Abstract
Germline mutations in PMS2 are associated with Lynch syndrome (LS), the most common known cause of hereditary colorectal cancer. Mutation detection in PMS2 has been difficult due to the presence of several pseudogenes, but a custom-designed long-range PCR strategy now allows adequate mutation detection. Many mutations are unique. However some mutations are observed repeatedly, across individuals not known to be related, due to the mutation being either recurrent, arising multiple times de novo at hot spots for mutations, or of founder origin, having occurred once in an ancestor. Previously, we observed 36 distinct mutations in a sample of 61 independently ascertained Caucasian probands of mixed European background with PMS2 mutations. Eleven of these mutations were detected in more than one individual not known to be related and of these, six were detected more than twice. These six mutations accounted for 31 (51%) ostensibly unrelated probands. Here we performed genotyping and haplotype analysis in four mutations observed in multiple probands and found two (c.137G>T and exon 10 deletion) to be founder mutations, one (c.903G>T) a probable founder, and one (c.1A>G) where founder mutation status could not be evaluated. We discuss possible explanations for the frequent occurrence of founder mutations in PMS2.
Keywords: colon cancer, founder mutation, genetic predisposition, PMS2
INTRODUCTION
Mutations in at least four mismatch repair genes cause Lynch syndrome (LS), a condition that predisposes to colorectal and endometrial cancer and to a lesser degree to a number of other cancers (1). Even though the proportion of all colorectal cancer caused by LS is a modest ~3% (but greater if diagnosis is at young age) there is a need for improved strategies to diagnose LS because clinical surveillance and prophylactic surgery can greatly reduce cancer morbidity and mortality (2–4). Different strategies have been devised to detect as many LS mutation carriers as possible as cost-effectively as possible and in many institutions, it is standard practice to perform immunohistochemistry (IHC) for the mismatch repair proteins as a first step in screening for Lynch syndrome (5–8).
While standard mutation detection methods apply well to MLH1, MSH2, and MSH6, testing for PMS2 gene mutations has been problematic due to the presence of numerous pseudogenes. The use of carefully-designed long-range PCR to avoid amplifying the pseudogenes has virtually solved the problem (9–12) so that presently, all four mismatch repair genes can be readily studied for mutations. Deletions in these mismatch repair genes are relatively common; therefore multiplex ligation-dependent probe amplification is also commonly used. Among the four mismatch repair genes, mutations in two (MLH1 and MSH2) cause high lifetime risks (penetrance) and together account for some 60–80% of all LS. Mutations in the other two genes (MSH6 and PMS2) have lower penetrance and each accounts for some 10–20% of all LS (13–18).
This communication deals with mutations in the PMS2 gene that were observed multiple times. Mutations that are observed in ostensibly unrelated individuals can be either recurrent (repeated spontaneous de novo occurrence; also known as “hot spot” mutations) or of founder type (inherited from a shared ancestor).
Material and Methods
Patients
This study is an extension of a previous study in which 99 probands with colon and/or endometrial cancer who demonstrated isolated absence of tumor staining for PMS2 by IHC were analyzed for PMS2 mutations (16). In total, 61 of the 99 probands (61%) had deleterious mutations (55 monoallelic; 6 biallelic). Of these, 36 were distinct mutations; 25 occurred in just one proband each, 5 occurred in 2 ostensibly unrelated probands and one (c.736_741del6ins11), a previously described ancient founder mutation (10), occurred in 12 ostensibly unrelated probands. The present paper describes four of the remaining five mutations which respectively occurred in seven, three, three and three ostensibly unrelated probands each (16). For this analysis, we have included an additional seven, previously unreported probands with these four PMS2 mutations (total 21 probands with four different mutations). For the fifth mutation, namely the complete gene deletion that occurred in 3 probands in Senter et al. (16) a DNA sample from only one patient was available; therefore this mutation was not studied.
