Abstract
Microtubules (MTs) are central to fundamental cellular processes including mitosis, polarization, and axon extension. A key issue is to understand how MT-associated proteins and therapeutic drugs, such as the anticancer drug paclitaxel, control MT self-assembly. To facilitate this research, it would be helpful to have automated methods that track the tip of dynamically assembling MTs as observed via fluorescence microscopy. Through a combination of digital fluorescence imaging with MT modeling, model-convolution, and automated image analysis of live and fixed MTs, we developed a method for MT tip tracking that includes estimation of the measurement error. We found that the typical single-frame tip tracking accuracy of GFP-tubulin labeled MTs in living LLC-PK1α cells imaged with a standard widefield epifluorescence digital microscope system is ~36 nm, the equivalent of ~4.5 tubulin dimer layers. However, if the MT tips are blunt, the tip tracking accuracy can be as accurate as ~15 nm (~2 dimer layers). By fitting a Gaussian survival function to the MT tip intensity profiles, we also established that MTs within living cells are not all blunt, but instead exhibit highly variable tapered tip structures with a protofilament length standard deviation of ~180 nm. More generally, the tip tracking method can be extended to track the tips of any individual fluorescently labeled filament, and can estimate filament tip structures both in vivo and in vitro with single-frame accuracy on the nanoscale.
Keywords: Microtubules, Tracking, Automation, MATLAB, Algorithm, LLC-PK1, Tips
INTRODUCTION
The cytoskeleton is a protein filament system in the cytoplasm of all cells, and it is responsible for cell shape and movement, as well as intracellular transport.1 MTs, one of the eukaryotic cytoskeletal components, are responsible for multiple functions within living cells, including positioning the nucleus, organizing the cytoplasm, transporting vesicles, and segregating chromosomes. The self-assembly dynamics of MTs are unusual, as MT plus ends undergo alternate phases of growth and shortening with stochastic switching between phases, a phenomenon known as “dynamic instability”.14
Video-enhanced differential interference contrast (DIC) light microscopy has revealed the dynamic instability behavior of single MTs.8,11,15,16,23 These studies allowed MT tip tracking using software developed by the Salmon lab with a frame rate of 30 Hz, but the program relied on manual hand-clicking on a video, such that the resulting MT length–time data was reported at ~1 Hz.23 The accuracy was not reported, although precision in newt lung epithelial cells was estimated to be better than 500 nm.7 Gildersleeve et al.11 also used a cursor overlay to manually track MTs that were visible on a video monitor. To assess the precision of their method, Gildesleeve et al.11 compared two independent operators and found a root-mean-squared (RMS) deviation between two independent operators of ~160 nm. More recently, automated analysis has been used to track the contours of fluorescently labeled MTs.5,6 Wan and co-workers recently used an error function fitting approach to determine the positions of ends of kinetochore MT fiber bundles, each of which contains numerous MTs.24 Hadjidemetriou et al.12 recently developed an automated fluorescent MT tip tracking algorithm, although the estimated accuracy with model-convolved images containing realistic noise was at least two pixels (260 nm) so that the resolution was comparable to manual tracking. Briefly, Hadjidemetriou et al. code tracks microtubules automatically after initial selection by user. At first, it uses the entire frame sequence to compute the region where the tip is expected to be; the tip feature is then calculated in that region for all time and used to form the tip trajectory. In general, automated methods have yet to be developed for tracking single fluorescent MT tips with better than diffraction-limited resolution. In addition, it is necessary to validate any automated tracking using fixed MTs and simulated MTs with realistic blur and noise. Also, it seems possible, at least in principle, to obtain higher order moments of the protofilament (PF) length distribution beyond the mean, such as the variance of the individual PF lengths.
Here we present a computational approach for high temporal and spatial resolution of MT assembly dynamics in living cells using digital fluorescence microscopy. Even though digital fluorescence microscopy lacks the ability to classically “resolve” structures that are smaller than half of the emission wavelength (~250 nm), we can now overcome such classical limitations through a combination of simulated imaging via model convolution and automated image analysis.10 In this work we describe a semi-automated MT tip-tracking algorithm that achieves tip position estimates in living cells with an accuracy of ~36 nm under standard imaging conditions, or ~6-fold less than the classical Rayleigh resolution criterion while maintaining temporal resolution of ~1 Hz. In addition, we are able to obtain quantitative information on MT tip structures, which establishes that digital fluorescence microscopy is capable of recovering structural information as the MT dynamically assembles and disassembles. Thus, these studies, validated by model-convolution and fixed MT imaging, demonstrate that the nanoscale dynamics of MT assembly and tip structure can be estimated in vivo.
MATERIALS AND METHODS
Cell Culture Preparation
Stably transfected GFP-tubulin LLC-PK1α cells17 (pig kidney epithelial cells) (see Fig. 1a) were seeded onto 35 mm glass bottom microwell dishes (MatTek Corporation, Ashland, MA) at densities around 200 cells/mm2 at 37 °C and 5% CO2 at least 24 h before observation to allow the cells sufficient time to adhere and spread. The media used for LLC-PK1α cells was Gibco®’s Opti-Mem® 1 (Invitrogen Corporation, Carlsbad, CA) with 10% fetal bovine serum (FBS).
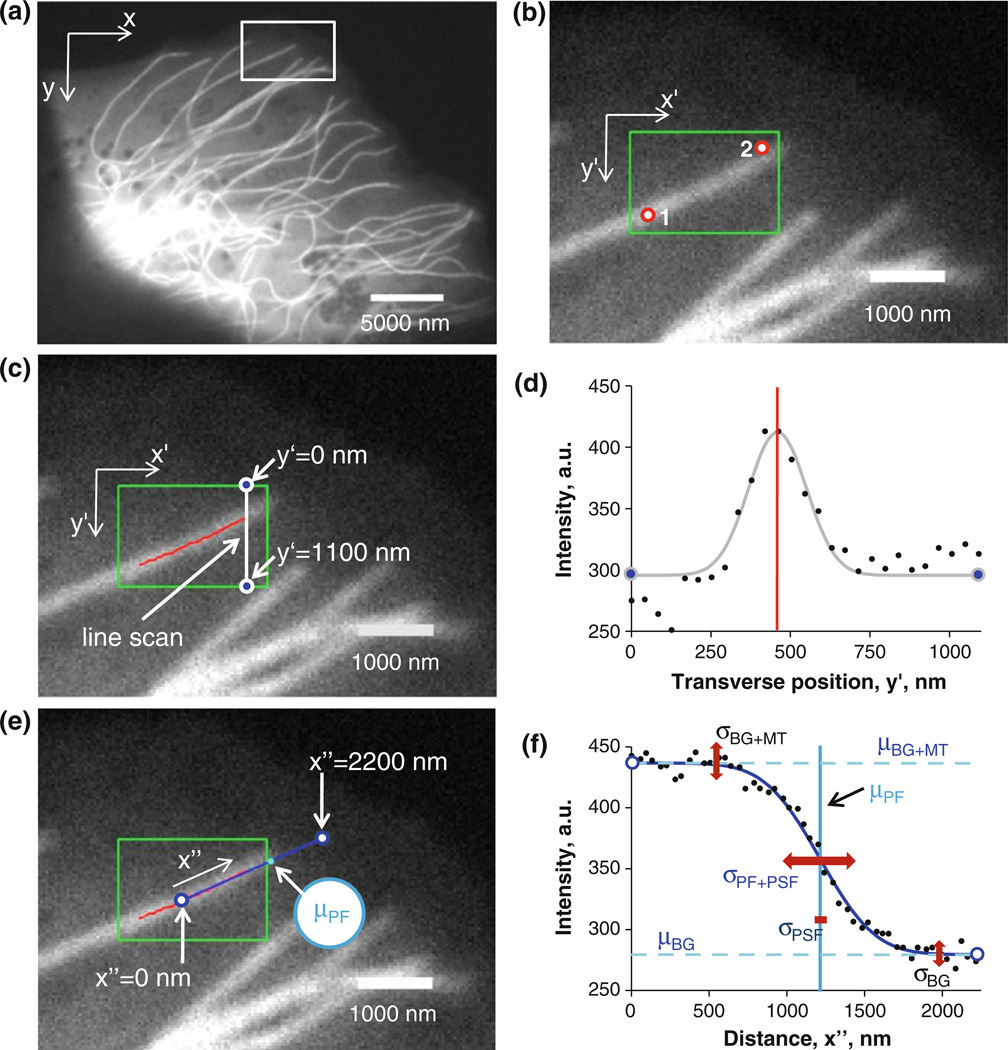
FIGURE 1. Overview of the MT tip tracking algorithm.
