Abstract
Herein we report a pressure-assisted capillary electrophoresis-mass spectrometric imaging (PACE-MSI) platform for peptide analysis. This new platform has addressed the sample diffusion and peak splitting problems that appeared in our previous groove design, and it enables homogenous deposition of the CE trace for high-throughput MALDI imaging. In the coupling of CE to MSI, individual peaks (m/z) can be visualized as discrete colored image regions and extracted from the MS imaging data, thus eliminating issues with peak overlapping and reducing reliance on an ultra-high mass resolution mass spectrometer. Through a PACE separation, 46 tryptic peptides from bovine serum albumin and 150 putative neuropeptides from the pericardial organs of a model organism blue crab Callinectes sapidus were detected from the MALDI MS imaging traces, enabling four to six-fold increase of peptide coverage as compared with direct MALDI MS analysis. For the first time, quantitation with high accuracy was obtained using PACE-MSI for both digested tryptic peptides and endogenous neuropeptides from complex biological samples in combination with isotopic formaldehyde labeling. Although MSI is typically employed in tissue imaging, we show in this report that, it offers a unique tool for quantitative analysis of complex trace-level analytes with CE separation. These results demonstrate a great potential of the PACE-MSI platform for enhanced quantitative proteomics and neuropeptidomics.
Keywords: capillary electrophoresis, CE, PACE, MALDI, mass spectrometric imaging, MSI, peptides, neuropeptides, quantitation
INTRODUCTION
Mass spectrometry (MS)-based characterization of trace-level peptides from complex samples, such as endogenous neuropeptide extracts, secreted peptides in biofluids, is of great challenge due to the extremely low concentration of analytes, background interference and limited sample amount. To achieve increased sensitivity, a variety of purification and separation techniques have been employed prior to MS analysis.1–8 Capillary electrophoresis (CE) is of great potential in MS-based proteomics and peptidomics.9–12 In addition to its high separation efficiency, it features low sample consumption (nano-liter volume) and fast separation, which makes the technique suitable for the analyses of complex samples with limited amounts.13–15 While both sheath flow-assisted and sheathless CE have been developed for coupling to electrospray ionization (ESI) MS,16–19 their offline coupling to matrix-assisted laser desorption/ionization (MALDI) MS provides an alternative way with better tolerance to impurities as well as increased flexibility.20 Recently, numerous research groups have reported the development of CE-MALDI MS for proteomics, peptidomics and metabolomics studies.21–24 In order to achieve enhanced MS signal, CE-MALDI interface is considered to be critical. A common offline coupling design employs discrete fraction collection, which causes loss of electrophoretic resolution.12 To address this limitation, some early studies attempted to reduce time intervals for fraction collection.25–27 Zhang et al. developed a continuous deposition method for offline CE-MALDI coupling.28 A CE trace was collected on a cellulose membrane pre-coated with matrix, and MALDI mass spectra were recorded by manually moving the MALDI target. This work represented the first attempt at continuous CE collection, but the data acquisition rate was low due to manual operation. In addition, since CE ground was connected on the target, current breakdown could be an issue due to poor connection. Karger and co-workers later reported off-line CE-MALDI interface using either tape29 or stainless-steel MALDI plate30. Their interface features a liquid junction to transfer CE samples to deposition capillary under a vacuum chamber. Fast separation and high sensitivity have been achieved, but the wide liquid junction could be a potential interference to CE separation, and fine control of vacuum is necessary to avoid morphology change and the formation of discrete spots on the stainless-steel plate which repels water.
Introduced by Caprioli et al. in 1997, MALDI mass spectrometric imaging (MSI) has been widely applied to investigate the molecular distribution of various compounds especially from biological tissues.31–33 With the micrometer-scale spatial resolution, MSI has an inherent great potential in coupling with micro-scale separation method such as CE. On one hand, imaging enables “continuous” collection of CE flow which preserves electropherogram resolution with the capability of data analysis based on selected mass or migration time. On the other hand, CE provides separation and quantitation ability for MSI. Based on early attempts to integrate separation techniques with MSI, 28, 34 two recent reports described the coupling of either CE or LC separation to MSI.35–36 Our group reported an original design of an offline CE-MSI interface with etched grooves on the MALDI plate that allow MALDI MSI experiments to be conducted.35 CE separation and quantitation have been demonstrated with 10 spiked peptide standards, showing the capability of CE-MSI in separation and quantitation of peptides. However, more in-depth studies in quantifying complex biological samples are limited by severe sample diffusion and peak splitting issues. These limitations were caused mainly by the poor homogeneity of the matrix trace in the etched grooves on the customized MALDI plate, and the way that the sheath flow was introduced via an un-protected fracture on the capillary which interrupted CE separation. Weidner and Falkenhagen reported another interface by coupling LC with MSI for the study of two polymers.36 A home-built electrospray deposition interface was applied to the capillary outlet in order to collect LC eluent spray on the MALDI plate, and matrix was introduced prior to the electrospray deposition interface by using a T-connector. Although intense imaging signal can be observed, samples were sprayed to a large area on the MALDI plate which greatly reduced spatial resolution that several-minute peak widths were observed, and the “matrix sheath flow” resulted in considerable interruption and diffusion. These potential drawbacks could limit its further application to quantitative analysis of low-abundance compounds of interest.
