Abstract
During nutrient excess, triglycerides are synthesized and stored to provide energy during times of famine. The presence of high glucose leads to the activation of carbohydrate response element binding protein (ChREBP), a transcription factor that induces the expression of a number of glycolytic and lipogenic enzymes. ChREBP is expressed in major metabolic tissues and while we have a basic understanding of ChREBP function in liver, in vivo genetic systems to study the function of ChREBP in other tissues are lacking. In this study, we characterized the role of the Drosophila homolog of ChREBP, Mlx interactor (Mio), in controlling fat accumulation in larvae and adult flies. In Mio mutants, high sugar-induced lipogenic enzyme mRNA expression is blunted and lowering Mio levels specifically in the fat body using RNA interference leads to a lean phenotype. A lean phenotype is also observed when the gene bigmax, the fly homolog of ChREBP’s binding partner Mlx, is decreased in the larval fat body. Interestingly, depleting Mio in the fat body results in decreased feeding providing a potential cause of the lowered triglycerides observed in these animals. However, Mio does not seem to function as a general regulator of hunger-induced behaviors as decreasing fat body Mio levels has no effect on sleep under fed or starved conditions. Together, these data implicate a role for Mio in controlling fat accumulation in Drosophila and suggests that it may act as a nutrient sensor in the fat body to coordinate feeding behavior with nutrient availability.
Keywords: Drosophila, fat storage, feeding, Mio/dChREBP, sleep
1. Introduction
After a meal, the oxidation of sugars and fats provides energy for basic cellular functions. Excess calories that are not used for energy are stored mainly as glycogen and triglycerides. This response was selected for over evolution as a means of preparing an organism for times of scarce food sources. However, in today’s western society where food is abundant and readily available, this ability of an animal to store surplus nutrients leads to excess fat accumulation and metabolic diseases such as obesity and diabetes. This effect is pronounced after a prolonged high sugar diet as these conditions lead to an acute increase in the activity of enzymes necessary for fat synthesis, the chronic production of lipogenic enzymes, and the concurrent synthesis and storage of fats [1].
A key regulator of this chronic response to high sugar intake and fat storage in mammals is the transcription factor, carbohydrate response element binding protein (ChREBP)1. In response to high glucose conditions, ChREBP translocates into the nucleus, where it activates the expression of pyruvate kinase and many lipogenic enzymes, including fatty acid synthase (FAS), acetyl-CoA carboxylase (ACC), and ATP citrate lyase (ATPCL), ultimately leading to increased fat accumulation [2,3]. In order to fully activate transcription, ChREBP must heterodimerize with another transcription factor called Max-like protein X (Mlx) [4]. ChREBP is expressed most highly in liver and adipose tissue, but significant expression is also observed in skeletal muscle, the intestine and kidney [5]. Conversely, Mlx has a relatively ubiquitous expression pattern [6]. While we have a basic understanding of ChREBP function in the liver, in vivo systems to study tissue-specific functions of ChREBP are lacking.
Drosophila is an excellent model system for investigating the tissue-specific control of metabolism [7]. Flies have specialized organ systems for nutrient uptake, storage, and metabolism that are functionally analogous to mammalian systems. The Drosophila midgut is the site of both digestion and nutrient uptake, while the fat body of the fly is the site of glycogen and triglyceride storage [8]. Many important metabolic genes and pathways in mammals are also highly conserved in flies (see table in [7]) and these genes can be manipulated easily using the genetic tools available in the Drosophila system [9,10], allowing the information identified to be applied to mammals.
The Drosophila genome contains a ChREBP-like gene named Mlx interactor (Mio) (also known as dmondo) and an Mlx-like gene called bigmax [11,12]. However, very little functional data exists for these gene products. Mio and bigmax are expressed throughout embryogenesis and are enriched in the fat body and malpighian tubules ([12]; Fly Atlas). Mio mRNA levels are also increased when larvae are fed 20% sucrose, a condition that also leads to increased expression of fat synthesis enzymes [13], suggesting a role for Mio in regulating high-sugar induced lipogenic gene expression. Therefore, we hypothesized that Mio and bigmax are involved in regulating fat storage in the fly. In this study, we found that the induction of lipogenic enzyme expression in response to high sugar is blocked in Mio mutant animals. Consistent with this finding, knocking down Mio and bigmax in the fly fat body leads to decreased fat storage. Further, overall food consumption is also blunted when Mio expression is decreased, providing a potential explanation for the observed lean phenotype. These data identify a novel role for Mio in the fat body to regulate the storage of triglycerides and overall food consumption and further supports the use of Drosophila as a model system for understanding tissue-specific control of metabolism and behavior.