(a) Periphery of an LLC-PK1α cell. x and y axes represent the camera coordinate system. The white box is a region of interest (ROI) containing at least one MT tip. (b) A MT tip is chosen within the region of interest. The tip selection is defined by two mouse clicks (red circles). x′ and y′ axes represent the ROI coordinate system. x′ and y′ are chosen so that the MT aligns primarily with the x′ axis. (c) Individual linescans at a given x′ position (delimited by blue dots) yields intensity vs. transverse position (i.e. along the y″ axis). A Gaussian fit to the line scans identifies the backbone of the MT (red points). (d) A single example linescan (black points) with the best-fit of Gaussian (gray). The mean of the Gaussian (red line) determines the location of the MT “backbone”. (e) The direction of the best fit line through MT backbone coordinates defines the MT tip axis x″ (blue line). (f) MT tip intensity profile (black dots) obtained by sampling pixel intensities along the MT tip axis. A Gaussian survival function is fit to the profile (blue line). Blue and dark blue parameters are used in the fit. μBG is the mean intensity value for the background, μBG+MT is the sum of mean signal and background intensity, σPF+PSF is the standard deviation of the Gaussian survival function. The μPF is the mean of the Gaussian survival function and is used to determine the MT tip location.
LLC-PK1α Cell Fixation
A fixative solution was prepared in a 50 mL centrifuge tube with PHEM buffer (60 mM Pipes, 25 mM Hepes, 5 mM EGTA, and 1 mM MgCl2), 0.25% glutaraldehyde, 3.7% paraformaldehyde, 3.7% sucrose, and 0.1% Triton X-100.26 The solution was then warmed to 37 °C in water bath. The media from plated cells was removed and cells were fixed by pipetting 300–400 µL of warm fixative solution onto the cells. The solution was removed after 10 min and a 3-step PBS (phosphate buffered saline) wash was performed. We then added ~300 µL of 50 mM ammonium chloride to each coverslip to create microscope slides and incubated them at room temperature for 10 min. A quick rinse of coverslips in DI water was then performed and excess liquid removed. A drop of antifade solution (SlowFade® Gold Antifade Reagent, Invitrogen™ Molecular Probes™, S36937; ~8–20 µL) was added to the cells and the coverslips placed on the microscope slides. We then air dried the slides under tin foil for 10 min and sealed the edges of coverslips with clear nail polish and allowed them to dry.
It must be emphasized that in order to minimize disturbing the cells to reduce deleterious effects of fixation on microtubules excess media was pipetted out from each dish and fixative was immediately and gently dropped onto a corner of the coverslip. These steps were performed at 37 °C without removing dishes from the incubator.
Digital Fluorescence Microscopy
All observations of LLC-PK1α cells were performed at 37 °C using a Nikon TE200 inverted microscope with a 60×, 1.49 NA Plan Apo objective in conjunction with a 2.5× projection lens resulting in a total magnification of 150×. To clearly identify single MTs, images were taken in the periphery of cells where the cell is thin enough to effectively confine the MTs to two dimensions (i.e. within a single focal plane, <500 nm in the axial direction). Since the depth of field of 1.49-NA lens is ~500 nm, it is logical to conclude that the MTs remain in focus if their z-coordinates remain within ~500 nm of each other, and such condition is adequately achieved in the periphery of LLCPK1 cells.4,5 The exposure time of image acquisition was 300 ms with a 100 W Hg arc lamp illumination system. Filters were 49002 ET-GFP (FITC/Cy2) manufactured by Chroma Technology Corporation (Bellows Falls, VT). The images were obtained using a CoolSnap HQ2 cooled CCD camera (Photometrics, Tucson, AZ) with ~60% quantum efficiency in the green region of the spectrum. As a result, our imaging system was able to collect images with a corresponding pixel size of ~42 nm in the specimen plane. The imaging system was controlled via Metamorph 7.2 software (Molecular Devices Corp., Downingtown, PA). A typical image is shown in Fig. 1a.
MT Tip Tracking Algorithm
The MT tip tracking algorithm was written in MATLAB® (The Mathworks, Natick, MA). The code builds upon the earlier work by Bicek et al.,5 where it was used to estimate the coordinates of MT backbone with subpixel accuracy. Here we used the previous code to determine coordinates of MT backbone at the tip to serve as an axis of growth/shortening, after which point we built a completely new portion to pinpoint tip position. Briefly, a user first defines which MT tips are of interest by enclosing those tip regions in separate rectangular regions (Fig. 1b). Such regions are defined by choosing two points with the computer mouse cursor. The two clicks must lie on the MT of interest with the second click being near the MT tip. Note that the user does not have to be precise in establishing these coordinates (see points 1 and 2 in Fig. 1b). In this way, not only is the rectangular box defined, but the directionality of the MT is also established from the first click toward the second, i.e. the program performs linescans transverse to the MT in the direction from the first click to the second toward the tip of the MT. Note that the two points are not exactly at the corners of the green box in Fig. 1b. Here an additional arbitrary padding is introduced to the boxed region to avoid cases with zero width or height (where the corners of the box are perfectly horizontal or perfectly vertical). After the region is defined, the program finds the center of mass of the rectangular box by giving brighter pixels more weight:
| (1) |
where mi and ri are pixel’s brightness and position, respectively. Depending on this center of mass, the rectangular region is either transposed or remains the same: if the line stretching from top left corner of the image to the center of mass of the image is more than 45° below the horizontal line, then the image matrix is transposed. If the opposite is true then the image matrix remains unaltered. This allows for the MT within the rectangular region to be optimally aligned with the axis of scanning (x′-axis). The region then undergoes a series of line-scans along the scanning axis (horizontal or vertical, depending on the center of mass). We avoided rotating the image regions to align MTs along the scanning direction to prevent the loss of information due to pixilation upon rotation by angles other than 90° (i.e. other than an exact matrix transposition operation on the image). For a given linescan, intensity vs. position data is obtained, to which the program fits a Gaussian curve to obtain the actual MT coordinate per line-scan (Figs. 1c and 1d). The fitting is done using
| (2) |
where I is a pixel intensity value at a given y′ coordinate within a line-scan step at x′, IBG is the mean background intensity, IMT is the mean signal above background, and σ is the Gaussian standard deviation (a measure of the point-spread function, PSF). The values for IBG, IMT, ym, and σ are initially determined from the first corner of the rectangular region and then the fit values from the previous line-scans are used as initial guesses for the subsequent line-scans. To avoid fitting to noise, the Gaussian fit width σ is constrained. The limits of the Gaussian fit width constraint are determined by fitting the Gaussian function to the theoretical point spread function of the microscope setup (Supplementary Fig. S1A, B). The resultant σPSF is used to define the interval [σPSF, 3σPSF], which then defines the acceptable range of the Gaussian width fit σ.
