Abstract
Quantitative proteomics analysis of cortical samples of ten Alzheimer’s disease (AD) brains versus ten normally aged brains was performed by following the accurate mass and time tag (AMT) approach with the high resolution LTQ Orbitrap mass spectrometer. More than 1400 proteins were identified and quantitated. A conservative approach of selecting only the consensus results of four normalization methods was suggested and used. A total of 197 proteins were shown to be significantly differentially abundant (p-values<0.05, corrected for multiplicity of testing) in AD versus control brain samples. Thirty seven of these proteins were reported as differentially abundant or modified in AD in the previous proteomics and transcriptomics publications. The rest to the best of our knowledge are new. Mapping of the discovered proteins with bioinformatic tools revealed significant enrichment with differentially abundant proteins of pathways and processes known to be important in AD, including signal transduction, regulation of protein phosphorylation, immune response, cytoskeleton organization, lipid metabolism, energy production, and cell death.
Keywords: Alzheimer’s disease, brain, cortical samples, proteomics, bioinformatics, normalization
Introduction
Alzheimer's disease (AD) is the most common form of dementia in the elderly affecting 10% of population aged over 65 years 1. Twin studies suggest that about 74% of the risk for late-onset AD is genetic 2. The remaining 26% is the result of the interplay between genetic variants and environmental factors 3. Proteomics directly addresses the level of gene products present in a given cell state and reflects the influence of both genetic and environmental factors and is therefore an important complement of genomics and transcriptomics in the study of the mechanism of late-onset AD.
Several reviews 4–11 summarize the results of the proteomics and transcriptomics studies of AD, where important findings of the differentially expressed and modified proteins in AD brain tissue, CSF, and serum have been reported. Review 4 summarizes proteomics of AD results from 1975 till 2006 and lists 96 proteins altered in AD, while the more recent clinical proteomics of AD review 6 summarizes the results of 43 2D-gel electrophoresis proteomics studies published since 1999 to 2010 and lists 93 proteins differentially expressed or modified in 13 different brain regions in mild cognitive impairment, early AD and AD. The 2011 review 9 of the systems biology of AD presents the list of 36 “overlapping” proteins affected in both human brain samples and in animal models of AD. Thirty of these proteins belong to 5 functional groups: cytoskeleton organization, energy production and metabolism, redox homeostasis, chaperons, and synaptic integrity.
Multiple groups looked at the biomarkers of AD in CSF, plasma, and serum, as reviewed in 8,10,11. The biomarker panels of 23, 17, and 15 proteins in CSF were suggested in 12–14 to distinguish AD patients from healthy elderly control subjects. Similarly, 5- and 18-protein biosignatures of AD were suggested for blood samples 15–16. Several groups focused on the studies of post-translational modifications in AD and studied Alzheimer’s brain phosphoproteome 17–19, oxidatively modified proteome 20–21, and brain proteins glycosylation 22.
The recently published whole transcriptome sequencing study 23 of the human brain samples of 2 AD patients versus 26 normal brain samples was performed by using the RNA-Seq next generation sequencing approach and demonstrated dramatic (up to plus 26-fold and minus 350-fold) over- and under-expression of multiple transcripts. Over 27 thousand transcripts were found in AD brain that were not observed in the normal, while 51 thousand found in normal were not present in AD. Both concordance and contradictions with the results of the previous microarray studies 24–25 of AD were reported, including the contradictions for the PP3GB, GRIA4, and GRIK1 genes.
There are several reasons for the relatively low overlap between the differentially expressed proteins and transcripts in AD discovered in different studies. First is the heterogeneity of AD reported by many authors 26–27 arguing that there exist several AD subtypes with both genetic and epigenetic factors contributing to the disease phenotype. With the relatively low number of AD brain samples analyzed both in proteomics and transcriptomics studies, this heterogeneity could lead to some contradicting results. Second relates to the comparison of proteomics and transcriptomics results which are typically relatively low correlated as reported by multiple studies 28–30 and is indicative of the numerous posttranslational modifications and regulations. Last but not least is the dramatic improvement both in transcriptomics and proteomics technology in the recent years which resulted in higher sensitivity and accuracy of quantitative analysis. Modern proteomics, based on the high resolution, high mass accuracy mass spectrometers, has “turned quantitative” and is ready for genome-wide comprehensive protein expression analysis, as recently summarized by the pioneers of the field 31–33. Studies 34–36 are some recent examples of successful use of modern LC-MS/MS based proteomics in psychiatry and neuroscience.
Here, we present the results of the quantitative proteomics study of temporal cortical samples of 10 brains affected by AD compared to 10 normally aged brains. Samples from the temporal lobe were selected for analysis, since this region is affected even at the early stages of AD 37. Analysis was performed using the accurate mass and time tag (AMT) proteomics approach 38–41 that was developed at PNNL about a decade ago and proved successful in multiple applications including the more recent quantitative proteomics studies of mouse brain 42–43. This approach incorporates the high mass measurement accuracy of Fourier transform ion cyclotron resonance (FTICR), Orbitrap, or other high mass accuracy MS instrument with accurate elution time measurements from liquid chromatography (LC) separations for peptide identification. The concept of the AMT tag strategy is based on generating a “look-up” table of tags based on LC-MS/MS peptide identifications in a limited but representative set of samples. This library of accurate masses and times for each peptide tag allows for the direct identification of peptides from LC-MS runs in the larger dataset without having to perform tandem mass spectrometry (MS/MS) on every sample. In mouse brain tissue AMT tag approach is able to identify and quantify upwards of one-thousand proteins 42–43. In this study, we enhanced and adapted the AMT approach to the analysis of multiple human brain samples by focusing on two stages: selection of the best available protein extraction protocol and improvement of the reliability of quantitation by using only the consensus results of four normalization methods.
Protein extraction
A prerequisite for any credible quantitative proteomics study is to get a good coverage of the proteome in the targeted sample. We evaluated three protocols for preparation of the tissue samples for LC-MS analysis. The main difference between the protocols is the type of detergent for solubilization of the tissue protein content:(i) 8M urea with 2% SDS, (ii) 8M urea, (iii) 6M guanidine. Three criteria were applied to assess the protocols, including the number of identified peptides and proteins, the percentage of identified peptides with missed cleavages, and the recovery of several pathology-related proteins. The details of the comparison are provided in the Supplementary Material Methods. Briefly, the 8M urea protocol resulted in the most comprehensive proteome coverage, the most efficient tryptic digestion with fewest missed cleavages identified, and the best recovery on the proteins of particular interests among the Alzheimer’s disease research community (see Suppl. Material Methods Figure S1). Although the addition of detergents, such as SDS, is usually preferred to help solubilize proteins from tissue samples, we found that the addition of SDS was not able to generate an overall satisfying proteomics result. Therefore, the 8M urea protocol was chosen to prepare all the samples in the main quantitative proteomics study presented in this paper.
Normalization
Quantitative proteomics requires normalization of protein abundances that compensates for systematic biases both in the sample preparation and in the sensitivity of LC-MS runs. This is especially important for label-free as opposed to quantitation approaches relying on isotopic labeling because of additional potential of biases incurred during the instrumental analysis. Label-free quantitation was initially proposed in the proteomics analysis of serum samples 44, where it was assumed that most of the proteins in the compared samples had similar abundances and only a relatively small percentage of proteins were differentially abundant. Based on this assumption, the median value of the ratios of abundances of the same proteins in the compared samples were used as normalization coefficients 44,45. However, the validity of this assumption in the analysis of samples that are expected to differ more radically is not evident. The obvious example is the comparison of proteomes of cancer and non-cancer cells where the substantial percentage of proteins can be affected. Alternatively, normalization can be based on the certain group of “housekeeping” proteins whose abundances are assumed not to be affected by cancer. However, as recently summarized 46, there is a growing body of evidence demonstrating that commonly used housekeeping proteins may not be adequate internal standards as they may be affected by a large number of factors including drug and experimental treatment, cell cycle phase, age, gender and stress conditions. Spiking of a standard reference material (exogenous proteins) may be useful as an additional control, but does not solve all the problems of the variability before spike-in and could create additional problems by ion suppression of endogenous proteins. Therefore, normalization in analysis of samples that are expected to differ substantially (as in AD) is an important and not yet solved problem both in quantitative proteomics and in microarray analysis as well 47–48. Below we suggest an approach for minimizing the effect of “normalization uncertainty” on the quantitation by using multiple normalization methods and selecting consensus results.
In this paper, we compared samples from AD and normally aged post-mortem brains that were expected to differ substantially. We identified and quantitated more than 1400 proteins in our analysis; abundances of many of them differ dramatically. We used 4 normalization methods (described in more detail in the Methods section), compared their results and selected for the consensus list only the proteins that were significantly (FDR<0.05, derived from the p-values corrected for multiple hypothesis testing)) differentially abundant across all normalization methods. The consensus list consisted of nearly two hundred significantly differentially abundant proteins including both known to be differentially abundant in AD and new ones.
Materials and Methods
Cortical Brain Samples
Frozen human brain tissues were obtained from the University of Michigan Alzheimer’s Disease Research Center’s brain bank. Our criteria for inclusion were as follows: self-defined ethnicity of European descent, neuropathologically confirmed LOAD or no neuropathology present, male gender and age of death greater than 65. Prior to selection for this study, the genotypes of all samples were additionally analyzed via the program STRUCTURE 49,50 as described in 51,52 and ethnic outliers (non-Caucasian) were removed from the study. Neuropathological diagnosis was defined by board-certified neuropathologists as per standard National Alzheimer’s Coordinating Center protocols. Sample properties (mean±s.d., range in parenthesis): Age (years): cases - 77.8 ± 3.3, (73–83), controls – 76.4±4.8, (69–83); post-mortem delay (hours): cases – 8.2 ± 3.2, (4.5–14), controls – 16.9 ±7.2, (6–28) ; Braak stage (mean and range): cases −5.4 (5–6), controls - 0.3 (0–2). Samples were de-identified before receipt, and the study met local human studies institutional review board and HIPPA regulations. This work is declared not human-subjects research and is IRB exempt under regulation 45 CFR 46.
Sample Preparation for Proteomic Analysis
The extraction and digestion of the proteins was performed using a commonly used protocol based on denaturation of protein in 8M urea followed by digestion with trypsin. The choice of this protein extraction protocol is justified in more detail in the Supplementary Material Methods. Briefly, approximately 5 mg of human brain tissue was resuspended in 100 µl denaturing solution (8M urea, 50 mM Tris-HCl pH 8.0 and 1 mM EDTA) and homogenized with a motorized pestle. The samples were sonicated for ~1 min in a bath sonicator (Branson, Danbury, CT) then the total protein content was measured by Bicinchoninic Protein Assay (BCA, Pierce, Rockford, IL). DTT was added to a concentration of 10 mM in sample, then to solubilize and unfold the proteins the samples were incubated for 60 min at 37°C with shaking. Cysteine residues were alkylated by adding iodoacetamide to 40 mM concentration and incubating for 1 hour at 37°C, with shaking, in the dark. For protein digestion the samples were diluted 10-fold with 50 mM ammonium bicarbonate (pH 7.8) and supplemented with 1 mM CaCl2. Trypsin was added in a 1:50 ratio (w: w trypsin: protein) and incubated for 3 hours at 37°C with shaking. The sample digests were purified with solid phase extraction using C18 columns (Discovery DSC-18, SUPELCO, 52601-U), lyophilized and resuspended in 25 mM ammonium bicarbonate pH 7.8. The peptide amounts were estimated with BCA assay.
LC-MS(/MS) Proteomics Analysis
Proteomics analysis was performed by following the well established accurate mass and time (AMT) tag approach 38–43 and by using the high resolution high mass accuracy mass spectrometer LTQ Orbitrap. The database of AMT tags was created by performing the LC/LC-MS/MS analysis (using SCX as a first dimension of separation) of the pooled 10 AD and pooled 10 control samples separately. The quantitation was performed by the LC-MS analysis of each of 20 individual samples. The details of AMT technology and software used for creation of AMT tag database and peptide identification and quantitation were described in 53–58 and are provided in the Supplementary Materials Methods. As shown in 59 the AMT tag analytical platform enables good level of reproducibility (correlation of peptide abundances of about R=0.94 across 9 technical replicates) and therefore allows avoiding technical replicates.
