Abstract
Both the PI3K→Akt→mTOR and MAPK signaling pathways are often deregulated in prostate tumors with poor prognosis. Here we describe a new genetically-engineered mouse model of prostate cancer in which PI3K-Akt-mTOR signaling is activated by inducible disruption of PTEN, and ERK1/2 MAP kinase signaling is activated by inducible expression of a BRAFV600E oncogene. These tissue-specific compound mutant mice develop lethal prostate tumors that are inherently resistant to castration. These tumors bypass cellular senescence and disseminate to lymph nodes, bone marrow and lungs where they form overt metastases in ~30% of the cases. Activation of PI3K→Akt→mTOR and MAPK signaling pathways in these prostate tumors cooperate to upregulate c-Myc. Accordingly, therapeutic treatments with Rapamycin and PD0325901 to target these pathways, respectively, attenuate c-Myc levels and reduce tumor and metastatic burden. Together, our findings suggest a generalized therapeutic approach to target c-Myc activation in prostate cancer by combinatorial targeting of the PI3K→Akt→mTOR and ERK1/2 MAPK signaling pathways.
Keywords: Akt/mTOR signaling, MAP kinase signaling, genetically engineered mouse models, Braf, Myc
Introduction
While the majority of prostate cancers can now be treated effectively with surgery or radiation therapy, a subset of highly aggressive tumors progress to lethal metastatic disease (1). Unlike other cancers, it has proven difficult to classify prostate tumors into molecular subtypes that distinguish those that are indolent from those that are aggressive. Nonetheless, certain genes and/or molecular pathways have been associated with disease progression and/or poor prognosis. Among those associated with disease progression, the MYC oncogene is somatically amplified I n a subset of advanced prostate tumors (2, 3), while nuclear MYC protein is up-regulated in cancer progression even in the absence of gene amplification (4), although the underlying mechanisms by which it is activated remain unresolved.
Among the key signaling pathways associated with disease progression and poor outcome, various components of both the PI3-kinase→Akt→mTOR and RAF→MEK→ERK MAP kinase pathways are frequently co-activated in advanced prostate tumors and associated with poor outcome (5-8). Moreover, studies in cell culture and in vivo have demonstrated the functional significance of co-activation of PI3-kinase→Akt→mTOR and MAP kinase signaling for prostate cancer tumor progression (9, 10). It is well-established that a driving force for activation of PI3-kinase→Akt→mTOR signaling is inactivation of PTEN, which is prevalent in prostate tumors (11, 12). However, despite the prevalence of MAP kinase activation in a majority of advanced prostate tumors (6), the underlying mechanisms are less clear (13). In particular, the major upstream activators of MAP kinase signaling, namely RAF kinases, are rarely mutated in prostate tumors (14-16); however, activation of the RAF signaling pathway occurs in >90% of prostate tumors (6), and at least one mechanism by which BRAF is activated is through chromosomal rearrangement (17).
In the current study, we have investigated the functional consequences of co-activation of PI3-kinase→Akt→mTOR and MAP kinase signaling for prostate tumorigenesis and metastasis in vivo by generating a genetically engineered mouse (GEM) model having conditional loss-of-function of Pten combined with activation of oncogenic Braf. These mice develop lethal prostate tumors that metastasize to lymph nodes and lungs in approximately 30% of the cases. Notably, these prostate tumors display robust activation of PI3-kinase→Akt→mTOR and MAP kinase signaling, coincident with Myc pathway activation. Therapeutic treatment with Rapamycin and PD0325901, which target PI3-kinase→Akt→mTOR and MAP kinase signaling respectively, results in reduced tumor and metastatic burden while suppressing Myc pathway activation. Taken together, these findings suggest that an important consequence of activation of PI3-kinase→Akt→mTOR and MAP kinase signaling in advanced prostate cancer is Myc pathway activation, which therefore may be a target of therapeutic intervention.
Materials and methods
Generation and analyses of genetically-engineered mice
All experiments using animals were performed according to protocols approved by the Institutional Animal Care and Use Committee at Columbia University Medical Center. The Nkx3.1CreERT2/+ allele (18), a conditional allele for Pten (Ptenflox/flox) (19) and a lox-stop-lox BrafV600E allele expressing an inducible Braf allele (20) have been described. Mice were maintained on a mixed strain C57BL/6 and 129/Sv, and FVB background and breed to generate the full spectrum of genotypic combinations. Primers for genotyping are listed in Supplementary Table S1. For tamoxifen induction of Cre activity, mice were administered tamoxifen (Sigma Cat #T5648) in corn oil by IP injection (225mg/kg) or oral gavage (100 mg/kg) once daily for 4 consecutive days, at 2-3 months of age as in (18). Control mice received only corn oil by the same delivery method. Note that unless otherwise indicated the ages of the mice analyzed refer to time following delivery of tamoxifen. Where indicated, mice were androgen-ablated by surgical castration at 1 month after tumor induction. Following tumor induction, mice were monitored on a daily basis for body condition (i.e., muscle tone and weight) and sacrificed when their body condition score was <1.5, as per guidelines of the Institutional Animal Care and Use Committee.
For phenotypic analyses, mice were sacrificed, prostate tissues were collected, photographed, and weights were determined. The prostatic lobes (anterior, dorsolateral, and ventral) were collected individually and fixed in 10% formalin and paraffin-embedded, cryopreserved in Optimal Cutting Temperature (OCT) compound, or snap-frozen in liquid nitrogen. Full necropsy was done on each mouse analyzed and, in addition to prostate, a spectrum of tissues was collected into 10% formalin for evaluation of the metastatic phenotype, which included kidneys, liver, lungs, pancreas, small intestine, heart, lumbar lymph node, caudal lymph node, and submandibular lymph nodes. Bone marrow was collected by macrodissection of the femur followed by flushing the marrow with 1 ml of PBS using a 26G needle. DNA was made from the flushed cells and then Platinum Genotype Tsp DNA polymerase (Invitrogen) was used to amplify fragments up to 500 bp therefore avoiding amplification of the non-deleted Ptenf/f allele.
Pathological grading was done on hematoxylin-eosin (H&E) stained paraffin sections using the classification of Park et al (21). Immunohistochemical and immunofluorescence staining was done on 3 μm paraffin sections as described in (5, 18). A list of antibodies is provided in Supplemental Table S2. Senescence Associated Beta-Galactosidase (SA-β-Gal) staining was done using the Cellular Senescence Assay Kit (Chemicon International) on tissue embedded directly in OCT. Images were photographed using a Nikon Eclipse E800 microscope equipped with a Nikon DXM1200 digital camera. For quantification of immunostaining, a minimum of 3 independent sections from 3 independent mice (i.e., 9 sections) from slides that had been immunostained with Ki67, AR, the indicated phospho-proteins, cytokeratin 5 or cytokeratin 8, were counted as done previously (22). Statistical analyses were performed using a two-tailed T-test. GraphPad Prism software (Version 5.0) was used for all statistical analysis and to generate data plots.