Here we report a significantly improved novel pressure-assisted CE-MSI (PACE-MSI) platform and the first successful quantitation of complex biological samples using CE-MSI. Compared with previously reported platforms, PACE-MSI addresses the problems of severe sample dilution and diffusion, peak splitting and low spatial resolution, and is able to generate highly homogenous CE trace for MSI analysis with excellent reproducibility. By integrating PACE and imaging, electrophoretic resolution is preserved which leads to improved quantitation accuracy. Furthermore, the overlapped peaks from the complex samples can be easily analyzed as separated colored regions on the MS images. These features result in enhanced MS signal and a significantly increased number of peptides being detected and accurately quantified from complex mixtures, highlighting the potential of the PACE-MSI platform for comparative proteomics and peptidomics studies.
EXPERIMENTAL SECTION
Chemical and materials
Acetic acid, ammonium hydroxide, acetone, acetonitrile, methanol, ammonium bicarbonate and urea were purchased from Fisher Scientific (Pittsburgh, PA). α-cyano-4-hydroxycinnamic acid (CHCA), trifluoroacetic acid (TFA), formic acid (FA), iodoacetamide (IAA) and bovine serum albumin (BSA) were from Sigma-Aldrich (St. Louis, MO). Sequencing grade modified trypsin and D/L-dithiothreitol (DTT) were from Promega (Madison, WI). Fused-silica capillary with 75 μm i.d. and 190 μm o.d. was purchased from Polymicro Technologies (Phoenix, AZ). Millipore C18 Ziptip column was used for sample cleaning, and all water used in this study was doubly distilled on a Millipore filtration system (Bedford, MA).
Animal dissection and sample preparation
The blue crabs (Callinectes sapidus) were purchased from a local grocery store and kept in an artificial seawater tank at 10–12 °C without food. The dissection procedure has been previously described.37 Briefly, the crabs were anesthetized with ice for 15–30 min before dissection. The pericardial organs (PO) were dissected in physiological saline, which consisted of 440 mM NaCl, 11mM KCl, 26 mM MgCl2, 13 mM CaCl2, 11 mM Trizma base, and 5 mM maleic acid in pH 7.45. The neuropeptides were extracted with cold acidified methanol, which consisted of methanol, water and acetic acid in the ratio of 90:9:1. Five POs were combined and homogenized in 50 μL acidified methanol in ice and centrifuged at 16,100×g for 15 min prior to harvesting the supernatants. The extraction was repeated for three times and the supernatants were then combined, dried and reconstituted with 10 μL of 0.1% TFA. Tryptic peptides were also prepared by digesting BSA following a previously reported procedure.38 Both the PO neuropeptide extracts and BSA tryptic peptides were desalted by Ziptip C18 column before isotopic formaldehyde labeling.
In solution formaldehyde labeling
After desalting, samples were eluted with 5 μL of 50% acetonitrile in 0.1% TFA. Two aliquots of 5 μL sample (BSA tryptic peptides or PO extract) with 1:1 or 1:10 concentration ratio were pairwise labeled with formaldehyde-H2 and formaldehyde-D2, respectively. Briefly, 1 μL of borane pyridine (C5H8BN, 120 mM in 10% methanol) was added to each aliquot, followed by 1 μL of formaldehyde-H2 (4% in water, v/v) or 1 μL of formaldehyde-D2 (4% in water, v/v). The vials were placed in 37 °C water bath for 15 min, followed by the addition of 1 μL of 0.2 M ammonium bicarbonate to quench the reaction. The two aliquots of the above labeled products were mixed, dried and reconstituted with 10 μL of CE buffer before analysis.
Fabrication and operation of PACE
A 65 cm long with 75 μm i.d./190 μm o.d. fused-silica capillary was used for PACE. In order to enable sheathless connection to high voltage, cellulose acetate membrane-coated porous joints were made 3 cm to the outlet ends of the capillary modified from our previous designs.37–38 For the inlet end, a 0.6 mL plastic vial was filled with the same CE buffer with positive electrode and capillary being inserted. It was raised 25 cm higher than the outlet end to initiate siphoning as the driving force for CE flow collection. CE power was supplied by TriSep-2100 HV power supplier from Unimicro Technologies (Pleasanton, CA). A high voltage of 9 kV was applied and lasted for 6 s to load sample, while 10 kV was employed for CE running. For regular offline PACE-fraction-MALDI MS, the CE flow was collected on the MALDI plate every 1 min for 25 min. For PACE-MSI, CE flow was collected through the PACE-MSI interface described below.