Protein Quantitation and Bioinformatics Analysis
Normalization
As discussed in the Introduction, normalization is not obvious in analysis of the samples that might have dramatic differences in abundances of numerous proteins. In this study, we used four normalization methods denoted VP, V01, V03 and Eigen MS and described below: (1) method VP is based on the assumption that the majority of proteins are not affected or the number of proteins approximately balanced between the up- or down-regulation; normalization coefficients are calculated as the median ratio of the abundances of peptides detected across all the samples 42; (2) method V01 introduced in 60 is based on the less strict assumption that not necessary the majority, but at least some proteins are not affected; normalization coefficients are calculated similar to above but based only on peptides whose relative abundances differ by not more than ±10% from the mode of relative peptide abundance distributions in all analyzed samples; (3) method V03 is the same as V01 except it considers as non-affected common peptides in the ±30% vicinity of the mode of the distributions; (4) method Eigen MS is based on the recently introduced 61 algorithm, which identifies the bias of arbitrary complexity by singular value decomposition (SVD) and removes it from the data.
Imputation
We imputed missing peptide abundance values by following two imputation methods. According to the first method we filtered out peptides with missing values present in both groups; we imputed the peptide abundance values only if they were missing in more than 7 out of 10 samples in one group and simultaneously present in all samples in the other group. Missing values were imputed with the minimum peptide abundance observed regardless of the group. This method leads to conservative estimate of the fold-change especially in the case of “complete informative missingness” 62, where the protein was not observed in all 10 samples in one group and was observed in all 10 samples in another group. Informative missingness is synonymous with censoring and occurs when a mass spectrometer is not able to detect peptides of abundances below some censoring cutoff. Such missing values are informative because we know that the values are below the lowest observed abundance for a peptide. This imputation method does not accurately predict the amplitude of the effect, but it correctly assigns the rank of the abundance value in the case of “informative missingness”. Thus, such imputation method is compatible with non-parametric rank-based statistical testing described below. This imputation method was used together with the normalization methods VP, V01, and V03. The second, imputation method uses a maximum likelihood model to determine if peptide abundance is missing completely at random or if it is censored as recently described in 62 and was used together with the Eigen MS normalization method. This method typically results in lower missing value peptide abundances and is therefore less conservative. After normalization and imputation the peptide abundance values were “rolled up” to protein abundance values, using reference roll-up algorithm 42,63.
Statistical analysis
Significance analysis was performed with Wilcoxon test, where the values of the normalized relative protein abundances served as an input data and ranking was done across the samples separately for each protein. The reason we choose Wilcoxon rank test over t-test is that we did not want to rely on assumption of the normality of protein abundance distribution in the human population and especially population affected by Alzheimer’s disease. Basically the hypothesis we tested is if there is overall increase or decrease in protein abundance using their ranks, but not necessarily the difference between the means of log-normal protein abundance distribution in the AD and control subjects. Correction for multi-testing, through calculation of false discovery rate (FDR) and q-values was performed following the algorithm introduced by J.D. Storey 64, as implemented in the mafdr.m function (Bioinformatics Toolbox, MATLAB 2008a, Mathworks, Inc., Natick, MA, USA). (Importantly, this FDR in determination of differentially abundant proteins in AD versus control samples should not be confused with the FDR of peptide identification that was discussed earlier.) Finally, to minimize the impact of normalization and imputation methods we selected as consensus significant only the proteins determined as significant according to all 4 normalization methods.
Pathway and Network Analysis
Enrichment analysis was performed with MetaCore 6.4 (GeneGo, Inc, St.Joseph, MI, USA). Thresholds were set at ±1.3-fold change and at 0.05 FDR levels. The total list of proteins detected in any of 20 samples was used as a background list. MetaCore’s enrichment analysis tool utilizes the hypergeometric model to determine statistical significance of enrichment with the differentially expressed genes of several groups of gene categories: GeneGo pathway maps, GO processes, GeneGo process networks, and GeneGo diseases. The above GeneGo categories are based on the GeneGo knowledge base, which reflects findings reported in the literature. GO processes are the categories defined in the Gene Ontology project 65.
Gene set enrichment analysis (GSEA) was performed following the algorithm introduced in 66 as implemented in Pathway Studio 7.1 (Ariadne Genomics, Rockville, MD, USA). Unlike MetaCore’s enrichment analysis, GSEA does not implement any thresholding. Instead, in GSEA, genes are ordered in a rank list according to their differential expression. The goal of GSEA is to check whether the members of a gene set tend to occur at the top or bottom of the list, in which case the gene set is correlated with the phenotypic class distinction. The gene set categories examined in Pathway Studio include: Ariadne Metabolic Pathways, Ariadne Signaling Pathways, Ariadne Ontology, as well as GO cellular component, molecular function, and biological process.
Verification method
Western Blot analysis for verification of differentially abundant proteins
Around 5mg of human normal and AD cortex samples were lysed in ice-cold RIPA buffer (20 mM Tris pH 7.5, 150 mM Nacl, 1% NP40, 0.5% Sodium deoxycholate, 1 mM EDTA and 0.1% SDS) by homogenization. Thereafter, cell lysates were placed on ice for 30 min and cleared by centrifugation at 3,000g for 15 min at 4 °C. The supernatants were collected and frozen at −80 °C until used for analysis. The protein concentrations of lysates were measured using the Bio-Rad protein assay kit.
Thirty µg of total protein from each sample was resolved on 10% sodium dodecyl sulfate-polyacrylamide gels (SDS-PAGE) and transferred onto nitrocellulose membranes (Protran, Schleicher & Schuell, Inc., Keene, NH). Membranes were blocked with 5% nonfat dry milk in Tris-buffered saline (20 mM Tris base (pH 7.5) and 150 mM NaCl) containing 0.05% Tween 20 (TBS-T), then probed with the primary antibody. The primary antibodies were diluted in 5% nonfat dry milk in TBS-T as indicated and used for immunoblotting: Anti-PKCγ (1:1000 dilution; sc-211 from Santa Cruz Biotechnology, Inc., Santa Cruz, CA); anti-NumbL (1:500 dilution; sc-135071 from Santa Cruz Biotechnology, Inc., Santa Cruz, CA) and anti-ENO2 (1:500 dilution; sc-51880 from Santa Cruz Biotechnology, Inc., Santa Cruz, CA). After washing in TBS-T, membranes were incubated with their appropriate horseradish peroxidase-conjugated secondary antibodies (Bio-Rad Laboratories, Inc.) and developed using an enhanced chemiluminescence detection system (Amersham Biosciences, Arlington Heights, IL) according to the instructions of the manufacturer and were exposed to X-Ray film (Phenix Research Products, Hayward, CA).
Densitometric analysis
To quantify the bands obtained via Western blot analysis, we applied ImageJ software based analysis (http://rsb.info.nih.gov/ij/). The area of the specific signal was corrected for the corresponding signal from the area of the loading control. Average normalized optical density (OD) values ± SE were used to plot matching diagrams.
Results
Normalization, quantitation, and significance analysis
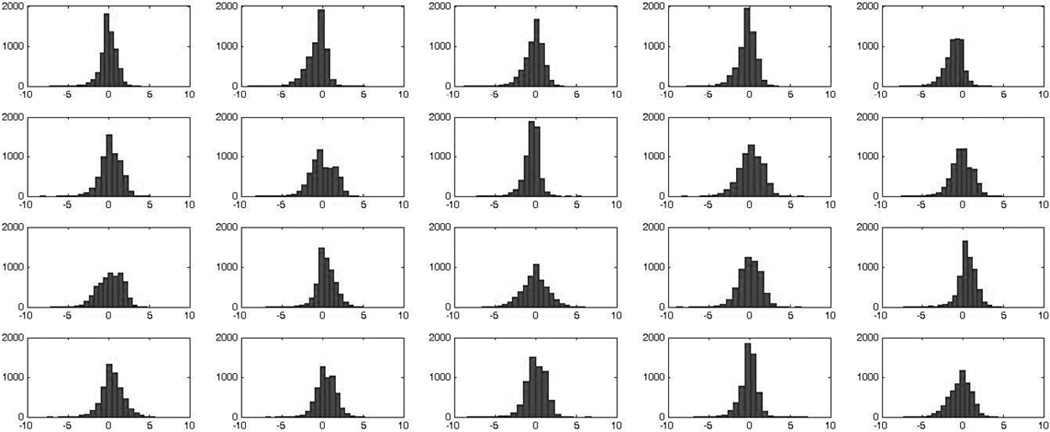
A total of 13,671 peptides attributed to 1408 proteins were detected and quantitated. Figure 1 presents the distributions of relative peptide abundances for the above 20 samples. In this figure, relative abundances of each peptide in each sample were calculated by dividing the individual peptide abundances by the average abundance for each peptide across all 20 samples followed by log2 transform. The range of relative peptide abundances is roughly 15,000, which indicates the presence of dramatic differences in peptide abundances across the samples. Therefore, as discussed in the Introduction, the choice of normalization method is not obvious and might be of importance. As described in the Methods section, we calculated protein abundances by using 4 normalization methods: VP, V01, V03, and Eigen MS. Table 1 presents the values of Pearson correlation of protein abundances calculated after normalization and imputation using the above 4 methods, while Supplementary Figure 1 presents the scatter plots of protein abundances calculated with these methods. The results for the methods VP, V01, V03 are similar (R≥0.993), while all three are noticeably different from Eigen MS (R≥0.908).
Figure 1.
Histograms of distributions of peptide abundances in 10 control (top two rows) and 10 AD (bottom two rows) brain samples. Vertical axis – number of peptides. Horizontal axis : log2(Iik/mIk), where mIk=ΣIik/N – mean abundance of the given peptide across all 20 samples.
Table 1.
Pearson Correlation (R) of Relative Protein Abundances Calculated with Four Normalization Methods
| Procedure | VP | V01 | V03 | Eigen MS |
|---|---|---|---|---|
| VP | 1 | 0.993 | 0.997 | 0.915 |
| V01 | 0.993 | 1 | 0.999 | 0.908 |
| V03 | 0.997 | 0.999 | 1 | 0.912 |
| Eigen MS | 0.915 | 0.908 | 0.912 | 1 |
Importantly, despite the above differences, a large group of 1049 (74% of total 1408) “consensus” proteins exists. For these proteins the sign of the fold change of mean abundance in AD samples versus the mean abundance in control samples does not depend on the normalization method chosen. Supplementary Table 1 presents the list of these consensus proteins with their log-changes and FDR levels calculated for each of the methods as well as mean values and standard deviations across 4 methods. In addition, the number of peptides used for quantitation of the given protein is presented. Note that the number of peptides used for protein quantitation (mean=4.4 peptides per protein; 55% proteins quantitated with two and more peptides) is decreased relative to the number of peptides used for protein identification (mean=6.9 peptides per protein; 72% proteins identified with 2 and more peptides). This is typical for large sample sets (the larger the number of the samples the higher the likelihood that some peptides are not detected in at least one of the samples of the group). Such peptides were discarded as described in the Supplementary Material Methods section. Using single peptide quantitation based on the measured abundance values is in our opinion more reliable than using double peptide quantitation based on imputation.