For western blot analyses, total protein extracts were prepared from the dorsolateral prostate or prostate tumors in 1X RIPA buffer (0.1% SDS, 1.0% Deoxycolate-Sodium Salt, 1.0% Triton X-100, 0.15 M NaCl, 10 mM Tris-HCl (pH 7.5), 1 mM EDTA) with fresh 1% protease inhibitor (Roche Basel, Switzerland #1697498) and 1% phosphatase inhibitor (Sigma-Aldrich St. Louis, Missouri #P2850). Protein lysates (10 mg per lane) were resolved by SDS page electrophoresis, immunoblotted with primary antibody followed by the appropriate secondary antibody and visualized using the ECL Plus Western Blotting Detection Kit (GE Healthcare/Amersham Biosciences Piscataway, New Jersey, #RPN2132). Western Blot data were quantified using ImageJ (http://rsb.info.nih.gov/ij/) (23). Details of antibodies are provided in Supplementary Table S2.
Pre-clinical analyses of genetically-engineered mice
Rapamycin was purchased from LC labs (Catalog #R-5000, lots #ASW-105 and #ASW-109); PD325901 was a generous gift from Pfizer (Batch U). Rapamycin was delivered via IP at 10 mg/kg in 5.2% Tween 80, 5.2% PEG400. PD325901 was delivered via oral gavage at 10 mg/kg in 0.05% hydroxy-propyl-methylcellulose, 0.02% Tween 80. Rapamycin and/or PD0325901 (or vehicle) were delivered for 5 consecutive days with 2 days off for a period of four weeks. Alternatively, mice were treated with Rapamycin and/or PD0325901 (or vehicle) for 4 days and dissected within 6 hours after the last treatment. Mice were weighed daily and observed for signs of distress following dosing. Notably, none of the treatments resulted in appreciable weight loss exceeding 10%.
Gene expression analysis
Gene expression profiling analysis was done using total RNA isolated from prostate tissues of Nkx3.1CE2/+; Ptenf/f (n = 6), or Nkx3.1CE2/+; Ptenf/f; BrafCA/+ mice treated with vehicle (n = 3), or Nkx3.1CE2/+; Ptenf/f; BrafCA/+ mice treated with RAP+PD (n = 3). RNA was isolated using the MagMAX-96 total RNA isolation kit (Ambion), which was reverse-transcribed and biotin-labeled using the Illumina TotalPrep RNA Amplification kit (Ambion). The cDNA (1.5 mg) was hybridized on mouseWG-6 v2 BeadArrays (Illumina) using an iScan BeadArray scanner (Illumina). Data were loaded and normalized using IlluminaExpressionFileCreator version 2 and Illumina normalizer with collapse mode using the maximum of all the probe values for each gene and without background subtraction. The resulting datasets were preprocessed to remove probesets whose difference between maximum and minimum values were less than 100. Data were log transformed for GSEA analysis.
The gene signatures of Nkx3.1CE2/+; Ptenf/f; BrafCA/+ versus Nkx3.1CE2/+; Ptenf/f mouse prostate tumors and RAP+PD-treated versus vehicle-treated Nkx3.1CE2/+; Ptenf/f; BrafCA/+ mouse prostate tumors were defined using the Welch T-test as a list of genes ranked by their differential expression. To identify pathways commonly deregulated in vehicle-treated and RAP+PD-treated Nkx3.1CE2/+; Ptenf/f; BrafCA/+ mouse prostate tumors, enrichment of the differentially expressed gene signatures in human pathways was evaluated using GSEA (24). Using pathways collected in manually drawn, peer-reviewed biological pathway maps including REACTOME (25), KEGG (26), and BioCarta (http://www.biocarta.com/genes/allpathways.asp) databases with 1,000 gene label permutations (gene-sets). Enrichment of Myc-related pathways by GSEA was done using six published gene sets (27-32). Selected genes were validated by quantitative real-time PCR using SYBR green reagent (Qiagen) in the Realplex2 machine (Eppendorf). Primer sequences for real-time PCR are provided in Supplementary Table S1.
Results
Cooperation of BrafV600E activation and Pten inactivation in a mouse model of advanced prostate cancer
To investigate the role of Braf pathway activation in conjunction with Pten loss-of-function, we used a tamoxifen-inducible Cre (CreERT2) under the control of the Nkx3.1 promoter (18), a prostate specific homeobox gene whose haploinsufficient loss-of-function leads to prostate intraepithelial neoplasia (PIN) (33). This Nkx3.1CreERT2 knock-in allele simultaneously inactivates Nkx3.1 and drives tamoxifen-dependent Cre-mediated recombination specifically in the prostate epithelium (Supplementary Fig. S1), including a relevant cell of origin of prostate cancer (18). We crossed the Nkx3.1CreERT2 allele with a Pten floxed allele (19), or with a conditionally-activatable Braf (BrafCA) allele, which expresses oncogenic BRAFV600E in response to Cre activity (20) (Fig. 1; Supplementary Fig. S1). Notably, our current model, which conditionally expresses activated Braf in prostatic luminal cells, is distinct from a previously reported transgenic model, which expresses a tetracycline-regulated Braf via the tyrosinase promoter resulting in prostate tumors (34).
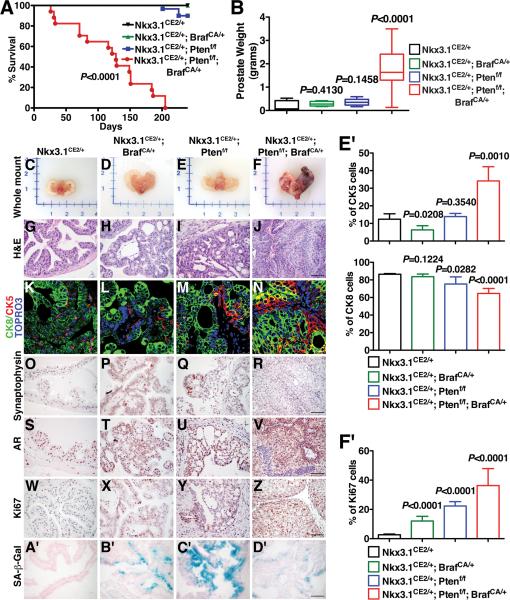
Figure 1.
