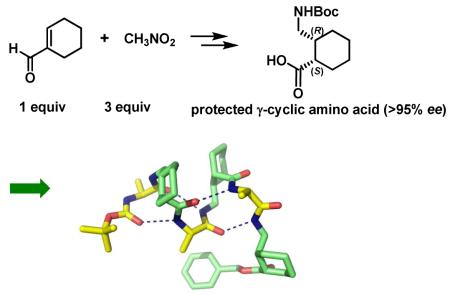
Abstract
An asymmetric synthesis of two new diastereomeric γ-amino acids is described. Both molecules contain a cyclohexyl ring to limit conformational flexibility about the Cα-Cβ bond; they differ in having cis vs. trans stereochemistry on the ring. Residues derived from the cis γ isomer are shown to support helical secondary structures in α/γ-peptide oligomers.
Peptidic oligomers that contain γ-amino acid residues have become increasingly popular as subjects for foldamer research.1,3 γ-amino acid building blocks in which a ring constrains at least one backbone bond are of particular interest because cyclic constraints have proven to be useful in stabilizing specific secondary structures among foldamers that contain β-amino acid residues.4 We previously found that a five membered ring incorporated via Cα and Cγ promotes a γ-peptide sheet secondary structure,5 and comparable observations have been reported by Smith et al. for γ-residues in which a cyclopropyl ring links Cα and Cβ.6 More recently we have shown that a γ residue of type I (Figure 1), with a cis cyclohexyl constraint at the Cβ-Cγ bond (θ torsion angle) supports helical secondary structures among γ- , α/γ- , β/γ- and α/β/γ-peptides.7 Identification of additional γ containing foldamers will require the development of a methodology to prepare new γ-amino acid building blocks with diverse conformational constraints.
Figure 1.
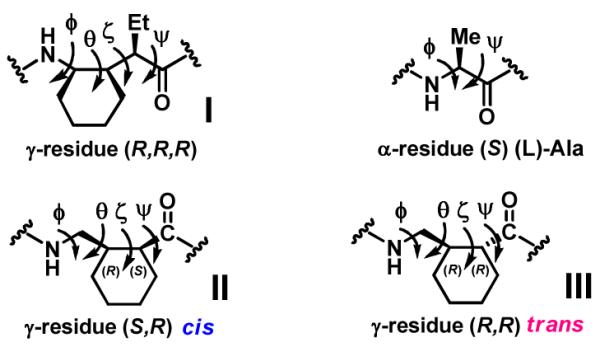
Structures of γ and α-amino acid residues
Here we describe asymmetric synthetic routes to γ-amino acids with a cyclohexyl constraint at the Cα-Cβ bond (ζ torsion angle). Michael addition of nitromethane to cyclohexene-1-carboxaldehyde facilitated via organocatalysis provides access to protected derivatives of the cis and trans γ-amino acid diastereomers, each in highly enantio enriched form.8 Ultimately, this route enables us to incorporate γ residues of types II and III into α/γ peptides. The cis residue (II) is shown to support a specific α/γ peptide helical secondary structure.
Our γ-amino acid synthesis efforts were motivated by results from Hayashi et al.9 and Wang et al.10; these groups simultaneously reported that pyrrolidine A catalyzes a highly enantioselective Michael addition of nitromethane to β-substituted propenal derivatives. Use of benzoic acid as a co-catalyst proved to be necessary for high yields, and simple alcohols were superior as reaction media to nonpolar or polar aprotic solvents. Most of the reported examples involved β-aryl propenals as the Michael acceptors; propenal derivatives bearing bulkier β-alkyl substituents required longer reaction times.9 It was unclear from these precedents whether cyclohexene-1-carboxaldehyde would be an effective Michael acceptor, since the enal unit bears both β-alkyl and α-alkyl substituents, which could sterically hinder the desired reactivity.