PACE-MSI interface
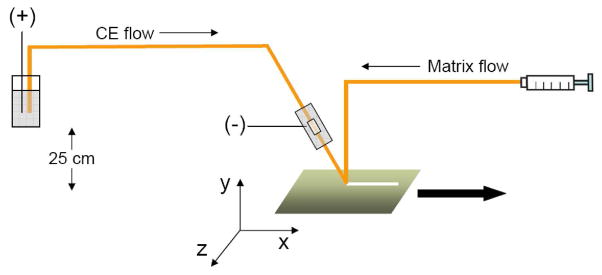
A commercially available ground stainless steel MALDI plate (Bruker Daltonics, Bremen, German) was used. Matrix flow (10 mg/mL CHCA in 50% ACN/0.1%TFA) was delivered with a 75 μm i.d./190 μm o.d. fused-silica capillary at 0.7 μL/min by using a syringe pump (Pump 11 Elite, Harvard Apparatus, Holliston, Massachusetts). The CE flow and matrix flow were mixed at the tips of two capillaries, which gently touched on the MALDI plate surface (Figure 1 and Figure 2A). With the MALDI plate moving along x-axis at a speed of 4.5 mm/min, which equaled to the distance between two spots on the MALDI plate, a straight and homogenous trace can be deposited on the plate surface. A lamp was used during fraction collection to facilitate the co-crystallization of CE fraction and matrix and reduce sample diffusion. A total of 23 minutes of CE fraction (from 2 min to 25 min after electrophoresis was initiated) was collected on the MALDI plate, which generated exactly 1 minute fraction between two spots on the MALDI plate.
Figure 1. The setup of the pressure-assisted capillary electrophoresis coupled to mass spectrometric imaging (PACE-MSI) interface.
Matrix flow is controlled by a syringe pump and delivered at a flow rate of 700 nL/min. CE flow rate is determined by the electroosmotic flow as well as height difference between capillary inlet and outlet along y-axis. Matrix and CE fraction are mixed at the tips of two capillaries, and collected immediately on the surface of the ground stainless steel MALDI plate. The MALDI plate is controlled mechanically and moves along the x-axis to the direction shown with arrow. A straight and uniform trace is deposited on the MALDI plate and is further analyzed by MALDI MS imaging.
Figure 2. The PACE-MSI interface and mixed trace on the MALDI sample plate.
A: CE flow and matrix flow are delivered separately and mixed at the capillary tips on the surface of a ground stainless steel MALDI plate. B and C: a 1:1 labeled peak pair at m/z 1356.6/1360.6 with identical peak intensities and migrations are observed in MS imaging when coupled with PACE.
MALDI MS Imaging
The MS images of separated BSA tryptic peptides and blue crab PO extracts were acquired with Autoflex III MALDI-TOF/TOF (Bruker Daltonics, Bremen, Germany) equipped with 200 Hz Smartbeam II laser. The positive reflectron mode was adopted with the following parameters: ion source 1 voltage 19.00 kV, ion source 2 voltage 16.62 kV, reflector 1 voltage 20.90 kV, reflector 2 voltage 9.64 kV and lens voltage 8.70 kV. For control samples by direct MALDI-TOF MS analysis, 500 shots were accumulated for each control spot. For MS imaging, approximately 0.8 mm× 60 mm regions of the CE trace, including all separated peptides, were selected. The step size for each pixel was set at 100 μm on the x-axis ×100 μm on the y-axis, and 200 shots were accumulated in a random mode for each pixel. The results were processed with the FlexImaging software from Bruker Daltonics.
RESULTS AND DISCUSSION
Evaluation of the PACE interface
We previously designed two CE setups for offline coupling with MALDI MS, 15,35 but the interface would either require negative mode with long separation time or need pressure from sheath flow as driving force that could result in loss of sensitivity and interruption to CE separation. Here, a new PACE setup is designed especially for coupling to MS imaging with three key features: (1) Sheath-free features with a membrane-covered fracture near the outlet end of the capillary, so that no sheath flow or liquid junction is needed as reported by other groups23, 29–30, and a narrow CE trace can be generated directly on MALDI plate without the need of spray or vacuum chamber30, 36. (2) Stable CE flow by employing siphoning as part of the driving force. We previously reported that siphoning, initiated by a pressure, can be used for precise control of the CE flow rate.15 It is estimated that a stable CE flow at approximately 500 nL/min can be achieved with a 25 cm height difference between capillary inlet and outlet, which is suitable for direct mixing with matrix at a similar flow rate. (3) Faster separation for trace-level peptides from complex samples compared to our previous CE-MALDI interface design. CE flow rate is determined by a combination of electroosmotic flow (EOF) and siphoning, which is controlled by adjusting voltage and height difference. It has been demonstrated that by using 10 kV as CE voltage, separation can be achieved when coupling to MSI with most neuropeptides being separated between 8–20 min, in contrast to 50 min separation time in our previous report.15