Samples from 21 subjects were studied. All research was conducted under approval of the Institutional Review Board (IRB) at The Ohio State University. Fourteen of these subjects were described previously (16) and five of these previously described subjects were accrued anonymously through research collaborations with the Australian Registry of the National Cancer Institute-funded Colon Cancer Family Registry (19). Anonymized samples from an additional 7 subjects were provided from the ARUP Laboratories (Salt Lake City, UT) (11). All subjects studied here had LS-associated tumors displaying absence of PMS2 protein with retention of MLH1, MSH2 and MSH6 protein. To determine the population frequency of the mutation-associated haplotypes we genotyped 80 control individuals. These samples were randomly drawn from Caucasians belonging to a collection of samples obtained from residents of central Ohio for the purpose of serving as controls for genetic studies.
Exon 10 deletion mutant breakpoint analysis
To determine breakpoints for patients with exon 10 deletions, patient DNA was first amplified by long-range PCR using TaKaRa LA Taq and primers specific for PMS2, spanning exon 8 to exon 11. Long-range amplicons were diluted 1:10 and used as template for nested PCR using primers spanning the breakpoint region. Amplicons were then sequenced using BigDye Terminator chemistry on the Applied Biosystems 3730 and compared to NC_000007.13, complement positions 6012870..6048737.
Genotyping
To characterize the haplotypes present in cases and controls we utilized 5 out of 6 microsatellite markers and 7 out of 9 SNPs previously reported (10) that span the PMS2 locus. The available subjects and 80 controls were typed for these PMS2 markers.
In order to prevent the amplification of pseudogenes, DNA samples were amplified using a previously described long-range PCR procedure (9, 11). Amplicons spanning exons 1–5 (long-range amplicon LR1) and 7–9 (LR2) were generated using the previously published primers. For the region encompassing exons 11–15, rather than generating two long-range products spanning exons 10–12 and 12–15, we used the forward primer located in exon 10 and the reverse primer located 3’ of exon 15. This generates an 18,341 bp product (LR3) (11). With this design modification, all of the long-range products have at least one primer anchored in an exon not present in any of the pseudogenes.
Using each of the long-range primer sets, 100 ng of DNA were amplified in 25 µl reactions containing 0.2 µM each primer, 1.25 U TaKaRa LA Taq (TaKaRa Bio Inc., Otsu, Shiga, Japan), 1 × LA PCR Buffer II, and 400 µM each dNTP. Cycling consisted of an initial denaturation at 94°C for 1 minute and 30 cycles of 10 seconds at 98°C and 10 or 18 minutes (for LR2 or LR3 respectively) at 68°C. Final elongation entailed 10 minutes at 72°C. The amplification result of long-range PCR was confirmed by gel electrophoresis and diluted (1 in 20) prior to marker-specific amplification.
Microsatellite markers were typed either by direct labeling of a PCR primer or by utilizing a labeled M13 primer in conjunction with an M13-tailed, amplicon-specific primer in a three primer PCR. Each 15 µl PCR reaction contained 7.5 µl AmpliTaq Gold master mix (PE Applied Biosystems, Foster City, CA), 100 ng genomic DNA, 10 pmol untailed primer, 5 pmol M13-tailed primer, and 10 pmol FAM-labeled M13 primer. Reactions were cycled using the following profile: 96°C for 10 min, 36 cycles of 96°C for 30 sec, 58°C for 30 sec, 72°C for 30 sec, and final extension at 72°C for 5 min. The PCR product was sized using an ABI 3730 DNA Analyzer.
For the genotyping of SNPs we used the same PCR conditions as above in the presence of 10 pmol forward and reverse primer with the appropriate long-range PCR product or genomic DNA used as template. The PCR product containing the SNP was subjected to the SNaPshot reaction (PE Applied Biosystems, Foster City, CA). The sequences of the primers used for microsatellite genotyping have been published (10) while sequences for the primers used in the SNaPshot reaction can be obtained upon request.
Haplotype construction
Genotyping data were used to construct haplotypes using the PHASE 2.0 program (20) according to the manual. Haplotypes associated with the mutation are shown.
RESULTS
Genotyping of control samples
We genotyped the controls and used the data to calculate the frequency of the alleles (see Fig. 1). The controls did not have any of the 4 mutations we discuss in this study. Using PHASE we constructed haplotypes in the controls. The haplotypes associated with each mutation were searched for in the controls and the number reported.