After the line-scans are performed, a set of coordinates x′ and y′ of the MT backbone at the tip is obtained (Fig. 1c, red line). The program then fits a straight line through x′–y′ coordinates with a line segment length that is set by user. This line segment serves to define an axis along which the MT tip grows (Fig. 1e, blue line), which we refer to as the x″ axis. The algorithm then “walks” along the x″ axis and samples the pixel intensity values. This sampling is done for every pixel-wide step, and it records intensities of five neighboring pixels (in the y′ direction) per step in the x″ direction: two pixels to the one side of the line, two pixels to the other, and the pixel lying directly on the MT axis (all five points lying along the y′ axis transverse to the MT). The resultant five intensity values per step are averaged to get the mean intensity value per step. Once the script accumulates intensity vs. position values, it fits a Gaussian survival function to this data as follows:
| (3) |
where IMT, σPF, σPF+PSF, and IBG are the intensity of MT without the background, mean protofilament length, combined standard deviation of protofilament lengths and the PSF, and background intensity, respectively (see Fig. 1f). Note that we define σPF+PSF as
| (4) |
where σPF is the standard deviation of the protofilament lengths.
The mean protofilament length is then converted, with sub-pixel resolution, to the tip position in the frame of reference of the original image (i.e. in the xy-coordinate frame of reference). Tip positions (σPF) and σPF+PSF of all selected MT tips are saved and the program moves onto the next movie frame where the process is repeated. Thus, at the end of the movie, a user can obtain a time series of tip positions (σPF), and standard deviations in protofilament lengths (σPF).
MT Simulation
The simulation of MT images built on our previous image simulation approach, which we call “model-convolution”.10 The overall approach was as follows: (1) define the MT, (2) give it a “speckle” pattern due to stochastic incorporation of labeled subunits25 convolve it with the microscope PSF, (3) map the image onto the camera pixel array (42 nm = 1 pixel), and (4) add noise to each pixel detector. Each step was highly constrained by our experimental observations and microscope specifications, so that the resulting simulated images looked very similar to our experimental images. In addition, the statistics of MT signal to noise were realistically constrained to match experiment. Overall the model-convolution provided us with a “ground truth” against which we could test the algorithm accuracy.
The details of the approach were as follows. First, an image of a white (pixel value = 1) blunt-ended line with perfectly black background (pixel value = 0) was generated with a pixel size of 2.5 nm (fine-grained image in Supplementary Fig. S1C). Next, each pixel of the line was given a probability of 5.2% for being labeled (a value of 1) or 94.8% of being not labeled (a value of 0) to accommodate the noise levels of pixel intensities observed experimentally for MTs in vivo (as in Fig. 1a) due to the fact that not every molecule of αβ-tubulin is labeled with eGFP (Supplementary Fig. S1D). The noise on MTs caused by the variability in tubulin labeling is called speckle noise.9 We call the parameter that determines the labeling in the simulation the speckle parameter (in this case, the speckle parameter is equal to 0.052 or 5.2%, as mentioned previously). For the methods of determining the magnitude of the speckle parameter, see the next section. Each pixel of the image then was multiplied by an appropriate factor so that after convolving the image with the PSF the experimentally observed signal level is matched (see Supplementary Fig. S1E; model convolution technique was applied according to our previous studies4,9,20). The signal levels on both simulated live and simulated fixed MTs were taken from averaging signal levels observed in the frames in the middle of the respective movies of experimental MTs. This was done to represent the average situation in the movies (i.e. our estimates are a worst case scenario for first half of the movies and are opposite for the latter half). After convolution, the image pixel size was increased to 42 nm to match our experimental imaging system described above (coarse-grained image). This was done by binning smaller pixels ((42 nm/2.5 nm) × (42 nm/2.5 nm) or 17 × 17) into bigger pixels and averaging their intensity values to assign intensities to bigger pixels (see Supplementary Fig. S1F). This coarse-grained image was then added to an image taken from a cellular background devoid of MTs. The resultant simulated image recapitulated experimentally obtained images in terms of their intensity values and noise levels, and had the same SNR observed experimentally. As a side note, the actual fraction of labeled tubulin dimers in LLC-PK1α cells is larger than 5.2%. Quantitative immunoblotting performed by Rusan et al.17 revealed eGFP-labeled fraction of tubulin dimers to be around 17%. The apparent discrepancy in these values is attributable to the fact that our fine-grained pixels do not match the molecule dimensions (a tubulin dimer is 8 nm long while our pixel size is 2.5 nm). However, given that the projected area of a tubulin dimer is ~8 nm × 4 nm = 32 nm2, and our pixel area of (2.5 nm)2 = 6.25 nm2, the resulting ratio of 32/6.25 = 5.1 provides reasonable agreement with the experimental expression level (5.2% × 5.1 = 26.5%, compared to the 17% observed experimentally by Rusan et al.17).
Speckle Parameter Determination for MT Tip Simulations
The image signal/noise ratio (SNR) was estimated from MT images using the ImageJ image processing and analysis software (NIH, Bethesda, Maryland, USA). A segmented line tool in ImageJ was used to sample pixel intensity values along experimental MTs. The average duration of live cell movies was 30 frames (0.5 s/frame), and we used the 15th frame to represent the typical situation with respect to SNR (almost all fixed cell movies were also 30 frames long, so the 15th frame was chosen as representative of the average SNR in fixed cell movies as well). This intensity vs. position data was then used to fit a line in order to estimate and eliminate any intensity gradients that arise from variability in cell thickness and intracellular structures. A mean intensity and its standard deviation were then calculated for several MTs (N = 32 for live cells, N = 10 for fixed cells). The procedure was also used to determine the SNR for the cellular background by using a segmented line tool parallel to, but offset from, the analyzed MTs. Thus, average values for background and signal intensity together with their standard deviations were collected. The average intensity values together with the background intensity standard deviation were then used for the MT tip simulation described in above. The simulation tested a range of speckle parameter values from 0.002 (0.2%) to 1 (100%) using increments of 0.002 (0.2%). All simulated MTs had similar starting parameters (i.e. mean signal and background intensities and standard deviation of the background intensity), but, due to a varying speckle parameter, they all had a different value for the standard deviation of the signal intensity along MTs. Thus, the plots of standard deviation of the signal intensity as a function of the speckle parameter were constructed (Supplementary Fig. S2). A power law function was then fit to the data, given by
| (5) |
where f(s) is the standard deviation of the signal intensity, s is the speckle parameter, k is a fitted scaling exponent, and a and b are fitted parameters. Since we measured the standard deviation of the signal on simulated MTs, we could then estimate the corresponding speckle parameter based on our standard deviation vs. speckle parameter plot. To increase our confidence in the estimated speckle parameter, we then simulated 100 MTs with the estimated speckle parameter to see if the average noise on the MT corresponded to the speckle levels of the simulation. This approach was validated by the fact that the speckle parameter is approximately proportional to the signal levels observed (i.e. the speckle parameter decreased from 0.052 to 0.02 as the intensity decreased from 87 a.u. to 47 a.u. as we moved from sampling live cell to fixed cell pixel intensity values). We note that this procedure for finding the speckle level needs to be performed for any new case/condition including: (1) different cell types, (2) different imaging systems, or (3) different movie lengths. For example, if we were to determine the speckle parameter at frame number 5 of live cell movies, we would have to estimate the mean intensity values of background and signal together with their standard deviations from the 5th frame of several live cell movies and use them in another round of the speckle parameter search as described above.