As many as 197 of these consensus proteins are significantly differentially abundant (statistical analysis described in Methods), in all four normalization/imputation methods. The list of these significant differentially abundant proteins together with their mean log-changes and max FDRs is presented in Supplementary Table 2. In addition, this table presents the comparison of the protein abundance changes with the published data (which is discussed in more detail in the Discussion section) and the information on the number of peptides used for the identification and quantitation of the above 197 proteins. Of these proteins, 188 (95.4%) were identified with two and more peptides, while 65 (33%) were quantitated with two and more peptides. The more stringent requirement for peptides used for quantitation arises from the need to have the peptide measurements across majority of the samples. Thus peptides with substantial portion of missing values were filtered out. Having single peptide for protein quantitation intuitively seems to reduce the precision and reliability of quantitation. However, less reliability should manifest itself in larger variance across the samples of the same group and therefore higher p-values. The proteins with the increased variability and therefore increased p-value have less chance to be called significant. It implies that quantitation with single peptide may have lower statistical sensitivity for detecting truly affected proteins, but likely does not compromise the reliability of the quantitation of proteins that pass the significance test. In our case, the mean of the corrected p-values for 131 proteins quantitated with single peptides is equal to 0.014 (min p=0.005, max p=0.0489). These proteins are flagged in the Supplementary Table 2 as quantitated with single peptides, but are considered reliably quantitated and included in the further discussion.
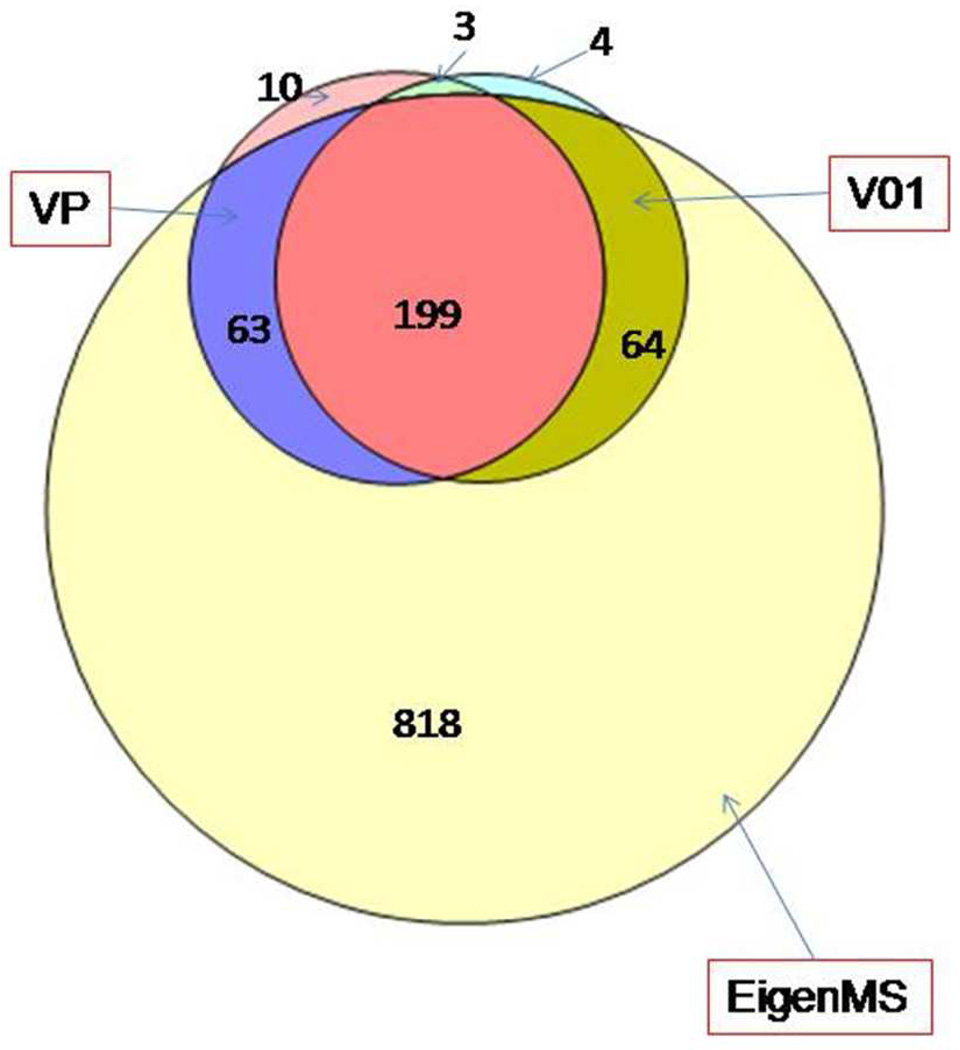
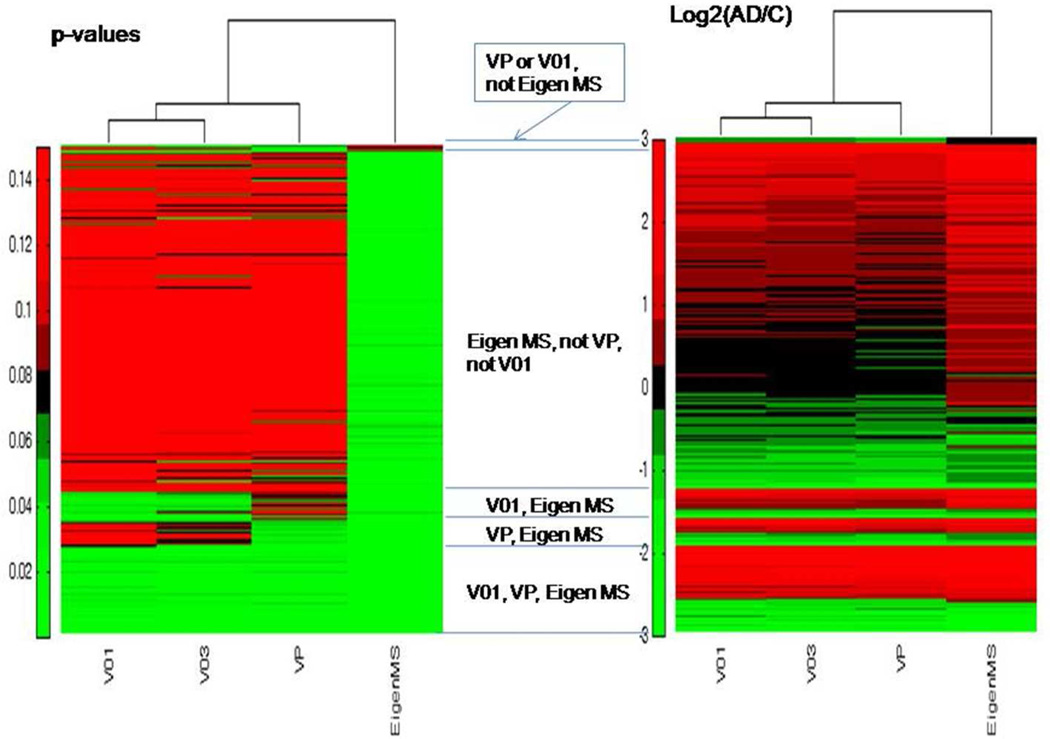
Figure 2 presents the Venn diagram of the proteins determined as significantly differentially abundant by using 3 normalization methods: VP, V01, and Eigen MS. The later one is rather inclusive and results in 1144 significant differentially abundant proteins which include practically all (326) proteins determined as significant either with VP or V01. The overlap between the lists of proteins significant according to VP and V01 is 199 – roughly 3/4 of each list. Results from V03 (not presented at the diagram for clarity) are similar to V01 and slightly reduce the overlap of 4 normalization methods to 197 consensus significantly differentially abundant proteins. Interestingly, though similar and highly correlated (see Table 1), normalization methods VP and V01 generate only 75% overlap of significant differentially abundant proteins. Figure 3 presents heat maps for the more detailed comparison of the corrected p-values and log2(AD/C) calculated with four normalization methods. The proteins are sorted, so that consensus significant ones are at the bottom of the map, followed by the proteins significant with two methods (Eigen MS and either VP or V01), then followed by significant only with Eigen MS, and finally proteins significant in either VP or V01, but not in Eigen MS. (Light green in the first heat map indicates FDR<0.05). The second heat map illustrates that abundance ratios are quite consistent across the normalization methods for consensus significant proteins. (See red and green color bands across the bottom part of the heat map). The level of agreement between the normalization methods for the values of abundance change in the 197 consensus significant differentially abundant proteins is rather high: relative standard deviation of log2(AD/C) values determined for the given protein with 4 normalization methods is less than 10% for 70 and less than 20% for 120 proteins. Note that 41 out of 197 proteins are flagged with “CIM” standing for “complete informative missingness”, meaning that they were observed in all 10 samples in one group and not observed in any of 10 samples of another group.
Figure 2.
Venn diagram comparison of proteins determined as significantly differentially abundant with 3 normalization methods: VP (275 proteins), V01 (270), Eigen MS (1144). Results for V03 (not shown) are similar to V01. See details in the text.
Figure 3.
Comparison of p-values corrected for multiplicity of testing (FDR) and abundance ratios calculated with four normalization methods. First heat map: FDR-values. Light green indicates FDR<0.05. Bottom part of the heat map – consensus significant proteins. Followed by proteins significant with two of normalization methods and then with single normalization method. Second heat map: log2(AD/C), green indicates proteins under-abundant in AD, red – over-abundant. Note that abundance ratios for consensus significant proteins are consistent across the normalization methods (green and red bands in the bottom of the map).
Below, we present the shortlist of the gene names of 10 most increased and 10 most decreased proteins in AD versus control with the log2 (AD/C) values in parenthesis (CIM proteins flagged). Ten most over-abundant proteins in AD samples: NBEAL1 (7.88, CIM), AUH (5.19), PICALM (4.63, CIM), TSN (4.35), PTBP2 (4.34, CIM), PTPN23 (4.15, CIM), MAPRE1 (4.12), C1QBP (4.08), PCYT1A (4.06), IQGAP1 (3.78). Ten most under-abundant proteins in AD samples: PPP1CB (−4.77, CIM), WARS2 (−4.22), FXYD7 (−4.18, CIM), HMGN2 (−3.69), FKBP1A (−3.64), PDE4DIP (−3.56), HSD24 (−3.34, CIM), NSMCE4A (−3.22), MYH15 (−3.07, CIM), CETN2 (−3.06). The given log ratios were calculated as median log ratios across all four normalization methods. Also note that in CIM cases the given log ratios are most likely conservative estimates, since conservative imputation procedures were used as explained in the Methods section.
