Abstract
Cancer, as well as other human disorders, has long been considered to result from the consequence of genetic mutations in key regulatory genes that reside in pathways controlling proliferation, cellular differentiation, DNA damage and repair. In the case of cancer, mutations are well documented to arise in key oncogenes and critically important tumor-suppressor genes as part of the disease progression process. In addition to more accepted, genetic mutations, a rapidly increasing body of evidence supports the general view that profound alterations also occur in ‘epigenes’, whose products serve to define the ‘epigenetic landscape’ of tumor cells. Aberrant changes in epigenetic mechanisms such as DNA methylation, histone modifications and expression of micro RNAs play an important role in cancer and contribute to malignant transitions. Here we review recent studies linking epigenetic mechanisms to epithelial-to-mesenchymal transition as defined in normal processes, as well as abnormal transitions that lead to oncogensis.
Keywords: Epithelial-to-mesenchymal transition (EMT), epigenetics, cancer, epigenetic therapy
Introducing Epigenetics and Epithelial-to-Mesenchymal transition (EMT)
Epigenetics can be defined as heritable alterations in states of gene expression that are not linked to changes in the DNA sequence [1, 2]. A wealth of emerging literature suggests that the precise organization of DNA in chromatin has important functional consequences. Essentially all DNA-templated processes such as transcription, replication, repair, recombination, and segregation are influenced by the complexity of the chromatin architecture. Chromatin states, whether in the broadest terms, active or silent, establish, maintain and propagate different patterns of gene expression during normal differentiation and development. Mistakes made in establishing these chromatin states, governed by chromatin remodeling activities, lead to mis-expression or improper silencing with far-reaching implications for human biology and human disease [3–8]. The fundamental and interrelated epigenetic events involved in gene regulation, development and tumor progression are DNA methylation, histone modifications, chromatin remodeling and micro-RNA expression. Recent studies on epigenetic mechanisms in cancer have demonstrated that epigenetic alterations also play important roles in epithelial-to-mesenchymal transition (EMT).
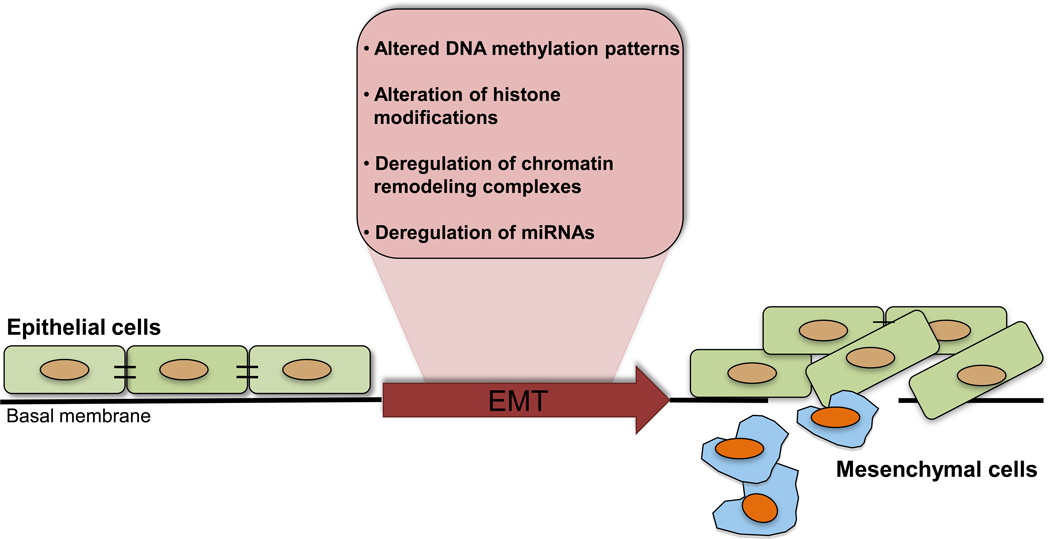
EMT is a crucial process during normal development. Several milestones in embryogenesis, including gastrulation, neural crest formation and heart morphogenesis, rely on dynamic transitions between epithelium and mesenchyme [9]. Typically, EMT involves loss of epithelial polarity, loss of adhesive properties, and acquisition of a fibroblastoid phenotype with increased cell motility. Also during metastasis, carcinoma cells transition to a fibroblast-like phenotype with drastically reduced cell-cell contact and increased migratory abilities. A collection of compelling evidence suggests that cancer progression is initiated via an EMT. These changes result in dispersed and isolated cells which are capable of invading the surrounding stroma, intravasating into the bloodstream, and eventually repopulating at distant sites as micrometastases [10–13]. Orchestration of complex and interlinked networks is necessary for cells to transition into a mesenchymal phenotype [14–16]. Molecular mechanisms underlying EMT were not analyzed until the early 1980s, but since then a large number of molecular differences between epithelial and mesenchymal cells have been described. Much more recently, researchers have learned that cancer cells have to acquire genetic as well as epigenetic changes to undergo EMT. In this review, we discuss how multiple, and likely, interconnected, mechanisms -- covalent DNA methylation, histone modifications, chromatin remodeling and miRNAs -- are or might be associated with EMT and cancer progression (Figure 1).
Figure 1.
Known epigenetic changes involved in epithelial-to-mesenchymal transition of tumor cells.