MT Taper Length Determination
The variance in protofilament (PF) length could potentially contribute significantly to the overall fitted standard deviation, σPF+PSF. We chose PF + PSF notation to emphasize the two sources of contribution to the Gaussian survival function parameters: (1) the MT’s protofilament tip distribution (PF), and (2) the PSF. By subtracting the PSF variance from the overall variance we are able to obtain an estimate of the variance in PF lengths. The tip is blunt if all 13 MT protofilaments are of length l. In practice, protofilaments at the tip are of different lengths, and so a gradual taper is produced, as is observed experimentally via cryo-electron microscopy of MTs in vitro.8,13 We assume that if the MT taper is of length l13−l1, then the 13 protofilaments of such a MT are distributed uniformly in length with the shortest protofilament having length l1 and the longest having the length of l13, which is approximately consistent with EM images of MTs in vitro (e.g. see Figs. 4b–e in Chretien et al.8).
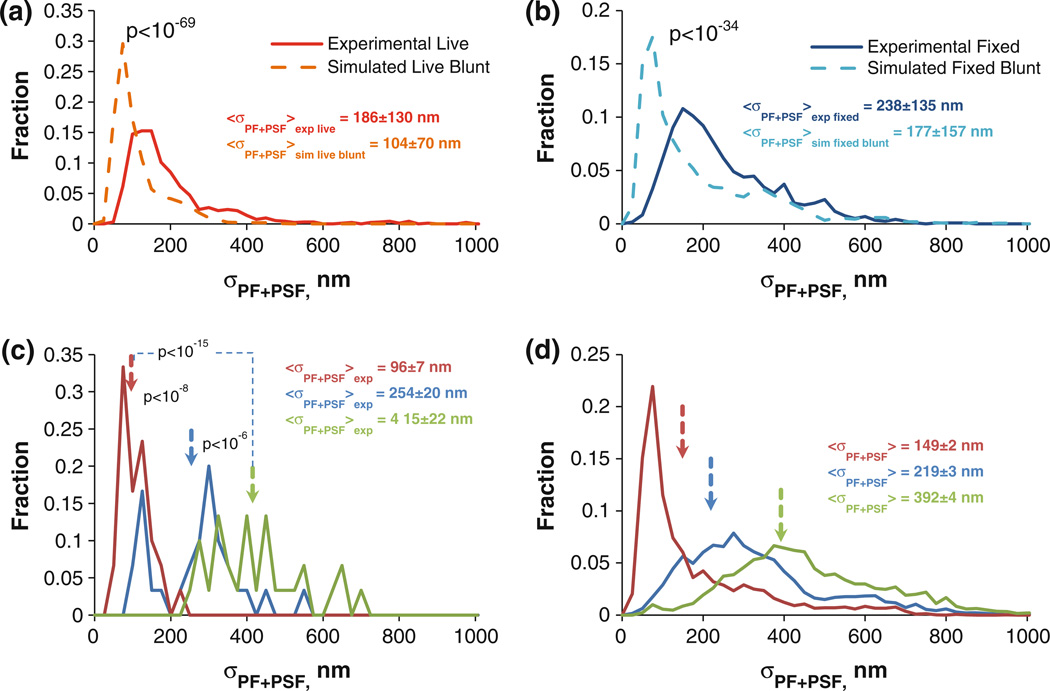
FIGURE 4. Determining MT taper and protofilament length variability.
(a) σPF+PSF distributions from live cell MTs (red) and blunt simulated live cell MTs (burnt orange). Uncertainties indicate the sample standard deviation of the distribution (Nexp. live = 930, Nsim. live blunt = 10230). (b) Same as a, except for fixed MTs (Uexp. fixed = 1185, Nsim. fixed blunt = 3000). (c) σPF+PSF distribution from three different experimental fixed MTs. (Uncertainties are 6SEM, N = 30). (d) σPF+PSF distributions of three simulated fixed cell MTs with N = 3000 for each distribution. The mean tapers from the three experimental MTs in (c) were used to generate the three distributions.
In order to see how σPF+PSF values change with taper dimensions, we increased taper lengths from 0 nm to 3000 nm in increments of 500 nm, for 20 stacks of 30 simulated MT image frames per taper length and checked the mean σPF+PSF value per taper (for more details on model convolution, see above). We then fit a line through input taper vs. σPF+PSF data to empirically determine the relationship between σPF+PSF and the corresponding taper length. This allowed us to construct empirical plots to serve as lookup tables for the analysis of the experimental data.
RESULTS
MT Tip Tracking Accuracy
Our first goal was to estimate the accuracy of our tip tracking algorithm. However, we found in our simulations that the accuracy was affected by the tip structure. The key fitting parameter used to characterize the tip structure is the standard deviation of the Gaussian survival function, σPF+PSF (Fig. 1f). Due to the resultant image arising from the convolution of the fluorescent object (i.e. the MT) with the system’s PSF, this standard deviation is a composite of the “object’s” standard deviation and the PSF’s standard deviation. In order for us to estimate variability in protofilament lengths (i.e. σPF), we need to know σPSF, the standard deviation of a PSF. Once we know σPSF and the measured σPF+PSF, we can calculate σPF, which allows us to gain information regarding the tip conformation. The standard deviation of the PSF, σPSF, can be approximated by the standard deviation of a Gaussian approximation to the theoretical Airy function (Supplementary Fig. S1B), estimated to be 72 nm for our system. One can see from Fig. 1f that the PSF’s standard deviation, σPSF, is in some cases negligible compared to the standard deviation of the Gaussian survival function, σPF+PSF. Therefore, the biggest contribution to the Gaussian survival function’s standard deviation in at least some cases comes from the actual object itself, and not the PSF. This fact was taken into account when simulating images of the MT tips and analyzing them in order to test the accuracy of the tip tracking algorithm.