Cooperation of Braf activation and Pten loss in prostate cancer. (A) Survival curve shows the percentage of mice of the indicated genotypes surviving after tumor induction. (B) Average wet weights of prostate tumors. (C-F) Whole mount images. (G-J) Representative H&E images of anterior prostate. (K-N) Immunofluorescence images show staining for cytokeratin 8 (CK8), which stains luminal epithelial cells, and cytokeratin 5 (CK5), which stains basal epithelial cells. Note that the prostate tumors are primarily luminal as evident by the robust staining for CK8, with more limited staining of CK5. (O-R) Immunohistochemical (IHC) staining for Synaptophysin. (S-V) IHC staining for AR in the intact mice. Note that the prostates from each of the models express nuclear AR. (W-Z) IHC staining for Ki67. (A’-D’) SA-β-Gal staining shows prominent senescence in the prostates from the Nkx3.1CE2/+; BrafCA/+ (NB) and Nkx3.1CE2/+; Ptenf/f (NP) mice, but not in the Nkx3.1CE2/+; Ptenf/f; BrafCA/+ (NPB) mice. (E’) Immunofluorescence staining index as measured by the percentage of the indicated CK5 and CK8 positive cells relative to total epithelial cells in the prostates of mice by genotypes as indicated. (F’) Average proliferation assessed by the number of Ki67-positive cells relative to total epithelial cells. Where indicated, p-values compare the experimental to the control (Nkx3.1CE2/+ prostate) and scale bars represent 100 μm, except in (K-N) where they represent 25 μm.
We found that mice lacking one allele of Nkx3.1, both alleles of Pten, and having an activatable Braf allele (i.e., Nkx3.1CreERT2/+; Ptenflox/flox; BrafCA/+ mice; hereafter denoted Nkx3.1CE2/+; Ptenf/f/; BrafCA/+ [NPB] mice) develop lethal prostate cancer 4 months after tamoxifen induction (i.e. by 6 months of age) (Fig. 1A; Supplementary Table S3). In contrast, mice lacking only Pten or having only an activatable Braf allele (denoted Nkx3.1CE2/+; BrafCA/+ [NB] mice or Nkx3.1CE2/+; Ptenf/f [NP] mice respectively) rarely develop lethal prostate tumors up to 2 years following tamoxifen induction (Fig. 1A; Supplementary Table S3). Indeed, by 4 months after tamoxifen induction, the prostate tumors from the NPB mice were large in size (~2-3 grams) and highly proliferative, compared with the NB or NP single mutants (Fig. 1B-F, W-Z, F’). Notably, cooperation of BRAFV600E activation and PTEN inactivation has also been observed in an analogous mouse model of melanoma (35, 36).
The progressive phenotype of the NPB mice was further evident by histopathological analyses, in which these mice displayed PIN by 2-4 weeks and invasive adenocarcinoma by 2-3 months after tamoxifen induction, which was considerably more aggressive than either the NB or NP single mutants (Fig. 1G-J; Supplementary Table S3). Nonetheless, the prostate tumors from the NPB mice, as well as the NB and NP single mutants, were primarily comprised of luminal epithelial cells, as evident by robust staining for cytokeratin 8, with significantly less staining for cytokeratin 5, a marker of basal epithelial cells and a few scattered neuroendocrine cells (Fig. 1K-R, E’). However, although the tumors in the NPB mice were comprised luminal cells, they displayed an expansion of CK5 positive cells NB or NP single mutants, as had also been observed in the Braf transgene model (34).
Considering that Braf pathway activation is most prominent in advanced prostate cancer in humans (6), we asked whether the NPB mice expressed androgen receptor and were sensitive to androgen depletion. We found that while the NPB prostates, as well as the NB and NP single mutants, each express nuclear androgen receptor (AR). (Fig. 1S-V). Furthermore, following androgen deletion by surgical castration, the NPB mice do not display significant tumor regression at 2 weeks following castration (Supplementary Fig. S2), suggesting that the tumors in the NPB mice are inherently resistant to castration. This contrasts with the NP mouse tumors, which are castration sensitive (37) as are the previously described Braf transgenic model, which is wild-type for Pten (34).
Consistent with this observation that the NPB tumors are inherently castration-resistant, another notable distinction of the NPB compound mutants relative to the NB and NP single mutants was the virtual absence in the NBP mice of SA-β-Gal staining, a marker of cellular senescence, which was in striking contrast to the robust SA-β-Gal staining in both single mutants (Fig. 1A’-D’). We have shown recently that one of the major difference among otherwise similar mouse prostate tumors is that the castration-resistant ones display little or no cellular senescence (37), therefore, this observation further supports the inherent castration-resistance of these NPB prostate tumors. As mentioned previously, the prostate tumors from the NPB mice were also much more highly proliferative than the NB or NP single mutants (Fig. 1WZ, F’), which is also consistent with their limited cellular senescence.
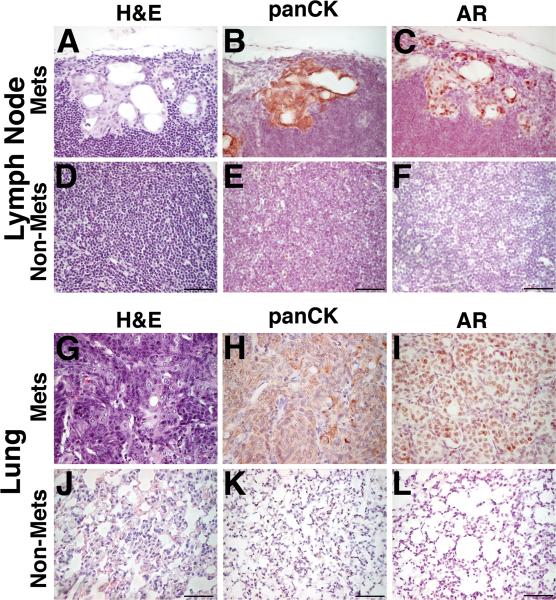
Finally, another striking distinction of the NPB prostate tumors is their propensity to metastasize to lymph nodes and lungs. In particular, NPB mice, but not NB or NP mice, displayed metastases to lungs as well as the lumbar lymph nodes in 30% of the cases (n=3/10) as evident by analyses of H&E as well as by staining for AR and Pan-Cytokeratin (Fig. 2; Supplementary Table S3). Notably, the NPB mice also displayed disseminated tumor cells in bone marrow (n=2/10), as detected by quantitative PCR (qPCR) (Supplementary Table S3). In summary, expression of oncogenic BrafV600E combined with Pten inactivation leads to advanced prostate cancer distinguished by its inherent castration-resistance, bypass of cellular senescence, and the occurrence of secondary metastases.
Figure 2.
Prostate tumors from Nkx3.1CE2/+; Ptenf/f; BrafCA/+ (NPB) mice form overt metastases to lungs and lymph nodes. (A-F) Representative H&E images show H&E staining and immunostaining with the indicated antibodies of lymph nodes metastasis as well as normal lymph nodes tissue. (G-L) Representative H&E images show H&E staining and immunostaining with the indicated antibodies of lung metastasis as well as normal lung tissue. The scale bars represent 100 μm.