Reaction of nitromethane and cyclohexene-1-carboxaldehyde (3:1 ratio) in the presence of 20 mol % A and 10 mol % benzoic acid provided a pair of isomeric γ-nitro aldehyde products in a ~4.5:1 ratio, based on NMR analysis (Scheme 1). The total yield was 90%. These aldehydes were immediately reduced to the corresponding trans and cis γ-nitro alcohols 1t and 1c; HPLC analysis indicated that each diastereomer was generated in > 95% ee. Reduction was employed to avoid epimerization adjacent to the aldehyde group. The catalytic mechanism underlying the Michael addition presumably involves formation of an iminium ion via condensation of starting aldehyde and A.
Scheme 1.
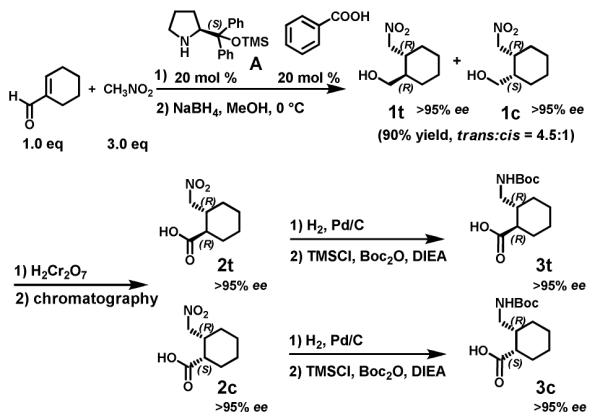
Diastereomers 1t and 1c were difficult to separate on a preparative scale, so the mixture of these δ-nitro alcohols was treated with H2Cr2O7 in acetone, which quantitatively generated the corresponding γ-nitro carboxylic acids 2t and 2c. These diastereomers could be separated by chromatography and crystallization (major isomer 2t is a while solid, while 2c is an oil). The absolute configuration of 2t was established as (R,R) via the crystal structure of L phenylalanine/γ dipeptide derivative 4 (Figure 2). We observed that the minor γ-nitro aldehyde stereoisomer generated by the Michael addition could be converted to the major γ-nitro aldehyde isomer by treatment with DBU, which suggests that γ-nitro carboxylic acid 2c has the S configuration at the ring carbon bearing the carboxyl group. This deduction was confirmed by oligomer crystal structures discussed below.
Figure 2.
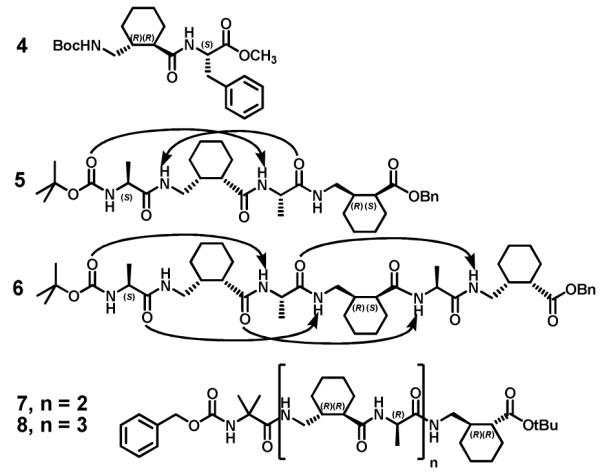
Structures of α/γ-peptides 4-8 (arrows indicate H-bonds in the crystal structures 5 and 6).