2. Materials and Methods
2.1 Fly Genetics
Flies were grown at 25°C on standard cornmeal-sugar-yeast medium unless otherwise stated. The following fly strains used in this study were obtained from the Bloomington Stock Center: UAS-GFP (#5194); UAS-bigmax-IR (#29325); Cg-Gal4 (#7011) and Miok05106/CyO (#10562). The yolk-Gal4 and to-Gal4 lines have been previously published [14,15]. The UAS-Mio-IR line (#52606), which knocks down Mio expression by producing a hairpin RNA, was obtained from the Vienna Drosophila RNAi Center [16]. The UAS-MiodsRNA line was made by cloning positions 1772–2679 of Mio-RB into pSymp-UAST [17] with subsequent P-element transformation performed by Duke University Model Systems Genomics. Control animals were cultured in the same vials as the experimental animals to account for crowding and other environmental conditions.
2.2 Triglyceride, Protein and DNA Measurements
Single wandering 3rd instar larvae, single adult female, or two adult male flies were homogenized in lysis buffer (140 mM NaCl, 50 mM Tris-HCl, pH 7.4, 0.1% Triton-X, 1X protease inhibitor cocktail (Roche Diagnostics) as described previously [18]. Triglyceride and protein concentrations were determined using the Stanbio Liquicolor (Fisher Scientific) and BCA Protein Assay (ThermoScientific) kits, respectively, according to manufacturer’s protocol. Total fat body DNA content was determined using the Quant-iT High Sensitivity DNA assay kit (Invitrogen) according to manufacturer’s instructions after homogenizing 3 female fat bodies in lysis buffer.
2.3 Gene Expression Analysis
Larvae 40–43 hr after egg deposition (AED) were fed either yeast paste or 20% sucrose on filter paper for 4 hr and then flash frozen with liquid nitrogen. Total RNA isolation, reverse transcription, and quantitative PCR were performed as previously described [19]. Primer sequences used for QPCR were: dFAS (sense 5′ CTGGCTGAGCAAGATTGTGTG 3′ and antisense 5′ TCGCACAACCAGAGCGTAGTA 3′), dACC (sense 5′ AGATGCAGAACGATGTCCGC 3′ and antisense 5′ CTCTTTGTGAAGCAGCTCCG 3′), dATPCL (sense 5′ CACGACAGATTGGTCCAAGCTC 3′ and antisense 5′ CTTGCTCTTCACGTCGGCTAAC 3′) and rp49 (sense 5′ GACGCTTCAAGGGACAGTATCTG 3′ and antisense 5′ AAACGCGGTTCTGCATGAG 3′).
2.4 Feeding Assay
Food consumption over a 24 hour period was measured by using a modified version of the Capillary Feeder (CAFE) Assay as described previously [20,21]. Briefly, three adult flies were placed in a vial with 1% agar as the only water source and 5% sucrose in a 5-μl glass micropipette (Fisher Scientific) as the sole food source. After 24 hours had elapsed, the amount of liquid consumed by the flies was measured and was corrected for any evaporation that occurred during the experiment.
2.5 Sleep Measurements
Flies were tested for starvation-induced sleep suppression as previously described [22]. Briefly, 3–5 day old female flies were anesthetized and loaded into glass tubes containing standard fly food. Following 16–24 hr of acclimation, baseline activity was recorded using the Drosophila Activity Monitor (DAM; Trikinetics, Waltham, MA) for 24 hr starting at lights on. Flies were then transferred to tubes containing 1% agar to measure sleep during starvation. Sleep was defined as 5 min of immobility as previously reported and analyzed using the Drosophila Sleep Counting Macro [23]. Change in sleep during starvation was calculated as ((% experimental-% baseline)/(% baseline))/100 as previously described [22].