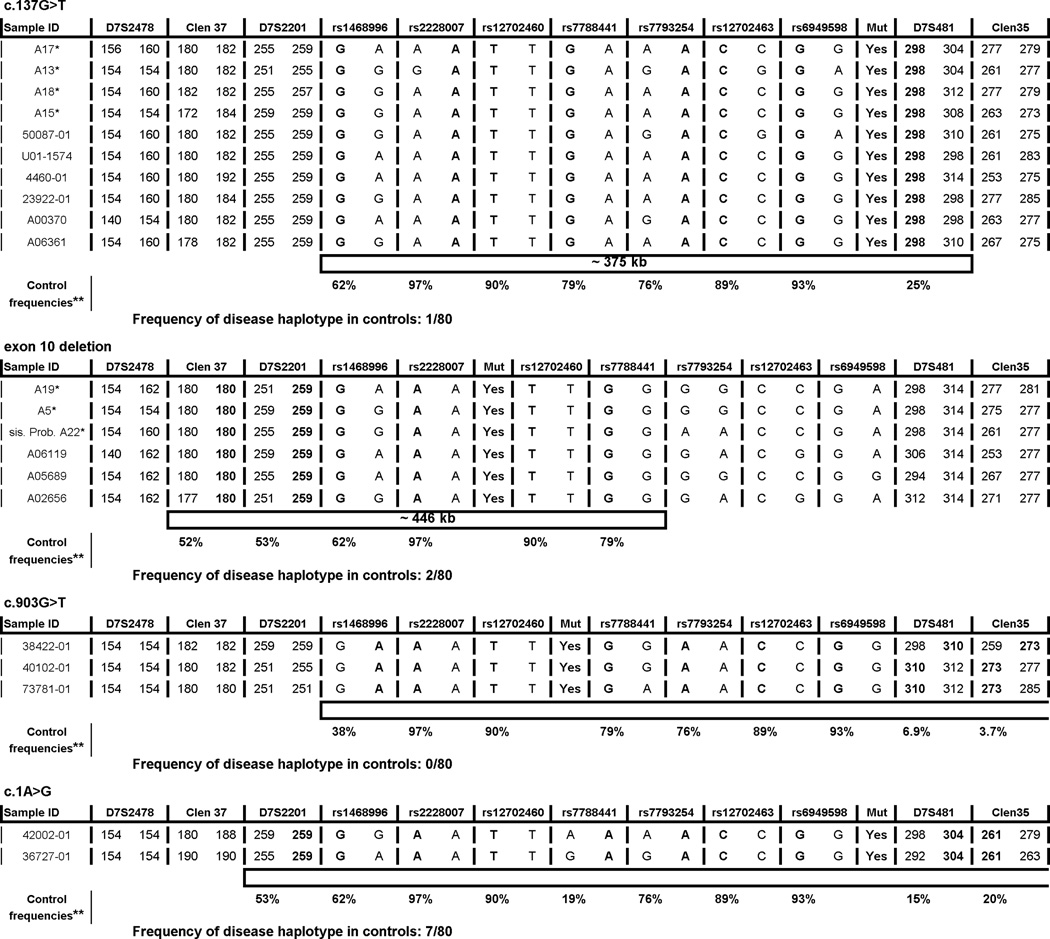
Fig. 1.
Genotype data spanning the PMS2 locus in probands carrying mutations c.137G>T, exon 10 deletion, c.903G>T or c.1A>G. The alleles associated with the mutation are bolded and the shared haplotypes are represented by the empty bars.
*the samples from the ARUP Laboratories; **frequencies in controls of the bolded alleles.
c.137G>T
We had access to samples from a total of 10 mutation-positive probands with c.137G>T, 6 from the original series (16) and an additional 4 from the ARUP collection. The results (Fig. 1) show that a disease-associated haplotype comprising some 375kb was shared by all subjects. The haplotype stretches from microsatellite D7S481 upstream of exon 1 to SNP rs1468996 downstream of exon 15. This 375kb shared haplotype was observed in one of 80 control individuals from the central Ohio area. All subjects were Caucasian and while ancestral information was not available for all probands, 3 probands reported ancestry in the United Kingdom and another reported Australian ancestry.