Western blot verification
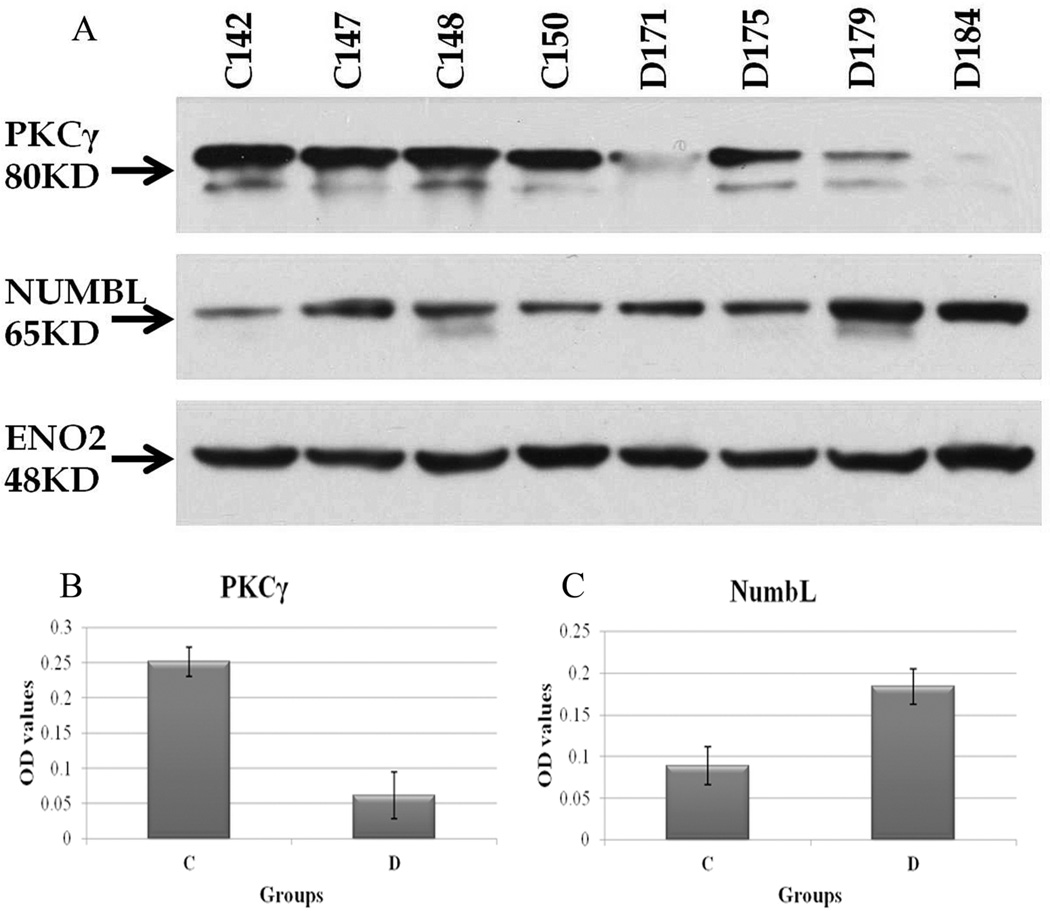
In order to validate the mass spectrometry results, western blot analysis was conducted on randomly selected normal and AD human cortex samples. Four out of ten of the available samples of each group were randomly selected. This insured adequate representation of the variability of protein expression across samples of the same group while preventing exhaustion of all of the precious sample cortices we had. In addition, we also randomly selected to probe for PRKCG, NUMBL and for ENO2 protein expression which were shown to be under-abundant, over-abundant and unchanged respectively in AD cortices versus normal patient cortices using mass spectrometry (see Supplementary Tables 1, 2) Indeed, densitometric analyses of western blot signals showed an average of 4.16 fold decrease and of 2.02 fold increase in PRKCG and NUMBL protein abundance respectively in AD cortices versus controls (Figure 4) which is in good concordance with our mass spectrometry based proteomics results (2.6-fold decrease for PRKCG and 3.2-fold increase for NUMBL). Also, ENO2 protein abundance levels remained practically unchanged in the two groups in concordance with mass spectrometry based quantitation results.
Figure 4.
Validation of expression of selected proteins by western blotting. (A) Western blot gels of PKC-gamma and NUMBL in normal and Alzheimer Disease human cortices. ENO2 was used as an internal control for equal loading. (B) and (C) Densitometric analysis of PKC-gamma and NUMBL protein expression in normal versus Alzheimer Disease human cortices (n = 4 samples per group). Average normalized OD values ± SE were used to plot respective diagrams. Control normal human cortex samples: C142, C147, C148, C150. Alzheimer’s Disease human cortex samples: D171, D175, D179, D184. OD: Optical density.
Pathway and network analysis
To explore the plausible biological relevance of the observed 197 significantly differentially abundant proteins, we performed enrichment analysis with MetaCore 6.4 (GeneGo, Inc) as described in Methods section. Supplementary Table 3 is an Enrichment Analysis Report presenting the lists of top 10 categories: GeneGo pathway maps, GO processes, GeneGo process networks, and GeneGo diseases enriched with differentially abundant proteins. Below, these categories are referred as increased (enriched amongst 124 over-abundant proteins) and decreased (enriched amongst 73 under-abundant proteins). Among the most significant are (p-values shown in parenthesis): (i) increased GeneGo pathways: signal transduction - cAMP signaling (0.016), regulation of lipid metabolism - stimulation of arachidonic acid production by ACM receptors (0.016), DNA damage - NHEJ mechanisms of DSBs repair (0.030); (ii) decreased GeneGo pathways: signal transduction - activation of PKC via G-Protein coupled receptor (0.0024), ubiquinone metabolism (0.015), immune response - NFAT in immune response (0.015); (iii) increased GO processes: negative regulation of phosphate metabolic process (0.0001), negative regulation of protein amino acid dephosphorylation (0.0007), response to corticosterone stimulus (0.002); (iv) decreased GO processes: branched chain family amino acid metabolic process (0.0024), positive regulation of interleukin-2 biosynthetic process (0.0027), positive regulation of cytokine biosynthetic process (0.008); (v) increased GeneGo process networks: DNA damage – core (0.005), DNA damage – checkpoint (0.006), apoptosis - apoptotic nucleus (0.019); (vi) decreased GeneGo process networks: transcription - mRNA processing (0.039), immune response -T helper cell differentiation (0.042). Among the enriched GeneGo diseases are: chromosome aberrations (4e-10), amyloidosis (5e-10), amyloid neuropathies (6e-9), memory disorders (2e-6).
To further explore the biological relevance of protein abundance changes observed in AD versus control samples we used the GSEA approach described in the Methods section. The 4 lists of relative protein abundances calculated with above 4 normalization methods were uploaded to Pathway Studio 7.1. Supplementary Table 4 presents the list of 75 gene sets (together with the names of the quantitated proteins) reported as enriched (mean p-value across all normalization methods below 0.05). Here, we present the shortlist of the most increased and most decreased gene sets with their mean log2 ratio (AD/Control) and p-value in parenthesis. Top increased gene sets: regulation of small GTPase mediated signal transduction (3.26, 0.008), NF-kappaB binding (2.99, 0.029), protein modification process (2.87, 0.002), transcription from RNA polymerase II promoter (2.49, 0.017), intermediate filament polymerization (2.32, 0.002), cell death (2.24, 0.0006), nucleosome (2.23, 0.023), telomere maintenance (2.20, 0.019), immune response (2.16, 0.011). Top decreased gene sets: phosphoprotein phosphatase inhibitor activity (−2.02, 0.022), transcription factors (−1.22, 0.009), guanyl-nucleotide exchange factor activity (−0.65, 0.048), cytoskeleton organization (−0.61, 0.041).
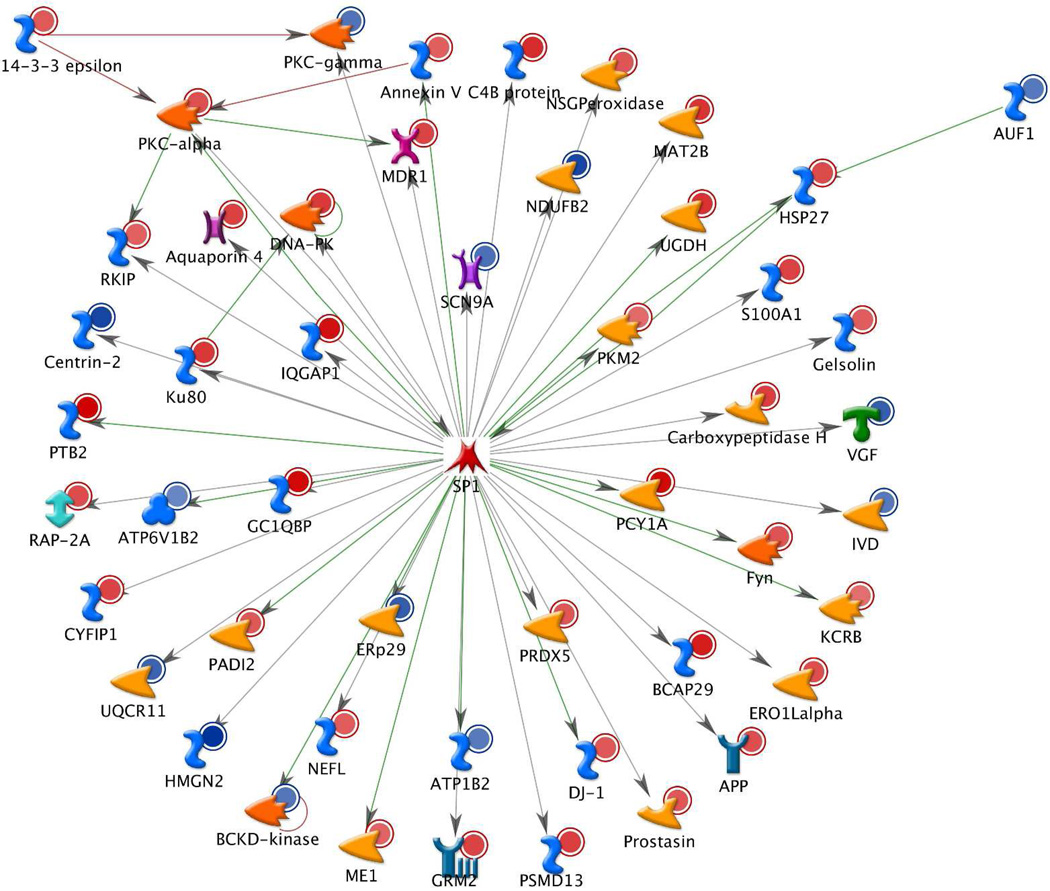
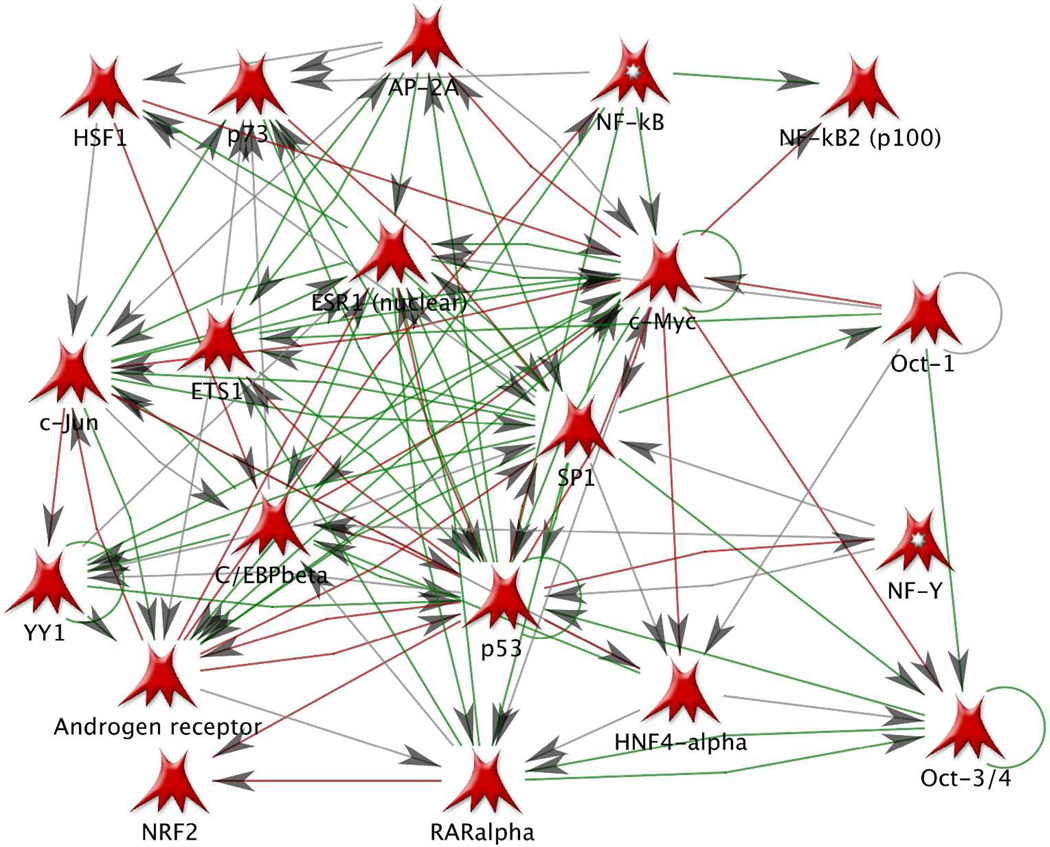
In search of the common regulators of the discovered 197 differentially abundant proteins we used the transcription regulation workflow in MetaCore 6.4 (GeneGo, Inc). The complete results of this analysis are presented in the Suppl. Report 1. Briefly, 20 transcription regulation networks were found to be significantly enriched (p-values within the range 7e-122 to 5e-17) with the above proteins. The top networks (with the number of affected differentially abundant proteins given in parenthesis) are: SP1 (49), c-Myc (36), HNF4-alpha (33), p53 (26), ESR1 (17). Figure 5 presents the SP1 network (red circles indicate over-abundant and blue circles under-abundant proteins). The rest of the networks are presented in the Suppl. Report 1. Importantly, these networks are interconnected: many proteins are affected by multiple transcription factors (TF) and itself are affecting other TFs. For example, APP is affected by the following TFs: SP1, c-Myc, p53, NF-kB, c-Jun, NF-Y, C/EBPbeta, oct-1. AP-2A regulates YY1, regulates and is affected by androgen receptor. PICALM is affected by c-Myc, p53, and ETS1. PKC-alpha regulates p53 and p73, affects and is affected by SP1 and ETS1. Figure 6 presents the direct interaction network of the above 20 transcription factors, which are connected through numerous transcription regulation and binding interactions, with both inhibition (red) and activation (green) effects. The high level of interconnectedness of the above transcription regulation networks adds confidence in the non-randomness and biological relevance of the discovered differentially abundant proteins.
