Abstract
Brain-derived neurotrophic factor (BDNF) promotes cell survival and differentiation in the central and peripheral nervous systems. Previously, we reported that BDNF is produced by salivary glands under acute immobilization stress in rats. However, expression of BDNF is poorly understood in humans, although salivary gland localization of BDNF in rodents has been demonstrated. In the present study, we investigated the expression and localization of BDNF in the human submandibular gland (HSG) using reverse transcription-polymerase chain reaction, western blot analysis, in situ hybridization (ISH), immunohistochemistry (IHC), and ELISA. BDNF was consistently localized in HSG serous and ductal cells, as detected by ISH and IHC, with reactivity being stronger in serous cells. In addition, immunoreactivity for BDNF was observed in the saliva matrix of ductal cavities. Western blotting detected one significant immunoreactive 14 kDa band in the HSG and saliva. Immunoreactivities for salivary BDNF measured by ELISA in humans were 40.76±4.83 pg/mL and 52.64±8.42 pg/mL, in men and women, respectively. Although salivary BDNF concentrations in females tended to be higher than in males, the concentrations were not significantly different. In conclusion, human salivary BDNF may originate from salivary glands, as the HSG appears to produce BDNF.
Keywords: brain-derived neurotrophic factor (BDNF), human, saliva, submandibular gland
I. Introduction
Brain-derived neurotrophic factor (BDNF) is a member of the neurotrophin (NT) family, which includes nerve growth factor (NGF), NT-3, -4/5, -6 and -7 [22]. BDNF is the most abundant NT in the central nervous system, and is closely involved in neural cell survival and maintenance, as well as in neural transmission [41]. In the hippocampus in particular, BDNF expression varies depending on stress [13], exercise [1], and learning [11]. Furthermore, BDNF plays an important role in facilitating the formation of neural networks. BDNF is also found in many peripheral tissues, such as the lachrymal glands [12], heart [43], and retina [44] in rodents, as well as in the testis [29], lung [35], and vascular endothelial cells [30] in human. Moreover, our previous study examined the effect of immobilization stress on BDNF expression in male rat submandibular glands [51]. Increased BDNF mRNA and protein expression were observed in duct cells as a result of immobilization stress. Furthermore, acute immobilization stress was observed to increase plasma BDNF levels, with a contribution from the submandibular glands [52].
The salivary glands consist of the major salivary glands, including the parotid, submandibular, and sublingual glands, as well as numerous minor salivary glands scattered throughout the oral cavity [15]. Their primary role is to secrete saliva, which is involved in food digestion, promotion of mastication, and antimicrobial activity. However, other roles for the salivary glands may also exist, such as in stress responses, and in producing biomarkers for tumors [45, 55]. Cell growth factors, such as epidermal growth factor (EGF) and NGF, in particular, are also produced in the rat submandibular gland [6, 7]. Mouse salivary gland tissue expresses a high level of NGF [6, 7], which is released into the bloodstream in large quantities from salivary glands during fighting [3], and plasma EGF levels are reduced after damage to the major salivary glands [17]. Hence, the salivary glands may play an important role in systemic health [50].
In humans, the presence of numerous growth factors in saliva has also been observed, including EGF [8], NGF [23], and hepatocyte growth factor (HGF) [49]. Recently, Mandel et al. used immunoblotting and enzyme digestion to demonstrate that pro- and mature BDNF are present in human saliva, and that a relationship exists between salivary BDNF concentrations and the presence of the Val66Met single nucleotide polymorphism (SNP) [24, 25]. These proteins play an essential role in the protection and repair of oral and gastric soft tissue, as well as in the maintenance of gustatory tissue. Numerous animal studies have reported that sialoadenectomy (removal of the salivary glands) results in decreased wound healing [5], gastric lesions [33], epithelial keratosis, and distinct changes in taste cell structure and number [27, 32]. Furthermore, conditions of decreased saliva production in humans, such as Sjogren’s syndrome, lead to increased incidence of oral infection and frequency of taste complaints [34, 54].
It is important to investigate whether BDNF and other growth factors are expressed in human submandibular gland (HSG), as the origin of salivary BDNF is not well understood and BDNF exhibits extensive function throughout the human body. In the present study, we investigated salivary BDNF in order to clarify expression patterns of BDNF in the HSG. To the best of our knowledge, this is the first study that describes the expression of BDNF in HSG.