The high levels of enantiopurity generated for both product diastereomers in the Michael addition allowed us to evaluate the resulting γ-amino acids as foldamer building blocks. Boc protected γ-amino acids 3t and 3c were readily prepared from γ-nitro acids 2t and 2c, respectively. We prepared α/γ-peptides with 1:1 α:γ alternation, 5-8 (Figure 2), because computational studies by Hofmann et al.3e and basic principles of conformational analysis suggested that such systems might display discrete folding propensities. Residues derived from γ-amino acid 3c (type II; as in α/γ-peptides 5 and 6) are expected to display gauche+ torsion angles about the θ and ζ backbone bonds. The computational studies predict that a local g+,g+ torsion angle sequence should promote formation of an α/γ peptide helix that contains C=O(i) H-N(i+3) H bonds. This "12 helix" (so named because the H bonds occur in 12 atom rings) has previously been documented among α/γ-peptides.3i,7a In contrast, residues derived from γ-amino acid 3t (type III; as in α/γ-peptides 7 and 8) are expected to display an anti torsion angle for θ and a gauche− torsion angle for ζ. The local a,g− torsion angle sequence, which is observed for the single γ residue of type III in dipeptide 4, is predicted to be consistent with an α/γ-peptide helix that contains C=O(i) H N(i-4) H bonds.3e This helix has never been documented experimentally. It should be noted that the absolute configuration is R at Cβ of each γ residue in α/γ-peptides 4-7, because all of these residues derive from Michael reactions catalyzed by A (S configuration). On the other hand, L α-amino acid residues were used for 5 and 6, but D α-amino acid residues were used for 7 and 8, based on the computational predictions of Hofmann et al.3e
High quality crystals of α/γ-peptides 5 and 6 were grown by slow evaporation of chloroform/heptanes/ether or chloroform/heptanes solutions, respectively, and the structures of these oligomers were solved based on X ray diffraction data. Unfortunately, we were unable to crystallize either 7 or 8, despite extensive effort. Thus, at present we have no insight on the secondary structural propensities of these α/γ-peptides containing γ residues with the trans cyclohexyl constraint (type III).
The solid state conformations of α/γ-peptides 5 and 6 revealed two different helical secondary structures (Figure 3), both of which were first proposed by Hofmann et al. based on computational studies.3e Tetramer 5 forms two intramolecular H bonds. The H bond involving the N terminal Boc carbonyl conforms to the expected C=O(i) H N(i+3) pattern; however, the other H bond has the pattern C=O(i) H N(i-1), a 10 membered ring interaction. This secondary structure has been designated the 12/10 helix; the different orientations of the 12 and 10 membered ring H bonds causes this helix to have only a small net dipole. The 12/10 helix has previously been proposed by Sharma, Kunwar et al. based on NMR analysis of α/γ-peptides containing acyclic γ residues.3f
Figure 3.
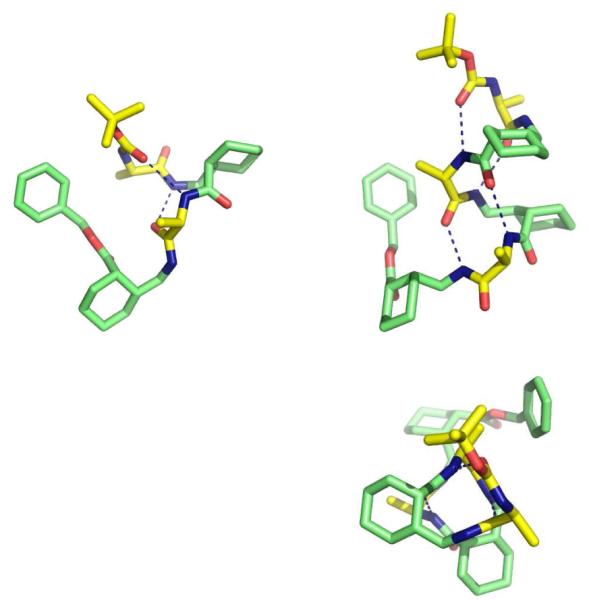
Crystal structures of 5 (left), 6 (right): (top) views perpendicular to helical axes; (bottom) views along the helical axes.