Figure 5.
The top significant transcription regulation network involving 49 out of 197 significantly differentially abundant proteins (AD versus control) determined in this study. Red circles – over-abundant proteins, blue circles - under-abundant proteins. Network map is generated with Build Network tool (option Transcription Regulation) of MetaCore 6.4 (GeneGo, Inc). See legend in Suppl . Fig. 2 for complete list of symbols. See Suppl. Report 1 for 19 more transcription networks regulating 197 differentially abundant proteins.
Figure 6.
Network of 20 transcription factors regulating 197 significant differentially abundant proteins. Network map is generated with Build Network tool (option Direct Interactions) of MetaCore 6.4 (GeneGo, Inc). Transcription factors are closely interconnected with transcription regulation and binding interactions. Green lines – activation, red – inhibition, grey – unspecified.
Discussion
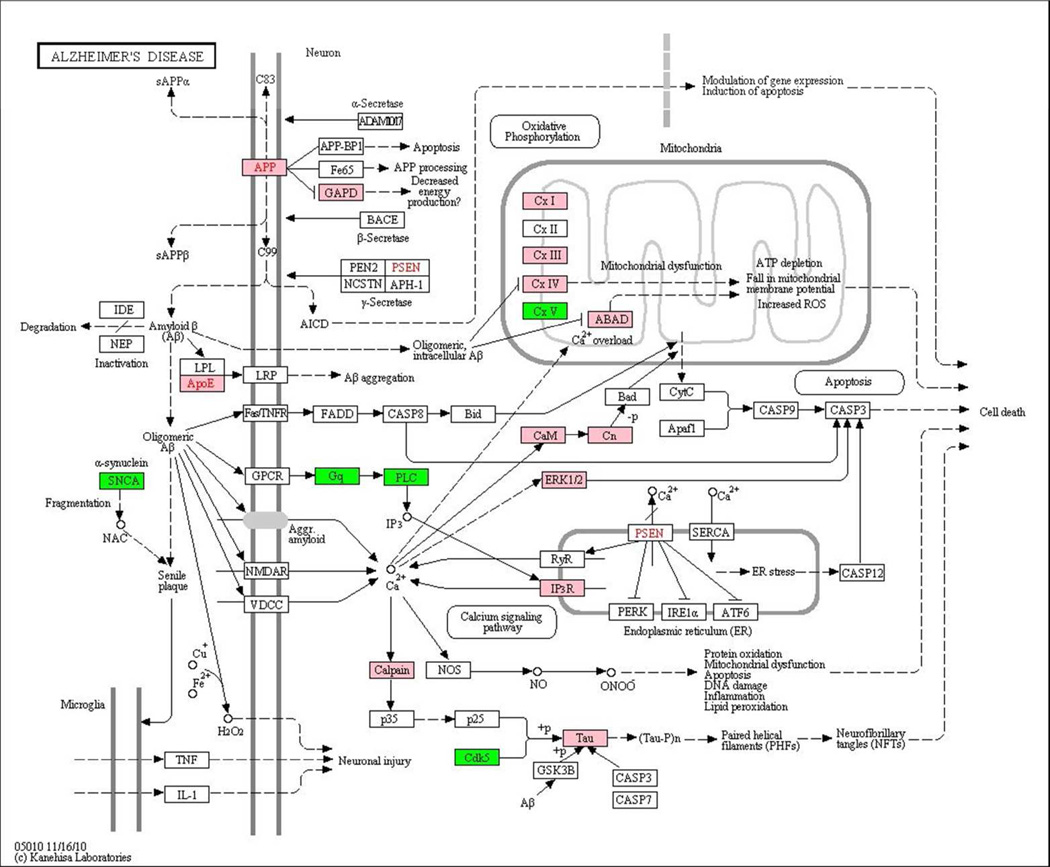
A total of 197 proteins were determined as significantly differentially abundant in the comparison of 10 AD and 10 control cortical brain samples. We compared these results with the published data on protein differential abundance in AD. The Supplementary Table 2 contains information on the published data related to the 197 significantly differentially abundant proteins determined in our study. Out of 197 proteins, 11 were mentioned as differentially abundant in AD in the 2006 omics review paper 4, 12 appeared in the list of genes with causal association with AD created with MetaCore 6.4 from the GeneGo database of publications, 21 were reported as differentially abundant in the recent proteomics and transcriptomics papers 6–23, 67–74, and 6 appeared in the AD pathway from KEGG 75. Figure 7 presents the results of mapping of the list of 1049 consensus proteins determined in our study on the AD pathway from KEGG. Over-abundant proteins are marked with pink and under-abundant as green. These KEGG AD related proteins are also flagged in the Supplementary Table 1. Importantly, the differentially abundant proteins observed in our study are present in three important branches of the AD pathway (APP, APOE and Tau).
Figure 7.
Alzheimer’s Disease pathway from KEGG. The list of 1049 consensus proteins determined in our study is mapped on the Alzheimer’s disease pathway from KEGG. Over-abundant proteins are marked with pink and under-abundant with green. The differentially abundant proteins observed in our study are present in three important branches of the KEGG AD pathway (APP, APOE and Tau).
A total of 37 proteins out of our 197 were mentioned in the above literature sources as differentially abundant or modified. Importantly, for the majority (28) of these proteins the sign of change in AD shown in our findings and reported in the literature coincides. (These concordant proteins are marked in Suppl. Table 2). Five of the proteins are mentioned in the literature as modified or affected (without indication of the sign of the change). While for other 4 (out of 37) proteins the sign of change differs in literature and our results. Specifically, p-glycoprotein (PGP) is over-abundant in AD in our study, while reported to be diminished in AD by Vogelgesang et al 76–77. Interestingly, however, PGP was also reported in 77 as over-abundant in the early stage of disease. Sulfotransferase 4A1is over-abundant in AD in our study while reported under-expressed in the cDNA study 74; protein cGMP-dependent 3’,5’-cyclic phosphodiesterase is 2-fold over-abundant in our study, while the transcript of this gene is under-expressed in the whole transcriptome study 23 ; cadherin 23 is 4-fold under-abundant in our study while the transcript of these gene is reported over-abundant in 23. Note that 3 out of 4 sign of change differences are observed in comparison of proteomics and transcriptomics measurements which are known to correlate poorly due to post-translational regulations. However, some of the transcriptomics and proteomics results are in good concordance, e.g. PICALM is at least 25-fold increased in AD in our study and the level of its mRNA is elevated in AD in 78, PCYT1A (choline-phosphate cytidylyltransferase A) is 16-fold increased in our study and 7-fold increased in the transcriptomics study 23, CAP2 (adenylyl cyclase-associated protein 2) is 2.4-fold decreased in our study and 2.6-fold decreased in the transcriptomics study 74.
In comparing our results with the recent proteomics of AD papers 71–72,21 interesting quantitative and qualitative similarities are worth mentioning. Specifically, in 71 the analysis of hippocampal samples was performed by 2D gel electrophoresis followed by MALDI TOF MS. As a result of the analysis, eighteen proteins were determined with significantly altered abundances in AD versus controls. Three of these proteins are in our list of 197 significantly differentially abundant proteins and are found to be over-abundant in both studies. The level of over-abundance, however, is higher in our study than in 71 for all three proteins (fold change found in our study versus fold change found in 71 is given in parenthesis): ferritin heavy chain (7.1-fold vs 1.23-fold), heat shock 70 protein (4.4-fold vs 1.14-fold), and glycerol-3-phosphate dehydrogenase (7-fold vs 1.28-fold).
Authors of another recent proteomics of AD paper 72 performed the 1D gel protein separation followed by in-gel digestion and high resolution LC-MS/MS study of the frontal cortex brain samples. The major difference from our approach is that they analyzed the detergent-insoluble subproteome in AD, i.e. were particularly interested in the thorough analysis of the neuropathological lesions associated with AD, while in our study we did not separate the plague from non-plague regions. Another important difference is that while in our study quantitation was based on the individual analysis of 10 AD and 10 control samples, in 72 analysis was performed by comparison of the pooled AD, control, and fronto-temporal degeneration (FTLD) samples. Eleven differentially abundant proteins were determined in AD versus FTLD and controls 72. Not surprisingly, amyloid beta was shown as over-abundant in both studies: 3.2-fold in our and 11-fold in 72. The members of the 14-3-3 protein family, known as important in neuronal development and abundant in brain tissue (1% wt of total soluble brain tissue protein)79, were also found as over-abundant in AD in both studies: 14-3-3 beta/alpha (3.3-fold) and 14-3-3 epsilon (2.5-fold) in our study, while two other isoforms 14-3-3 eta (3.7-fold) and 14-3-3 zeta (3.3-fold) were found in 72.
Several redox proteomics studies summarized in 21 reported a number of oxidatively modified brain proteins belonging or associated with mitochondrial proteome both in AD and in mild cognitive impairment. Interestingly, among our 197 significant differentially abundant proteins 14 are directly related to the mitochondrial proteome. Four of these proteins are over-abundant, while ten are under-abundant in AD in our study. Among them are: AUH (methylglutaconyl-CoA hydratase, mitochondrial) 36-fold over-abundant (FDR=0.0022), WARS2 (Tryptophanyl-tRNA synthetase, mitochondrial) 18-fold under-abundant (FDR=0.007), which to the best of our knowledge were not previously reported as affected in AD. Although our study does not examine oxidative protein modifications, it demonstrates the dramatic changes in the mitochondrial proteome and therefore is in concordance with the previous observations 21.
PICALM was recently in the center of attention of AD community due to GWAS studies that demonstrated association of AD risk with PICALM and CLU SNPs 80–82. Elevation of PICALM mRNA in AD brain tissue recently was studied with real time PCR and shown to happen predominantly in the endothelial cells 78. In our proteomics study PICALM was observed in all 10 AD samples and was not detected in any of 10 control samples, thus confirming its significant over-abundance in AD. Clusterin (CLU) is present as over-abundant (1.5-fold) in our consensus protein list (Suppl. Table 1), but didn’t make it to the significant protein list (mean FDR =0.09). Another protein of particular interest, ubiquitin-like modifier-activating enzyme 6 (UBA6) is shown in our study to be significantly (FDR=0.02) and strongly (6-fold) over-abundant in AD. UBA6 was recently shown to activate not only ubiquitin but also FAT10 (human leukocyte antigen F-associated transcript 10) which represents the new layer of regulation of ubiquitin conjugation system 83–84. The important role of ubiquitin-proteosome system in AD pathogenesis is reflected in reviews 85–86.