BrafV600E activation combined with Pten inactivation is associated with elevated Myc activity
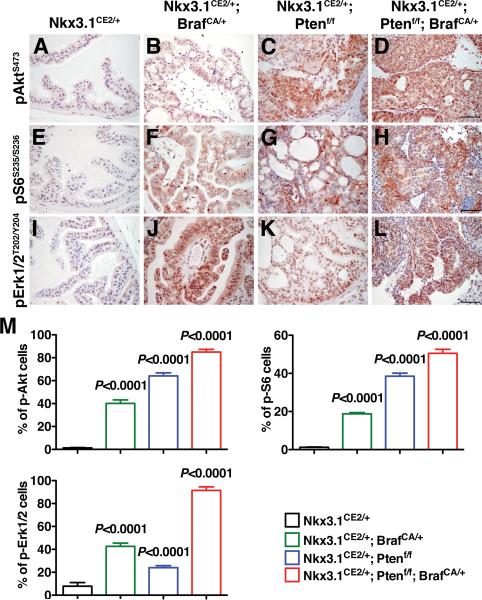
Next, we pursued biochemical and computational analyses to identify molecular pathways that contribute to the advanced prostate cancer phenotype of the NPB mice. Using immunohistochemical and western blotting analyses, we found that prostate tumors from these mice displayed prominent activation of multiple components of the PI3-kinase→Akt→mTOR and MAP kinase signaling pathways, as well as activated Ras signaling (Fig. 3; Fig. 4). In most cases, the NPB prostates displayed more robust of pathway activation relative to either NB or NP single mutant, as exemplified by activation of the Ras pathway markers, p38 and p-GSK3 (Fig. 4C). However, some notable trends in the single mutants provide insights into the relative contribution of Braf activation versus Pten loss for prostate tumorigenesis. For example, phosphorylation of 4E-BP1(Ser65) is abundant in the prostate tumors of the NB single mutant as well as the NPB mice, suggesting that Braf may directly influence this mTOR pathway component.
Figure 3.
Activation of kinase pathways in mouse prostate tumors. (A-L) Immunohistochemical staining for the indicated phospho-proteins. (M) Immunohistochemical staining index as measured by the percentage of the indicated phospho-proteins positive cells relative to total epithelial cells in the prostates of mice by genotypes as indicated. Note that the Nkx3.1CE2/+; Ptenf/f (NP) and Nkx3.1CE2/+; Ptenf/f; BrafCA/+ (NPB) prostates display activation (phosphorylation) of Akt, while the Nkx3.1CE2/+; BrafCA/+ (NB) and Nkx3.1CE2/+; Ptenf/f; BrafCA/+ (NPB) have activation of Erk. Where indicated, p-values compare the experimental to the control (Nkx3.1CE2/+ prostate) and scale bars represent 100 μm.
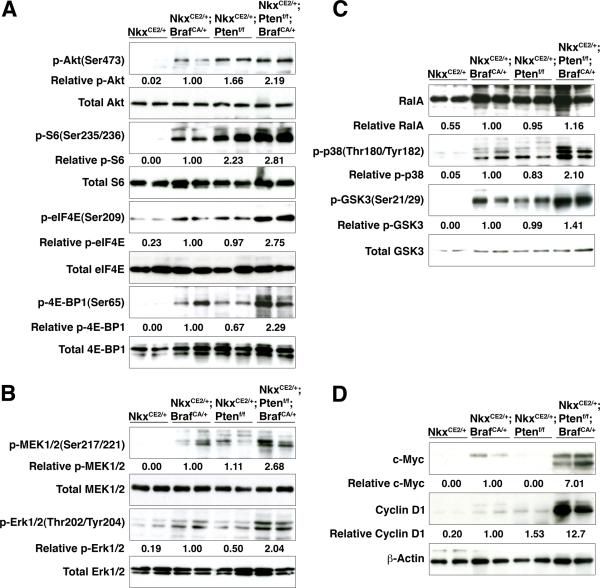
Figure 4.
Pathway analyses of mouse prostate tumors. Western blot analyses were done to assess activation of the PI3-kinase→Akt→mTOR pathway (A), MAP kinase signaling (B), RAS pathway activation (C), and expression of Myc and Cyclin D1 (D). Western blots were performed using total protein extracts prepared from dorsolateral prostate or prostate tumors from the indicated genotypes. Western Blots were quantified using ImageJ.
Notably, we also observed that c-Myc and Cyclin D1 were robustly up-regulated in the prostate tumors from the NPB mice, compared to those of the single mutants (Fig. 4D). Notably, Myc has been shown to be a target of Raf activation in other contexts (38-41), and its expression has been associated with bypass from cellular senescence (42, 43). Thus, the progressive phenotype of the NPB mice is associated with the activation of PI3-kinase→Akt→mTOR and MAP kinase pathways, as well as up-regulation of Myc and Cyclin D1.
We further investigated the molecular pathways that distinguish the NPB phenotype from NP phenotype using Gene Set Enrichment Analysis (GSEA) to identify biological processes that are altered in these prostate tumors by comparing the NPB mice with the NP single mutants (Supplementary Table S4, S5A, S5B). Among the significantly enriched categories identified were those corresponding to “cell cycle” (p < 0.001), “G1 pathway” (p = 0.001), “pathway in cancer” (p < 0.001) and “ERK pathway” (p = 0.004) (Supplementary Fig. S3; Supplementary Table S5A), consistent with our phenotypic analyses (see Fig. 1). Additional categories that were significantly enriched included “focal adhesion” (p < 0.001) and “extracellular receptor interaction” (p < 0.001) (Supplementary Fig. S3; Supplementary Table S5A).