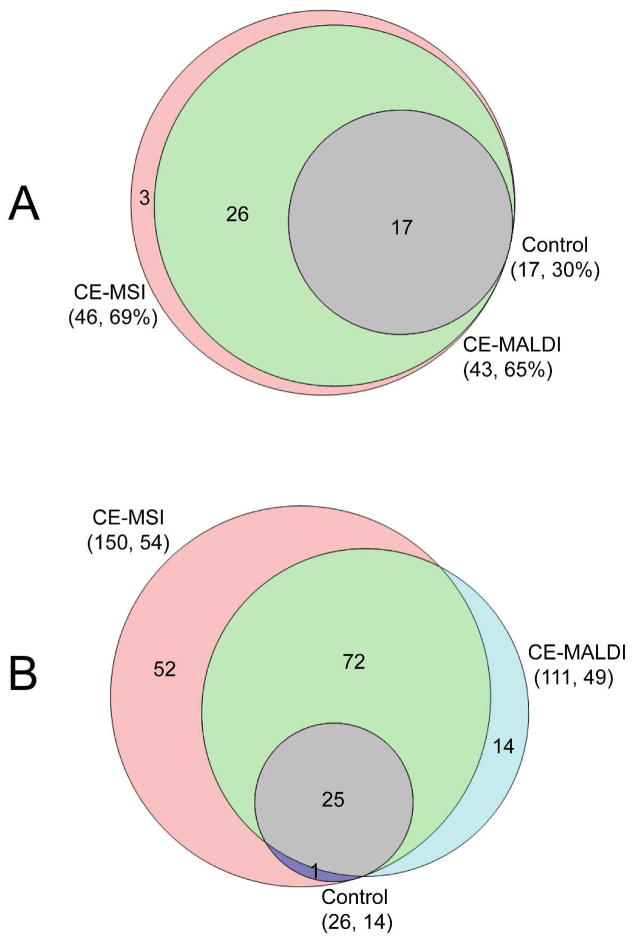
We have coupled PACE to MALDI-TOF/TOF with discrete spot collection for evaluation of the PACE system. BSA tryptic peptides and the blue crab PO extracts were firstly labeled with isotopic formaldehyde in a ratio of 1:1 (formaldehyde-H2: formaldehyde-D2), and were then loaded onto the CE capillary. Individual mass spectra collected from 13 to 17 min, with tryptic peptides being effectively separated after PACE, were shown in Figure S1. It was found that compared with the un-separated control sample by direct MALDI MS analysis, PACE gave rise to much higher peak intensities and significantly enhanced signal-to-noise ratios (S/N) in mass spectra, resulting in an increased number of peptides being detected. As shown in Figure 3, by coupling PACE with MALDI MS, 43 BSA tryptic peptides and 111 putative peptides were detected, highlighting the capability of PACE system for complex peptide mixtures in addition to the unique features for coupling with MSI.
Figure 3. Comparison of the number of peptides detected via MALDI MS by PACE-MSI, regular CE and un-separated control sample.
A: Number of BSA tryptic peptides detected in the mass range from m/z 500 to 2500. The performance is shown as numbers of peptide detected and protein sequence coverage in the parentheses. B: Number of putative neuropeptides from the blue crab PO detected in the mass range from m/z 500 to 2500. The performance is shown as the number of peptides detected and the number of peptides previously identified in parentheses.
Design and Evaluation of the PACE-MSI interface
When coupling CE with MSI, it is critical that the matrix and CE flow shall not interfere with each other, and the CE trace on the MALDI plate surface should be continuous and homogenous to achieve high separation resolution while minimizing diffusion. As shown in Figure 2A, in the PACE-MSI system, CE flow and matrix flow were individually delivered without interference, and they were mixed and collected on the surface of a ground stainless steel MALDI plate directly without grooves. With a continuous and highly uniform trace, 1:1 isotopically labeled peptide peak pairs show identical intensities and distributions, which enable accurate quantitative analysis (Figure 2B and 2C). We were able to set the step size of MALDI imaging to as small as 50 μm ×50 μm, though 100 μm ×100 μm pixel sizes were utilized to shorten experimental time. With the MALDI plate moving at 4.5 mm/min, each pixel covers 1.3 seconds of CE fraction along x-axis. Thus for a 30–40 s wide CE peak, which is commonly observed for peptides being separated from complex biological samples, more than 20 pixels can be covered for each peak. For the y-axis, 5–6 rows of pixels can be covered with track width at around 0.8 mm, which enables a comprehensive coverage of spatial distribution of complex peptides.
The sensitivity of the PACE-MSI system was tested with peptide standards. Two standard peptides (m/z 1046.54 DRVYIHPF and m/z 1533.86 PPPPPPPPPPPPPPR) were dissolved in CE buffer at 125 fmol, and approximately 20 nL (2.5 fmol) was loaded for separation. As shown in Figure S2, each peptide peak is covered by more than 100 imaging pixels, which indicates that on average lower than 25 amol of peptide can be detected from each pixel. Different from regular CE-MALDI coupling, current MSI software will only give imaging signal without a “real” peak or electropherogram. However, the image positions accurately reflect CE migration times and image intensities can be used for relative quantitation.
We also validated the reproducibility of the PACE-MSI interface by investigating the migration time of representative peptides as reflected in MS images. Nine BSA tryptic peptide peaks with representative migration times were selected from the mass spectra generated by both regular PACE-fraction-MALDI MS and PACE-MSI analysis, and 4 repeating experiments were performed in 4 days for PACE-MSI with the same sample and CE condition as used for regular PACE-fraction-MALDI MS analysis. As shown in Table S1, similar migration times among 4 repeating experiments were observed with RSDs among 1.3%–3.5%, indicating good reproducibility of the PACE-MSI interface.