3. Results
3.1 Mio is essential for high sugar-induced expression of lipogenic enzymes
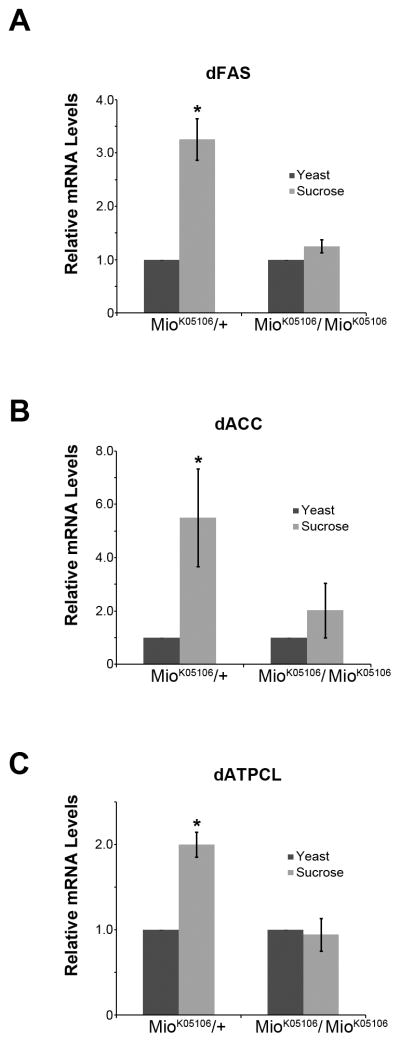
When fed high sugar diets, animals increase triglyceride production by increasing the activity of certain lipogenic enzymes such as acetyl-CoA carboxylase (ACC), fatty acid synthase (FAS), or ATP citrate lyase (ATPCL) [1]. In addition, under chronic high sugar feeding, the expression of these same lipogenic enzymes is induced and ChREBP has been implicated in this response [1,5]. Feeding 2nd instar Drosophila larvae a high sugar diet of 20% sucrose for only 4 hr leads to an increase in the mRNAs of some lipogenic genes. The mechanisms controlling this induction, however, are not fully understood [13]. To test whether Mio contributes to regulating this increase in lipogenic gene expression in response to high sugar, we used a P-element insertion into the Mio gene previously shown to be a strong hypomorphic, loss of function Mio allele [11]. We fed these Mio mutant larvae a high sugar diet (20% sucrose) for 4 hr and then performed quantitative PCR for the Drosophila homologs of the lipogenic enzymes ACC, FAS and ATPCL. As expected, in control animals fed high sugar, dACC, dFAS, and dATPCL mRNA levels were increased compared to control animals fed yeast paste (Fig. 1; [13]). Interestingly, this increase in the expression of key lipogenic enzymes was blunted in Mio mutant larvae fed high sugar (Fig. 1). These data suggest that under high sugar conditions, Mio is necessary for the proper expression of lipogenic enzymes.
Figure 1. Mio is necessary for proper expression of lipogenic enzymes under high sugar conditions.
Heterozygous (Miok05106/+) and homozygous (Miok05106/Miok05106) Mio mutant larvae 40–43 hr AED were fed yeast paste (control) or 20% sucrose (high sugar) for 4 hr. Larvae were collected, total RNA isolation was performed followed by quantitative RT-PCR for the following genes: (A) Fatty acid synthase (dFAS), (B) Acetyl-CoA carboxylase (dACC), and (C) ATP citrate lyase (dATPCL) genes. mRNA levels of flies fed yeast were set to 1.0 and mRNA levels of flies fed 20% sucrose were then normalized to their appropriate yeast control for each genotype. Each experiment was performed at least three times and the values represent means ± SEM. *, P <0.05 by unpaired Student’s t test comparing 20% sucrose to the yeast control within each genotype.
3.2 Mio expression promotes triglyceride storage in Drosophila larvae and adults
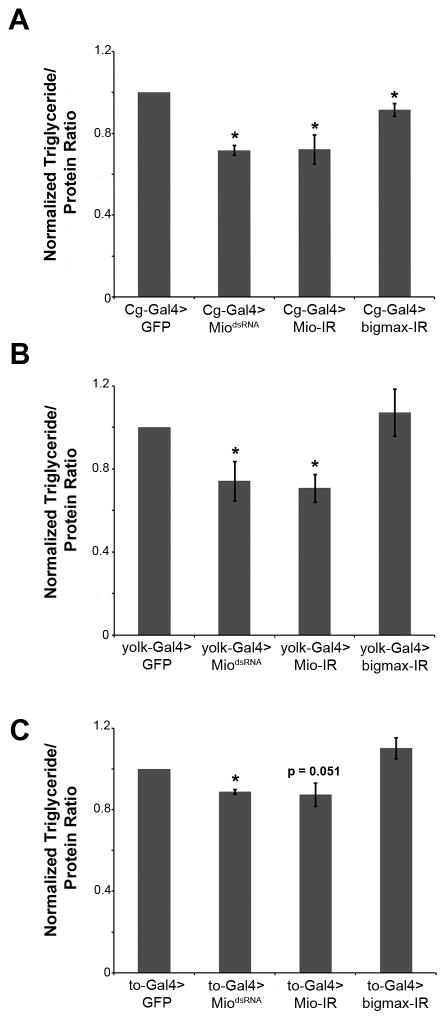
Since high sugar-induced lipogenic gene expression was altered in Mio mutants, we hypothesized that triglyceride storage would also be affected in these animals. However, we were unable to measure triglycerides in the Mio mutants as they die during the pupal stage of development (data not shown; [11]). Therefore, in order to test whether Mio regulates triglyceride storage, we performed experiments in which RNA interference (RNAi) was used to decrease Mio expression specifically in the fat body (the fly liver and adipose tissue equivalent). We expressed two different transgenes to inhibit Mio function, MiodsRNA and Mio-IR, as well as a transgene to decrease bigmax, in both wandering third instar larvae and adults using the Gal4-upstream activating sequence (UAS) system. Cg-Gal4, yolk-Gal4, and to-Gal4 drivers were used as they express highly in the larval, adult female, and adult male fat bodies, respectively. Decreasing Mio and bigmax levels in the fat body of wandering third instar larvae resulted in lowered triglyceride levels compared to controls (Fig. 2A). A similar phenotype was observed in adults with fat body-specific Mio knockdown; however, decreasing bigmax in the adult fat body had no effect on triglyceride accumulation (Fig. 2B and 2C). These results implicate Mio and bigmax in regulating lipid storage in the fat body of Drosophila.