Hexamer 6 formed the expected 12 helix conformation in the solid state; all four possible C=O(i) H N(i+3) H bonds were formed. The crystal contains three independent molecules, but all three conformations are very similar to one another. Balaram et al. have previously proposed a 12 helical conformation for α/γ-peptides in which the γ residues are derived from gabapentin (spiro cyclohexyl ring at Cβ), based on NMR data.3i We have recently provided crystallographic evidence for the 12 helix in α/γ-peptides that contain γ residues with a cis cyclohexyl constraint on the Cβ Cγ bond.7a
Table 1 compares backbone torsion angles for the α and γ residues observed in crystal structures of the α/γ-peptides 5 and 6, which contain cis γ residues (II), and the analogous data for the lone trans γ residue (III) in dimer 4. For comparison, backbone torsion angles are given for a previously reported 1:1 α/γ-peptide crystal structure that contains γ residues of type I;7a this example contains the (R,R,R) γ residue and D alanine. Each of the γ residues in 6 displays a g+,g+ local conformation about the Cα-Cβ (ζ) and Cβ Cγ (θ) bonds, and ψ and ϕ torsion angles for the α residues near 120°, with a somewhat wider distribution for the latter torsion angle. In contrast, the previously described γ residues of type I display g−,g− local conformations. For both type I and type II γ residues, the crystal structures are consistent with the C=O(i) H N(i+3) H bonded helical conformation for 1:1 α/γ-peptides predicted by Hofmann et al.3f The α residue ψ torsion angles in our crystal structures (near 30° for L Ala) are similar to those observed for a canonical 310 helix, which shares the C=O(i) H N(i+3) H bonding pattern.
Table 1.
Backbone Torsion Angles (deg) for α/γ-Peptides
| peptides | residue | ϕ | θ | ζ | ψ |
|---|---|---|---|---|---|
| 1:1 α/γ-tetramer 5 (γ-residue II) |
α | −74.7 | 27.0 | ||
| γ | 64.5 | 47.5 | 45.0 | −118.2 | |
| α | −94.0 | 152.2 | |||
| γ | −98.0 | 59.7 | 60.2 | −169.0 | |
|
| |||||
| 1:1 α/γ-hexamer 6 (γ-residue II) |
α | −65.9 | 35.9 | ||
| γ | −120.1 | 54.7 | 55.3 | −129.7 | |
| α | −70.3 | −27.2 | |||
| γ | −138.3 | 58.6 | 56.2 | −112.1 | |
| α | −83.8 | −28.5 | |||
| γ | 110.5 | 167.9 | −54.6 | −64.4 | |
|
| |||||
| (L)-Phe/γ-dimer (γ-residue III) |
α | −89.4 | 142.5 | ||
| γ | 105.5 | −177.9 | −54.1 | 108.6 | |
|
| |||||
| 1:1 α/γ-hexamer7a (R,R,R)-γ residue I |
α | 75.5 | 22.8 | ||
| γ | 134.3 | −57.7 | −50.5 | 109.4 | |
| α | 56.3 | 41.1 | |||
| γ | 134.7 | −56.7 | −55.7 | 109.8 | |
| α | 67.2 | 45.9 | |||
| γ | 162.1 | −55.4 | −51.3 | 116.0 | |
|
| |||||
| computational study By Hofmann3e |
α | 72.3 | 28.8 | ||
| γ | 123.4 | −52.6 | −62.3 | 124.9 | |
| α | 69.8 | 29.1 | |||
| γ | 123.3 | −52.1 | −62.7 | 122.6 | |
| α | 69.6 | 30.6 | |||
| γ | 122.8 | −53.8 | −64.0 | 129.8 | |
Average parameters for the C=O(i) H N(i+3) H bonded helices formed by 1:1 α/γ-peptides were derived from the structural data as described previously11 and are presented in Table 2. Each helical parameter was calculated from the α carbons of four consecutive residues (for γ residues, Cβ was used as an imaginary α carbon). Non helical residues at C termini were excluded from these calculations. Table 2 includes for comparison parameters for two other helices that contain C=O(i) H N(i+3) hydrogen bonds: the 1:1 a/γ-peptide 12 helix containing formed by γ residues of type I and the α-peptide 310 helix. The parameters for all three helices are quite similar, although it is intriguing that the 12 helix containing γ residues of type II has a somewhat smaller rise per turn than the 12 helix containing γ residues of type I.
Table 2.