Exon 10 deletion
The exon 10 deletion was found in three unrelated probands from Australia (16). For this study we had access to an additional 2 probands and a mutation-carrying sister of the third proband, all from the ARUP collection. Breakpoints were confirmed (c.989-296_144+706del) and were identical to breakpoints previously reported in individuals with exon 10 deletions (21, 22). A shared haplotype extending from rs7788441 in intron 7 to microsatellite marker Clen37 downstream of exon 15 was observed in six probands (Fig. 1). The same haplotype occurred in 2 of 80 controls. These data are consistent with a relatively short shared ancestral haplotype. All subjects were Caucasian with unknown ancestral origin.
c.903G>T
The c.903G>T mutation leads to the skipping of exon 8 (16). We studied all 3 probands and detected a shared haplotype spanning from microsatellite marker Clen35 upstream of exon 1 to SNP rs1468996 some 280kb downstream of PMS2. This haplotype was seen in 0 of the 80 controls. All subjects were Caucasian and two of three probands reported ancestry from Austria, Hungary, and Germany.
c.1A>G
Of the 3 probands originally detected carrying this mutation, 2 were available for study. A shared haplotype was seen from microsatellite marker Clen35 upstream of exon 1 to D7S2201 some 390kb downstream of PMS2. This haplotype was seen in 7/80 controls. Ancestral information is known for only one of the two subjects, being mixed Irish, French, and Native American.
DISCUSSION
The existence of numerous pseudogenes has made it more difficult to search for mutations in PMS2 than in the other three MMR genes. As a consequence, data on the proportion of all Lynch syndrome that is caused by PMS2 mutations are scarce. Moreover, the documented low penetrance of PMS2 mutations relative to the penetrance of the MMR genes MLH1 and MSH2 (see below) means that PMS2 mutations will be underdiagnosed in the clinical setting where mutation analyses typically are applied to individuals displaying the “high risk” features of strong family history of early onset Lynch syndrome cancers. For these reasons the proportion of LS caused by mutations in PMS2 can best be estimated by studies in which unselected cases of Lynch syndrome-associated cancers are screened. The data summarized in a large review (7) show MLH1 and MSH2 involvement in 32% and 39% respectively while PMS2 and MSH6 were reported to be present in 15% and 14% respectively of all Lynch syndrome cases. An additional population-based study of 500 CRC cases (23) disclosed 18 LS probands with a similar distribution of mutations. Thus, as an overall conclusion MSH2 and MLH1 account for ~70% of diagnosed LS while MSH6 and PMS2 together account for the remaining ~30% (7). From a practical point of view these numbers suggest that to adequately assess the presence of LS all 4 genes must be considered.
This study focuses on those mutations in PMS2 that occurred repeatedly in a series of 99 probands whose tumors did not stain for PMS2 protein by IHC (16). The subjects emanated from numerous institutions mainly in Northern Europe, North America and Australia, being mostly Caucasians of European origin. It is important to bear in mind that we cannot therefore make inference about other ethnicities or nationalities. Moreover, because the initial series of 99 ostensibly unrelated probands contained at least 24 population-based probands while at least 19 probands were from high risk clinics (exact numbers are not available), there may be a bias in favor of higher rather than lower penetrance mutations if such exist. Nevertheless, with these limitations our series of subjects is by far the largest of its kind and therefore allows at least some tentative conclusions of population relevance.
We show that repeated mutations in PMS2 are common and whenever feasible to assess, are likely to be of founder nature. Among the 61 probands, 31 carried a mutation seen in at least three probands and one mutation was observed in seven probands. In addition, as shown in Table 1 several of these mutations have been seen and published in patients who were not part of the initial series described in Senter et al. (16). Thus it appears that approximately half of all PMS2 mutations occur repeatedly in the Caucasian population.
Table 1.
PMS2 mutations found in two or more of 61 probands studied by Senter et al. (16) and number of probands with the same mutations reported in the literature.
| Mutation | # Probands | ||
|---|---|---|---|
| Senter et al. (16) | Literature | Total | |
| c.736_741del6ins11 (P246CfsX3)* | 12 | 11,37 | 16 |
| c.137G>T (S46I) | 7 | 11,37−40 | 15 |
| Deletion exon 10 (c.989– 296_1144+706del) | 3 | 11,21,22 | 7 |
| c.903G>T (skips exon 8) | 3 | - | 3 |
| c.1A>G (5' truncation) | 3 | - | 3 |
| Complete gene deletion | 3 | 22,37 | 5 |
| c.1840A>T (K614X) | 2 | - | 2 |
| c.1831_1832insA (1611NfsX2) | 2 | 11 | 3 |
| c.2113G>A (E705K) | 2 | - | 2 |
| c.949C>T (Q317X)** | 2 | - | 2 |
| Deletion of exons 5, 6, 7 | 2 | - | 2 |
(Clendenning et al. (10))
This mutation occurred monoallelic in one proband and homozygous biallelic in one proband. In the latter case the parents were first cousins and each was heterozygous for the mutation.