The PACE-MSI interface has been applied to the analysis of both BSA tryptic peptides and neuropeptides extracted from the blue crab PO. As shown in Figure 3, a regular PACE-MALDI MS with discrete spot collection greatly increased MS peak intensities and S/N ratios especially for neuropeptides from complex samples compared with direct MALDI MS analysis. However, PACE-MSI features continuous collection and results in the greatest number of peptides being detected in both BSA tryptic digests and the blue crab PO extracts compared with regular PACE-MALDI coupling. When tested with BSA tryptic peptides, on average 45±2 peptides were detected with PACE-MSI, compared with 42±2 peptides being identified from PACE-MALDI TOF/TOF through discrete fraction collection (n=3). Lists of all BSA tryptic peptides and neuropeptides identified using the above methods can be found in Table S2 and Table S3 in the supporting information. Although the current imaging software can not provide peak information against migration time to generate electropherogram, we have manually reconstructed an electropherogram by recording the change of image intensities among pixels. As shown in Figure S3, the peaks of 10 representative neuropeptides from PO extracts have been simulated and reconstructed in an electropherogram.
Evaluation of PACE-MSI for peptide quantitation
Formaldehyde labeling has been considered to be a highly efficient mass-difference quantitation with fast reaction and low cost. 39 This technique labels samples with light formaldehyde (FH2, +28.03 Da for each incorporated label) and heavy formaldehyde (FD2, +32.05 Da for each incorporated label), generating a 4.02 Da mass difference between each peak pair. Here we report the first use of CE-MS imaging to quantify trace-level analytes from biological samples. Both BSA tryptic peptides and neuropeptides extracted from the blue crab PO were labeled in 1:1 ratio of concentration (FH2:FD2), and combined for PACE-MSI, PACE-MALDI MS with discrete spot collection and direct MALDI MS analysis. Peak areas from MALDI-TOF/TOF were compared to the calculated FH2:FD2 ratios. For MS imaging, however, the FlexImaging software cannot provide peak information such as intensity or area. Ratios were estimated by calculating peak areas as triangles for the first three isotopic peaks. It has been found that in both cases for coupling to MALDI MS via discrete spot collection and coupling to MSI, PACE enabled accurate relative quantitation with less than 15% error compared to theoretical ratio of 1:1 when applied to complex peptides with wide dynamic ranges. An example of BSA tryptic peptide RHPEYAVSVLLR (m/z 1439.81), extracted from MS image signal, is shown in Figure S4A. In addition to 1:1 labeling, we also tested 0.1:1 labeling, in which circumstances errors were easily introduced to the system due to the addition of an extremely low amount of light-labeled sample. Although 1:6.0 to 1:6.5 ratios were calculated from un-separated controls, we have observed higher quantitation accuracy with light: heavy ratios ranging from 1:7.0 to 1:8.5 with PACE due to the reduction of interferences. An example for peptide RHPEYAVSVLLR (m/z 1439.81) is shown in Figures S4B and S4C. For 0.1:1 labeling experiment, a region of interest (ROI, highlighted in Figure S4C) is selected covering signals from both light (m/z 1467.8) and heavy (m/z 1471.8) labeled peaks. By measuring averaged peak areas, a ratio of 1:7.3 is estimated, compared with 1:7.2 by PACE-MALDI MS with discrete spot collection and 1:6.4 by direct MALDI MS analysis. Development of imaging software is highly desirable for extending the current quantitative analysis by PACE-MSI to even larger scale analysis.
Analysis of trace-level analytes from complex samples with PACE-MSI