Average Parameters for i, i+3 H-bonded Helices
| backbone | helix type |
res/turn n |
rise/turn p (Å) |
rise/res d (Å) |
radius r (Å) |
|---|---|---|---|---|---|
| 1:1 α/γ-peptides (γ-residues II) |
12 | 2.9 | 5.4 | 1.9 | 2.5 |
| 1:1 α/γ-peptides (γ-residues I) |
12 | 3.0 | 6.1 | 2.0 | 2.5 |
| α-peptides | 310 | 3.2 | 5.8 | 1.8 | 2.0 |
We have developed a short, asymmetric route to γ-amino acids that features a cyclohexyl constraint on the Cα-Cβ bond. Both cis and trans diastereomers can be produced in highly enantiopure forms. Preliminary evaluation of the foldamer potential of these γ-amino acids was carried out via synthesis and examination of oligomers with a 1:1 α:γ backbone. Crystal structures revealed a strong helix forming propensity for the cis γ residue, but no insights could be gleaned regarding the trans γ residue. Further use of the cis γ residues for fundamental foldamer research would be facilitated by improvements in the synthetic route, since the required protected γ-amino acid is derived from the minor diastereomer produced in the key Michael addition. We anticipate that incorporation of the new γ residues into other types of heterogeneous peptidic backbones will give rise to new types of foldamers.
Supplementary Material
Acknowledgments
This research was supported by NSF (CHE 0848847). NMR spectrometers were purchased with partial support from NIH and NSF.
Footnotes
Supporting Information Available. Full experimental details, characterization data for all compounds including 1H and 13C NMR and crystallographic data for 5 and 6 (CIF). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1(a).Recent reviews: Guichard G, Huc I. Chem. Commun. 2011;47:5933. doi: 10.1039/c1cc11137j. Hecht S, Huc I, editors. Foldamers: Structure, Properties and Applications. Wiley VCH Weinheim; Germany: 2007. Seebach D, Beck AK, Bierbaum DJ. Chem. Biodivers. 2004;1:1111. doi: 10.1002/cbdv.200490087.
- 2(a).Selected γ-peptide examples: Hanessian S, Luo X, Schaum R, Michnick S. J. Am. Chem. Soc. 1998;120:8569. Hintermann T, Gademann K, Jaun B, Seebach D. Helv. Chim. Acta. 1998;81:983. Seebach D, Brenner M, Rueping M, Schweizer B, Jaun B. Chem. Commun. 2001:207. Baldauf C, Gunther R, Hofmann HJ. Helv. Chim. Acta. 2003;86:2573. Farrera Sinfreu J, Zaccaro L, Vidal D, Salvatella X, Giralt E, Pons M, Albericio F, Royo MA. J. Am. Chem. Soc. 2004;126:6048. doi: 10.1021/ja0398621. Claudon P, Violette A, Lamour K, Decossas M, Fournel S, Heurtault B, Godet J, Mely Y, Gregoire B. Jamart, Petit M. C. Averlant, Briand JP, Duportail G, Monteil H, Guichard G. Angew. Chem., Int. Ed. 2010;49:333. doi: 10.1002/anie.200905591.
- 3(a).Selected examples with γ residues in heterogeneous backbones: Hagihara M, Anthony NJ, Stout TJ, Clardy J, Schreiber SL. J. Am. Chem. Soc. 1992;114:6568. Karle IL, Pramanik A, Banerjee A, Bhattacharjya S, Balaram P. J. Am. Chem. Soc. 1997;119:9087. Ananda K, Vasudev PG, Sengupta A, Raja KMP, Shamala N, Balaram P. J. Am. Chem. Soc. 2005;127:16668. doi: 10.1021/ja055799z. Qureshi MKN, Smith M. Chem. Commun. 2006:5006. doi: 10.1039/b611882h. Baldauf C, Gunther R, Hofmann HJ. J. Org. Chem. 2006;71:1200. doi: 10.1021/jo052340e. Sharma GVM, Jadhav VB, Ramakrishna KVS, Narsimulu K, Subash V, Kunwar AC. J. Am. Chem. Soc. 2006;128:14657. doi: 10.1021/ja064875a. Baruah PK, Sreedevi NK, Gonnade R, Ravindranathan S, Damodaran K, Hofmann HJ, Sanjayan GJ. J. Org. Chem. 2007;72:636. doi: 10.1021/jo062032w. Vasudev PG, Ananda K, Chatterjee S, Aravinda S, Shamala N, Balaram P. J. Am. Chem. Soc. 2007;129:4039. doi: 10.1021/ja068910p. Chatterjee S, Vasudev PG, Raghothama S, Ramakrishnan C, Shamala N, Balaram P. J. Am. Chem. Soc. 2009;131:5956. doi: 10.1021/ja900618h. Araghi RR, Jackel C, Cofen H, Salwiczek M, Vokel A, Wagner SC, Wieczorek S, Baldauf C, Koksch B. ChemBioChem. 2010;11:335. doi: 10.1002/cbic.200900700. Araghi RR, Baldauf C, Gerling UIM, Cadicamo CD, Koksch B. Amino Acids. 2011;41:733. doi: 10.1007/s00726-011-0941-z.