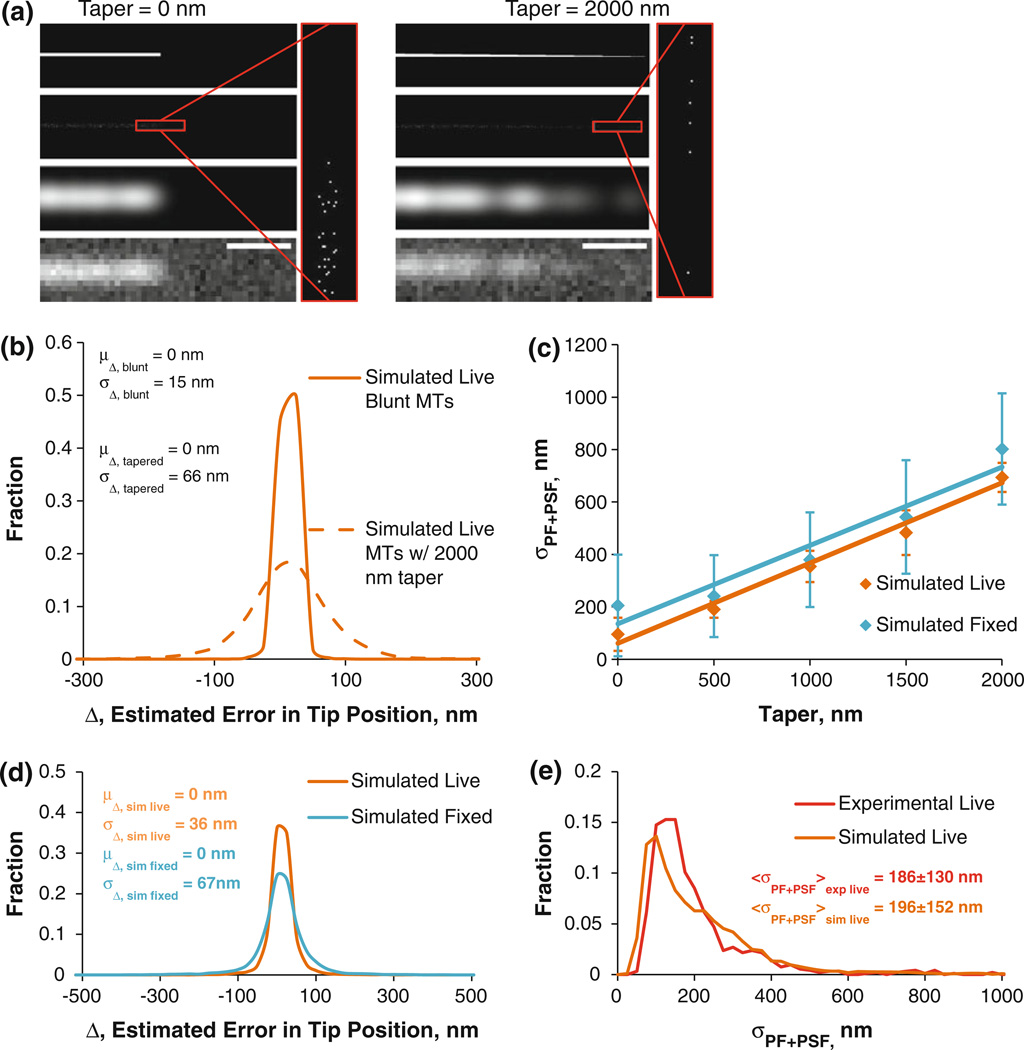
When simulated live blunt MT tips were analyzed, the accuracy of the tip tracking algorithm was ~15 nm, equivalent to ~2 tubulin dimers (Figs. 2a and 2b). However, as can be seen from Fig. 1f, the standard deviation of the Gaussian survival function, σPF+PSF, is often much greater than the standard deviation of the PSF, σPSF. Therefore, it is reasonable to assume that the actual dimensions of the object will affect the fit to the intensity profile data, and, consequently, the accuracy of the tip tracking program. Indeed, when simulated live MT tips were given a taper size of 2000 nm, which is at the upper extreme of taper length evident from cryo-EM data by Chrétien et al.,8 the accuracy worsened to 66 nm (Figs. 2a and 2b). Therefore, since the accuracy of the tip tracking algorithm depends on taper length, a realistic distribution of taper lengths was needed for the simulations.
FIGURE 2. Simulation of blunt and tapered MTs and estimated accuracy in tracking.
(a) Leftmost panels: steps in simulating a blunt MT; top left: starting unspeckled blunt MT; upper middle left: speckled MT tip (red inset enlarges the tip); lower middle left: model-convolved speckled MT tip; bottom left: final image of speckled blunt MT tip coarse-grained to match the microscope setup with the noise levels resembling experimental values. Rightmost panels: same as in leftmost panels, except for a tapered MT tip. Scale bars = 500 nm. (b) A histogram of tip tracking accuracy of blunt and tapered MT tips given by the histogram of errors in tip tracking. (c) Plots of σPF+PSF vs. simulated MT taper, used to convert σPF+PSF values into taper lengths and vice versa. (d) Accuracy in MT tip tracking with simulated live/fixed cell taper distributions. (e) Tip structure distribution, as measured by the σPF+PSF distributions of experimental (red) and simulated (burnt orange) live cell MTs.
To account for the effect of taper on tip position accuracy, we converted the observed σPF+PSF values of all MT tips at all time points of MT movies to taper sizes using an empirically-determined linear relation between simulated MT taper and σPF+PSF (shown in Fig. 2c). These realistic taper lengths were then used to generate and analyze simulated MT tips in order to determine the accuracy of the tip tracking (Figs. 2d and 2e). The result of this investigation yielded average single time point MT tip tracking accuracy of ~36 nm. We need to mention that due to photobleaching, the signal worsens toward the end of the movies, reducing not only tip position accuracy but also the accuracy in estimation of σPF+PSF. Since the signal is the strongest during the first frames of MT movies and the weakest during the last frames, we used the 15th frame of each movie to determine SNR and speckle parameter values, such that the average situation is represented.