Overall, the level of overlap and concordance of our results and previously published data is similar to that of any new “omics” study of AD. Historically, some proteins were reported as affected in AD by the majority of the studies, some were novel in each next study which employed higher sensitivity approach. Later, some of them were confirmed and joined the core AD proteome, and some remained non-confirmed mainly due to the heterogeneity of AD and low sample size of the studies. The record high numbers (thousands) of new transcripts affected in AD were reported in the recent whole transcriptome study 23. Although not yet confirmed by alternative methods, these novel transcripts provide an important resource for future studies. We expect the same to be true to our 160 novel differentially abundant in AD proteins. Importantly, the overlap between the studies is higher at the level of biological processes; for instance, signal transduction, regulation of protein phosphorylation, immune response, cytoskeleton organization, lipid metabolism, energy production, and cell death were reported as affected in AD both in our and many other studies including 5, 9, 19, 21, 23. The affiliation of the novel differentially abundant proteins to the biological processes known to be affected in AD increases the confidence in the relevance of these proteins.
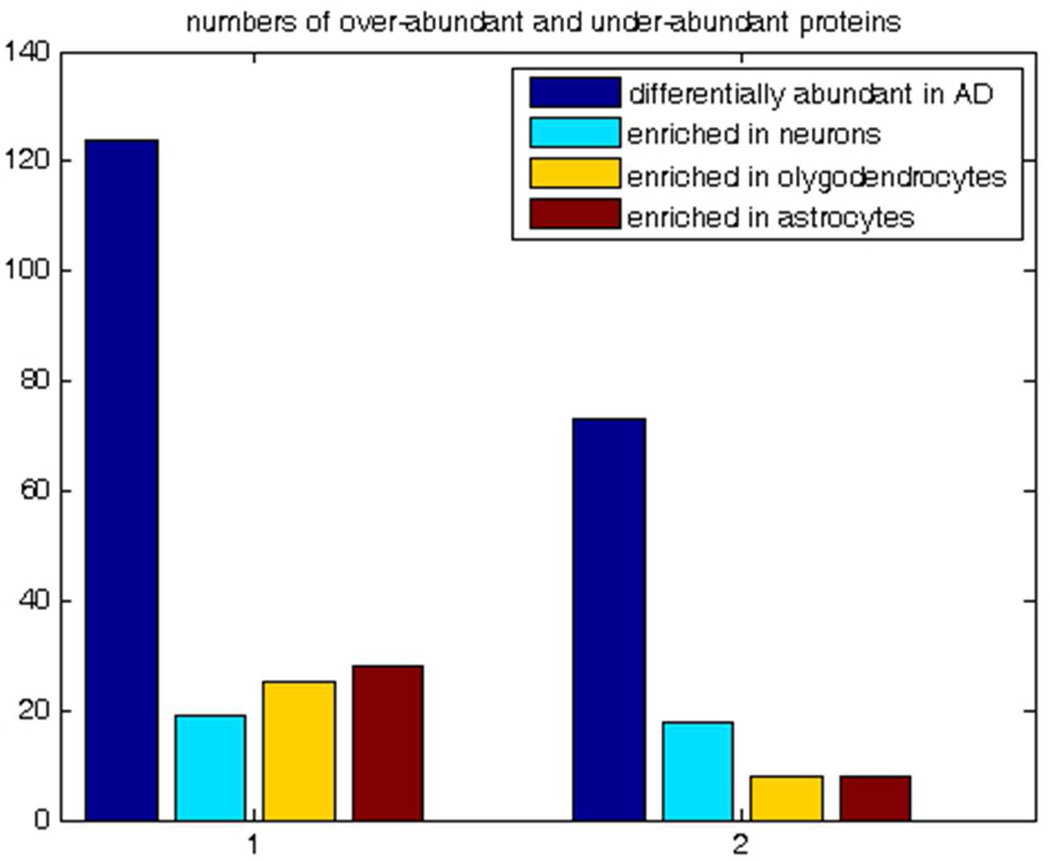
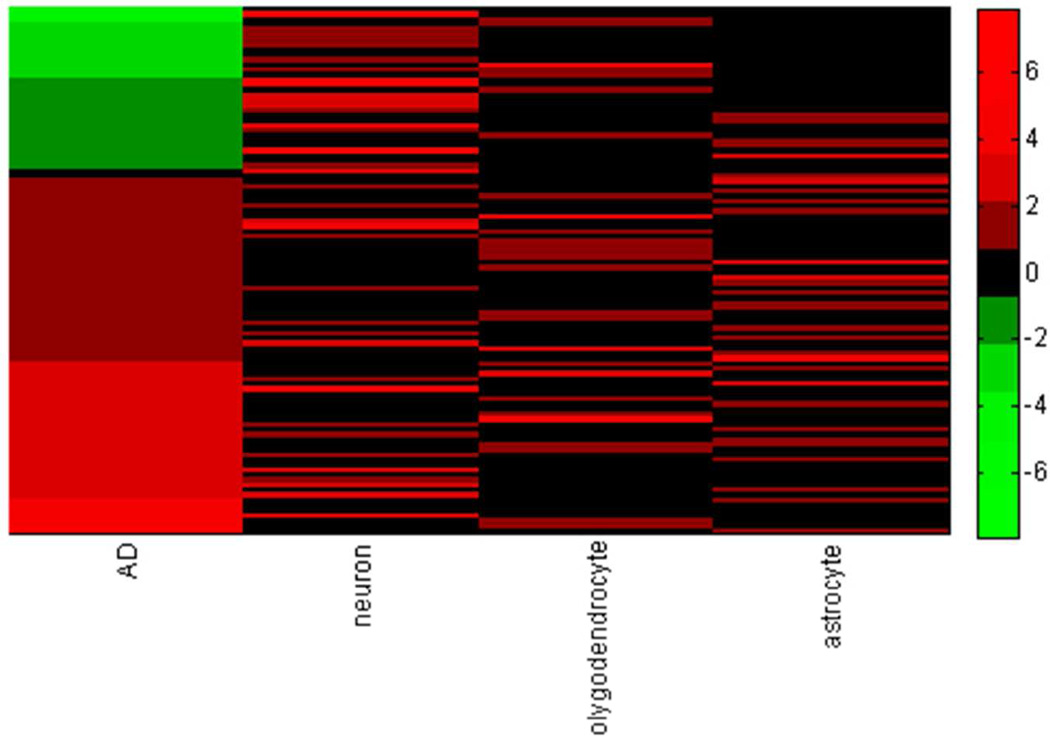
The interpretation of the observed differential abundance of 197 proteins is complicated by the fact that AD is characterized by the massive neuropathological alterations and shifts in cell population, e.g. formation of plaques and tangles, neuronal loss, gliosis, etc 87. With this in mind, we avoided using terms “up-regulated” and “down-regulated” and referred to the observed differentially abundant proteins as “over-abundant” and “under-abundant”. Here, we make an attempt to roughly estimate the relative role of cell population shift and intracellular process regulation in the observed differential abundance of the 197 proteins of this paper. We mapped our 197 differentially abundant proteins on the recently published database of transcripts enriched in astrocytes, neurons, and oligodendrocytes of mouse forebrain 88; to the best of our knowledge no such whole transcriptome cell type enrichment data exist for the human brain cells. By using the MammalHom tool (http://depts.washington.edu/l2l/aboutmammalhom.html) 184 mouse homologues of our 197 proteins were found. The results of the mapping are presented in Figure 8A. Of 124 proteins observed as over-abundant in AD in our study, 19 are enriched in neurons, 25 in oligodendrocytes, and 28 in astrocytes; of 73 under-abundant in AD, 18 are enriched in neurons, 8 in oligodendrocytes, and 8 in astrocytes; 93 of our differentially abundant proteins are not shown to be enriched in any of three cell types. Figure 8B presents the heat map of the differential abundance and cell type enrichment for 104 of our 197 proteins that overlap with the cell type enrichment database (Suppl. Fig 3 provides the zoomable version of this heat map with the names of the proteins). Table 2 presents Pearson correlation of differential abundance and cell type enrichment. As expected, differential abundance in AD is negatively correlated (−0.25) with enrichment in neurons and positively correlated with enrichment in astrocytes (0.10), which agrees with the fact of neuronal loss in AD. However, this correlation is not strong. With all the reservations in mind about the differences between human and mouse brains, we think that this result together with the fact that 93 of the differentially abundant proteins are not enriched in any of three cell types indicates that the observed differential abundance of our 197 proteins represents the combined effect of both the regulation of the intracellular processes and the alternations in the cell type composition of the AD brain. The more detailed quantitative discrimination of the effects of the intracellular regulation and of the shift of cell population would require laser capture microdissection and separate proteomics analysis of several cell types and is beyond the scope of this study.
Figure 8.
Mapping of the 197 proteins significantly differentially abundant in AD onto the mouse CNS cell type enrichment database. Fig 8A. Proteins over-abundant and under-abundant in AD and their cell type enrichment. Fig 8B. Heat map of abundance and cell type enrichment for 104 proteins common for the list of 197 significantly differentially abundant in AD and cell type enrichment database 88, where transcripts were considered enriched if they were at least 1.5-fold over-expressed and statistically different by significance analysis of microarrays with false discovery threshold of 1%.
Table 2.
Pearson Correlation(R) of differential abundance in AD and cell type enrichment
| Abundance/enrichment | AD | neurons | Oligodendrocytes | Astrocytes |
|---|---|---|---|---|
| AD | 1 | −0.2535 | 0.012 | 0.1052 |
| Neurons | −0.2535 | 1 | −0.2884 | −0.3296 |
| oligodendrocytes | 0.012 | −0.2884 | 1 | −0.23 |
| Astrocytes | 0.1052 | −0.3296 | −0.23 | 1 |
Conclusion
We generated a list of 197 proteins differentially abundant in AD temporal cortical samples versus normally aged controls, by using the high sensitivity high resolution mass spectrometry based proteomics with the AMT label-free quantitation approach together with a conservative procedure of selecting only the proteins determined as significant (FDR<0.05) in four normalization/imputation methods. The sign of the change in abundance is confirmed by the published data for 28 of these proteins, for 3 it is in contradiction with the existing transcriptomics data, and only for one in contradiction with existing proteomics data. The majority (160 out of 197) proteins were not previously reported as differentially abundant or modified in AD.
We performed verification of our findings by analyzing with Western blot two (PRKCG and NUMBL) randomly selected differentially abundant proteins out of these 160. For both of them Western blot and mass spectrometry based proteomics results are in concordance: PRKCG – 4-fold and 2.6-fold decrease, NUMBL – 2-fold and 3.2-fold increase. Our findings will be further validated with the highly sensitive targeted proteomics approach 89 to quantify the above 197 differentially abundant proteins. The targeted proteomics quantitation will serve as an independent method of validation that is more practical in the case of the numerous proteins than Western blot or ELISA validation of the total list of differentially abundant proteins.
Mapping of the discovered differentially abundant proteins revealed significant enrichment of gene sets, pathways, and processes known to be important in AD thus further confirming the relevance of our findings. Based on the maturity of the LC-MS based proteomics, conservative consensus selection of the differentially abundant proteins used in the current study, and the concordance of our results with the published data for a large group of proteins, we believe it is important to publish the list of discovered 197 differentially abundant proteins (Suppl. Table 2) The present study offers a valuable data resource for data mining and generation of hypotheses for follow-up studies of Alzheimer’s disease etiology.
Supplementary Material
Acknowledgements
We are grateful to the donors and their families. This work is supported by National Institute on Aging EUREKA grant R01-AG-034504 to Dr. Myers. The University of Michigan Alzheimer’s Disease Research Center’s brain bank is supported by NIA grant P50-AG08671. Proteomic analyses were supported by the NIH National Center for Research Resources (RR18522 to R.D.S.) and were performed in the Environmental Molecular Sciences Laboratory, a US Department of Energy (DOE) national scientific user facility located at the Pacific Northwest National Laboratory (PNNL) in Richland, Washington. PNNL is a multi-program national laboratory operated by Battelle Memorial Institute for the DOE under Contract DE-AC05-76RL01830.
Footnotes
Conflict of Interest
Authors declare no conflict of interest.