Compared with regular offline CE-fraction-MALDI MS, a unique feature of PACE-MSI is the ability to convert mass spectral peaks to image signals, which enables data analysis based on targeted mass or migration time for studying overlapped peaks in mass spectra of complex samples. By selecting different imaging regions, separated peaks can be displayed in individual mass spectra for qualitative and quantitative analysis when overlapping peaks cannot be fully resolved by a mass spectrometer, reducing reliance on ultra-high resolution mass analyzer for complex sample analysis. Alternatively, this improvement in separation of overlapping peaks from complex mixtures could yield even greater peak capacity and peptidome coverage when high resolution mass spectrometer is available.
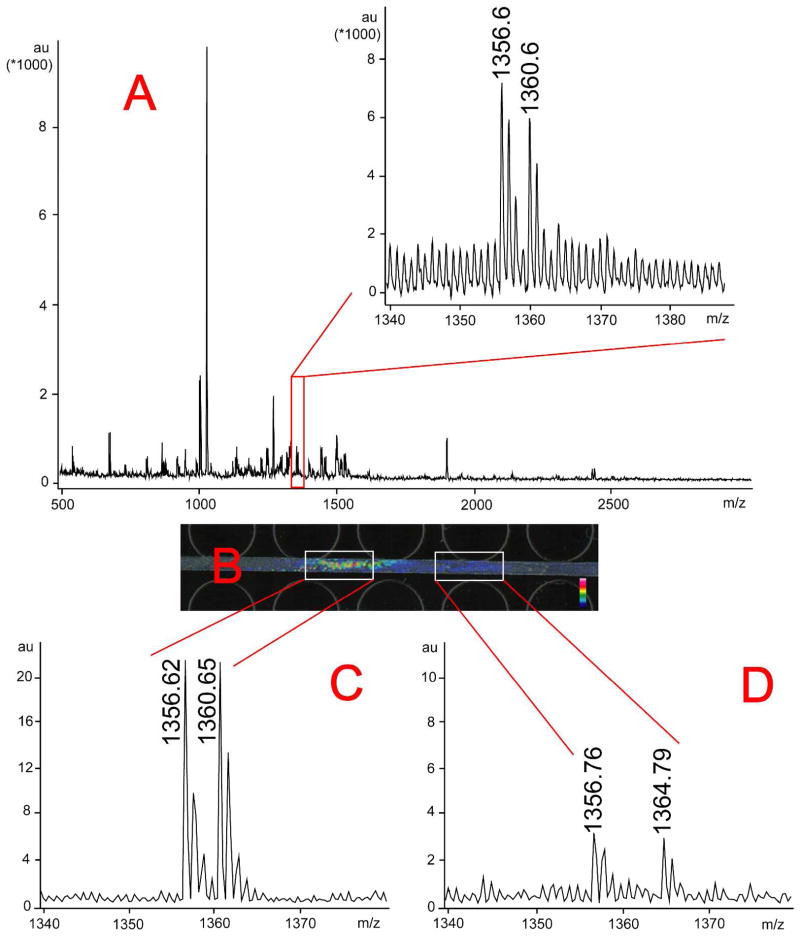
Figure 4 shows how this feature can be utilized for the analysis of overlapped peaks from a complex sample. With direct analysis of un-separated, isotopically labeled (in 1:1 ratio) blue crab PO extract using MALDI-TOF/TOF, a peak pair with m/z around 1356.6/1360.6 was observed in the mass spectrum (Figure 4A). This peak pair was considered as a neuropeptide candidate based on the mass difference, but its S/N ratio was low and their labeled ratio was higher than 1:1. With PACE-MSI, by selecting the mass range from m/z 1356.1 to m/z 1357.1, we were able to observe two separated imaging signal regions with different signal intensities as shown in Figure 4B. By selecting each region, we were able to display the mass spectrum for the selected region and observed two different peak pairs m/z 1356.62/1360.65 and m/z 1356.76/1364.79. The peak intensity for m/z 1356.76/1364.79 was too low to be detected from un-separated control sample, and the overlapped light labeled peaks caused the observed peak intensity ratio of m/z 1356.7/1360.7 being higher than expected 1:1 ratio in the mass spectrum from direct analysis. However, with PACE-MSI we were able to distinguish the two peak pairs with highly accurate quantitation at almost 1:1 ratios (ratios as shown in Figures 4C and 4D), which provides a promising tool for the analysis of complex samples. Compared with regular offline CE-fraction-MALDI MS, no manual inspection of all mass spectra is needed, and enhanced MS signal is observed with PACE-MSI.
Figure 4. PACE-MSI analysis of peak pair M/Z 1356.6/1360.6 from complex neuropeptide extracts of the blue crab PO.
A: Mass spectrum of the blue crab PO extract by direct MALDI MS analysis. A zoom-in inset showed peak pair m/z 1356.6/1360.6. B: Images of mass range m/z 1356.1–1357.1, with two regions being highlighted with white boxes. The two regions were separated from each other and exhibited significantly different signal intensities. C and D: Mass spectra corresponding to each region. Two different peak pairs, including m/z 1356.6/1360.6 and m/z 1356.7/1364.7, were detected from each region.
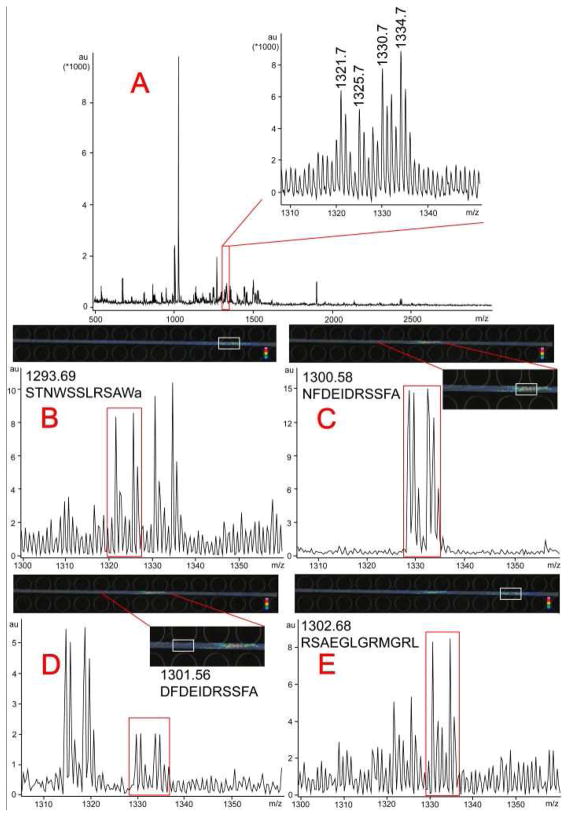
In addition to analyzing a single peak, it is more convenient to analyze peak clusters with low S/N ratios using PACE-MSI compared to CE-MALDI MS with discrete spot collection. As shown in Figure 5A, there are many low-intensity peaks with low S/N ratios in the mass range from m/z 1310 to m/z 1340. We could roughly identify two peak pairs at m/z 1321.7/1325.7 and 1330.7/1334.7, but they had unusual isotopic distributions and their peak intensity ratios deviated from the expected 1:1 ratio after labeling. Instead of searching mass spectra from regular offline CE-MALDI MS, by scanning the mass range of 1310–1340 with 1 Da wide mass filter, we could readily observe 4 different colored regions which were separated from background with PACE-MSI (Figures 5B–5E), including a B type allatostatin neuropeptide (STNWSSLRSAWamide, m/z 1293.69, for peak pair 1321.7/1325.7 in Figure 5B), a CPRP family neuropeptide (RSAEGLGRMGRL, m/z 1302.68, for peak pair 1330.7/1334.7 in Figure 5E), an orcokinin family peptide NFDEIDRSSFA (m/z 1300.58, for peak pair 1328.6/1332.6 in Figure 5C) and another orcokinin DFDEIDRSSFA (m/z 1301.56, for peak pair 1329.6/1333.6 in Figure 5D). Compared with regular CE-MALDI with discrete fraction collection, PACE-MSI makes it possible to analyze complex samples based on either selected mass or migration time, thus providing great benefits for peak identification and structure elucidation of analytes from complex samples in addition to its capability for more accurate quantitation.