II. Materials and Methods
Tissue samples
Normal HSG tissues were obtained by neck dissections (n=12) at Kanagawa Dental College (Kanagawa, Japan). For staining with in situ hybridization (ISH) (n=4), tissue specimens were fixed in 0.01 M phosphate-buffered saline (PBS) containing 4% paraformaldehyde (pH 7.4) at room temperature for 16 hr, embedded in paraffin and serial 3-µm sections were cut. For staining with hematoxylin and eosin (HE) and immunohistochemistry (IHC) (n=8), tissues specimens were fixed in 10% buffered formalin at room temperature for 18 hr, embedded in paraffin and serial 3-µm sections were cut. Normal paraffin-embedded HSG tissues were used for staining. Normal human hippocampal tissues, ready-made paraffin-embedded or frozen tissue slides (Cosmo Bio, Tokyo, Japan), were used as positive controls for immunostaining and ISH. Total RNA from HSG tissue (n=4) was used for mRNA analysis and Ultrapure total hippocampal RNA (n=1) (Cosmo Bio) was used as a control for mRNA analysis. Total protein from the single HSG tissue was also used for western blot analysis of normal HSG. All patient materials used in this study were obtained following fully informed consent regarding the nature and the aims of the study in accordance with the Ethics Committee of the Kanagawa Dental College.
Participants and saliva collection
Fifty healthy volunteers (26 male and 24 female) from Kanagawa Dental College in Kanagawa, Japan, participated in this study. Participants had a mean age of 27±6.4 years. Participants had not consumed any food or drink, nor brushed their teeth, for 2 hr before sample collection. They were instructed not to consume alcoholic beverages for the 24 hr prior to sample collection. All participants were non-medicated non-smokers. Information about age, general and oral health was also collected. All samples were collected between 9 and 10 a.m., within a 10-min period, to minimize any possible effect of diurnal variation. All saliva samples were collected using the Salivette (Sarstedt Co. Ltd., Nümbrecht, Germany) absorbent method. The Salivette samples were collected according to the manufacturer’s instructions. Briefly, participants were instructed to chew on a cotton roll for 2 min, or until the cotton was fully saturated with saliva, and then expectorate the cotton into the Salivette tube. Participants were asked not to handle the cotton roll in order to prevent possible contamination. All saliva samples were stored on ice until handling (approximately 1 hr), at which point the tubes were centrifuged at 2000 rpm for 15 min at 4°C and the samples aliquotted. All samples were stored at –80°C until use. Upon thawing, the samples were centrifuged once more to ensure complete debris removal [46].
cDNA synthesis and reverse transcription-polymerase chain reaction (RT-PCR)
Total RNA from the HSG tissues (n=4) and Ultrapure total hippocampal RNA (n=1) (Cosmo Bio), stored at −80°C until use, was reverse transcribed at 50°C for 30 min, 99°C for 5 min, and 5°C for 5 min using a single-strand cDNA synthesis kit (Roche Diagnostics, Ltd., Lewes, UK) according to the manufacturer’s instructions [51]. Following the reverse transcription (RT) reaction, cDNA products were stored at −20°C until use. RT-polymerase chain reaction (RT-PCR) was performed using the RNA LA PCRTM Kit (AMV) Ver.1.1 according to the manufacturer’s instructions (TaKaRa, Tokyo, Japan). RT was primed using random 9-mers and cDNA synthesis was conducted using the following cycle conditions: 30°C for 10 min, 42°C for 20 min, 99°C for 5 min and 5°C for 5 min. Oligonucleotide primers were designed to amplify a 167-bp fragment of human BDNF. Primer sequences were 5'-CAGGGGCATAGACAAAAG-3' (sense) and 5'-CTTCCCCTTTTAATGGTC-3' (antisense) [21]. RT products were amplified using Tag DNA polymerase after denaturation for 10 min at 95°C, followed by 25 cycles of denaturation at 94°C for 30 sec, primer annealing at 60°C for 30 sec and product extension at 72°C for 30 sec, and a final extension at 72°C for 5 min. Primers for detecting the internal control marker, a 260-bp fragment of the β-actin housekeeping gene, were 5'-CCTGTATGCCTCTGGTCGTA-3' (sense) and 5'-CCATCTCTTGCTCGAAGTCT-3' (antisense) [42]. PCR products were electrophoresed on 1.5% TBE agarose gels and stained using ethidium bromide.