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Evans DA, Funkenstein HH, Albert MS, Scherr PA, Cook NR, Chown MJ, Hebert LE, Hennekens CH, Taylor JO. Prevalence of Alzheimer’s disease in a community population of older persons. Higher than previously reported. JAMA. 1989;262:2551–2556. [PubMed] [Google Scholar]
- 2.Gatz M, Pedersen NL, Berg S, Johansson B, Johansson K, Mortimer JA, Posner SF, Viitanen M, Winblad B, Ahlbom A. Heritability for Alzheimer’s disease: the study of dementia in Swedish twins. J Gerontol A Biol Sci Med Sci. 1997;52:M117–M125. doi: 10.1093/gerona/52a.2.m117. [DOI] [PubMed] [Google Scholar]
- 3.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 4.Papassotiropoulos A, Fountoulakis M, Dunckley T, Stephan DA, Reiman EM. Genetics, transcriptomics and proteomics of Alzheimer’s disease. J Clin Psychiatry. 2006;67:652–670. doi: 10.4088/jcp.v67n0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lovestone S, Guntert A, Hye A, Lynham S, Thambisetty M, Ward M. Proteomics of Alzheimer’s disease: understanding mechanisms and seeking biomarkers. Expert Rev Proteomics. 2007;4:227–238. doi: 10.1586/14789450.4.2.227. [DOI] [PubMed] [Google Scholar]
- 6.Korolainen MA, Nyman TA, Aittokallio T, Pirttila T. An update on clinical proteomics of Alzheimer’s research. J.Neurochem. 2010;112:1386–1414. doi: 10.1111/j.1471-4159.2009.06558.x. [DOI] [PubMed] [Google Scholar]
- 7.Cocabelos R, Martinez-Bouza R. Genomics and Pharmacogenomics of Dementia. CNS Neuroscience & Therapeutics. 2011;17:566–576. doi: 10.1111/j.1755-5949.2010.00189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cummings JL. Biomarkers in Alzheimer’s disease drug development. Alzheimer’s & Dementia. 2011;7:e13–e44. doi: 10.1016/j.jalz.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 9.Juhasz G, Foldi I, Penke B. Systems biology of Alzheimer’s disease: How diverse molecular changes result in memory impairment in AD. Neurochem Int. 2011;58:739–750. doi: 10.1016/j.neuint.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 10.Davinelli S, Intrieri M, Russo C, Di Constanzo A, Zella D, Bosco P, Scapagnini G. The “Alzheimer’s disease signature”; potential perspectives for novel biomarkers. Immunity & Ageing. 2011;8:7–17. doi: 10.1186/1742-4933-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Domenico F, Coccia R, Butterfield DA, Perluigi M. Circulating biomarkers of protein oxidation for Alzheimer’s disease: Expectations within limits. Biochimica et Biophysica Acta. 2011;1814:1785–1795. doi: 10.1016/j.bbapap.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 12.Finehout EJ, Franck Z, Choe LH, Relkin N, Lee KH. Cerebrospinal fluid proteomic biomarkers for Alzheimer’s disease. Ann Neurol. 2007;61:120–129. doi: 10.1002/ana.21038. [DOI] [PubMed] [Google Scholar]
- 13.Simonsen AH, McGuire J, Hansson O, Zetterberg H, Podust VN, Davies HA, et al. Novel panel of cerebrospinal fluid biomarkers for the prediction of progression to Alzheimer dementia in patients with mild cognitive impairment. Arch Neurol. 2007;64:366–370. doi: 10.1001/archneur.64.3.366. [DOI] [PubMed] [Google Scholar]
- 14.Simonsen AH, McGuire J, Podust VN, Hagnelius NO, Nilsson TK, Kapaki E, et al. A novel panel of cerebrospinal fluid biomarkers for the differential diagnosis of Alzheimer’s disease versus normal aging and frontotemporal dementia. Dement Geriatr Cogn Disord. 2007;24:434–440. doi: 10.1159/000110576. [DOI] [PubMed] [Google Scholar]
- 15.Ray S, Britschgi M, Herbert C, Takeda-Uchimura Y, Boxer A, Blennow K, et al. Classification and prediction of clinical Alzheimer’s diagnosis based on plasma signaling proteins. Nat Med. 2007;13:1359–1362. doi: 10.1038/nm1653. [DOI] [PubMed] [Google Scholar]
- 16.Palmer JC, Baig S, Kehoe PG, Love S. Endothelin-converting enzyme-2 is increased in Alzheimer’s disease and up-regulated by Abeta. Am J Pathol. 2009;175:262–270. doi: 10.2353/ajpath.2009.081054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xia Q, Cheng D, Duong DM, Gearing M, Lah JJ, Levey AI, Peng J. Phosphoproteomic analysis of human brain by calcium phosphate precipitation and mass spectrometry. J Proteome Res. 2008;7:2845–2851. doi: 10.1021/pr8000496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rudrabhatla P, Jaffe H, Pant HC. Direct evidence of phosphorylated neuronal intermediate filament proteins in neurofibrillary tangles (NFTs): phosphoproteomics of Alzheimer’s NFTs. FASEB J. 2011;11:3896–3905. doi: 10.1096/fj.11-181297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Di Domenico F, Sultana R, Barone E, Perluigi M, Cini C, Mancuso C, Cai J, Pierce WM, Butterfield DA. Quantitative proteomics analysis of phosphorylated proteins in the hippocampus of Alzheimer’s disease subjects. J Proteomics. 2011;74:1091–1103. doi: 10.1016/j.jprot.2011.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reed T, Perluigi M, Sultana R, Pierce WM, Klein JB, Turner DM, Coccia R, Markesbery WR, Butterfield DA. Redox proteomic identification of 4-hydroxy-2-nonenal-modified brain proteins in amnestic mild cognitive impairment: Insight into the role of lipid peroxidation in the progression and pathogenesis of Alzheimer’s disease. Neurobiology of Disease. 2008;30:107–120. doi: 10.1016/j.nbd.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 21.Sultana R, Butterfield DA. Oxidatively modified, mitochondria-relevant brain proteins in subjects with Alzheimer’s disease and mild cognitive impairment. J Bioenerg Biomembr. 2009;41:441–446. doi: 10.1007/s10863-009-9241-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Charlwood J, Dingwalls C, Matico R, Hussain I, Johanson K, Moore S, Powel DJ, Skehel JM, Ratcliffe S, Clarke B, Trill J, Sweitzer S, Camilleri P. Characterization of the glycosylation profiles of Alzheimer’s beta-secretase protein asp-2 expressed in variety of cell lines. J Biol Chem. 2001;276:16739–16748. doi: 10.1074/jbc.M009361200. [DOI] [PubMed] [Google Scholar]
- 23.Twine NA, Janitz K, Wilkins MA, Janitz M. Whole transcriptome sequencing reveals gene expression and splicing differences in brain regions affected by Alzheimer’s disease. PloS One. 2011;6:e16266. doi: 10.1371/journal.pone.0016266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hata R, Masumura M, Akatsu H, Li F, Fujita H, et al. Up-regulation of calcineurin Abeta mRNA in the Alzheimer’s disease brain: assessment by cDNA microarray. Biochem Biophys Res Commun. 2001;284:310–316. doi: 10.1006/bbrc.2001.4968. [DOI] [PubMed] [Google Scholar]
- 25.Ginsberg SD, Crino PB, Hemby SE, Weingarten JA, Lee VM, et al. Predominance of neuronal mRNAs in individual Alzheimer’s disease senile plaques. Ann Neurol. 1999;45:174–181. [PubMed] [Google Scholar]
- 26.Larner AJ, Doran M. Clinical phenotypic heterogeneity of Alzheimer’s disease associated with mutations of the preselin-1 gene. J Neurol. 2006;253:139–158. doi: 10.1007/s00415-005-0019-5. [DOI] [PubMed] [Google Scholar]
- 27.Folstein MF. Heterogeneity of Alzheimer’s disease. Nerobiology of Aging. 1989;5:434–435. doi: 10.1016/0197-4580(89)90086-9. [DOI] [PubMed] [Google Scholar]
- 28.Gygi SP, Rochon Y, Franza BR, et al. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hack CJ. Integrated transcriptome and proteome data: the challenges ahead. Brief Funct Genomic Proteomic. 2004;3:212–219. doi: 10.1093/bfgp/3.3.212. [DOI] [PubMed] [Google Scholar]
- 30.Chen G, Gharib TG, Huang C-C, Taylor JMG, Misek DE, Kardia SLR, Giordano TJ, Iannettoni MD, Orringer MB, Hanash SM, Beer DG. Discordant protein and mRNA expression in lung adenocarcinomas. Mol Cellular Proteomics. 2002;1:304–313. doi: 10.1074/mcp.m200008-mcp200. [DOI] [PubMed] [Google Scholar]
- 31.Ong SE, Mann M. Mass spectrometry based proteomics turns quantitative. Nat Chem Biol. 2005;1:252–262. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- 32.Cox J, Mann M. Is proteomics a new genomics? Cell. 2007;130:395–398. doi: 10.1016/j.cell.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 33.Ahrens CH, Brunner E, Qeli E, Basler K, Aebersold R. Generating and navigating proteome maps using mass spectrometry. Nature Rev Mol Cell Biol. 2010;11:789–801. doi: 10.1038/nrm2973. [DOI] [PubMed] [Google Scholar]
- 34.Levin Y, Wang L, Schwartz E, Koethe D, Leweke FM, Bahn S. Global proteomics profiling reveals altered proteomic signature in schizophrenia serum. Molecular Psychiatry. 2010;15:1088–1100. doi: 10.1038/mp.2009.54. [DOI] [PubMed] [Google Scholar]
- 35.Chan MK, Tsang TM, Harris LW, Guest PC, Holmes E, Bahn S. Evidence for disease and antipsychotic medication effects in post mortem brain from schizophrenia patients. Molecular Psychiatry. 2010;15:1–14. doi: 10.1038/mp.2010.100. [DOI] [PubMed] [Google Scholar]
- 36.Michaelevski I, Medzihradszky KF, Lynn A, Burlingame AL, Fainzilber M. Axonal transport proteomics reveals mobilization of translation machinery to the lesion site in injured sciatic nerve. Mol Cell Proteomics. 2010;9:976–987. doi: 10.1074/mcp.M900369-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burton EJ, Barber R, Mukaetova-Ladinska EB, Robson J, Perry RH, Jaros E, Kalaria RN, O’Brien JT. Medial temporal lobe atrophy on MRI differentiates Alzheimer’s disease from dementia with Lewy bodies and vascular cognitive impairment: a prospective study with pathological verification of diagnosis. Brain. 2009;132:195–203. doi: 10.1093/brain/awn298. [DOI] [PubMed] [Google Scholar]
- 38.Conrads TP, Anderson GA, Veenstra TD, Pasa-Tolic L, Smith RD. Utility of accurate mass tags for proteome-wide protein identification. Anal Chem. 2000;72:3349–3354. doi: 10.1021/ac0002386. [DOI] [PubMed] [Google Scholar]
- 39.Smith RD, Anderson GA, Lipton MS, Pasa-Tolic L, Shen Y, Conrads TP, Veenstra TD, Udseth HR. An accurate mass tag strategy for quantitative and high-throughput proteome measurements. Proteomics. 2002;2:513–523. doi: 10.1002/1615-9861(200205)2:5<513::AID-PROT513>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 40.Smith RD, Anderson GA, Lipton MS, Masselon CD, Pasa-Tolic L, Shen Y, Udseth HR. The Use of accurate mass tags for high-throughput microbial proteomics. OMICS. 2002;6:61–90. doi: 10.1089/15362310252780843. [DOI] [PubMed] [Google Scholar]
- 41.Jaitly N, Monroe ME, Petyuk VA, Clauss TRW, Adkins JN, Smith RD. Robust algorithm for alignment of liquid chromatography-mass spectrometry analyses in an accurate mass and time tag data analysis pipeline. Anal Chem. 2006;78:7397–7409. doi: 10.1021/ac052197p. [DOI] [PubMed] [Google Scholar]