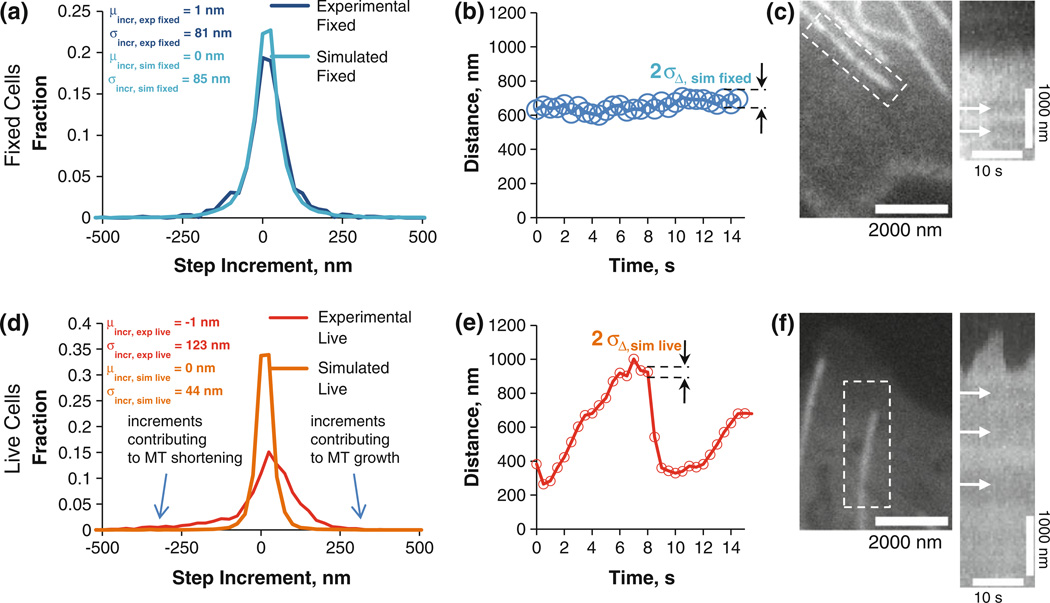
Unfortunately we do not know the absolute MT tip location in our experiments, so using this measurement to validate our live MT tip tracking is not an option. However, to estimate the tip tracking accuracy we can use incremental changes in tip position per time step as an alternative. We refer to this measure as a “step increment” in this context. Live LLCPK1α cells have MTs that constantly grow and shorten, as opposed to our simulations, where all MTs are static. If we try to compare step increment distributions between experimental live and simulated live MT tip tracking, most likely they will not match. Therefore, to remove MT dynamics from the picture, we decided to use fixed LLCPK1α cells with no MT activity, which allows us to establish an experimental control in the absence of assembly dynamics.18 Using the mid-frame SNR and speckle parameters, we simulated fixed cell MTs similarly to the live MT simulation (for resultant σPF+PSF distribution, see Supplementary Fig. S3A) the standard deviation of step increment distribution for fixed MTs was ~85 nm (Fig. 3a, cyan blue plot). This result is very close to the experimentally obtained value of ~81 nm (Fig. 3a, dark blue plot) and shows that our validation of the tip tracking algorithm by the means of simulation is adequate. The corresponding estimated single time point accuracy for tracking MT tips in fixed cells is 67 nm (Figs. 2d and 3b). An example of a fixed cell microtubule is given in Fig. 3c with the corresponding kymograph to the right. Arrows show the presence of stationary speckles indicating the absence of MT transport.
FIGURE 3. Method validation using fixed MTs and subsequent application to live MTs.
(a) Histogram of the step increments, dark blue plot showing the experimental fixed cell analysis, while light blue represents the simulated fixed cell data. The excellent agreement demonstrates that the simulation faithfully predicts the experimentally measured step increment distribution. (b) distance vs. time for the tip position. Circle diameter is equal to two times sD,sim fixed at each time point to demonstrate algorithm accuracy; (c) Fixed MT tip; kymograph of the fixed MT tip with arrows indicating speckles and lack of MT transport; (d–f) same as a–c, except for a live MT tip. Live MTs have increments (d, red) that are much more variable than expected from measurement error alone (d, orange).
Using the same approach as described above, we then simulated MT tips as were observed in living cells. The SNR parameters used in the simulation were found by analyzing the 15th frame of MT movies (out of 30 frames). We found that the standard deviation of the step increment distribution for living cell movies was 44 nm (Fig. 3d, burnt orange), with a single time point accuracy of 36 nm (Fig. 3e), as was previously mentioned (Figs. 2d with the experimental and simulated σPF+PSF distributions shown in Fig. 2e). When compared to the standard deviation of the step increment in experimental living cell MTs, our simulated value is significantly smaller (44 nm vs. 123 nm, Fig. 3d). This increase in step increment standard deviation in the living cell MTs shows that live cell MTs are highly dynamic due to stochastic net addition or loss of tubulin between observations. Based on these results, we conclude that the single time point tip tracking accuracy in living cells is 36 nm on average, at least when using our imaging system with LLCPK1α epithelial cells. This means that we can track assembly dynamics in vivo under these specific conditions with single time point accuracy equivalent to ~4.5 tubulin dimer layers. An example of a live cell microtubule is on Fig. 3f with the corresponding kymograph to the right. Arrows show the presence of stationary speckles to emphasize the absence of MT transport which, otherwise, would have affected tip tracking results.
Estimation of MT Tip Structure
Because σPF+PSF was generally larger than σPSF, we then estimated the tip structures in fixed and live cells. Comparing σPF+PSF distributions for live and fixed cells we found that they are qualitatively similar to each other, indicating that MT tip structures were not altered significantly upon cell fixation (compare Figs. 4a and 4b, solid plots). Using σPF+PSF data for experimental live and fixed cells, as well as the linear conversion in Fig. 2c, we can also approximate mean taper lengths in live and fixed cells. Thus, converting the average σPF+PSF values of live and fixed cells into tapers yielded mean tapers of 418 nm (with standard deviation of 420 nm) and 387 nm (with standard deviation of 420 nm), respectively. We also determined σPF to be ~171 nm for live cells and ~227 nm for fixed, estimated by:
| (6) |
where σPSF = 72 nm for our imaging system (see Figs. 4a and 4b, solid plots).
Since we identified all the components contributing to σPF+PSF values (variability in protofilament lengths and the standard deviation of the PSF), we then used this measurement to test various assumptions about the state of MT tips. One hypothesis that we can test statistically is whether all tips in living or fixed cells are blunt. In order for us to test this hypothesis, we first collected live and fixed cell MT σPF+PSF values. We then used our MT simulation algorithm to generate 10230 live cell simulated MTs whose tips are blunt. The resultant σPF+PSF distribution could then be compared to σPF+PSF distributions of experimental live cell MTs, and found to be statistically different (p-value < 10−69 see Fig. 4a). Similarly, fixed cell MTs were also found to be inconsistent with the hypothesis that all MT plus ends are blunt (p-value < 10−34, see Fig. 4b).
We then determined whether we could discriminate taper lengths between individual fixed MTs. The average taper lengths described above were obtained by using all data points across all MT movies. However, one can use one or several frames in order to estimate whether or not taper structures differ from MT to the next, or from time point to time point. Using our algorithm, we estimated σPF+PSF values from three different fixed cell MTs, found their mean, and used the calibration plot for the fixed cells in Fig. 2c to determine their respective taper values. Thus, for one MT the average value of σPF+PSF was 96 nm, and the corresponding taper length was ~0 nm. For a second MT the average σPF+PSF was 254 nm, and the corresponding taper was 438 nm. Finally, for a third MT, the average σPF+PSF was 415 nm, yielding a taper of 940 nm. Despite having only 30 data points per MT tip, the three distributions were clearly distinct from one another. To better approximate the true distributions of σPF+PSF at these three taper values, we also simulated 3000 fixed cell MTs for each of the three tapers. As shown in Fig. 4d, we found that these distributions recapitulated the experimentally observed variability, and yet were still distinct from each other. Thus, we are able to detect statistically significant differences in tip structures between individual MTs.