Figure 2. Mio expression in the fat body is necessary for normal triglyceride storage in Drosophila larvae and adults.
(A) Normalized triglyceride/protein ratios in Cg-Gal4>MiodsRNA, Cg-Gal4>Mio-IR, and Cg-Gal4>bigmax-IR wandering instar larvae compared to Cg-Gal4>GFP controls. (B) Normalized triglyceride/protein ratios in yolk-Gal4>MiodsRNA, yolk-Gal4>Mio-IR, and yolk-Gal4>bigmax-IR 5–8 day old females compared to yolk-Gal4>GFP controls. (C) Normalized triglyceride/protein ratios in to-Gal4>MiodsRNA, to-Gal4>Mio-IR, and to-Gal4>bigmax-IR 5–8 day old males compared to to-Gal4>GFP controls. Each experiment was performed at least three times and the values represent means ± SEM. *, P <0.05 by unpaired Student’s t test comparing each experimental genotype to its appropriate GFP control.
3.3 Mio controls triglyceride levels by regulating the amount of fat per cell in the adult fat body
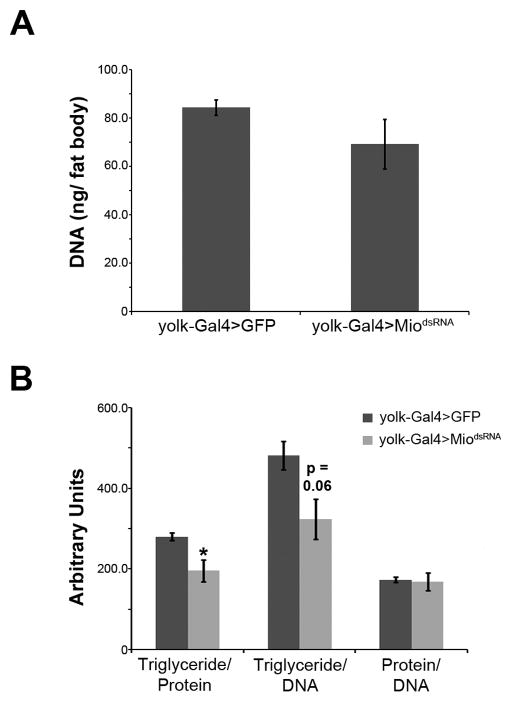
Modulation of total triglyceride levels in an animal’s adipose tissue requires alterations in the number of fat cells, the amount of fat per cell, or a combination of the two [24]. To determine whether Mio regulates fat body triglycerides by affecting fat cell number or the amount of fat per cell, we dissected fat bodies from animals with decreased Mio levels and measured total DNA content, a measurement previously shown to correlate directly with cell number [18]. While fat body-specific knockdown of Mio has no significant effect on DNA levels (Fig. 3A), Mio knockdown reduces triglyceride/protein and triglyceride/DNA ratios (Fig. 3B), suggesting that Mio regulates fat levels by controlling the amount of fat per cell rather than changing fat cell number.
Figure 3. Mio regulates the amount of fat stored in each cell of the adult fat body.