Figure 5. PACE-MSI analysis of mass range M/Z 1320 to 1335 from complex neuropeptide extracts from the blue crab PO.
A: Mass spectrum of the blue crab PO extract by direct MALDI MS analysis. A zoom-in inset showed mass range 1320–1335 with peak pairs at m/z 1321.7/1325.7 and m/z 1330.7/1334.7 which were difficult to differentiate. B–E: By scanning the MS images of mass range m/z 1320 to 1335 with PACE-MSI, four neuropeptides as reflected in different image regions were observed with their corresponding mass spectra shown. They were well-separated with enhanced MS signals, and were accurately quantified with light: heavy labeled peak ratios around 1:1.
CONCLUSIONS
A newly established PACE-MSI platform has been applied for the first time to quantitative analysis of complex trace-level peptide extract. By integrating CE with MS imaging, overlapped peaks in a single mass spectrum can be accurately quantified and visualized in the images as separate regions, providing a novel method for analysis of complex analytes. Compared with regular offline CE-MALDI MS using discrete fraction collection, significantly enhanced MS signals and increased number of peptides have been observed for the new PACE-MSI platform with the ability of analyzing complex samples based on selected mass and migration time. These results demonstrate the potential of employing PACE-MSI in proteomics and peptidomics analysis. Based on this initial development, further optimizations of the system as well as new applications of PACE-MSI are currently underway including integrating it to multi-dimensional separations and applying PACE-MSI to the analysis of more complex samples beyond neuropeptides. Further improvements of MS imaging software will greatly facilitate the large-scale data analysis including peak identification and quantitation by this platform.
Supplementary Material
Acknowledgments
This work is supported in part by the National Science Foundation grant (CHE-0967784), and National Institutes of Health grants (1R01DK071801, 1R56DK071801). The authors thank Bruker Daltonics for graciously loaning the Autoflex III MALDI TOF/TOF mass spectrometer. We also thank PNNL and the OMICS.PNL.GOV website for providing Venn Diagram Plotter software. L. Li acknowledges an H.I. Romnes Faculty Research Fellowship. Z. Zhang thanks Claire Schmerberg from the Li lab for thoughtful suggestions on the CE-MSI interface design.
Footnotes
Supporting Information. Additional information as noted in the text. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Gatlin CL, Eng JK, Cross ST, Detter JC, Yates JR. Anal Chem. 2000;72:757–763. doi: 10.1021/ac991025n. [DOI] [PubMed] [Google Scholar]
- 2.Wang T, Ma J, Zhu G, Shan Y, Liang Z, Zhang L, Zhang Y. J Sep Sci. 2010;33:3194–3200. doi: 10.1002/jssc.201000324. [DOI] [PubMed] [Google Scholar]
- 3.Perry M, Li Q, Kennedy RT. Anal Chim Acta. 2009;653:1–22. doi: 10.1016/j.aca.2009.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fang X, Wang W, Yang L, Chandrasekaran K, Kristian T, Balgle BM, Lee CS. Electrophoresis. 2008;29:2215–2223. doi: 10.1002/elps.200700609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lapainis T, Sweedler JV. J Chromatogr A. 2008;1184:144–158. doi: 10.1016/j.chroma.2007.10.098. [DOI] [PubMed] [Google Scholar]
- 6.Wang F, Han G, Yu Z, Jiang X, Sun S, Chen R, Ye M, Zou H. J Sep Sci. 2010;33:1879–1887. doi: 10.1002/jssc.200900718. [DOI] [PubMed] [Google Scholar]
- 7.McLaren DG, Chen DDY. Anal Chem. 2004;76:2298–2305. doi: 10.1021/ac0350460. [DOI] [PubMed] [Google Scholar]
- 8.Zhao L, Qin H, Wu R, Zou H. J Chromatogr A. 2012;1228:193–204. doi: 10.1016/j.chroma.2011.09.051. [DOI] [PubMed] [Google Scholar]
- 9.Kasicka V. Electrophoresis. 2010;31:122–146. doi: 10.1002/elps.200900442. [DOI] [PubMed] [Google Scholar]
- 10.Herrero M, Ibanez E, Cifuentes A. Electrophoresis. 2008;29:2148–2160. doi: 10.1002/elps.200700404. [DOI] [PubMed] [Google Scholar]