Epigenetic mechanisms in cancer cells
DNA methylation
One of the more extensively described epigenetic modification in humans is the methylation of cytosine. DNA methylation is believed to be a mechanism of stable gene silencing, which is crucial for regulating gene expression and chromatin states, in an interplay with histone modifications and chromatin-associated proteins. In mammalian cells, DNA methylation is mostly found within CpG dinucleotides, which tend to form clusters known as CpG islands, and in regions of large repetitive sequences, such as retrotransposon elements and centromeric repeats[17–19]. CpG islands are mostly located at the 5’ end of genes and mark approximately 60% of human gene promoters [19, 20]. DNA methylation has long been associated with gene silencing and is especially important for genomic imprinting, wherein one of the two parental alleles is hypermethylated to ensure monoallelic expression, and for inactivation of the X-chromsome in females [21, 22]. Furthermore, repetitive genomic sequences are heavily methylated to maintain chromosomal integrity by preventing chromosomal rearrangements, translocations and gene disruption through the reactivation of transposable elements [5, 17, 23, 24]. DNA-methylation is also a mechanism to control expression of germline-specific genes, like MAGE (Melanoma antigen-encoded gene) family members. It is further used to silence tissue-specific genes, such as MASPIN (mammary serine protease inhibitor, also known as SERPIN5B), in tissues were they should not be expressed [25–27]. Extensive DNA methylation changes, probably induced by cell differentiation, have recently also been described to occur at CpG island “shores”, which are areas of relatively low CpG density close to CpG islands [28, 29]. The recent discovery that 5-methylcytosine can be converted into 5-hydroxymethylcytosien by the 2-oxoglutarate- and Fe (II)-dependent oxygenases TET1, TET2 and TET3 indicates that there is much more complexity to the relationship between DNA methylation and gene expression than first believed [30–34]. More insight is necessary to understand the role of 5-hydroxymethylcytosine, detected in ES cells and Purkinje neurons, to get a much clearer understanding of this new modification and its potential contribution to the cancer epigenome.
Gene silencing through DNA methylation is achieved by a variety of mechanisms. For example, it can inhibit binding of transcription factors to target sites, or alternatively, function as docking sites for methyl-binding domain proteins (e.g. MBD proteins, MeCP2), which induce gene silencing through the recruitment of histone deacetylases (HDACs) [35, 36]. In normal cells, CpG islands in expressed genes are often found to be unmethylated. Cancer cells, however, acquire distorted methylation patterns, which display an inversion of the pattern found in normal tissues. In fact, changes in DNA methylation pattern were the first epigenetic alterations described in cancer cells [37, 38]. Cancer cells are characterized by a general loss of DNA methylation (approximately 20–60% less cytosine methylation) [39]. On the other hand, site-specific hypermethylation at CpG islands is frequently observed at the same time [5]. While the underlying mechanisms that initiate these global changes are still under investigation, recent studies indicate that changes occur very early in cancer development and may contribute to cancer initiation [40].
Covalent histone modifications
Clearly, DNA-methylation is one critical layer in a complex mechanism that is responsible for establishing chromatin states, a layer which in some organisms is well-documented to be influenced by histone modifications. The fundamental packaging element of chromatin is the nucleosome, which consists of 147 pairs of DNA wrapped around an octamer of eight globular histone proteins (two each of H2A, H2B, H3 and H4) [41, 42]. The N-and C-termini of the histones protrude from the nucleosome into the nuclear milieu, were they can be highly decorated with a diverse set of post-translational modifications (PTMs) that are recognized to govern the structure and function of chromatin. These modifications include acetylation, phosphorylation, methylation, citrullination, ADP-ribosylation, and ubiquitylation [43, 44]. PTMs such as these can also occur within the globular domains of the histone proteins, providing a staggering degree of complexity to the ‘language’ of covalent histone modifications that may contribute to a histone or epigenetic “code” that remains under active investigation (see below).
A rapidly emerging body of literature indicates that epigenetic alterations are fundamental for normal development, cell differentiation and also play an important role in abnormal human pathologies, such as cancer. Modulation of chromatin by covalent histone marks is one fundamental way of regulating DNA accessibility during processes such as gene transcription, DNA replication and DNA damage repair. For example trimethylation of lysines (K) 4, 36 or 79 on histone 3 (H3K4me3, H3K36me3, H3K79me3), and acetylation of H3K9 and H3K14 (H3K9ac, H3K14ac, and in some developmentally-regulated enhancer elements, even H3K27ac) most often correlate with transcriptional activation, whereas histone modifications like di- or trimethylation of H3K9 (H3K9me2 and H3K9me3) and trimethylation of H3K27 (H3K27me3) generally mediate gene repression [45–47]. The histone code postulates that the state of chromatin, active or repressed, depends on the combination of histone modifications, which regulate critical downstream events by providing a signalling platform to recruit “readers” or “effector” proteins [48–50]. Alterations in this “code” either through changes in the “writer” and “erasers” of these covalent marks, or the effector protein complexes that read them, are closely linked with oncogenesis [7, 8, 18]. For example, a wide-ranging study of histone-modifying enzymes even allowed the discrimination between cancer samples and their normal counterparts and grouped the tumor samples by cell type [51]. Recently the mechanisms of chromatin remodeling gained increased attention in cancer research because of the clear links to human biology and human malignant diseases. For example, the histone variant H3.3 and one of its dedicated chaperone systems DAXX/ATRX were linked to the formation of malignant tumors. Even though H3.3 differs from its canonical counterparts by only 4–5 amino acids, H3.3 is enriched at highly dynamic regions of chromatin which are subject to high rates of nucleosome turnover, including telomeres [52–56]. A number of recent studies reported mutations in DAXX/ATRX in patients with pancreatic cancer, acute myeloid leukemia, and pediatric glioblastoma and even described alterations in H3.3 as “driving mutations” in pediatric glioblastoma [57–59]. These “game-changing” studies provoke the intriguing, but unexpected, possibility that mutated histone variants, notably H3.3, might function as oncogenes. To what extent, for example, do the point mutations in histone proteins, alter any of the PTMs described above, thereby bringing about mis-expression of critically important downstream genes? Clearly, this will be an exciting area for future investigations.
Remarkable progress has been made in describing molecular mechanisms that introduce variation into the chromatin template, however, identifying the physiologically-relevant combinations of chromatin modifications in normal and pathological states remains a daunting challenge. As well, until mechanisms of inheritance for histone modifications are better understood, the extent to which histones are true carriers of epigenetic information will be open for debate and future experimentation.
Deregulation of miRNAs
MicroRNAs (miRNAs) are non-coding RNA molecules consisting of approximately 21–23 nucleotides that regulate gene expression through post-transcriptional silencing of target genes. They are differentially expressed, depending on the tissue and the developmental context, and some estimates suggest that approximately 30% of messenger RNA transcripts are regulated by miRNAs [60]. Sequence-specific miRNAs pair with the 3’ untranslated regions of target messenger RNA within a RNA-induced silencing complex and conclude in target messenger RNA degradation or inhibition of translation [61]. MicroRNAs have been shown to regulate various important physiological and pathological processes by altering the expression levels of their target mRNAs that play major roles in diverse cellular processes, such as differentiation, proliferation, migration, invasion and survival. Like normal genes, the expression of miRNAs can be altered by various mechanisms, e.g. transcription factor binding, chromosomal rearrangements, genetic and also epigenetic alterations [62–64].