(A) Total DNA content of fat bodies dissected from 4- to 7-day-old adult yolk-Gal4>MiodsRNA females or yolk-Gal4>GFP controls. (B) Triglyceride/protein, triglyceride/DNA, and protein/DNA ratios of fat bodies dissected from yolk-Gal4>MiodsRNA females or yolk-Gal4>GFP controls. Each experiment was performed at least three times and the values represent means ± SEM. *, P <0.05 by unpaired Student’s t test.
3.4 Mio functions in the fat body to promote feeding
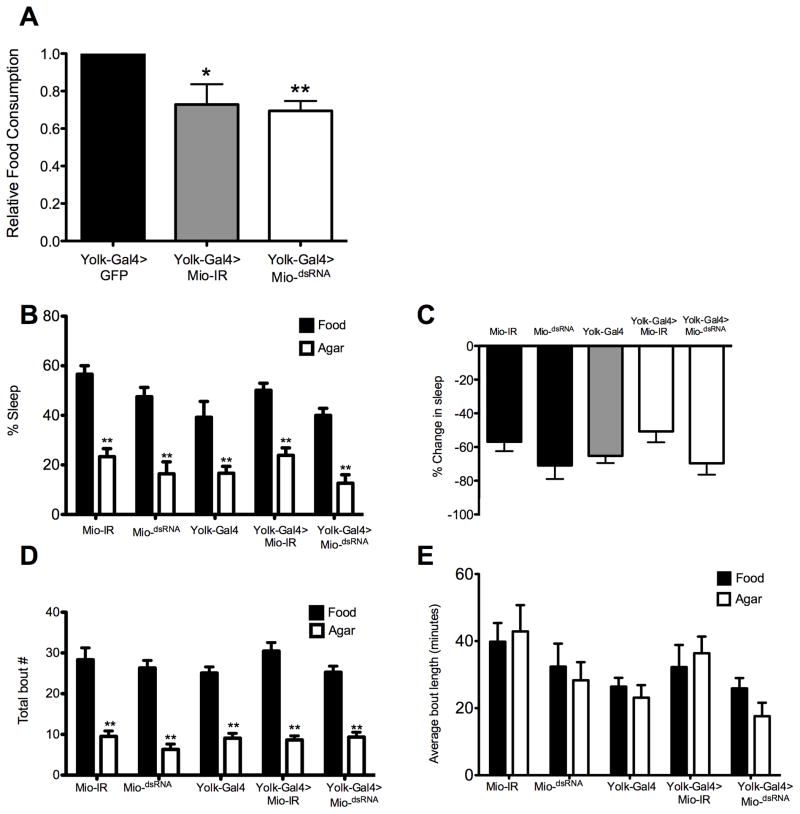
Nutrient stores and molecules that signal energy availability potently modulate hunger-dependent behaviors including feeding and sleep [25]. To determine whether Mio functions in the fat body to modulate feeding, we measured food consumption over a 24 hr period in flies using the CAFE assay [21]. Fat body-specific knockdown of Mio using yolk-Gal4 resulted in reduced food consumption, suggesting that Mio acts in the fat body to promote feeding (Fig. 4A). This result led us to question whether Mio generally promotes hunger-dependent behaviors or specifically controls feeding. One such hunger-dependent behavior is starvation-induced sleep suppression [26]. Flies with mutations in metabolic genes that result in altered triglyceride levels have marked changes in overall sleep homeostasis, and flies that are starved sleep less in order to forage for food [22,27]. To determine whether Mio selectively controls feeding behavior or generally regulates hunger-dependent behaviors, we tested whether knockdown of Mio in the fat body alters starvation-induced sleep suppression. To do this, sleep was measured during 24 hr on food and then 24 hr on starvation medium as previously described [22]. Flies with Mio knockdown in the fat body suppressed sleep similarly to control flies, suggesting that Mio is not required for metabolic regulation of sleep (Fig. 4B, C). Starvation-induced sleep suppression results from decreased sleep bout number rather than shortened sleep bout length [25]. We, therefore, examined the architecture of sleep in Mio knockdown flies and found that they suppress sleep by decreasing sleep bout number similarly to controls, fortifying the notion that Mio does not modulate starvation-induced sleep suppression (Fig. 4D, E). Taken together, these findings suggest that Mio regulates feeding behavior but not metabolic regulation of sleep, indicating that Mio specifically controls food consumption and does not act as a general regulator of hunger-driven behaviors.
Figure 4. Mio acts in the fat body to promote feeding.