- 11.Fonslow BR, Yates JR. J Sep Sci. 2009;32:1175–1188. doi: 10.1002/jssc.200800592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simpson D, Smith RD. Electrophoresis. 2005;26:1291–1305. doi: 10.1002/elps.200410132. [DOI] [PubMed] [Google Scholar]
- 13.Hu S, Dovichi NJ. Anal Chem. 2002;74:2833–2850. doi: 10.1021/ac0202379. [DOI] [PubMed] [Google Scholar]
- 14.Kraly J, Fazal MA, Schoenherr RM, Bonn R, Harwood MM, Turner E, Jones M, Dovichi NJ. Anal Chem. 2006;78:4097–4110. doi: 10.1021/ac060704c. [DOI] [PubMed] [Google Scholar]
- 15.Wang J, Ma M, Chen R, Li L. Anal Chem. 2008;80:6168–6177. doi: 10.1021/ac800382t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moini M. Anal Bioanal Chem. 2002;373:466–480. doi: 10.1007/s00216-002-1283-1. [DOI] [PubMed] [Google Scholar]
- 17.Ding J, Vouros P. Anal Chem. 1999;71:378A–385A. doi: 10.1021/ac9904415. [DOI] [PubMed] [Google Scholar]
- 18.Moini M. Anal Chem. 2001;73:3497–3501. doi: 10.1021/ac010189c. [DOI] [PubMed] [Google Scholar]
- 19.Edwards JL, Chisolm CN, Shackman JG, Kennedy RT. J Chromatogr, A. 2006;1106:80–88. doi: 10.1016/j.chroma.2005.08.082. [DOI] [PubMed] [Google Scholar]
- 20.Williams BJ, Russell WK, Russell DH. Anal Chem. 2007;79:3850–3855. doi: 10.1021/ac062395w. [DOI] [PubMed] [Google Scholar]
- 21.Snovida SI, Chen VC, Krokhin O, Perreault H. Anal Chem. 2006;78:6556–6563. doi: 10.1021/ac060738k. [DOI] [PubMed] [Google Scholar]
- 22.Amantonico A, Urban PL, Zenobi R. Analyst. 2009;134:1536–1540. doi: 10.1039/b907039g. [DOI] [PubMed] [Google Scholar]
- 23.Page JS, Rubakhin SS, Sweedler JV. Anal Chem. 2002;74:497–503. doi: 10.1021/ac0156621. [DOI] [PubMed] [Google Scholar]
- 24.Wang J, Zhang Y, Xiang F, Zhang Z, Li L. J Chromatogr, A. 2010;1217:4463–4470. doi: 10.1016/j.chroma.2010.02.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vannatta MW, Whitmore CD, Dovichi NJ. Electrophoresis. 2009;30:4071–4074. doi: 10.1002/elps.200900414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Busnel JM, Josserand J, Lion N, Girault HH. Anal Chem. 2009;81:3867–3872. doi: 10.1021/ac900128q. [DOI] [PubMed] [Google Scholar]
- 27.Ojima N, Shingaki T, Yamamoto T, Masujima T. Electrophoresis. 2001;22:3478–3482. doi: 10.1002/1522-2683(200109)22:16<3478::AID-ELPS3478>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 28.Zhang H, Stoeckli M, Andren PE, Caprioli RM. J Mass Spectrom. 1999;34:377–383. doi: 10.1002/(SICI)1096-9888(199904)34:4<377::AID-JMS778>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 29.Preisler J, Hu P, Rejtar T, Karger BL. Anal Chem. 2000;72:4785–4795. doi: 10.1021/ac0005870. [DOI] [PubMed] [Google Scholar]
- 30.Rejtar T, Hu P, Juhasz P, Campbell JM, Vestal ML, Preisler J, Karger BL. J Proteome Res. 2002;1:171–179. doi: 10.1021/pr015519o. [DOI] [PubMed] [Google Scholar]
- 31.Caprioli RM, Farmer TB, Gile J. Anal Chem. 1997;69:4751–4760. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- 32.Heeren RMA, Smith DF, Stauber J, Kükrer-Kaletas B, MacAleese L. J Am Soc Mass Spectrom. 2009;20:1006–1014. doi: 10.1016/j.jasms.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 33.Amstalden van Hove ER, Smith DF, Heeren RMA. J Chromatogr A. 2010;1217:3946–3954. doi: 10.1016/j.chroma.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 34.Zhang H, Caprioli RM. J Mass Spectrom. 1996;31:1039–1046. doi: 10.1002/(SICI)1096-9888(199609)31:9<1039::AID-JMS398>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 35.Wang JH, Ye H, Zhang Z, Xiang F, Girdaukas G, Li L. Anal Chem. 2011;83:3462–3469. doi: 10.1021/ac200708f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weidner SM, Falkenhagen J. Anal Chem. 2011;83:9153–9158. doi: 10.1021/ac202380n. [DOI] [PubMed] [Google Scholar]
- 37.Hui L, Cunningham R, Zhang Z, Cao W, Jia C, Li L. J Proteome Res. 2011;10:4219–4229. doi: 10.1021/pr200391g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Z, Wang J, Hui L, Li L. J Chromatogr, A. 2011;1218:5336–5343. doi: 10.1016/j.chroma.2011.05.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ning Z, Zhou H, Wang F, Abu-Farha M, Figeys D. Anal Chem. 2011;83:4407–4426. doi: 10.1021/ac200857t. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.