Additionally, miRNAs can in turn regulate intracellular epigenetic mechanisms by modulating enzymes involved in modifying histones (enhancer of zeste-EZH2) and in methylating DNA (DNA methyltransferases DNMT3A and DNMT3B) [65–67]. Studies from recent years accumulate evidence that widespread changes of microRNA expression patterns are associated with tumor progression [68]. Some miRNAs are overexpressed in cancer cells, whereas others are downregulated and, depending on their target genes, miRNAs can function as tumor suppressors or oncogenes [69, 70]. Deregulation of miRNA expression has also been linked to EMT, e.g. by targeting repressors of E-cadherin [71, 72].
Considering the fundamental role of epigenetic mechanisms in the maintenance of gene regulation, it is not surprising that aberrations of these integrated patterns are found in cancer cells. The fine tuned cross-talk of the epigenetic machinery is disturbed in cancer cells and leads to a distorted epigenetic landscape promoting tumor progression. The main epigenetic alterations found in human cancer tissues are aberrant hypermethylation of tumor-suppressor genes, global DNA-hypomethylation and changes in histone modifications [5, 6, 18, 24]. The study of epigenetic mechanisms in cancer during the last decade has provided extensive information about the mechanisms that contribute to the neoplastic phenotype and changes in DNA-methylation pattern, histone modifications and miRNA expression are now accepted as common hallmarks of cancer.
Epigenetic control of genes associated with EMT
Epithelial and mesenchymal cells differ in various functional and phenotypic characteristics and complex signaling networks are necessary to orchestrate EMT [15, 16]. The study of epigenetics in tumor progression during recent years has provided intriguing clues and some insights into mechanisms that promote EMT through the regulation of expression of genes critical to relevant transformation pathways. Cells undergoing EMT show complex alterations in gene expression patterns and a proportion of these alterations could be explained by disrupted epigenetic mechanisms, such as histone modifications and DNA-methylation [24, 73]. Cancer cells almost certainly have specific epigenomes distinct from their parental cell type of origin, but little is known as to when and how alterations in epigenetic landscapes take place along the transformation process. While changes in gene expression patterns during EMT have been well described, the function of DNA methylation in this transition is less clear. However, several studies, described below, document aberrant promoter hypermethylation of genes that are associated with the regulation of EMT. These genes involve cadherin genes, laminins, thrombospondins, estrogen signaling and axon guidance molecules, to mention a few.
The silencing of the E-cadherin gene (CDH1) by aberrant promoter CpG island hypermethylation is one of the most informative examples. E-cadherin is an important “caretaker” of the epithelial phenotype and downregulation of E-cadherin leads to several intra-and inter-cellular changes that are of direct relevance for EMT [15, 16]. Limited E-cadherin levels result in the loss of E-cadherin-dependent intercellular epithelial cell-cell contact complexes, and E-cadherin-regulated sequestering of β-catenin in the cytoplasm is inhibited. Consequently, β-catenin localizes to the nucleus and promotes the Wnt signaling pathway by activating transcriptional regulation through LEF/TCF4 (lymphoid enhancer-binding factor/T-cell factor 4). In addition, other epigenetic mechanisms, such as recruitment of repressive chromatin-remodeling complexes and changes in histone modifications have been reported to be involved in the silencing of the E-cadherin gene. Here, the transcriptional repressors Snail and Slug bind to the CDH1 gene and recruit histone deacetylase (HDAC) containing complexes, such as the Sin3A/HDAC1/HDAC2 complex [74, 75]. Another example is the silencing of CDH1 by Snai1 in breast cancer cells lacking estrogen receptor alpha. In the presence of estrogen receptor alpha (ERα), Snai1 is repressed by a MTA3/Mi-2/NuRD remodeling complex binding to its promoter. The absence of ERα abolishes the formation of this repressive complex and leads to expression Snai1, which in turn represses the epithelial marker protein E-cadherin [76]. Snai1 has also been shown to physically interact with lysine-specific demethylase 1 LSD1 (KDM1A), on epithelial gene promoters, such as E-cadherin. LSD1 removes dimethylation of lysine 4 on histone H3 (H3K4m2), a histone mark often associated with active chromatin, leading to a more repressive chromatin state. LSD1 is essential for Snai1-regulated gene repression and for maintenance of the silencing of Snai1 target genes in invasive cancer cells [77].
Another well-described and illustrative example for epigenetic silencing of an EMT-related gene, is the repression of the estrogen receptor alpha (ESR1) gene. Estrogen receptor alpha (ERα) is an important regulator of proliferation and differentiation in mammary epithelial cells. In a ligand-dependant fashion, ERα induces gene expression by binding to its transcriptional targets directly but als through secondary effects mediated by biological activities of direct target genes [78–80]. The loss of ERα is a marker for poor prognosis in a significant proportion of breast cancer patients [81], and in many cases this repression is a result of the hypermethylation of CpG islands within the ESR1 promoter [82, 83]. Hypermethylation within this promoter leads to estrogen-independent tumor growth and subsequently to cancers with increased aggressiveness and failure to endocrine therapy. Other studies have shown that DNA-methyltransferases and HDACs play important roles in the silencing of ESR1. Co-treatment of ERα-negative breast cancer cells with the methyltransferase inhibitor 5-aza-2-deoxycytidine (5-aza-dC) and HDAC-inhibitors, such as TSA can synergistically re-induce ESR1 gene expression, indicating that DNA-methyltransferases and HDACs function through a common mechanistic pathway to repress transcription [82]. Other epigenetic mechanisms besides aberrant hypermethylation have also been shown to be involved in repression of ESR1 expression. For example, the repressive transcription factor Snail binds to the ESR1 promoter in ERα-positive MCF-7 breast cancer cells, leading to the recruitment of HDAC1/2 complexes, which in turn, leads to a decrease in histone H3 lysine 9 acetylation levels. Although the recruitment of these complexes does not result in permanent silencing, the binding of Snail and concerted loss of histone acetylation bring about drastic downregulation of ESR1 gene expression [84].