(A) Total food consumption was measured in the CAFE assay over 24 hrs in yolk-Gal4>GFP, yolk-Gal4>MiodsRNA, and yolk-Gal4>Mio-IR 5–8 day old females. Mio knockdown in the fat body significantly reduced feeding compared to GFP controls. (B) Flies were tested for sleep 24 hr on food and 24 hr on agar. Flies expressing Mio-IR and MiodsRNA using yolk-Gal4 significantly reduced sleep during starvation similarly to yolk-Gal4, UAS-Mio-IR and UAS-MiodsRNA controls. (C) Analysis of starvation-induced sleep suppression revealed no significant differences between Mio knockdown flies (yolk-Gal4>Mio-IR and yolk-Gal4>MiodsRNA) and yolk-Gal4, UAS-Mio-IR and UAS-MiodsRNA controls. (D, E) yolk-Gal4, UAS-Mio-IR, UAS-MiodsRNA, yolk-Gal4>Mio-IR, and yolk-Gal4>MiodsRNA all displayed significantly reduced bout number (D), and no changes in average bout length (E), indicating that Mio does not alter sleep architecture in fed or starved flies. Each experiment was performed at least two times and the values represent means ± SEM. *, P <0.05, ** P <0.01 by unpaired Student’s t test.
4. Discussion
In this study, we have shown that decreasing Mio levels specifically in the fat body led to lower triglycerides in larvae and adult flies. This suggests that Mio plays a role in lipid accumulation in these animals and supports the hypothesis that Mio acts to regulate fat storage in Drosophila. These findings are in agreement with data from a study where ChREBP knockout mice were found to have lower triglyceride levels in their adipose tissue compared to wild type control mice [5]. ChREBP acts with the myc-family transcription factor Mlx to activate transcription [4]. We have shown that decreasing the expression of the Drosophila homolog of Mlx, bigmax, in the larval fat body results in decreased triglycerides (Fig. 2A), suggesting that Mio and bigmax may act together to control fat metabolism. Further biochemical analysis of these two proteins is necessary to determine whether they bind to each other in order to activate the transcription of target genes.
Previous studies have shown that a high sugar diet leads to changes in expression of multiple genes involved in fat synthesis [13]. The factors involved in up-regulating the transcription of these genes are, however, unknown. Mio is a likely regulator of the transcription of some of these lipogenic genes as its mammalian homolog is regulated in response to high sugar [3]. Data presented here showing that high sugar-induced lipogenic gene expression is blunted in Mio mutants (Fig. 1) provides support for this hypothesis. However, the targets described here are probably only a small subset of Mio-regulated genes, and further experimentation is necessary in order to identify the full complement of genes that are regulated by Mio.
The Drosophila fat body has been implicated in the regulation of both feeding and sleep [28]. Here, we show that decreased levels of Mio in the fat body led to decreased food consumption, but not altered sleep. Mio knockdown flies were tested for sleep during fed and starved states and did not differ from wildtype under either condition. These findings raise the possibility that Mio selectively acts to regulate feeding behavior. Testing Mio-deficient flies in additional hunger-dependent assays such as appetitive memory and sucrose-yeast food choice would address this question [29,30].
The decreased feeding in Mio knockdown flies could be responsible for the decreased fat per cell and overall lower triglyceride levels observed in these Mio mutants. One question from this study that remains unanswered is how Mio functions in the fat body to control feeding. The findings that Mio expression in the fat body is necessary for normal feeding suggests that Mio acts in the fat body as a sensor capable of detecting the status of the body’s energy reserves and conveying that information to the brain to control feeding patterns accordingly. An inherent ability of the fat body to release peptides into the hemolymph of the fly could explain this proposed communication between these organs. It is possible that Mio may activate the transcription of a factor secreted from the fat body that acts as the messenger to the brain. In order to identify whether such a Mio-responsive factor exists, the full complement of Mio target genes needs to be identified. It is also possible that the fat body may be communicating with the brain through a direct neuronal connection. In mammals, white adipose tissue is directly innervated by the sympathetic nervous system [31]. This connection is thought to be a major stimulus for initiating the mobilization of lipid stores. A similar connection between the fat body and the brain may be present in Drosophila, but evidence supporting this claim is lacking.
In summary, the data presented in this study shows that Mio, the Drosophila homolog of mammalian ChREBP, functions in the fat body to promote both lipid storage as well as feeding behavior. These data provide support for Mio acting as a nutrient sensor in the Drosophila fat body to coordinate metabolism and behavior in response to changes in nutrient abundance. This study also describes a genetic system for identifying and understanding the genes and mechanisms involved in controlling feeding and metabolism under high sugar conditions.
Highlights.
Mio is the Drosophila homolog of ChREBP.
Mio is necessary for high sugar-induced lipogenic gene expression.