The most common mutation described in Senter et al. (16), c.736_741del6ins11, has been studied in detail previously (10). The second most common mutation described in Senter et al. (16), c.137G>T (Ser46Ile) has the characteristics of a deleterious missense change (24,25). Our data allow us to conclude that this mutation is inherited from a single shared ancestor. The haplotype is short suggesting that the mutation occurred many generations ago, but with the limited number of affected individuals available for study we are not able to estimate the age of the mutation with any degree of precision. Additionally the exon 10 deletion (the second mutation examined in our study) very likely is of a founder type, due to the presence of shared breakpoints and a shared haplotype and the same shared haplotype occurred only in a very small number (2/80) of controls.
The third mutation investigated in this study, namely c.903G>T, that leads to the skipping of exon 8 (16) showed somewhat longer shared haplotype. For this shared haplotype we were unable to determine the upstream start. Since this haplotype was never detected in controls and although we have studied only 3 probands we conclude that this is a probable founder mutation.
The last mutation analyzed (c.1A>G) disrupts the first translation initiation codon leading to 5’ truncation of the putative protein. Only 2 samples were available from subjects with this mutation and they shared an even longer haplotype which was present in 7/80 controls. In this case too we were unable to determine the upstream start of the shared haplotype. Thus, while shared ancestry is a distinct possibility, this mutation could also be the one that recurs frequently de novo. We note here that de novo mutations are rare in the mismatch repair genes (26).
Founder mutations are not unique to PMS2. Founder mutations are well known in MLH1 (27–31), and MSH2 (32–34). At least one recurrent “hot spot” mutation is widespread worldwide. This is the intronic MSH2 c.942+3A>T splice site mutation that apparently arises frequently de novo as a result of meiotic misalignment at a stretch of 26 adenines in the 5’ region of intron 5 (35). Are founder or recurrent mutations less common in MLH1, MSH2 and MSH6 than in PMS2? The large multicenter study on MSH6 by Baglietto et al. (18) identified 74 distinct mutations in a total of 113 probands. Among the 74 mutations, 22 were observed in more than one proband (range = 2 to 6 probands). In total, 29/113 families displayed mutations seen more than twice, as compared to the 31/61 noted by us for PMS2. Thus it is possible that the two MMR genes with the lowest penetrance (PMS2 and MSH6) also share the property of having frequent recurrent or founder mutations, but they may be more abundant in PMS2 than MSH6. It is documented that the penetrance of cancer is lower in PMS2 (lifetime risk of CRC ~20%) than in MLH1 and MSH2 (lifetime risk of CRC ~ 40–60%) (Senter et al. (16) and references cited within). Unfortunately, data establishing the proportion of repeated mutations in MLH1 and MSH2 are not readily available. We are not aware of publications in which the occurrence of mutations has been determined in large numbers of probands from panmixing (as opposed to isolated; geographically distinct) populations. Nevertheless, population-based studies reviewed in Palomaki et al. (7) list the MMR mutations found in altogether only 82 probands with Lynch syndrome. These data are too few to conclude anything with certainty about the proportion of founder mutations in the two most prevalent MMR genes compared to PMS2 and MSH6. We suggest that further, much larger population-based studies are desirable to shed light on this question.
Founder mutations are believed to become enriched by at least 2 alternative mechanisms. First, if a rare mutation is introduced into an isolated population that subsequently expands without significant influx of genes, it can become enriched simply by genetic drift. (More often however, it can decrease or become extinct from genetic drift.) This mechanism is believed to account for those numerous examples of frequent founder mutations seen in Icelanders, Finns, Ashkenazi Jews, French Canadians, and other typical isolated founder populations. This mechanism does not readily apply to our findings in PMS2 which are derived from large panmixing Caucasian populations. Another well known cause by which a particular mutation can become enriched occurs when its effect carries an advantage (positive selection). This mechanism is well known e.g. from the hemoglobin gene where heterozygosity for the most common sickle cell anemia mutation confers protection against malaria (36). We are unaware of any evidence about positive selection of mutations in PMS2.