Recently, another epigenetic modulator, the lysine methyltransferase SET8, was described to be involved in EMT of breast cancer cells. Yang and colleagues demonstrated that SET8 interacts and cooperates with TWIST, a transcription factor known to play an important role in promoting EMT, to regulate expression levels of the E-cadherin and N-cadherin genes. SET8 is recruited by TWIST to these two gene promoters where it acts as a dual epigenetic modifier through its H4K20 monomethylation activity. H4K20me1 has generally been associated with gene silencing, but also with transcriptional activation of single genes [85]. Expression of SET8 was found to be positively associated with expression of N-cadherin and TWIST, promoting EMT, and negatively correlated with E-cadherin levels in MCF-7 breast adenocarcinoma cells [86].
The above examples describe the epigenetic mechanisms involved in the regulation of specific EMT-associated genes and demonstrate that a fine tuned interplay of DNA-methylation, histone modifications and chromatin remodeling complexes is necessary to control gene expression. As might be expected, genome-wide changes in epigenetic landscapes are now being reported to support the above view. In AML12 mouse hepatocytes undergoing EMT following TGFβ-treatment, McDonald and colleagues found DNA-methylation patterns to be unchanged during EMT [79]. However, a global loss of the heterochromatin mark H3K9me2 was observed, together with an increase in euchromatin marks, H3K4me3 and H3K36me3. Intriguingly, these changes were dependent on expression of LSD1, since loss of LSD1 and its activity drastically affected cell migration and chemoresistance of the cells driven into EMT. Mapping of the genome-wide changes in histone modifications revealed that these alterations were mainly specific to large-organized domains of heterochromatin enriched in H3K9 methylation, so called LOCKs [79]. These results suggest that like stem cell differentiation, germ cell development and malignant transformation, EMT is characterized by widespread changes in chromatin modifications [6, 40, 87, 88]. It will be interesting to investigate if these changes are a general characteristic of EMT by performing similar studies in other cell systems and in other physiological contexts.
Genome-wide investigations of epigenetic regulation in EMT is still in its early stages, but the discovery of new genes involved in this malignant transition will be of great importance for the treatment of aggressive cancer types. In addition, the discovery of EMT-associated miRNAs will further our understanding of the multilayer epigenetic events that control EMT.
Emerging literature describes changes in miRNA expression patterns in primary human tumors relative to normal tissues and it conceivable that dysregulation of miRNAs plays a role in tumorigenesis [68–70]. Several miRNAs have been described to enhance tumor progression. Examples for miRNAs that promote cell migration in breast cancer are miR-10b, miR-373 and miR520c [89, 90]. The latter two were shown to promote tumor progression by suppression of CD44 [89]. The well-described oncogenic miR-21 also contributes to EMT by promoting cell metastasis by inhibiting tumor suppressor genes like TPM1, MASPIN and PDCD4, genes that heve been linked to tumor cell migration and invasion [91–93]. On the other hand, miRNAs can also function as suppressors of EMT and metastasis, like the miRNAs miR-126 and miR-335, which inhibit tumor cell proliferation and migration in breast cancer [94, 95].
Another well-described group of micro-RNAs involved in EMT is the miR-200 family. Members of this miRNA family are known to be downregulated in human tumors and are important for the inhibition of EMT by targeting known repressors of E-cadherin, such as ZEB1, ZEB2, SIP1 and transcription factor 8 [75, 96–99]. Interestingly, in cancer cells miRNAs are also used to modify epigenetic patterns in cells undergoing EMT. For example, miR-138 inhibits EMT in squamous cell carcinoma cell lines by regulating the expression of ZEB and EZH2 [100]. Also, miR-101 has been described to regulate the histone methyltransferase responsible for H3K27 methylation, EZH2. The targeting of EZH2 by miR-101 leads to a degradation of EZH2 and therefore increased expression of tumor suppressor genes due to the down-regulation of this repressive chromatin mark [66, 67, 101]. MiR-101 is found to be downregulated in several cancer types, which consequently brings about higher expression levels of EZH2.
In addition, the expression of miRNAs can reciprocally be changed by epigenetic mechanisms, such as CpG island hypermethylation and alterations in histone modifications. Epigenetic silencing of miRNAs plays an important role in the acquisition of an invasive cell phenotype and the development of metastasis [71]. MiRNA expression studies with the DNA methylation inhibitor 5-aza-2′-deoxycytidine of metastatic cells from lymph node metastasis of different human cancers revealed a DNA-methylation pattern characteristic of cancer cells, specifically for the CpG islands in the promoter regions of miR-148a, miR-9 family and miR-34b/c.
Epigenetic silencing of these miRNAs activates genes such as c-myc, CDK6 and E2F3 targeted by miR-34b/c downregulation, and TGIF2 (transforming growth factor-β-induced factor-2), which are known to play important roles in EMT programming and cancer progression [71, 102]. Changes in expression levels of these miRNAs have been found in several cancer types such as breast [103], ovarian [104] and pancreatic cancer [105]. Interestingly, miR-9 has also been described as a microRNA with oncogenic character, due to its involvement in the silencing of E-cadherin in hepatocellular carcinomas promoting cell invasion [106]. This finding suggests that miRNAs, depending on the tissue, potentially play a double role in either promoting or abolishing tumor progression. In addition, hypermethylation of the miR-9 family has also been detected in hematopoietic malignancies [107] and renal cell carcinoma [108]. Studies within the last two years also report silencing of some miR-200 familiy members by DNA-hypermethylation-associated in cancer cells [109–113]. As mentioned above, the miR-200 family plays an important role in regulating epithelial–mesenchymal transitions [75, 97, 99]. Importantly, new findings indicate that DNA-methylation-associated silencing of the miR-200 family is a dynamic process that mediates evolving epithelial–mesenchymal transition phenotypes in colorectal and breast tumors [114]. Thus, current studies collectively demonstrate, that miRNAs can both modulate the epigenetic machinery and be regulated through epigenetic alterations. These inter-connected mechanisms constitute a fine-tuned control loop, which should be further explored for epigenetic therapy in the future.