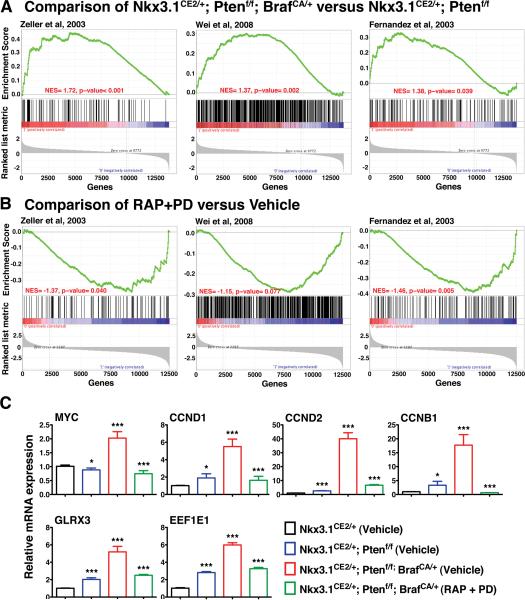
Considering our observation that Myc is up-regulated in the NPB relative to the NP mice (see Fig. 4D), we asked whether the gene signature corresponding to the NPB versus NP prostates was associated with up-regulation of the Myc pathway activity (Fig. 5A-C; Supplementary Table S6A). In particular, we compared the mouse prostate signature with six independent Myc pathway signatures, which were as follows: (i) genes up-regulated and promoters bound by MYC in human B-cells (32); (ii) genes whose promoters contain E-box motifs and whose expression is induced by Myc in neuroblastoma cells (30); (iii) genes identified as Myc target by ChIP analyses in human cancer cells (28); (iv) genes differentially expressed in human cancer cells expressing Myc (27); (v) genes up-regulated in B cell lymphoma tumors expressing MYC (31); and (vi) genes up-regulated in Burkitt's lymphoma cells expressing MYC (29). In comparison with each of these independent Myc signatures, the gene signature corresponding to the NPB versus NP mice was significantly enriched for up-regulation of the Myc pathway genes, as evident from the positive enrichment score in each case (Fig. 5A; Supplementary Table S6A). These observations provide further support our finding that Myc is activated in the NPB mouse prostate tumors.
Figure 5.
Enrichment of Myc pathway activation. (A) Gene Set Enrichment Analysis (GSEA) showing comparison the gene signature from the NPB (Nkx3.1CE2/+; Ptenf/f; BrafCA/+) versus the NP (Nkx3.1CE2/+; Ptenf/f) tumors with three independent human Myc pathway signatures (28, 30, 32). (Three additional Myc pathways signatures are summarized in Supplementary Table S6A.) The positive enrichment scores for the curve indicate an enrichment of the Myc pathway genes in the mouse signature genes. (B) GSEA comparing the drug response signature (i.e., NPB mice treated with RAP+PD versus vehicle) with three independent human Myc pathway signatures (28, 30, 32). The negative enrichment scores indicate enrichment of the under-expressed part of the drug response signature in the mouse prostate. (C) Real-time PCR validation of selected genes. Data are expressed as the fold change of mRNA relative to that of Nkx3.1CE2/+. The values are the means ± SD; ***P < 0.0001, ** P < 0.001, * P < 0.01. The * on Nkx3.1CE2/+; Ptenf/f; BrafCA/+ (RAP + PD) indicates the comparison between Nkx3.1CE2/+; Ptenf/f; BrafCA/+ (RAP + PD) and Nkx3.1CE2/+; Ptenf/f; BrafCA/+ (Vehicle). All other * indicate the comparisons with Nkx3.1CE2/+ (Vehicle).
Combination targeted therapy reduces tumor and metastatic burden, and alleviates Myc activation
Based on these phenotypic and molecular analyses, we reasoned that the NPB mice would provide an excellent in vivo model to study the consequences of therapeutic targeting of the PI3-kinase→Akt→mTOR plus MAP kinase signaling pathways, and to assess their relevance for Myc pathway activation. Toward this end, we performed preclinical studies using the combination of Rapamycin (RAP) to target mTOR signaling together with a MEK inhibitor (PD0325901) (PD) to target MAP kinase signaling. We have previously found that this RAP+PD combination was highly effective for treatment of prostate cancer in a mouse model based on germline inactivation of Nkx3.1 and Pten (5). Furthermore, combinatorial targeting of PI3-kinase→Akt→mTOR and MAP kinase signaling pathways has shown promising results in preclinical studies and clinical trials for several cancers, including prostate as well as lung and breast (5, 44-46), while targeting tumors driven by Braf activation combined with Pten loss using the same or similar agents has shown promise in preclinical studies of melanoma and thyroid cancer (35, 47).
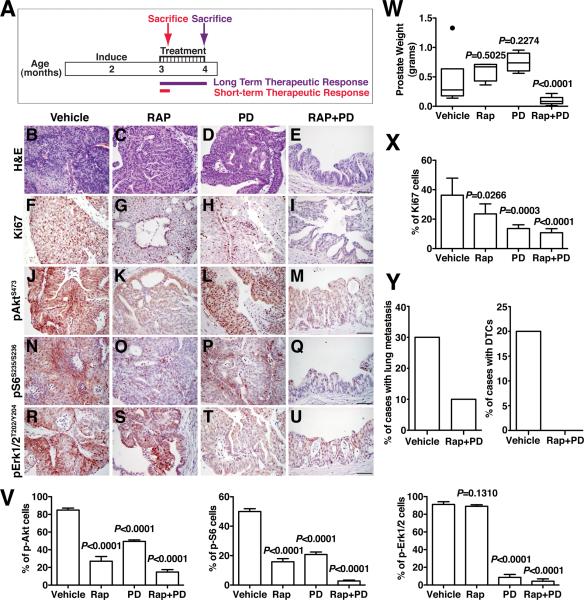
Therefore, we evaluated the consequences of RAP and/or PD for therapeutic intervention in the NPB mice in preclinical studies (Fig. 6A; Supplementary Table S7). We found that treatment with both RAP+PD had a significantly impacted on tumor burden as evident by inspection of several independent endpoints. In particular, the NPB mice displayed a significant (p < 0.0001) reduction in tumor weight after treatment (n = 10), as well as histological evidence of reduced tumor growth (n = 6) and a significant (p < 0.0001) reduction in cellular proliferation (n = 5) (Fig. 6B-I, W, X; Supplementary Table S7). In contrast to the robust efficacy of the RAP+PD combination, the single agents were only modestly effective, as we have observed previously (5), although PD0325901 alone displayed significant efficacy for inhibiting proliferation (Fig. 6F-I, X). Notably, in addition to the effects on tumor burden, the RAP+PD treatment also decreased metastatic burden, as evident by the reduced incidence of lung metastases (n=1/10) and disseminated tumor cells in the bone marrow (n=0/10) (Fig. 6Y; Supplementary Table S7).
Figure 6.
Combination therapy reduces tumor burden. (A) Design of preclinical studies utilizing combination therapy with Rapamycin (RAP) and PD0325901 (PD). Nkx3.1CE2/+; Ptenf/f; BrafCA/+ (NPB) mice were induced with tamoxifen at 2 months of age and drug treatment was initiated 1 month later and continued for 1 month, during which time mice were treated with vehicle or RAP and/or PD. Following cessation of treatment, mice were sacrificed (Long-term Response). For evaluation of immediate response to drug action, a cohort of mice were treated with vehicle or RAP and/or PD for 4 days and sacrificed within 6 hours after the last treatment (Short-term Response). Unless otherwise indicated the analyses show data for the Long-term Response. (B-E) Representative H&E images of mice treated with vehicle, RAP and/or PD as indicated. (F-I) Representative Ki67 immunostaining of prostate tissues treated as indicated. (J-U) Immunostaining for the indicated phospho-proteins. (V) Immunostaining index as measured by the percentage of the indicated phospho-proteins positive cells relative to total epithelial cells in the prostates of mice treated as indicated. (W) Average prostate weight for mice treated as indicated. (X) Proliferation index as measured by the percentage of Ki67 positive cells relative to total epithelial cells in the prostates of mice treated as indicated. (Y) The penetrance of lung metastasis and DTCs of mice treated as indicated. Where indicated, p-values compare the drug-treated to vehicle-treated groups, and scale bars represent 100 μm.