Mio controls triglyceride storage in the fly fat body.
Mio acts in the fat body to regulate food consumption.
This study implicates Mio in tissue-specific regulation of metabolism and feeding.
Acknowledgments
We would like to thank the Bloomington Stock Center and the Vienna Drosophila RNAi Center for fly stocks used in this study. This study was supported by NIH grants R21DK089391 to MJB and P20RR016464 to ACK and internal funds from Hofstra University and University of Nevada at Reno to JRD and ACK, respectively.
Footnotes
The abbreviations used are: ChREBP, carbohydrate response element binding protein; FAS, fatty acid synthase; ACC, acetyl-CoA carboxylase; ATPCL, ATP citrate lyase; Mlx, Max-like protein X; Mio, Mlx interactor; AED, after egg deposition; DAM, Drosophila Activity Monitor; RNAi, RNA interference; UAS, upstream activating sequence.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Towle HC, Kaytor EN, Shih HM. Regulation of the expression of lipogenic enzyme genes by carbohydrate. Annual review of nutrition. 1997;17:405–433. doi: 10.1146/annurev.nutr.17.1.405. [DOI] [PubMed] [Google Scholar]
- 2.Benhamed F, Denechaud PD, Lemoine M, Robichon C, Moldes M, Bertrand-Michel J, Ratziu V, Serfaty L, Housset C, Capeau J, Girard J, Guillou H, Postic C. The lipogenic transcription factor ChREBP dissociates hepatic steatosis from insulin resistance in mice and humans. The Journal of clinical investigation. 2012;122:2176–2194. doi: 10.1172/JCI41636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Postic C, Dentin R, Denechaud PD, Girard J. ChREBP, a transcriptional regulator of glucose and lipid metabolism. Annual review of nutrition. 2007;27:179–192. doi: 10.1146/annurev.nutr.27.061406.093618. [DOI] [PubMed] [Google Scholar]
- 4.Stoeckman AK, Ma L, Towle HC. Mlx is the functional heteromeric partner of the carbohydrate response element-binding protein in glucose regulation of lipogenic enzyme genes. The Journal of biological chemistry. 2004;279:15662–15669. doi: 10.1074/jbc.M311301200. [DOI] [PubMed] [Google Scholar]
- 5.Iizuka K, Bruick RK, Liang G, Horton JD, Uyeda K. Deficiency of carbohydrate response element-binding protein (ChREBP) reduces lipogenesis as well as glycolysis. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:7281–7286. doi: 10.1073/pnas.0401516101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Billin AN, Eilers AL, Queva C, Ayer DE. Mlx, a novel Max-like BHLHZip protein that interacts with the Max network of transcription factors. The Journal of biological chemistry. 1999;274:36344–36350. doi: 10.1074/jbc.274.51.36344. [DOI] [PubMed] [Google Scholar]
- 7.Baker KD, Thummel CS. Diabetic larvae and obese flies-emerging studies of metabolism in Drosophila. Cell metabolism. 2007;6:257–266. doi: 10.1016/j.cmet.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leopold P, Perrimon N. Drosophila and the genetics of the internal milieu. Nature. 2007;450:186–188. doi: 10.1038/nature06286. [DOI] [PubMed] [Google Scholar]
- 9.Matthews KA, Kaufman TC, Gelbart WM. Research resources for Drosophila: the expanding universe. Nature reviews Genetics. 2005;6:179–193. doi: 10.1038/nrg1554. [DOI] [PubMed] [Google Scholar]
- 10.Venken KJ, Bellen HJ. Emerging technologies for gene manipulation in Drosophila melanogaster. Nature reviews Genetics. 2005;6:167–178. doi: 10.1038/nrg1553. [DOI] [PubMed] [Google Scholar]
- 11.Billin AN, Ayer DE. The Mlx network: evidence for a parallel Max-like transcriptional network that regulates energy metabolism. Current topics in microbiology and immunology. 2006;302:255–278. doi: 10.1007/3-540-32952-8_10. [DOI] [PubMed] [Google Scholar]
- 12.Peyrefitte S, Kahn D, Haenlin M. New members of the Drosophila Myc transcription factor subfamily revealed by a genome-wide examination for basic helix-loop-helix genes. Mechanisms of development. 2001;104:99–104. doi: 10.1016/s0925-4773(01)00360-4. [DOI] [PubMed] [Google Scholar]