- 4(a).Raguse TL, Lai JR, Gellman SH. J. Am. Chem. Soc. 2003;125:5592. doi: 10.1021/ja0341485. [DOI] [PubMed] [Google Scholar]; (b) Schmitt MA, Choi SH, Guzei IA, Gellman SH. J. Am. Chem. Soc. 2005;127:13130. doi: 10.1021/ja0536163. [DOI] [PubMed] [Google Scholar]; (c) Horne WS, Gellman SH. Acc. Chem. Res. 2008;41:1399. doi: 10.1021/ar800009n. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Horne WS, Johnson LM, Ketas TJ, Klasse PJ, Lu M, Moore JP, Gellman SH. Proc. Natl. Acad. Sci. U.S.A. 2009;106:14751. doi: 10.1073/pnas.0902663106. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Price JL, Hadley EB, Steinkruger JD, Gellman SH. Angew. Chem., Int. Ed. 2010;49:368. doi: 10.1002/anie.200904714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woll MG, Lai JR, Guzei IA, Taylor SJC, Smith MEB, Gellman SH. J. Am. Chem. Soc. 2001;123:11077. doi: 10.1021/ja011719p. [DOI] [PubMed] [Google Scholar]
- 6.Qureshi MKN, Smith M. Chem. Commun. 2006:5006. doi: 10.1039/b611882h. [DOI] [PubMed] [Google Scholar]
- 7(a).Guo L, Chi Y, Almeida AM, Guzei IA, Parker BK, Gellman SH. J. Am. Chem. Soc. 2009;131:16018. doi: 10.1021/ja907233q. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Guo L, Almeida AM, Zhang W, Reidenbach AG, Choi SH, Guzei IA, Gellman SH. J. Am. Chem. Soc. 2010;132:7868. doi: 10.1021/ja103233a. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Sawada T, Gellman SH. J. Am. Chem. Soc. 2011;133:7336. doi: 10.1021/ja202175a. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Guo L, Zhang W, Reidenbach AG, Giuliano JW, Guzei IA, Spenser LC, Gellman SH. Angew. Chem., Int. Ed. 2011;50:5843. doi: 10.1002/anie.201101301. [DOI] [PMC free article] [PubMed] [Google Scholar]; For an altertnative synthesis of γ-amino acids of type I, see: Nodes WJ, Nutt DR, Chippindale AM, Cobb AJA. J. Am. Chem. Soc. 2009;131:16016. doi: 10.1021/ja9070915.
- 8.For racemic synthesis of γ-amino acids corresponding to residues II and III, see: Johnson MR, Gauuan JF, Guo C, Guzzo PR, Le VD, Shenoy RA, Hamby J, Roark H, Stier M, Mangette JE. Synth. Comm. 2011;41:2769.
- 9.Gotoh H, Ishikawa H, Hayashi Y. Org. Lett. 2007;9:5307. doi: 10.1021/ol702545z. [DOI] [PubMed] [Google Scholar]
- 10.Zu L, Xie H, Li H, Wang J, Wang W. Adv. Synth. Catal. 2007;349:2660. [Google Scholar]
- 11(a).Barlow DJ, Thorton JM. J. Mol. Biol. 1988;5:601. doi: 10.1016/0022-2836(88)90641-9. [DOI] [PubMed] [Google Scholar]; (b) Sugeta H, Miyazawa T. Biopolymers. 1967;5:673. [Google Scholar]; (c) Kahn PC. Comput. Chem. 1989;13:185. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.