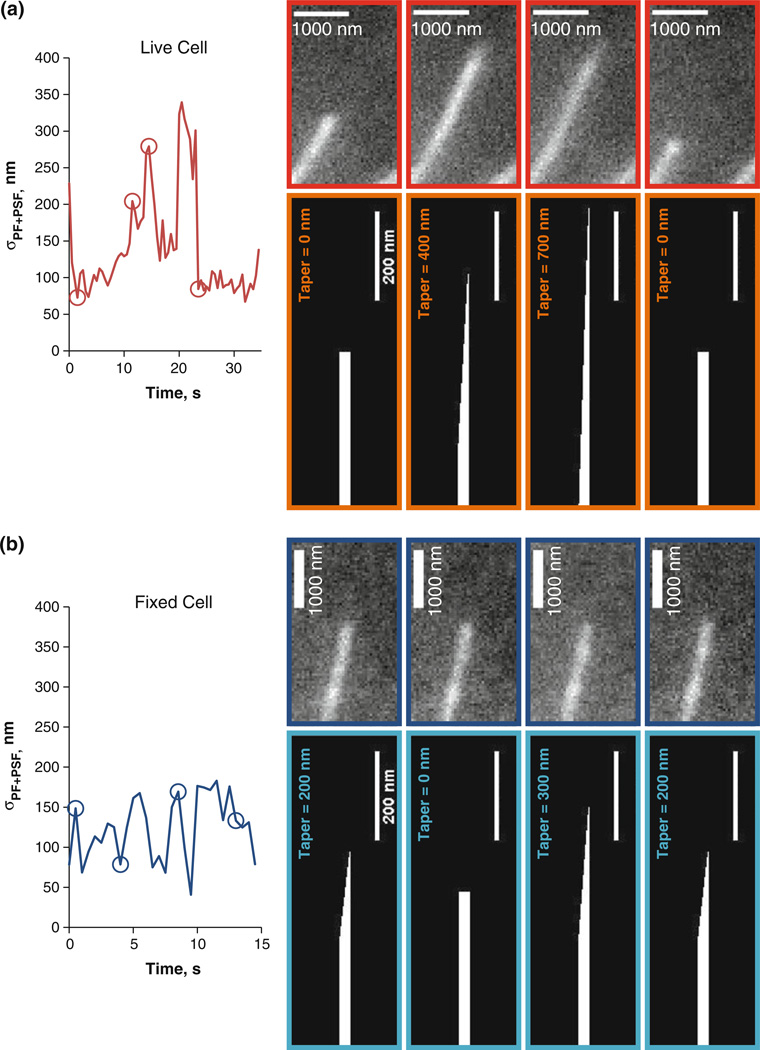
Finally, the algorithm allowed us to estimate the conformational dynamics of MT tips by recording σPF+PSF via a Gaussian survival function fitted to intensity vs. distance during the tracking (see Fig. 5a). Thus, the tip tracking algorithm can potentially be used to determine the effect of a therapeutic drug (e.g. taxol) on dynamic instability parameters and the MT tip configuration (compare non-dynamic fixed cell MT in Fig. 5b to live cell MT in Fig. 5a). Notice that even though the analyzed MT is taken from a fixed cell in Fig. 5b, the presence of photon counting noise creates variability in the estimated tip structures. However, the variability is less than that seen in live cell MTs, indicating the ability to track live cell MT tip structure dynamics.
FIGURE 5. MT Tip Structure Dynamics.
(a) Left plot: Time series of σPF+PSF for a single live cell MT. Panels outlined in red show the MT tip at four different times, denoted by red circles on the plot to the left. The panels in burnt orange show predicted MT tip tapers at those four time points. (b) Same as (a), except for a fixed MT.
DISCUSSION
Currently, there are several approaches like 3D-SIM, PALM, STORM, and others that are able to reveal highly detailed information on the interior of cells.2,3,19 These methods yield images with detail down to 5–10 nm resolution; however, obtaining combined high temporal and spatial resolution in live cells remains an objective that is yet to be achieved. Although such innovative methods can obtain nanometer-scale information, they require fixing the specimen (3D-SIM, PALM) and they take ~minutes to hours to image (STORM), so if live cell imaging were to ever be implemented with such methods, the ability to resolve temporal details that are especially important when dealing with such dynamic structures as MTs would be lost. By utilizing live cell imaging with the tip tracking algorithm presented here, we can avoid such obstacles.
The goal of this work was to improve the quality of MT tip tracking using conventional digital fluorescence microscopy combined with semi-automated image analysis. While developing the algorithm, we discovered a relationship between the accuracy of the program and the state of MT tips. As a result, the final version of the tip tracking algorithm allowed us to track MT tips at single time points with sub-pixel precision (~36 nm RMS error in live LLCPK1-a cells), surpassing the typical Rayleigh resolution limit on eGFP-tubulin imaging by 6-fold. In addition, the algorithm allowed us to estimate the conformational dynamics of MT tips. Thus, our tip tracking algorithm allows for near-molecular measurement accuracy using conventional imaging systems. This method compares favorably with the methods described earlier, and is suitable for studying dynamic and temporally-evolving MT tips. For example, in the case of manual hand-tracking with a mouse-driven cursor overlaid onto video monitor projection of DIC images, the precision was found to be <500 nm7 and ~160 nm.11 Neither study used model-convolution or fixed MTs to assess the accuracy of the measurements. Subsequently, manual tracking has been used to analyze fluorescence images of MTs, including for GFP-MTs in LLCPK1α cells.17 Rusan et al. did not report the accuracy and precision, although our own visual assessment of their time series trajectories suggests submicrometer precision. The method of Hadjidemetriou et al. used high curvature of the isointensity contours of tip points to track MT tips and is described in detail in section 2.6 of their work.12 They estimated the accuracy to be ~260 nm, which is comparable to the previous hand-tracking approaches but has the advantage of being semi-automated. Our present study describes a semi-automated method that has accuracy of ~36 nm, improving on the previous methods by several-fold. In the future it will be important to assess the accuracy under less ideal conditions, including cases where MTs do not lie in the focal plane.
After we found that we could evaluate MT tip structure with our algorithm, we tested whether MT tips in living cells are blunt. When we compared σPF+PSF distributions from simulated live-cell blunt MTs and the MTs of experimental live cells, the p-value from a two-sample t-test was <10−69. Thus, we conclude that not all MTs in our LLCPK1α cells have blunt-ended tips. Rather, we estimate that they have an average standard deviation in their protofilament lengths of ~171 nm. This result gives us insight into the magnitude of single tubulin dimer bonding energies, an issue addressed in works by VanBuren et al.21,22 A strong lateral bond relative to the longitudinal bond will result in tubulin dimers forming stronger associations with their lateral neighbors, thereby making the MT tip relatively blunt. Alternatively, if longitudinal bonds are much stronger that the lateral ones, individual protofilaments would favor longitudinal rather than lateral additions, which would result in dynamic growth with non-blunt tips predominating. This second case is what seems to be the situation with live LLC-PK1α cell MTs since blunt tips are relatively rare compared to tips with dispersed protofilament lengths. The extent to which various MT-associated proteins are responsible for the relative strength of the longitudinal bonds will be an important area of future investigation.
Finally, it must be mentioned that there is always a possibility of MTs being affected by cell fixation. We used fixed cell MTs to compare their tip tracking to the tip tracking on the simulated images. Our MT simulations did not include MT dynamics so in order to truly compare “apples to apples” we needed to have non-dynamic experimental MTs which we achieved with cell fixation. We applied our algorithm to simulated as well as experimental fixed cell MTs and found that their step change distributions were nearly identical, which indicated that our model convolution approach for tip tracking validation is adequate. However, if one needs to qualitatively determine effects of fixation altered MT tip structures in any way, one can compare σPF+PSF distributions from live and fixed cells. In our case, cell fixation did not have significant effect on MT tips since the distributions were similar (see Figs. 4a and 4b).