- 13.Zinke I, Schutz CS, Katzenberger JD, Bauer M, Pankratz MJ. Nutrient control of gene expression in Drosophila: microarray analysis of starvation and sugar-dependent response. The EMBO journal. 2002;21:6162–6173. doi: 10.1093/emboj/cdf600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dauwalder B, Tsujimoto S, Moss J, Mattox W. The Drosophila takeout gene is regulated by the somatic sex-determination pathway and affects male courtship behavior. Genes & development. 2002;16:2879–2892. doi: 10.1101/gad.1010302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Georgel P, Naitza S, Kappler C, Ferrandon D, Zachary D, Swimmer C, Kopczynski C, Duyk G, Reichhart JM, Hoffmann JA. Drosophila immune deficiency (IMD) is a death domain protein that activates antibacterial defense and can promote apoptosis. Developmental cell. 2001;1:503–514. doi: 10.1016/s1534-5807(01)00059-4. [DOI] [PubMed] [Google Scholar]
- 16.Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, Couto A, Marra V, Keleman K, Dickson BJ. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 17.Giordano E, Rendina R, Peluso I, Furia M. RNAi triggered by symmetrically transcribed transgenes in Drosophila melanogaster. Genetics. 2002;160:637–648. doi: 10.1093/genetics/160.2.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DiAngelo JR, Birnbaum MJ. Regulation of fat cell mass by insulin in Drosophila melanogaster. Molecular and cellular biology. 2009;29:6341–6352. doi: 10.1128/MCB.00675-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DiAngelo JR, Bland ML, Bambina S, Cherry S, Birnbaum MJ. The immune response attenuates growth and nutrient storage in Drosophila by reducing insulin signaling. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:20853–20858. doi: 10.1073/pnas.0906749106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DiAngelo JR, Erion R, Crocker A, Sehgal A. The central clock neurons regulate lipid storage in Drosophila. PloS one. 2011;6:e19921. doi: 10.1371/journal.pone.0019921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ja WW, Carvalho GB, Mak EM, de la Rosa NN, Fang AY, Liong JC, Brummel T, Benzer S. Prandiology of Drosophila and the CAFE assay. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:8253–8256. doi: 10.1073/pnas.0702726104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keene AC, Duboue ER, McDonald DM, Dus M, Suh GS, Waddell S, Blau J. Clock and cycle limit starvation-induced sleep loss in Drosophila. Current biology: CB. 2010;20:1209–1215. doi: 10.1016/j.cub.2010.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pfeiffenberger C, Lear BC, Keegan KP, Allada R. Processing sleep data created with the Drosophila Activity Monitoring (DAM) System. Cold Spring Harbor protocols. 2010;2010 doi: 10.1101/pdb.prot5520. pdb prot5520. [DOI] [PubMed] [Google Scholar]
- 24.Cristancho AG, Lazar MA. Forming functional fat: a growing understanding of adipocyte differentiation. Nature reviews Molecular cell biology. 2011;12:722–734. doi: 10.1038/nrm3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McDonald DM, Keene AC. The sleep-feeding conflict: Understanding behavioral integration through genetic analysis in Drosophila. Aging. 2010;2:519–522. doi: 10.18632/aging.100181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horne J. REM sleep, energy balance and ‘optimal foraging’. Neuroscience and biobehavioral reviews. 2009;33:466–474. doi: 10.1016/j.neubiorev.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 27.Thimgan MS, Suzuki Y, Seugnet L, Gottschalk L, Shaw PJ. The perilipin homologue, lipid storage droplet 2, regulates sleep homeostasis and prevents learning impairments following sleep loss. PLoS biology. 2010;8 doi: 10.1371/journal.pbio.1000466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu K, Zheng X, Sehgal A. Regulation of feeding and metabolism by neuronal and peripheral clocks in Drosophila. Cell metabolism. 2008;8:289–300. doi: 10.1016/j.cmet.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krashes MJ, Waddell S. Drosophila memory: will Orb(2) predict the future? Current biology: CB. 2008;18:R74–76. doi: 10.1016/j.cub.2007.11.053. [DOI] [PubMed] [Google Scholar]
- 30.Ribeiro C, Dickson BJ. Sex peptide receptor and neuronal TOR/S6K signaling modulate nutrient balancing in Drosophila. Current biology: CB. 2010;20:1000–1005. doi: 10.1016/j.cub.2010.03.061. [DOI] [PubMed] [Google Scholar]
- 31.Bartness TJ, Bamshad M. Innervation of mammalian white adipose tissue: implications for the regulation of total body fat. The American journal of physiology. 1998;275:R1399–1411. doi: 10.1152/ajpregu.1998.275.5.R1399. [DOI] [PubMed] [Google Scholar]