Epigenetic Therapy
As described in this review, cells undergoing EMT are characterized by multiple, distinct epigenetic changes, such as DNA hypermethylation and histone modifications inducing chromatin remodeling. In many cases, hypermethylation and histone deacteylation are biomarkers of tumor progression and therefore associated with poor prognosis for cancer patients [102]. However, the reversible nature of drastic epigenetic alterations in cancer cells led to novel targets for anti-cancer drugs and the development of “epigenetic therapy” [115]. The aim of these treatment options is the reconstitution of the epigenome of normal cells together with the re-expression of epigenetically mis-silenced genes. In recent years successful drugs have been discovered that effectively reverse DNA methylation and aberrant histone modifications, such as DNA-methylation inhibitors zebularine, 5-azacytidine and 5-aza-2’-deoxycytidine and HDAC-inhibiting drugs, including SAHA (suberoylanilide hydroxamid acid), valproic acid (VPA) and trichostatin A (TSA) [116]. Several of these drugs have significant anti-tumor activity and some of these drugs were approved by the US Food and Drug Administration (FDA) for the treatment of cancer patients [117–122].
Combinatorial use of epigenetic drugs involving DNA methylation and HDAC inhibitors together, has proven to be especially effective in several types of cancer. For example, co-treatment of estrogen-receptor negative breast cancer cells with DNMT and HDAC inhibitors, such as TSA, can synergistically induce estrogen-receptor re-expression. At the same time, treated breast cancer cells show reduced soluble DNMT1 expression and DNMT activity, leading to partial demethylation of ER promoter regions, and increased acetylation of histones H3 and H4 [123]. Other preclinical studies of combinatorial HDAC and DNMT inhibitor treatment in colon and lung cancer models showed successful re-expression of silenced genes, decreased tumorigenesis in lung cancers, and increased apoptosis [124]. Besides the development of new, potentially less toxic DNA methylation and HDAC inhibitors, drugs inhibiting histone methyltransferases (HMT) and demethylases are also being actively pursued. For example, the HMT inhibitor DZNep was shown to target EZH2, a protein of the polycomb repressive complex 2 and to effectively induce apoptosis in breast cancer cells [125]. Also polyamine-based LSD1-inhibtors like 2d or PG-11144 were used to reactivate epigenetically silenced tumor suppressor genes in cancers [126, 127]. One proof of principle that so called “reader” proteins are a promising target for epigenetic drug therapy is the development of small molecule inhibitors that block the bromodomains of BET-family of chromatin adaptors, like Brd4. Translocation and/or overexpression of Brd4 have been linked to a master regulator role in lethal forms of childhood epithelial cancer and AML by leading to overactivation of the MYC oncogene in these cancer types. The inhibition of BET proteins with small-molecule bromodomain inhibitors decreased the activity of the MYC oncogene and inhibited tumor cell proliferation in vitro and in vivo [128–132]
In addition, miRNAs are also attractive targets for epigenetic therapy. A study using 5-aza-CdR and 4-phenylbutyric acid demonstrated the successful reactivation of anti-oncogenic miR-127 [64]. Also, synthetic miRNAs, which mimic tumor suppressor miRNAs, have proven to be effective in repressing tumorigenesis [133]. However, the development of efficient and targeted delivery vehicles of synthetic miRNAs is still necessary for this treatment option to be more generally successful.
Conclusion
Although we are just starting to understand the involvement and the importance of epigenetic mechanisms in the process of EMT, we should expect major steps forward over the next few years. Many studies have already identified epigenetically modified genes and miRNAs known to play major roles during EMT.
Despite the fact that an understanding of epigenetics in EMT still needs to be translated into clinical treatment options, we should promote future studies of novel EMT-associated genes and miRNAs that are modulated by the epigenetic machinery in cancer cells. New fundamental insights into the epigenetic regulatory mechanisms and advances in powerful technologies will enable us to discover more EMT-related modifications and therefore potential predictive biomarkers for clinical outcome and targets for novel epigenetic therapeutics.
Acknowledgment
This work was supported by a NIH grant R01GM098870 to C.D.A.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev. 2009;23:781–783. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Chi P, Allis CD, Wang GG. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010;10:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 5.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 6.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang GG, Allis CD, Chi P. Chromatin remodeling and cancer, Part II: ATP-dependent chromatin remodeling. Trends Mol Med. 2007;13:373–380. doi: 10.1016/j.molmed.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang GG, Allis CD, Chi P. Chromatin remodeling and cancer, Part I: Covalent histone modifications. Trends Mol Med. 2007;13:363–372. doi: 10.1016/j.molmed.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 9.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 10.Hay ED, Zuk A. Transformations between epithelium and mesenchyme: normal, pathological, and experimentally induced. Am J Kidney Dis. 1995;26:678–690. doi: 10.1016/0272-6386(95)90610-x. [DOI] [PubMed] [Google Scholar]
- 11.Grunert S, Jechlinger M, Beug H. Diverse cellular and molecular mechanisms contribute to epithelial plasticity and metastasis. Nat Rev Mol Cell Biol. 2003;4:657–665. doi: 10.1038/nrm1175. [DOI] [PubMed] [Google Scholar]
- 12.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 13.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 14.Nieto MA. The Ins and Outs of the Epithelial to Mesenchymal Transition in Health and Disease. Annu Rev Cell Dev Biol. 2011 doi: 10.1146/annurev-cellbio-092910-154036. [DOI] [PubMed] [Google Scholar]