SDS-PAGE and western blot analysis
Total proteins (40 µg) from the single HSG tissue and whole saliva (n=5) were boiled for 3 min with Laemmli sample buffer (Wako, Tokyo, Japan) and allowed to cool to room temperature. Samples were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 15% gels with 2.5 ng of recombinant BDNF (rBDNF) (R&D Systems, Minneapolis, MN, USA) used as a positive control. The Precision Plus All Blue Standard (Bio-Rad, Tokyo, Japan) molecular weight standard was used to monitor protein migration during electrophoresis. Proteins were transferred onto PVDF membranes (Millipore Corp., Bedford, MA, USA) in transfer buffer (25 mM Tris, 190 mM glycine, 20% MeOH) for 1 hr at 100 V and 4°C, then blocked for 1 hr at room temperature in blocking buffer (PBS, 0.1% Tween-20, 1% NP-40) with 5% non-fat dry milk (NFDM; w/v). For immunodetection, PVDF membranes were incubated overnight at 4°C with anti-BDNF rabbit polyclonal antibody (1:1000; molecular weight 14 kDa; sc-546, Santa Cruz Biochemistry, Santa Cruz, CA, USA) in blocking buffer with 5% NFDM. After washing in Tween-phosphate-buffered saline (PBST, 0.1% Tween 20, 50 mM Tris, pH 7.6, 150 mM NaCl), membranes were incubated in anti-rabbit IgG conjugated with HRP (Dako Cytomation, Glostrup, Denmark), diluted 1:2000 in blocking buffer with 5% NFDM, for 1 hr at room temperature. Membranes were washed again and the ECL Plus Chemiluminescence system (Amersham Biotech, Piscataway, NJ, USA) was used for detection. The specificity of the primary antibody was tested both by peptide neutralization and by determining cross-reactivity to 2.5 ng of recombinant NGF, NT-3, and NT-4 (R&D Systems). Negative controls using only the secondary antibody were also run. The specific signals on immunoblotted membranes were computer-captured with the Multi Gauge Version 3.1 of Science Lab 2005 (Fujifilm, Tokyo, Japan) [40].
BDNF in situ hybridization (ISH)
Complementary RNA (cRNA) probes were produced by in vitro transcription of linearized pGEM-T Easy Vector (Promega Co., Madison, WI, USA). The human BDNF oligonucleotide primers used are as described above. Digoxigenin (DIG)-11-UTP-labeled single-stranded cRNA probes for human BDNF were prepared using a DIG labeling kit SP6/T7 (Roche) according to the manufacturer’s instructions. Procedures for ISH were as described previously [19, 38]. ISH was performed on 3-µm paraffin sections digested with 1 µg/mL proteinase K for 20 min at 37°C. Hybridization was performed at 50°C for 17 hr using DIG-11-UTP-labeled single-stranded cRNA probes dissolved in hybridization medium (Wako). After hybridization, mRNA was detected colorimetrically using a DIG-non-radioactive nucleic acid detection kit (Roche) [18].
BDNF immunohistochemistry (IHC)
Immunohistochemical analysis was performed using Simple Stain MAX-PO (Nichirei, Tokyo, Japan). Slides were pre-incubated in 3% H2O2 for 5 min and sections were subsequently incubated with anti-human BDNF monoclonal antibody (1:100, Techne, Minneapolis, MN, USA) for 1 hr at room temperature. After washing with PBS, sections were incubated with horseradish peroxidase (HRP)-labeled anti-rabbit IgG with amino acid polymer (Nichirei), for 30 min at room temperature. Color was developed using 0.02% 3,3'-diaminobenzidine-tetrahydrochloride (DAB) containing 0.0003% H2O2 in Tris-buffered saline (TBS) for 5 min, and sections were subsequently counterstained with hematoxylin. For negative control experiments, non-immunized rabbit or mouse IgG was used instead of the primary antibody. To determine the binding specificity, a competitive assay was also conducted using rBDNF (R&D Systems) [20, 39].
BDNF ELISA analysis
Human BDNF was detected by sandwich ELISA according to the manufacturer’s instructions (CYT306; Millipore Corp.). All assays were performed in F-bottom 96-well plates (Nunc, Wiesbaden, Germany). Tertiary antibodies were conjugated to horseradish peroxidase (HRP) and color was developed with tetramethylbenzidine (TMB) and measured at 450/570 nm. BDNF content was quantified against a standard curve with a detection limit was <4 pg/mL that was calibrated with known amounts of BDNF. All samples were tested twice, and the mean was calculated. Cross-reactivity to related neurotrophins (NGF, NT-3, and NT-4) was less than 3%. Intra- and inter-assay coefficients of variation were 3.7% and 8.5%, respectively. Concentrations were expressed as pg/mL [14].