CONCLUSIONS
As confirmed by our simulations, the MT tip tracking algorithm introduced here allows for not only sub-pixel accuracy (36 nm in live cells), but also is able to yield information regarding the tip state of a MT by estimating σPF at any given frame. Even though a single time point estimate of MT taper is noisy, one can collect several σPF+PSF estimates from single fixed cell MTs to improve accuracy of the estimate. The algorithm can also be applied to other fluorescently labeled linear structures, and may also work with imaging methods differing from digital fluorescence microscopy. With this method we have demonstrated unprecedented accuracy in MT tip tracking in vivo, and the ability to acquire information about MT tip structure at the nanoscale. This algorithm will facilitate molecular-scale understanding of MT-associated proteins and therapeutic drugs that alter MT dynamics in normal and diseased cells.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dominique Seetapun for technical assistance. The research was conducted with support from NIH (GM071522 and GM076177), and NSF (MCB-0615568).
Footnotes
ELECTRONIC SUPPLEMENTARY MATERIAL
The online version of this article (doi:10.1007/s12195-010-0155-6) contains supplementary material, which is available to authorized users.
REFERENCES
- 1.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. New York: Garland Science; 2007. [Google Scholar]
- 2.Bates M, Huang B, Dempsey GT, Zhuang X. Multicolor super-resolution imaging with photo-switchable fluorescent probes. Science. 2007;317:1749–1753. doi: 10.1126/science.1146598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 4.Bicek D, Tüzel E, Kroll DM, Odde DJ. Analysis of microtubule curvature. Methods Cell Biol. 2007;83:237–268. doi: 10.1016/S0091-679X(07)83010-X. [DOI] [PubMed] [Google Scholar]
- 5.Bicek AD, Tuzel E, Demtchouk A, Uppalapati M, Hancock WO, Kroll DM, Odde DJ. Anterograde microtubule transport drives microtubule bending in LLC-PK1 epithelial cells. Mol. Biol. Cell. 2009;20:2943–2953. doi: 10.1091/mbc.E08-09-0909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brangwynne CP, Koenderink GH, Barry E, Dogic Z, MacKintosh FC, Weitz DA. Bending dynamics of fluctuating biopolymers probed by automated high-resolution filament tracking. Biophys. J. 2007;93:346–359. doi: 10.1529/biophysj.106.096966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cassimeris L, Pryer NK, Salmon ED. Real-time observations of microtubule dynamic instability in living cells. J. Cell Biol. 1988;107:2223–2231. doi: 10.1083/jcb.107.6.2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chrétien D, Fuller SD, Karsenti E. Structure of growing microtubule ends: two-dimensional sheets close into tubes at variable rates. J. Cell. Biol. 1995;129:1311–1328. doi: 10.1083/jcb.129.5.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gardner MK, Odde DJ, Bloom K. Hypothesis testing via integrated computer modeling and digital fluorescence microscopy. Methods. 2007;41:232–237. doi: 10.1016/j.ymeth.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 10.Gardner M, Sprague B, Pearson C, Cosgrove B, Bicek A, Bloom K, Salmon E, Odde D. Model convolution: a computational approach to digital image interpretation. Cell. Mol. Bioeng. 2010;3:163–170. doi: 10.1007/s12195-010-0101-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gildersleeve RF, Cross AR, Cullen KE, Fagen AP, Williams RC. Microtubules grow and shorten at intrinsically variable rates. J. Biol. Chem. 1992;267:7995–8006. [PubMed] [Google Scholar]
- 12.Hadjidemetriou S, Toomre D, Duncan J. Motion tracking of the outer tips of microtubules. Med. Image Anal. 2008;12:689–702. doi: 10.1016/j.media.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 13.Mandelkow EM, Mandelkow E, Milligan RA. Microtubule dynamics and microtubule caps: a time-resolved cryo-electron microscopy study. J. Cell Biol. 1991;114:977–991. doi: 10.1083/jcb.114.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- 15.Odde DJ, Cassimeris L, Buettner HM. Kinetics of microtubule catastrophe assessed by probabilistic analysis. Biophys. J. 1995;69:796–802. doi: 10.1016/S0006-3495(95)79953-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Odde DJ, Buettner HM, Cassimeris L. Spectral analysis of microtubule assembly dynamics. AIChE J. 1996;42:1434–1442. [Google Scholar]
- 17.Rusan NM, Fagerstrom CJ, Yvon AC, Wadsworth P. Cell cycle-dependent changes in microtubule dynamics in living cells expressing green fluorescent protein-{alpha} Tubulin. Mol. Biol. Cell. 2001;12:971–980. doi: 10.1091/mbc.12.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schek HT, III, Gardner MK, Cheng J, Odde DJ, Hunt AJ. Microtubule assembly dynamics at the nanoscale. Curr. Biol. 2007;17:1445–1455. doi: 10.1016/j.cub.2007.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schermelleh L, Carlton PM, Haase S, Shao L, Winoto L, Kner P, Burke B, Cardoso MC, Agard DA, Gustafsson MGL, Leonhardt H, Sedat JW. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science. 2008;320:1332–1336. doi: 10.1126/science.1156947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sprague BL, Pearson CG, Maddox PS, Bloom KS, Salmon E, Odde DJ. Mechanisms of microtubule-based kinetochore positioning in the yeast metaphase spindle. Biophys. J. 2003;84:3529–3546. doi: 10.1016/S0006-3495(03)75087-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.VanBuren V, Odde DJ, Cassimeris L. Estimates of lateral and longitudinal bond energies within the microtubule lattice. Proc. Nat. Acad. Sci. USA. 2002;99:6035–6040. doi: 10.1073/pnas.092504999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.VanBuren V, Cassimeris L, Odde DJ. Mechanochemical model of microtubule structure and self-assembly kinetics. Biophys. J. 2005;89:2911–2926. doi: 10.1529/biophysj.105.060913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walker RA, O’Brien ET, Pryer NK, Soboeiro MF, Voter WA, Erickson HP, Salmon ED. Dynamic instability of individual microtubules analyzed by video light microscopy: rate constants and transition frequencies. J. Cell Biol. 1988;107:1437–1448. doi: 10.1083/jcb.107.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wan X, O’Quinn RP, Pierce HL, Joglekar AP, Gall WE, DeLuca JG, Carroll CW, Liu S, Yen TJ, McEwen BF, Stukenberg PT, Desai A, Salmon E. Protein architecture of the human kinetochore microtubule attachment site. Cell. 2009;137:672–684. doi: 10.1016/j.cell.2009.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waterman-Storer CM, Salmon E. How microtubules get fluorescent speckles. Biophys. J. 1998;75:2059–2069. doi: 10.1016/S0006-3495(98)77648-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Witte H, Neukirchen D, Bradke F. Microtubule stabilization specifies initial neuronal polarization. J. Cell Biol. 2008;180:619. doi: 10.1083/jcb.200707042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.