In summary, founder mutations appear to be common in PMS2. As more PMS2 mutations are identified through population-based screening of colon and/or endometrial cancers using IHC followed by appropriate germline genetic testing, more PMS2 mutation carriers are likely to be identified and could provide much more detailed estimates of the prevalence of these mutations. It is possible that if certain mutations are identified in a significant number of patients, standard methodology of PMS2 mutation detection could be altered by testing for common mutations before sequencing the entire gene.
Acknowledgements
This work was supported by the National Cancer Institute, National Institutes of Health under RFA # CA-95-011 and through cooperative agreements with members of the Colon Cancer Family Registry (Colon CFR) and P.I.s. The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the CFRs, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government or the CFR.” Samples and corresponding data were provided by “Australasian Colorectal Cancer Family Registry (U01 CA097735).
We thank Shuying Sun, Jennifer Panescu, James Mackay, Joy Larsen Haidle, Marc Greenblatt, Daniel Buchanan, Graham Giles, Graham Suthers, Graham Young and Jack Goldblatt for help and Paul Fuerst for helpful discussion.
Grant sponsor: National cancer institute; grant number: CA16058 and CA67941.
Footnotes
Conflict of interest: Authors declare no conflict of interest.
REFERENCES
- 1.Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med. 2003;348:919–932. doi: 10.1056/NEJMra012242. [DOI] [PubMed] [Google Scholar]
- 2.Järvinen HJ, Aarnio M, Mustonen H, et al. Controlled 15-year trial on screening for colorectal cancer in hereditary nonpolyposis colorectal cancer families. Gastroenterology. 2000;118:829–834. doi: 10.1016/s0016-5085(00)70168-5. [DOI] [PubMed] [Google Scholar]
- 3.Schmeler KM, Lynch HT, Chen LM, et al. Prophylactic surgery to reduce the risk of gynecologic cancers in the Lynch syndrome. N Engl J Med. 2006;354:261–269. doi: 10.1056/NEJMoa052627. [DOI] [PubMed] [Google Scholar]
- 4.Järvinen HJ, Renkonen-Sinisalo L, Aktán-Collán K, Peltomäki P, Aaltonen LA, Mecklin JP. Ten years after mutation testing for Lynch syndrome: cancer incidence and outcome in mutation-positive and mutation-negative family members. J Clin Oncol. 2009;27:4793–4797. doi: 10.1200/JCO.2009.23.7784. [DOI] [PubMed] [Google Scholar]
- 5.Vasen HF, Watson P, Mecklin JP, Lynch HT. New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC. Gastroenterology. 1999;116:1453–1456. doi: 10.1016/s0016-5085(99)70510-x. [DOI] [PubMed] [Google Scholar]
- 6.Umar A, Boland CR, Terdiman JP, et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palomaki GE, McClain MR, Melillo S, Hampel HL, Thibodeau SN. EGAPP supplementary evidence review: DNA testing strategies aimed at reducing morbidity and mortality from Lynch syndrome. Genet Med. 2009;11:42–65. doi: 10.1097/GIM.0b013e31818fa2db. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boland CR, Shike M. Report from the Jerusalem workshop on Lynch syndrome-hereditary nonpolyposis colorectal cancer. Gastroenterology. 2010;138:2197, e1–e7. doi: 10.1053/j.gastro.2010.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clendenning M, Hampel H, LaJeunesse J, et al. Long-range PCR facilitates the identification of PMS2-specific mutations. Hum Mutat. 2006;27:490–495. doi: 10.1002/humu.20318. [DOI] [PubMed] [Google Scholar]
- 10.Clendenning M, Senter L, Hampel H, et al. A frame-shift mutation of PMS2 is a widespread cause of Lynch syndrome. J Med Genet. 2008;45:340–345. doi: 10.1136/jmg.2007.056150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vaughn CP, Robles J, Swensen JJ, et al. Clinical analysis of PMS2: mutation detection and avoidance of pseudogenes. Hum Mutat. 2010;31:588–593. doi: 10.1002/humu.21230. [DOI] [PubMed] [Google Scholar]