- 15.Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 16.Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147:275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 18.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2009;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci U S A. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Leung FC. An evaluation of new criteria for CpG islands in the human genome as gene markers. Bioinformatics. 2004;20:1170–1177. doi: 10.1093/bioinformatics/bth059. [DOI] [PubMed] [Google Scholar]
- 21.Csankovszki G, Nagy A, Jaenisch R. Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. J Cell Biol. 2001;153:773–784. doi: 10.1083/jcb.153.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, et al. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429:900–903. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- 23.Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300:455. doi: 10.1126/science.1083557. [DOI] [PubMed] [Google Scholar]
- 24.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 25.Bodey B. Cancer-testis antigens: promising targets for antigen directed antineoplastic immunotherapy. Expert Opin Biol Ther. 2002;2:577–584. doi: 10.1517/14712598.2.6.577. [DOI] [PubMed] [Google Scholar]
- 26.Futscher BW, Oshiro MM, Wozniak RJ, Holtan N, Hanigan CL, Duan H, et al. Role for DNA methylation in the control of cell type specific maspin expression. Nat Genet. 2002;31:175–179. doi: 10.1038/ng886. [DOI] [PubMed] [Google Scholar]
- 27.Illingworth R, Kerr A, Desousa D, Jorgensen H, Ellis P, Stalker J, et al. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 2008;6:e22. doi: 10.1371/journal.pbio.0060022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doi A, Park IH, Wen B, Murakami P, Aryee MJ, Irizarry R, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, et al. Dynamic regulation of 5- hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 32.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 36.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 37.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 38.Riggs AD, Jones PA. 5-methylcytosine, gene regulation, and cancer. Adv Cancer Res. 1983;40:1–30. doi: 10.1016/s0065-230x(08)60678-8. [DOI] [PubMed] [Google Scholar]
- 39.Goelz SE, Vogelstein B, Hamilton SR, Feinberg AP. Hypomethylation of DNA from benign and malignant human colon neoplasms. Science. 1985;228:187–190. doi: 10.1126/science.2579435. [DOI] [PubMed] [Google Scholar]
- 40.Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 41.Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 42.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 43.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 44.Khorasanizadeh S. The nucleosome: from genomic organization to genomic regulation. Cell. 2004;116:259–272. doi: 10.1016/s0092-8674(04)00044-3. [DOI] [PubMed] [Google Scholar]
- 45.Hebbes TR, Thorne AW, Crane-Robinson C. A direct link between core histone acetylation and transcriptionally active chromatin. Embo J. 1988;7:1395–1402. doi: 10.1002/j.1460-2075.1988.tb02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 47.Liang G, Lin JC, Wei V, Yoo C, Cheng JC, Nguyen CT, et al. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc Natl Acad Sci U S A. 2004;101:7357–7362. doi: 10.1073/pnas.0401866101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gardner KE, Allis CD, Strahl BD. Operating on chromatin, a colorful language where context matters. J Mol Biol. 2011;409:36–46. doi: 10.1016/j.jmb.2011.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 50.Turner BM. Histone acetylation and an epigenetic code. Bioessays. 2000;22:836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 51.Ozdag H, Teschendorff AE, Ahmed AA, Hyland SJ, Blenkiron C, Bobrow L, et al. Differential expression of selected histone modifier genes in human solid cancers. BMC Genomics. 2006;7:90. doi: 10.1186/1471-2164-7-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Drane P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 2010;24:1253–1265. doi: 10.1101/gad.566910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lewis PW, Elsaesser SJ, Noh KM, Stadler SC, Allis CD. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc Natl Acad Sci U S A. 2010;107:14075–14080. doi: 10.1073/pnas.1008850107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tang J, Wu S, Liu H, Stratt R, Barak OG, Shiekhattar R, et al. A novel transcription regulatory complex containing death domain-associated protein and the ATR-X syndrome protein. J Biol Chem. 2004;279:20369–20377. doi: 10.1074/jbc.M401321200. [DOI] [PubMed] [Google Scholar]
- 56.Xue Y, Gibbons R, Yan Z, Yang D, McDowell TL, Sechi S, et al. The ATRX syndrome protein forms a chromatin-remodeling complex with Daxx and localizes in promyelocytic leukemia nuclear bodies. Proc Natl Acad Sci U S A. 2003;100:10635–10640. doi: 10.1073/pnas.1937626100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heaphy CM, de Wilde RF, Jiao Y, Klein AP, Edil BH, Shi C, et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011;333:425. doi: 10.1126/science.1207313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiao Y, Shi C, Edil BH, de Wilde RF, Klimstra DS, Maitra A, et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331:1199–1203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 60.Filipowicz W, Jaskiewicz L, Kolb FA, Pillai RS. Post-transcriptional gene silencing by siRNAs and miRNAs. Curr Opin Struct Biol. 2005;15:331–341. doi: 10.1016/j.sbi.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 61.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 62.Berdasco M, Esteller M. Aberrant epigenetic landscape in cancer: how cellular identity goes awry. Dev Cell. 2011;19:698–711. doi: 10.1016/j.devcel.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 63.Deng S, Calin GA, Croce CM, Coukos G, Zhang L. Mechanisms of microRNA deregulation in human cancer. Cell Cycle. 2008;7:2643–2646. doi: 10.4161/cc.7.17.6597. [DOI] [PubMed] [Google Scholar]
- 64.Saito Y, Jones PA. Epigenetic activation of tumor suppressor microRNAs in human cancer cells. Cell Cycle. 2006;5:2220–2222. doi: 10.4161/cc.5.19.3340. [DOI] [PubMed] [Google Scholar]
