, Kuc J (1991) beta-1,3-Glucanase, chitinase, and peroxidase activities in tobacco issues resistant and susceptible to blue mould as related to flowering, age and sucker development. Physiol Mol Plant Pathol 39: 433–440 [Google Scholar]
- Yapalni N, Shulaev V, Raskin I (1993) endogenous salicylic levels correlate with accumulation of pathogenesis-related proteins and virus resistance in tobacco. Phytopathology 83: 702–708 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.