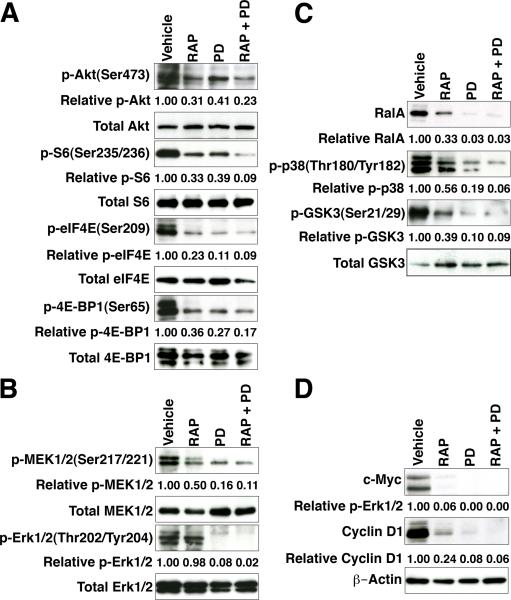
These phenotypic changes in tumor and metastatic burden were accompanied by reduced expression of the relevant targets, namely pAKT(Ser473), pS6(Ser235/236) and pERK(Thr202/Tyr204) (Fig. 6J-V). Furthermore, following just 4 days of RAP and/or PD treatment, we observed a significant reduction in activation of multiple components of the PI3-kinase→Akt→mTOR, MAP kinase, and Ras signaling pathways (Fig. 7A, B, and C). In particular, PI3-kinase→Akt→mTOR pathway components were reduced following RAP treatment and after the RAP+PD combination, while components of the MAP kinase and Ras pathways were down-regulated by treatment with PD as well as with the RAP+PD combination.
Figure 7.
Combination therapy suppresses Myc activation. Western blot analyses of tissues from Short-term Response mice show expression of PI3-kinase→Akt→mTOR (A), MAP kinase (B), and Ras (C) pathway markers, as well as Myc and Cyclin D1 (D) following drug treatment. Western blots were performed using total protein extracts prepared from dorsolateral prostate or prostate tumors from the indicated drug treatments. Western Blots were quantified using ImageJ.
Notably, expression of Myc and Cyclin D1 were reduced by treatment with the individual agents and were abolished by the RAP+PD combination (Fig. 7D), suggesting that inhibition of PI3-kinase→Akt→mTOR and MAP kinase signaling led to Myc down-regulation. To confirm this finding, we assessed whether genes associated with Myc activation that were up-regulated in the NPB mice (see Fig. 5A) were down-regulated in response to RAP+PD treatment. For this purpose, we generated expression profiles from prostate tumors from NPB mice that had been treated with RAP+PD or vehicle (Supplementary Table S8) and then used GSEA analyses to compare this signature to the same Myc gene signatures we had found to be enriched in the NPB versus NP prostate tumors (see Fig. 5A). In contrast to the strong positive enrichment of the Myc pathway signatures identified in the transition from NPB to NP mice, we found an inverse negative enrichment of these Myc pathway signatures when comparing the RAP+PD-treated to the vehicle-treated samples (compare Fig. 5A and Fig. 5B, Supplementary Table S6B). This observation was further confirmed by qPCR validation, focusing on Myc target genes involved in cell cycle progression, such as Cyclin D1, Cyclin D2, Cyclin B1 and glutaredoxin 3 (Fig. 5C). In particular, Myc targets that were up-regulated in the NPB prostate tumors were correspondingly down-regulated following treatment with RAP+PD (Fig. 5C). In summary, these findings show that combination therapy using Rapamycin and PD0325901 to target PI3-kinase→Akt→mTOR signaling and MAP kinase signaling results in a profound reduction in tumor burden, reduces the occurrence of metastases, and reverts Myc pathway activation.
Discussion
Here we describe a new mouse model of advanced prostate cancer based on inducible Pten inactivation combined with inducible expression of oncogenic BrafV600E, which leads to dysregulation of both PI3-kinase→Akt→mTOR and MAP kinase signaling. Our findings suggest that cancer progression that results from activation of Braf signaling together with loss of Pten is the consequence of several important features that distinguish these advanced tumors. In particular, the prostate tumors in the NPB mice are inherently castration-resistant and express robust levels of AR, are highly proliferative, display a bypass of cellular senescence, and most notably, metastases to lungs and the lymph node. Furthermore, our analyses have revealed that Myc is a major downstream effector of PI3-kinase→Akt→mTOR and MAP kinase signaling in these NPB prostate tumors, which can be suppressed by therapeutic interventions targeting these signaling pathways. Thus, our findings provide a new preclinical model for evaluating agents that target the PI3-kinase→Akt→mTOR and MAP kinase signaling pathways, as well as novel insights regarding Myc as a target for therapeutic intervention.
Since up-regulation of ERK1/2 MAP kinase signaling is prevalent in advanced prostate cancer, the role of RAF kinases, which are the major upstream effectors, has remained controversial. Although point mutation of BRAF does not play a major role in prostate tumorigenesis (14-16), activation of RAF→MEK→ERK signaling occurs in a majority of prostate tumors with poor prognosis (6) and at least one mechanism for its activation is via chromosomal translocation of the BRAF gene (17). Indeed, the absence of BRAF mutations in prostate cancer is perhaps not surprising, since prostate tumors are not typically characterized by mutational activation and are generally more indolent relative to other cancer types. Thus, a major future challenge will be to define how RAF→MEK→ERK signaling is activated in prostate cancer so that the appropriate targets for therapeutic intervention can be identified.
It is interesting to compare the consequences of PTEN inactivation combined with oncogenic BRAFV600E expression in melanoma versus prostate cancer. Similar to prostate cancer, combinatorial activation of PI3-kinase→Akt→mTOR signaling and MAP kinase signaling occurs frequently in melanoma and is associated with poor outcome (14, 48, 49), and genetically-engineered mouse models in which Pten and Braf are perturbed in melanoma have revealed their functional collaboration (35, 36). However, in striking contrast to prostate cancer, BRAF mutations are highly prevalent in melanoma (14, 50), and the progression of melanoma is generally much more aggressive than is the case for prostate cancer. Thus, while the molecular consequences of BRAFV600E expression and PTEN inactivation (i.e., activation of PI3-kinase→Akt→mTOR signaling and MAP kinase signaling) may be similar in melanoma and prostate cancer, the molecular evolution of the tumors is likely to be distinct. Further comparison of the molecular features of prostate cancer and melanoma driven by BRAFV600E expression and PTEN inactivation may provide insights into their common versus tissue-specific molecular mechanisms of cancer progression, which can be facilitated by the availability of complementary mouse models for these tumor sites.