- 65.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Friedman JM, Jones PA, Liang G. The tumor suppressor microRNA-101 becomes an epigenetic player by targeting the polycomb group protein EZH2 in cancer. Cell Cycle. 2009;8:2313–2314. doi: 10.4161/cc.8.15.9168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Friedman JM, Liang G, Liu CC, Wolff EM, Tsai YC, Ye W, et al. The putative tumor suppressor microRNA-101 modulates the cancer epigenome by repressing the polycomb group protein EZH2. Cancer Res. 2009;69:2623–2629. doi: 10.1158/0008-5472.CAN-08-3114. [DOI] [PubMed] [Google Scholar]
- 68.Ventura A, Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell. 2009;136:586–591. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 70.Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lujambio A, Esteller M. How epigenetics can explain human metastasis: a new role for microRNAs. Cell Cycle. 2009;8:377–382. doi: 10.4161/cc.8.3.7526. [DOI] [PubMed] [Google Scholar]
- 72.Mongroo PS, Rustgi AK. The role of the miR-200 family in epithelial-mesenchymal transition. Cancer Biol Ther. 2010;10:219–222. doi: 10.4161/cbt.10.6312548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 74.Bolos V, Peinado H, Perez-Moreno MA, Fraga MF, Esteller M, Cano A. The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressors. J Cell Sci. 2003;116:499–511. doi: 10.1242/jcs.00224. [DOI] [PubMed] [Google Scholar]
- 75.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 76.Fujita N, Jaye DL, Geigerman C, Akyildiz A, Mooney MR, Boss JM, et al. MTA3 and the Mi-2/NuRD complex regulate cell fate during B lymphocyte differentiation. Cell. 2004;119:75–86. doi: 10.1016/j.cell.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 77.Lin T, Ponn A, Hu X, Law BK, Lu J. Requirement of the histone demethylase LSD1 in Snai1-mediated transcriptional repression during epithelial-mesenchymal transition. Oncogene. 2010;29:4896–4904. doi: 10.1038/onc.2010.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Couse JF, Korach KS. Estrogen receptor null mice: what have we learned and where will they lead us? Endocr Rev. 1999;20:358–417. doi: 10.1210/edrv.20.3.0370. [DOI] [PubMed] [Google Scholar]
- 79.McDonald OG, Wu H, Timp W, Doi A, Feinberg AP. Genome-scale epigenetic reprogramming during epithelial-to-mesenchymal transition. Nat Struct Mol Biol. 2011;18:867–874. doi: 10.1038/nsmb.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nilsson S, Gustafsson JA. Estrogen receptor transcription and transactivation: Basic aspects of estrogen action. Breast Cancer Res. 2000;2:360–366. doi: 10.1186/bcr81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lapidus RG, Nass SJ, Davidson NE. The loss of estrogen and progesterone receptor gene expression in human breast cancer. J Mammary Gland Biol Neoplasia. 1998;3:85–94. doi: 10.1023/a:1018778403001. [DOI] [PubMed] [Google Scholar]
- 82.Giacinti L, Claudio PP, Lopez M, Giordano A. Epigenetic information and estrogen receptor alpha expression in breast cancer. Oncologist. 2006;11:1–8. doi: 10.1634/theoncologist.11-1-1. [DOI] [PubMed] [Google Scholar]
- 83.Lo PK, Sukumar S. Epigenomics and breast cancer. Pharmacogenomics. 2008;9:1879–1902. doi: 10.2217/14622416.9.12.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dhasarathy A, Kajita M, Wade PA. The transcription factor snail mediates epithelial to mesenchymal transitions by repression of estrogen receptor-alpha. Mol Endocrinol. 2007;21:2907–2918. doi: 10.1210/me.2007-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schotta G. H4K20 monomethylation faces the WNT. Proc Natl Acad Sci U S A. 2011;108:3097–3098. doi: 10.1073/pnas.1100147108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yang F, Sun L, Li Q, Han X, Lei L, Zhang H, et al. SET8 promotes epithelial-mesenchymal transition and confers TWIST dual transcriptional activities. Embo J. 2012 doi: 10.1038/emboj.2011.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hawkins RD, Hon GC, Ren B. Next-generation genomics: an integrative approach. Nat Rev Genet. 2010;11:476–486. doi: 10.1038/nrg2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, Nair S, et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol. 2008;10:202–210. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- 90.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 91.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 92.Han M, Liu M, Wang Y, Mo Z, Bi X, Liu Z, et al. Re-expression of miR-21 contributes to migration and invasion by inducing epithelial-mesenchymal transition consistent with cancer stem cell characteristics in MCF-7 cells. Mol Cell Biochem. 2011 doi: 10.1007/s11010-011-1195-5. [DOI] [PubMed] [Google Scholar]
- 93.Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350–359. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- 94.Png KJ, Halberg N, Yoshida M, Tavazoie SF. A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature. 2011;481:190–194. doi: 10.1038/nature10661. [DOI] [PubMed] [Google Scholar]
- 95.Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 97.Hurteau GJ, Carlson JA, Spivack SD, Brock GJ. Overexpression of the microRNA hsa-miR-200c leads to reduced expression of transcription factor 8 and increased expression of E-cadherin. Cancer Res. 2007;67:7972–7976. doi: 10.1158/0008-5472.CAN-07-1058. [DOI] [PubMed] [Google Scholar]
- 98.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liu X, Wang C, Chen Z, Jin Y, Wang Y, Kolokythas A, et al. MicroRNA-138 suppresses epithelial-mesenchymal transition in squamous cell carcinoma cell lines. Biochem J. 2011;440:23–31. doi: 10.1042/BJ20111006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695–1699. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lujambio A, Calin GA, Villanueva A, Ropero S, Sanchez-Cespedes M, Blanco D, et al. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci U S A. 2008;105:13556–13561. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lehmann U, Hasemeier B, Christgen M, Muller M, Romermann D, Langer F, et al. Epigenetic inactivation of microRNA gene hsa-mir-9-1 in human breast cancer. J Pathol. 2008;214:17–24. doi: 10.1002/path.2251. [DOI] [PubMed] [Google Scholar]
- 104.Laios A, O'Toole SA, Flavin R, Martin C, Ring M, Gleeson N, et al. An integrative model for recurrence in ovarian cancer. Mol Cancer. 2008;7:8. doi: 10.1186/1476-4598-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Omura N, Goggins M. Epigenetics and epigenetic alterations in pancreatic cancer. Int J Clin Exp Pathol. 2009;2:310–326. [PMC free article] [PubMed] [Google Scholar]