- 12.Vaughn CP, Hart KJ, Samowitz WS, Swensen JJ. Avoidance of pseudogene interference in the detection of 3' deletions in PMS2. Hum Mutat. 2011;32:1063–1071. doi: 10.1002/humu.21540. [DOI] [PubMed] [Google Scholar]
- 13.Hampel H, Frankel WL, Martin E, et al. Screening for the Lynch Syndrome (Hereditary Nonpolyposis Colorectal Cancer) New Engl J Med. 2005;352:1851–1860. doi: 10.1056/NEJMoa043146. [DOI] [PubMed] [Google Scholar]
- 14.Hampel H, Stephens JA, Pukkala E, et al. Cancer risk in Hereditary Nonpolyposis Colorectal Cancer syndrome: later age of onset. Gastroenterology. 2005;129:415–421. doi: 10.1016/j.gastro.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 15.Truninger K, Menigatti M, Luz J, et al. Immunohistochemical analysis reveals high frequency of PMS2 defects in colorectal cancer. Gastroenterology. 2005;128:1160–1171. doi: 10.1053/j.gastro.2005.01.056. [DOI] [PubMed] [Google Scholar]
- 16.Senter L, Clendenning M, Sotamaa K, et al. The clinical phenotype of Lynch syndrome due to germ-line PMS2 mutations. Gastroenterology. 2008;135:419–428. doi: 10.1053/j.gastro.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall G, Clarkson A, Shi A, et al. Immunohistochemistry for PMS2 and MSH6 alone can replace a four antibody panel for mismatch repair deficiency screening in colorectal adenocarcinoma. Pathology. 2010;42:409–413. doi: 10.3109/00313025.2010.493871. [DOI] [PubMed] [Google Scholar]
- 18.Baglietto L, Lindor NM, Dowty JG, et al. Risks of Lynch syndrome cancers for MSH6 mutation carriers. J Natl Cancer Inst. 2010;102:193–201. doi: 10.1093/jnci/djp473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newcomb PA, Baron J, Cotterchio M, et al. Colon Cancer Family Registry. Colon Cancer Family Registry: an international resource for studies of the genetic epidemiology of colon cancer. Cancer Epidemiol Biomarkers Prev. 2007;16:2331–2343. doi: 10.1158/1055-9965.EPI-07-0648. [DOI] [PubMed] [Google Scholar]
- 20.Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van der Klift H, Wijnen J, Wagner A, et al. Molecular characterization of the spectrum of genomic deletions in the mismatch repair genes MSH2, MLH1, MSH6, and PMS2 responsible for hereditary nonpolyposis colorectal cancer (HNPCC) Genes Chromosomes Cancer. 2005;44:123–138. doi: 10.1002/gcc.20219. [DOI] [PubMed] [Google Scholar]
- 22.Overbeek LI, Kets CM, Hebeda KM, et al. Patients with an unexplained microsatellite instable tumour have a low risk of familial cancer. Br J Cancer. 2007;96:1605–1612. doi: 10.1038/sj.bjc.6603754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hampel H, Frankel WL, Martin E, et al. Feasibility of screening for Lynch syndrome among colorectal cancer patients. J Clin Oncol. 2008;26:5783–5788. doi: 10.1200/JCO.2008.17.5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakagawa H, Lockman JC, Frankel WL, et al. Mismatch repair gene PMS2: disease-causing germline mutations are frequent in patients whose tumors stain negative for PMS2 protein, but paralogous genes obscure mutation detection and interpretation. Cancer Res. 2004;64:4721–4727. doi: 10.1158/0008-5472.CAN-03-2879. [DOI] [PubMed] [Google Scholar]
- 25.Jackson CC, Holter S, Pollett A, et al. Café-au-lait macules and pediatric malignancy caused by biallelic mutations in the DNA mismatch repair (MMR) gene PMS2. Pediatr Blood Cancer. 2008;50:1268–1270. doi: 10.1002/pbc.21514. [DOI] [PubMed] [Google Scholar]