Finally, our current findings have provided new insights into the relationship of Myc up-regulation to activation of PI3-kinase→Akt→mTOR and MAP kinase signaling in prostate cancer. In particular, our findings demonstrate that Myc, whose up-regulation in prostate cancers is associated with disease progression and outcome (1, 51), is downstream of P I 3-kinase→Akt→mTOR and MAP kinase signaling and can be down-regulated by agents that affect these pathways. In the case of melanoma, a mouse model based on Pten loss and Braf activation has revealed the involvement of β-catenin signaling via GSK3 (36), which may also ultimately affect Myc signaling. These observations further underscore the potential value of comparative analyses of molecular pathways of prostate cancer and melanoma through studies of the corresponding mouse models, as well as the significance of Myc as a key downstream target of these signaling pathways.
Supplementary Material
Acknowledgments
This work was supported by grants CA115717 (to CAS), CA084294 (to CAS, MMS and AC), U54 CA121852 (to AC, CAS, MMS), CA154293 (to MMS and CAS), DK076602 (to MMS), and Silico Research Centre of Excellence NCI-caBIG, SAIC 29XS192 (to AC). AA is a recipient of a Marie Curie International Outgoing Fellowship (PIOF-GA-2009-253290), co-sponsored with the Catalan Institute of Oncology-Bellvitge Institute for Biomedical Research, Barcelona, Spain. TK was supported by post-doctoral training grants from the American Urological Association Foundation and the American Association for Cancer Research. AM is a recipient of a National Science Foundation Post-doctoral Training Grant (#0937060). CAS is an American Cancer Society Research Professor supported in part by a generous gift from the F.M. Kirby Foundation.
Footnotes
Disclosure: None.
References
- 1.Shen MM, Abate-Shen C. Molecular genetics of prostate cancer: new prospects for old challenges. Genes Dev. 2010;24:1967–2000. doi: 10.1101/gad.1965810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jenkins RB, Qian J, Lieber MM, Bostwick DG. Detection of c-myc oncogene amplification and chromosomal anomalies in metastatic prostatic carcinoma by fluorescence in situ hybridization. Cancer Res. 1997;57:524–31. [PubMed] [Google Scholar]
- 3.Sato K, Qian J, Slezak JM, Lieber MM, Bostwick DG, Bergstralh EJ, et al. Clinical significance of alterations of chromosome 8 in high-grade, advanced, nonmetastatic prostate carcinoma. Journal of the National Cancer Institute. 1999;91:1574–80. doi: 10.1093/jnci/91.18.1574. [DOI] [PubMed] [Google Scholar]
- 4.Gurel B, Iwata T, Koh CM, Jenkins RB, Lan F, Van Dang C, et al. Nuclear MYC protein overexpression is an early alteration in human prostate carcinogenesis. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2008;21:1156–67. doi: 10.1038/modpathol.2008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kinkade CW, Castillo-Martin M, Puzio-Kuter A, Yan J, Foster TH, Gao H, et al. Targeting AKT/mTOR and ERK MAPK signaling inhibits hormone-refractory prostate cancer in a preclinical mouse model. J Clin Invest. 2008;118:3051–64. doi: 10.1172/JCI34764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gioeli D, Mandell JW, Petroni GR, Frierson HF, Jr., Weber MJ. Activation of mitogen-activated protein kinase associated with prostate cancer progression. Cancer Res. 1999;59:279–84. [PubMed] [Google Scholar]
- 8.Kremer CL, Klein RR, Mendelson J, Browne W, Samadzedeh LK, Vanpatten K, et al. Expression of mTOR signaling pathway markers in prostate cancer progression. Prostate. 2006;66:1203–12. doi: 10.1002/pros.20410. [DOI] [PubMed] [Google Scholar]
- 9.Gao H, Ouyang X, Banach-Petrosky WA, Gerald WL, Shen MM, Abate-Shen C. Combinatorial activities of Akt and B-Raf/Erk signaling in a mouse model of androgen-independent prostate cancer. Proc Natl Acad Sci U S A. 2006;103:14477–82. doi: 10.1073/pnas.0606836103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uzgare AR, Isaacs JT. Enhanced redundancy in Akt and mitogen-activated protein kinase-induced survival of malignant versus normal prostate epithelial cells. Cancer Res. 2004;64:6190–9. doi: 10.1158/0008-5472.CAN-04-0968. [DOI] [PubMed] [Google Scholar]
- 11.Nardella C, Carracedo A, Salmena L, Pandolfi PP. Faithfull modeling of PTEN loss driven diseases in the mouse. Curr Top Microbiol Immunol. 2010;347:135–68. doi: 10.1007/82_2010_62. [DOI] [PubMed] [Google Scholar]
- 12.Salmena L, Carracedo A, Pandolfi PP. Tenets of PTEN tumor suppression. Cell. 2008;133:403–14. doi: 10.1016/j.cell.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 13.Gioeli D. Signal transduction in prostate cancer progression. Clin Sci (Lond) 2005;108:293–308. doi: 10.1042/CS20040329. [DOI] [PubMed] [Google Scholar]
- 14.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 15.Cho NY, Choi M, Kim BH, Cho YM, Moon KC, Kang GH. BRAF and KRAS mutations in prostatic adenocarcinoma. Int J Cancer. 2006;119:1858–62. doi: 10.1002/ijc.22071. [DOI] [PubMed] [Google Scholar]
- 16.Shen Y, Lu Y, Yin X, Zhu G, Zhu J. KRAS and BRAF mutations in prostate carcinomas of Chinese patients. Cancer Genet Cytogenet. 2011;198:35–9. doi: 10.1016/j.cancergencyto.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 17.Palanisamy N, Ateeq B, Kalyana-Sundaram S, Pflueger D, Ramnarayanan K, Shankar S, et al. Rearrangements of the RAF kinase pathway in prostate cancer, gastric cancer and melanoma. Nat Med. 2010;16:793–8. doi: 10.1038/nm.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, Kruithof-de Julio M, Economides KD, Walker D, Yu H, Halili MV, et al. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature. 2009;461:495–500. doi: 10.1038/nature08361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lesche R, Groszer M, Gao J, Wang Y, Messing A, Sun H, et al. Cre/loxP-mediated inactivation of the murine Pten tumor suppressor gene. Genesis. 2002;32:148–9. doi: 10.1002/gene.10036. [DOI] [PubMed] [Google Scholar]