- 106.Tan HX, Wang Q, Chen LZ, Huang XH, Chen JS, Fu XH, et al. MicroRNA-9 reduces cell invasion and Ecadherin secretion in SK-Hep-1 cell. Med Oncol. 2010;27:654–660. doi: 10.1007/s12032-009-9264-2. [DOI] [PubMed] [Google Scholar]
- 107.Roman-Gomez J, Jimenez-Velasco A, Agirre X, Castillejo JA, Navarro G, San Jose-Eneriz E, et al. Epigenetic regulation of human cancer/testis antigen gene, HAGE, in chronic myeloid leukemia. Haematologica. 2007;92:153–162. doi: 10.3324/haematol.10782. [DOI] [PubMed] [Google Scholar]
- 108.Hildebrandt MA, Gu J, Lin J, Ye Y, Tan W, Tamboli P, et al. Hsa-miR-9 methylation status is associated with cancer development and metastatic recurrence in patients with clear cell renal cell carcinoma. Oncogene. 2010;29:5724–5728. doi: 10.1038/onc.2010.305. [DOI] [PubMed] [Google Scholar]
- 109.Ceppi P, Mudduluru G, Kumarswamy R, Rapa I, Scagliotti GV, Papotti M, et al. Loss of miR-200c expression induces an aggressive, invasive, and chemoresistant phenotype in non-small cell lung cancer. Mol Cancer Res. 2010;8:1207–1216. doi: 10.1158/1541-7786.MCR-10-0052. [DOI] [PubMed] [Google Scholar]
- 110.Chen J, Wang L, Matyunina LV, Hill CG, McDonald JF. Overexpression of miR-429 induces mesenchymal-to-epithelial transition (MET) in metastatic ovarian cancer cells. Gynecol Oncol. 2011;121:200–205. doi: 10.1016/j.ygyno.2010.12.339. [DOI] [PubMed] [Google Scholar]
- 111.Neves R, Scheel C, Weinhold S, Honisch E, Iwaniuk KM, Trompeter HI, et al. Role of DNA methylation in miR-200c/141 cluster silencing in invasive breast cancer cells. BMC Res Notes. 2010;3:219. doi: 10.1186/1756-0500-3-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Vrba L, Jensen TJ, Garbe JC, Heimark RL, Cress AE, Dickinson S, et al. Role for DNA methylation in the regulation of miR-200c and miR-141 expression in normal and cancer cells. PLoS One. 2010;5:e8697. doi: 10.1371/journal.pone.0008697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wiklund ED, Bramsen JB, Hulf T, Dyrskjot L, Ramanathan R, Hansen TB, et al. Coordinated epigenetic repression of the miR-200 family and miR-205 in invasive bladder cancer. Int J Cancer. 2010;128:1327–1334. doi: 10.1002/ijc.25461. [DOI] [PubMed] [Google Scholar]
- 114.Davalos V, Moutinho C, Villanueva A, Boque R, Silva P, Carneiro F, et al. Dynamic epigenetic regulation of the microRNA-200 family mediates epithelial and mesenchymal transitions in human tumorigenesis. Oncogene. 2011 doi: 10.1038/onc.2011.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kelly TK, De Carvalho DD, Jones PA. Epigenetic modifications as therapeutic targets. Nat Biotechnol. 2011;28:1069–1078. doi: 10.1038/nbt.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yoo CB, Jones PA. Epigenetic therapy of cancer: past, present and future. Nat Rev Drug Discov. 2006;5:37–50. doi: 10.1038/nrd1930. [DOI] [PubMed] [Google Scholar]
- 117.Esteller M. DNA methylation and cancer therapy: new developments and expectations. Curr Opin Oncol. 2005;17:55–60. doi: 10.1097/01.cco.0000147383.04709.10. [DOI] [PubMed] [Google Scholar]
- 118.Fiskus W, Wang Y, Joshi R, Rao R, Yang Y, Chen J, et al. Cotreatment with vorinostat enhances activity of MK-0457 (VX-680) against acute and chronic myelogenous leukemia cells. Clin Cancer Res. 2008;14:6106–6115. doi: 10.1158/1078-0432.CCR-08-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kaminskas E, Farrell AT, Wang YC, Sridhara R, Pazdur R. FDA drug approval summary: azacitidine (5-azacytidine, Vidaza) for injectable suspension. Oncologist. 2005;10:176–182. doi: 10.1634/theoncologist.10-3-176. [DOI] [PubMed] [Google Scholar]
- 120.Kuendgen A, Lubbert M. Current status of epigenetic treatment in myelodysplastic syndromes. Ann Hematol. 2008;87:601–611. doi: 10.1007/s00277-008-0477-9. [DOI] [PubMed] [Google Scholar]
- 121.Lane AA, Chabner BA. Histone deacetylase inhibitors in cancer therapy. J Clin Oncol. 2009;27:5459–5468. doi: 10.1200/JCO.2009.22.1291. [DOI] [PubMed] [Google Scholar]
- 122.Scuto A, Kirschbaum M, Kowolik C, Kretzner L, Juhasz A, Atadja P, et al. The novel histone deacetylase inhibitor, LBH589, induces expression of DNA damage response genes and apoptosis in Phacute lymphoblastic leukemia cells. Blood. 2008;111:5093–5100. doi: 10.1182/blood-2007-10-117762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yang X, Ferguson AT, Nass SJ, Phillips DL, Butash KA, Wang SM, et al. Transcriptional activation of estrogen receptor alpha in human breast cancer cells by histone deacetylase inhibition. Cancer Res. 2000;60:6890–6894. [PubMed] [Google Scholar]
- 124.Belinsky SA, Klinge DM, Stidley CA, Issa JP, Herman JG, March TH, et al. Inhibition of DNA methylation and histone deacetylation prevents murine lung cancer. Cancer Res. 2003;63:7089–7093. [PubMed] [Google Scholar]
- 125.Tan J, Yang X, Zhuang L, Jiang X, Chen W, Lee PL, et al. Pharmacologic disruption of Polycombrepressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21:1050–1063. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Huang Y, Greene E, Murray Stewart T, Goodwin AC, Baylin SB, Woster PM, et al. Inhibition of lysinespecific demethylase 1 by polyamine analogues results in reexpression of aberrantly silenced genes. Proc Natl Acad Sci U S A. 2007;104:8023–8028. doi: 10.1073/pnas.0700720104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Huang Y, Stewart TM, Wu Y, Baylin SB, Marton LJ, Perkins B, et al. Novel oligoamine analogues inhibit lysine-specific demethylase 1 and induce reexpression of epigenetically silenced genes. Clin Cancer Res. 2009;15:7217–7228. doi: 10.1158/1078-0432.CCR-09-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci U S A. 2011;108:16669–16674. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Schwartz BE, Hofer MD, Lemieux ME, Bauer DE, Cameron MJ, West NH, et al. Differentiation of NUT midline carcinoma by epigenomic reprogramming. Cancer Res. 2011;71:2686–2696. doi: 10.1158/0008-5472.CAN-10-3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Iorio MV, Casalini P, Piovan C, Braccioli L, Tagliabue E. Breast cancer and microRNAs: therapeutic impact. Breast. 2011;20(Suppl 3):S63–S70. doi: 10.1016/S0960-9776(11)70297-1. [DOI] [PubMed] [Google Scholar]