- 26.Win AK, Jenkins MA, Buchanan DD, et al. Determining the frequency of de novo germline mutations in DNA mismatch repair genes. J Med Genet. 2011;48:530–534. doi: 10.1136/jmedgenet-2011-100082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nyström-Lahti M, Kristo P, Nicolaides NC, et al. Founding mutations and Alu-mediated recombination in hereditary colon cancer. Nat Med. 1995;1:1203–1206. doi: 10.1038/nm1195-1203. [DOI] [PubMed] [Google Scholar]
- 28.Moisio AL, Sistonen P, Weissenbach J, de la Chapelle A, Peltomäki P. Age and origin of two common MLH1 mutations predisposing to hereditary colon cancer. Am J Hum Genet. 1996;59:1243–1251. [PMC free article] [PubMed] [Google Scholar]
- 29.Caluseriu O, Di Gregorio C, Lucci-Cordisco E, et al. A founder MLH1 mutation in families from the districts of Modena and Reggio-Emilia in northern Italy with hereditary non-polyposis colorectal cancer associated with protein elongation and instability. J Med Genet. 2004;41:e34. doi: 10.1136/jmg.2003.013714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borràs E, Pineda M, Blanco I, et al. MLH1 founder mutations with moderate penetrance in Spanish Lynch syndrome families. Cancer Res. 2010;70:7379–7391. doi: 10.1158/0008-5472.CAN-10-0570. [DOI] [PubMed] [Google Scholar]
- 31.Tomsic J, Liyanarachchi S, Hampel H, et al. An American founder mutation in MLH1. Int J Cancer. 2012;130:2088–2095. doi: 10.1002/ijc.26233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wagner A, Barrows A, Wijnen JT, et al. Molecular analysis of hereditary nonpolyposis colorectal cancer in the United States: high mutation detection rate among clinically selected families and charactrization of an American founder genomic deletion of the MSH2 gene. Am J Hum Genet. 2003;72:1088–1100. doi: 10.1086/373963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lynch HT, Coronel SM, Okimoto R, et al. A founder mutation of the MSH2 gene and hereditary nonpolyposis colorectal cancer in the United States. JAMA. 2004;291:718–724. doi: 10.1001/jama.291.6.718. [DOI] [PubMed] [Google Scholar]
- 34.Clendenning M, Baze MD, Sun S, et al. Origins and prevalence of the American founder mutation of MSH2. Cancer Res. 2008;68:2145–2153. doi: 10.1158/0008-5472.CAN-07-6599. [DOI] [PubMed] [Google Scholar]
- 35.Desai DC, Lockman JC, Chadwick RB, et al. Recurrent germline mutation in MSH2 arises frequently de novo. J Med Genet. 2000;37:646–652. doi: 10.1136/jmg.37.9.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ashley-Koch A, Yang Q, Olney RS. Sickle hemoglobin (HbS) allele and sickle cell disease: a HuGE review. Am J Epidemiol. 2000;151:839–845. doi: 10.1093/oxfordjournals.aje.a010288. [DOI] [PubMed] [Google Scholar]
- 37.Herkert JC, Niessen RC, Olderode-Berends MJ, et al. Pediatric intestinal cancer and polyposis due to bi-allelic PMS2 mutations: case series, review and follow-up guidelines. Eur J Cancer. 2011;47:965–982. doi: 10.1016/j.ejca.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 38.Leenen C, Geurts-Giele W, Dubbink H, et al. Pitfalls in molecular analysis for mismatch repair deficiency in a family with biallelic pms2 germline mutations. Clin Genet. 2011;80:558–565. doi: 10.1111/j.1399-0004.2010.01608.x. [DOI] [PubMed] [Google Scholar]
- 39.Giunti L, Cetica V, Ricci U, et al. Type A microsatellite instabililty in pediatric gliomas as an indicator of Turcot syndrome. Eur J Hum Genet. 2009;17:919–927. doi: 10.1038/ejhg.2008.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Auclair J, Leroux D, Desseigne F, et al. Novel biallelic mutations in MSH6 and PMS2 genes: gene conversion as a likely cause of PMS2 gene inactivation. Hum Mutat. 2007;28:1084–1090. doi: 10.1002/humu.20569. [DOI] [PubMed] [Google Scholar]