- 20.Dankort D, Filenova E, Collado M, Serrano M, Jones K, McMahon M. A new mouse model to explore the initiation, progression, and therapy of BRAFV600E-induced lung tumors. Genes & development. 2007;21:379–84. doi: 10.1101/gad.1516407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park JH, Walls JE, Galvez JJ, Kim M, Abate-Shen C, Shen MM, et al. Prostatic intraepithelial neoplasia in genetically engineered mice. Am J Pathol. 2002;161:727–35. doi: 10.1016/S0002-9440(10)64228-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhatia-Gaur R, Donjacour AA, Sciavolino PJ, Kim M, Desai N, Young P, et al. Roles for Nkx3.1 in prostate development and cancer. Genes Dev. 1999;13:966–77. doi: 10.1101/gad.13.8.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- 24.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Croft D, O'Kelly G, Wu G, Haw R, Gillespie M, Matthews L, et al. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 2011;39:D691–7. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–7. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 28.Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J, et al. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17:1115–29. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schuhmacher M, Kohlhuber F, Holzel M, Kaiser C, Burtscher H, Jarsch M, et al. The transcriptional program of a human B cell line in response to Myc. Nucleic Acids Res. 2001;29:397–406. doi: 10.1093/nar/29.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei JS, Song YK, Durinck S, Chen QR, Cheuk AT, Tsang P, et al. The MYCN oncogene is a direct target of miR-34a. Oncogene. 2008;27:5204–13. doi: 10.1038/onc.2008.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu D, Cozma D, Park A, Thomas-Tikhonenko A. Functional validation of genes implicated in lymphomagenesis: an in vivo selection assay using a Myc-induced B-cell tumor. Ann N Y Acad Sci. 2005;1059:145–59. doi: 10.1196/annals.1339.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeller KI, Jegga AG, Aronow BJ, O'Donnell KA, Dang CV. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol. 2003;4:R69. doi: 10.1186/gb-2003-4-10-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abate-Shen C, Shen MM, Gelmann E. Integrating differentiation and cancer: the Nkx3.1 homeobox gene in prostate organogenesis and carcinogenesis. Differentiation. 2008;76:717–27. doi: 10.1111/j.1432-0436.2008.00292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeong JH, Wang Z, Guimaraes AS, Ouyang X, Figueiredo JL, Ding Z, et al. BRAF activation initiates but does not maintain invasive prostate adenocarcinoma. PLoS One. 2008;3:e3949. doi: 10.1371/journal.pone.0003949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dankort D, Curley DP, Cartlidge RA, Nelson B, Karnezis AN, Damsky WE, Jr., et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nature genetics. 2009;41:544–52. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Damsky WE, Curley DP, Santhanakrishnan M, Rosenbaum LE, Platt JT, Gould Rothberg BE, et al. beta-Catenin Signaling Controls Metastasis in Braf-Activated Pten-Deficient Melanomas. Cancer Cell. 2011;20:741–54. doi: 10.1016/j.ccr.2011.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Floc'h N, Kinkade CW, Kobayashi K, Aytes A, Lefebvre C, Mitrofanova A, et al. Dual targeting of the Akt/mTOR signaling pathway inhibits castration-resistant prostate cancer in a genetically engineered mouse model. Cancer Res. doi: 10.1158/0008-5472.CAN-12-0283. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kerkhoff E, Houben R, Loffler S, Troppmair J, Lee JE, Rapp UR. Regulation of c-myc expression by Ras/Raf signalling. Oncogene. 1998;16:211–6. doi: 10.1038/sj.onc.1201520. [DOI] [PubMed] [Google Scholar]
- 39.Aziz N, Cherwinski H, McMahon M. Complementation of defective colony-stimulating factor 1 receptor signaling and mitogenesis by Raf and v-Src. Mol Cell Biol. 1999;19:1101–15. doi: 10.1128/mcb.19.2.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang W, Liu HT. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 2002;12:9–18. doi: 10.1038/sj.cr.7290105. [DOI] [PubMed] [Google Scholar]
- 41.Zhu J, Blenis J, Yuan J. Activation of PI3K/Akt and MAPK pathways regulates Myc-mediated transcription by phosphorylating and promoting the degradation of Mad1. Proc Natl Acad Sci U S A. 2008;105:6584–9. doi: 10.1073/pnas.0802785105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reimann M, Lee S, Loddenkemper C, Dorr JR, Tabor V, Aichele P, et al. Tumor stroma-derived TGF-beta limits myc-driven lymphomagenesis via Suv39h1-dependent senescence. Cancer Cell. 2010;17:262–72. doi: 10.1016/j.ccr.2009.12.043. [DOI] [PubMed] [Google Scholar]
- 43.Zhuang D, Mannava S, Grachtchouk V, Tang WH, Patil S, Wawrzyniak JA, et al. C-MYC overexpression is required for continuous suppression of oncogene-induced senescence in melanoma cells. Oncogene. 2008;27:6623–34. doi: 10.1038/onc.2008.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14:1351–6. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carracedo A, Ma L, Teruya-Feldstein J, Rojo F, Salmena L, Alimonti A, et al. Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer. J Clin Invest. 2008;118:3065–74. doi: 10.1172/JCI34739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wee S, Jagani Z, Xiang KX, Loo A, Dorsch M, Yao YM, et al. PI3K pathway activation mediates resistance to MEK inhibitors in KRAS mutant cancers. Cancer Res. 2009;69:4286–93. doi: 10.1158/0008-5472.CAN-08-4765. [DOI] [PubMed] [Google Scholar]
- 47.Jin N, Jiang T, Rosen DM, Nelkin BD, Ball DW. Synergistic action of a RAF inhibitor and a dual PI3K/mTOR inhibitor in thyroid cancer. Clin Cancer Res. 2011;17:6482–9. doi: 10.1158/1078-0432.CCR-11-0933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsao H, Goel V, Wu H, Yang G, Haluska FG. Genetic interaction between NRAS and BRAF mutations and PTEN/MMAC1 inactivation in melanoma. J Invest Dermatol. 2004;122:337–41. doi: 10.1046/j.0022-202X.2004.22243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Curtin JA, Fridlyand J, Kageshita T, Patel HN, Busam KJ, Kutzner H, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353:2135–47. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 50.Dhomen N, Marais R. BRAF signaling and targeted therapies in melanoma. Hematol Oncol Clin North Am. 2009;23:529–45, ix. doi: 10.1016/j.hoc.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 51.Koh CM, Bieberich CJ, Dang CV, Nelson WG, Yegnasubramanian S, De Marzo AM. MYC and Prostate Cancer. Genes Cancer. 2010;1:617–28. doi: 10.1177/1947601910379132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.