- 42.Petyuk VA, Qian W-J, Chin MH, Wang H, Livesay EA, Monroe ME, Adkins JN, Jaitly N, Anderson DJ, Camp DG, Smith DJ, Smith RD. Spatial mapping of protein abundances in the mouse brain by voxelation integrated with high-throughput liquid chromatography-mass spectrometry. Genome Research. 2007;17:328–336. doi: 10.1101/gr.5799207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petyuk VA, Qian W, Smith RD, Smith DJ. Mapping protein abundance patterns in the brain using voxelation combined with liquid chromatography and mass spectrometry. Methods. 2010;50:77–84. doi: 10.1016/j.ymeth.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang W, Zhou H, Lin H, Roy S, Shaler TA, Hill LR, Norton S, Kumar P, Anderle M, Becker CH. Quantification of proteins and metabolites by mass spectrometry without isotopic labeling or spiked standards. Anal. Chem. 2003;75:4818–4826. doi: 10.1021/ac026468x. [DOI] [PubMed] [Google Scholar]
- 45.Silva JC, Denny R, Dorschel CA, Gerenstein M, Kass IJ, Li G-Z, McKenna T, Nold MJ, Richardson K, Young P, Geromanos S. Quantitative proteomic analysis by accurate mass retention time pairs. Anal. Chem. 2005;77:2187–2200. doi: 10.1021/ac048455k. [DOI] [PubMed] [Google Scholar]
- 46.Ferguson RE, Carroll HP, Harris A, Maher ER, Selby PJ, Banks RE. Housekeeping proteins: a preliminary study illustrating some limitations as useful references in protein expression studies. Proteomics. 2005;5:566–571. doi: 10.1002/pmic.200400941. [DOI] [PubMed] [Google Scholar]
- 47.Lim WK, Wang K, Lefebvre C, Califano A. Comparative analysis of microarray normalization procedures: effects on reverse engineering gene networks. Bioinformatics. 2007;23:i282–i288. doi: 10.1093/bioinformatics/btm201. [DOI] [PubMed] [Google Scholar]
- 48.Stafford P. Methods in Microarray Normalization. Boca Raton, USA: CRC Press; 2008. [Google Scholar]
- 49.Pritchard JK, Stephens M, Rosenberg NA, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Myers AJ, Gibbs JR, Webster JA, Rohrer KC, Zhao AS, Marlowe L, et al. A survey of genetic human cortical gene expression. Nat Genet. 2007;39:1494–1499. doi: 10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- 52.Webster JA, Gibbs JR, Clarke J, Ray M, Zhang W, Holmans P, et al. Genetic control of human brain transcript expression in Alzheimer’s disease. Am J Hum Genet. 2009;84:445–458. doi: 10.1016/j.ajhg.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mayampurath AM, Jaitly N, Purvine SO, Monroe ME, Auberry KJ, Adkins JN, Smith RD. DeconMSn: a software tool for accurate parent ion monoisotopic mass determination for tandem mass spectra. Bioinformatics. 2008;24:1021–1023. doi: 10.1093/bioinformatics/btn063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Petyuk VA, Mayampurath AM, Monroe ME, Polpitiya AD, Purvine SO, Anderson GA, Camp DG, 2nd, Smith RD. DtaRefinery, a software tool for elimination of systematic errors from parent ion mass measurements in tandem mass spectra data sets. Mol Cell Proteomics. 2010;9:486–496. doi: 10.1074/mcp.M900217-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Elias JE, Gygi SP. Target-decoy search strategy for mass spectrometry-based proteomics. Methods Mol Biol. 2010;604:55–71. doi: 10.1007/978-1-60761-444-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Petyuk VA, Qian WJ, Smith RD, Smith DJ. Mapping protein abundance patterns in the brain using voxelation combined with liquid chromatography and mass spectrometry. Methods. 2010;50:77–84. doi: 10.1016/j.ymeth.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jaitly N, Mayampurath A, Littlefield K, Adkins JN, Anderson GA, Smith RD. Decon2LS: An open-source software package for automated processing and visualization of high resolution mass spectrometry data. BMC Bioinformatics. 2009;10:87. doi: 10.1186/1471-2105-10-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Monroe ME, Tolic N, Jaitly N, Shaw JL, Adkins JN, Smith RD. Viper: an advanced software package to support high throughput LC-MS peptide identification. Bioinformatics. 2007;23:2021–2023. doi: 10.1093/bioinformatics/btm281. [DOI] [PubMed] [Google Scholar]
- 59.Qian WJ, Jacobs JM, Liu T, Camp DG, 2nd, Smith RD. Advances and challenges in liquid chromatography-mass spectrometry-based proteomics profiling for clinical applications. Mol Cell Proteomics. 2006;5:1727–1744. doi: 10.1074/mcp.M600162-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Andreev VP, Dwivedi RC, Paz-Filho G, Krokhin OV, Wong M-L, Wilkins JA, Licinio J. Dynamics of plasma proteome during leptin replacement therapy in genetically-based leptin deficiency. Pharmacogenomics J. 2011;11:162–173. doi: 10.1038/tpj.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Karpievitch YV, Taverner T, Adkins JN, Callister SJ, Anderson GA, Smith RD, Dabney AR. Normalization of peak intensities in bottom-up MS-based proteomics using singular value decomposition. Bioinformatics. 2009;25:2573–2580. doi: 10.1093/bioinformatics/btp426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Karpievitch YV, Stanley J, Taverner T, Huang J, Adkins JN, Ansong C, Heffron F, Metz TO, Qian W-J, Yoon H, Smith RD, Dabney AR. A statistical framework for protein quantitation in bottom-up MS-based proteomics. Bioinformatics. 2009;25:2028–2034. doi: 10.1093/bioinformatics/btp362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Polpitiya AD, Qian WJ, Jaitly N, Petyuk VA, Adkins JN, Camp DG, 2nd, Anderson GA, Smith RD. DAnTE: a statistical tool for quantitative analysis of -omics data. Bioinformatics. 2008;24:1556–1558. doi: 10.1093/bioinformatics/btn217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Storey JD. A direct approach to false discovery rates. J. Royal Statistical Society, Series B. 2002;64:479–498. [Google Scholar]
- 65.The Gene Ontology Consortium. Gene Ontology: tool for the unification of biology. Nature Genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Subramanian A, Tamayoa P, Moothaa VK, Mukherjeed S, Eberta BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. PNAS. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang J, Goodlett DR, Peskind ER, Quinn JF, Zhou Y, Wang Q, et al. Quantitative proteomic analysis of age-related changes in human cerebrospinal fluid. Neurobiol Aging. 2005;26:207–227. doi: 10.1016/j.neurobiolaging.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 68.Zhang J, Goodlett DR, Quinn JF, Peskind E, Kaye JA, Zhou Y, et al. Quantitative proteomics of cerebrospinal fluid from patients with Alzheimer disease. J Alzheimers Dis. 2005;7:125–133. doi: 10.3233/jad-2005-7205. [DOI] [PubMed] [Google Scholar]
- 69.Abdia F, Quinn JF, Jankovic J, McIntosh M, Leverenz JB, Peskind E, et al. Detection of biomarkers with a multiplex quantitative proteomic platform in cerebrospinal fluid of patients with neurodegenerative disorders. J Alzheimerss Dis. 2006;9:293–348. doi: 10.3233/jad-2006-9309. [DOI] [PubMed] [Google Scholar]
- 70.Ho L, Sharma N, Blackman L, Festa E, Reddy G, Pasinetti GM. From proteomics to biomarker discovery in Alzheimer’s disease. Brain Research Rev. 2005;48:360–369. doi: 10.1016/j.brainresrev.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 71.Sultana R, Boyd-Kimball D, Cai J, Pierce WM, Klein JB, Merchant M, et al. Proteomics analysis of the Alzheimer’s disease hippocampal proteome. J Alzheimers Dis. 2007;11:153–164. doi: 10.3233/jad-2007-11203. [DOI] [PubMed] [Google Scholar]
- 72.Gozal YM, Duong DM, Gearing M, Cheng C, Hanfelt JJ, Funderburk C, et al. Proteomics analysis reveals novel components in the detergent-insoluble subproteome in Alzheimer’s disease. J Proteome Res. 2009;8:5069–5079. doi: 10.1021/pr900474t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tsuji T, Shiozaki A, Kohno R, Yoshizato K, Shimohama S. Proteomic profiling and neurodegeneration in Alzheimer’s disease. Neurochemical Research. 2002;27:1245–1253. doi: 10.1023/a:1020941929414. [DOI] [PubMed] [Google Scholar]
- 74.Emilsson L, Saetre P, Jazin E. Alzheimer’s disease: mRNA expression profiles of multiple patients show alterations of genes involved with calcium signaling. Neurobiology of Disease. 2006;21:618–625. doi: 10.1016/j.nbd.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 75. http://www.genome.jp/kegg/
- 76.Vogelgesang S, Cascorbi I, Schroeder E, Pahnke J, Kroemer HK, Siegmund W, et al. Deposition of Alzheimer's beta-amyloid is inversely correlated with P-glycoprotein expression in the brains of elderly non-demented humans. Pharmacogenetics. 2002;12:535–541. doi: 10.1097/00008571-200210000-00005. [DOI] [PubMed] [Google Scholar]
- 77.Vogelgesang S, Warzok RW, Cascorbi I, Kunert-Keil C, Schroeder E, Kroemer HK, et al. The role of P-glycoprotein in cerebral amyloid angiopathy; implications for the early pathogenesis of Alzheimer's disease. Curr Alzheimer Res. 2004;1:121–125. doi: 10.2174/1567205043332225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Baig S, Joseph SA, Tayler H, Abraham R, Owen MJ, Williams J, et al. Distribution and expression of picalm in Alzheimer’s disease. J Neuropathol Exp Neurol. 2010;69:1071–1077. doi: 10.1097/NEN.0b013e3181f52e01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fu H, Subramanian RR, Masters SC. 14-3-3proteins: structure, function, and regulation. Annu. Rev. Pharmacol. Toxicol. 2000;40:617–647. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- 80.Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat. Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat. Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 82.Corneveaux JJ, Myers AJ, Allen AN, Pruzin JJ, Ramirez M, et al. Association of CR1 CLU, and PICALM with Alzheimer’s disease in a cohort of clinically characterized and neuropathologically verified individuals. HMG Advance Access. doi: 10.1093/hmg/ddq221. published June 9, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chiu YH, Sun Q, Chen ZJ. E1-L2 activates both ubiquitin and FAT10. Mol. Cell. 2007;27:1014–1023. doi: 10.1016/j.molcel.2007.08.020. [DOI] [PubMed] [Google Scholar]
- 84.Groettrup M, Pelzer C, Schmidtke G, Hofmann K. Activating the ubiquitin family: UBA6 challenges the field. TIBS. 2008;33:230–237. doi: 10.1016/j.tibs.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 85.Upadhya SC, Hegde AN. Role of the ubiquitin proteasome system in Alzheimer's disease. BMC Biochem. 2007;8(Suppl 1):S12–S19. doi: 10.1186/1471-2091-8-S1-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Song S, Jung Y-K. Alzheimer’s disease meets the ubiquitin-proteosome system. Trends in Molecular Medicine. 2004;10:565–570. doi: 10.1016/j.molmed.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 87.Serrano-Pozo A, Frosch MP, Masliah E, Hyman BT. Neuropathological alterations in Alzheimer’s disease. In: Selkoe DJ, Mandelkow E, Holtzman DM, editors. The Biology of Alzheimer’s Disease. Cold Spring Harbor, NY: CSH Press; 2012. pp. 43–65. [Google Scholar]
- 88.Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J. Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kiyonami R, Schoen A, Prakash A, Peterman S, Zabrouskov V, Picotti P, Aebersold R, Huhmer A, Domon B. Increased selectivity, analytical precision, and throughput in targeted proteomics. Mol Cell Proteomics. 2011;10:1–11. doi: 10.1074/mcp.M110.002931. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.