Statistical analysis
Statistical analyses were carried out using the SPSS (Version 17.0; SPSS Inc., Chicago, IL, USA) statistics program. Student’s t-test was used to assess differences in BDNF concentrations between men and women (expressed as mean±SD). A probability level of 0.05 or less was accepted as significant.
III. Results
RT-PCR measurement of BDNF gene expression in HSG
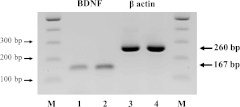
Amplified products corresponding to human BDNF transcripts were detected in RT-PCR samples derived from human hippocampus and HSG (lanes 1 and 2, respectively; Fig. 1). Human β-actin transcripts were also detected in RT-PCR samples derived from human hippocampus and HSG (lanes 3 and 4, respectively; Fig. 1). The sizes of the amplified fragments for BDNF and β-actin were 167 and 260 bp, respectively. In addition, BDNF gene expression was observed in all cases.
Fig. 1.
RT-PCR for BDNF in normal human hippocampus and submandibular glands. Expression of BDNF transcripts is observed in lanes 1 (hippocampus) and 2 (submandibular gland). Expression of β-actin transcripts is observed in lanes 3 (hippocampus) and 4 (submandibular gland). Lane M shows φ×174 HaeIII digest marker.
Western blot analysis of BDNF in saliva and HSG
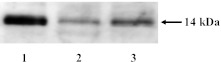
Immunoblotting of rBDNF, as a positive control, with antiserum against human BDNF revealed the expected specific mature full-length 14 kDa band (Fig. 2). In HSG and whole saliva, specific immunoreactive bands were also detected at 14 kDa (Fig. 2). Full-length BDNF protein expression was observed in all cases.
Fig. 2.
Western blot analysis of BDNF in human saliva and HSG. Western blot analysis with BDNF antiserum detected recombinant BDNF (lane 1) and endogenous BDNF in whole saliva (lane 2) and HSG (lane 3). The 14 kDa protein band observed in saliva and HSG is consistent with the expected size of mature full-length BDNF, and is confirmed by the use of recombinant BDNF.
BDNF in situ hybridization
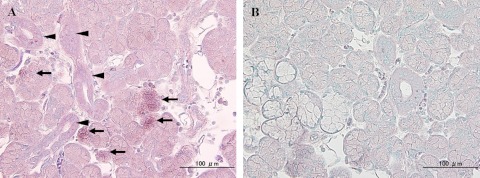
In normal HSG tissue, hybridization signals were predominantly identified in serous cells rather than mucous cells (Fig. 3A). Ductal cells were weakly positive to antisense probes for BDNF mRNA (Fig. 3A) and sense probes failed to produce hybridization signals (Fig. 3B). Hybridization signals for BDNF mRNA were predominantly observed in serous cells, and weakly expressed in ductal cells in all of cases. These results corresponded well with the immunohistochemical distribution profiles.
Fig. 3.
Expression and localization of BDNF mRNA in HSG. (A) Secretory serous cells display a granular pattern (arrows) for BDNF mRNA, whereas ductal cells are only weakly positive for BDNF mRNA (arrowheads). (B) Negative hybridization signals in serous, mucous and ductal cells using sense BDNF cRNA probe.
BDNF immunohistochemistry
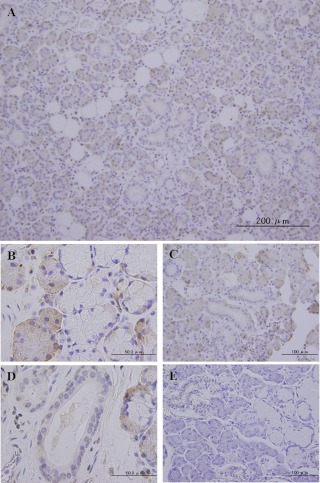
In normal HSG tissue, BDNF was mainly localized in serous cells (Fig. 4A), whereas most mucous cells were immunonegative for BDNF (Fig. 4A). Furthermore, distribution of BDNF protein was more intense in serous cells than in mucous cells (Fig. 4B). Ductal epithelia, including intercalated, striated (Fig. 4C) and interlobular, intralobular (Fig. 4D) and excretory ducts, were weakly immunopositive. Saliva matrix secreted into ductal cavities was also BDNF immunopositive (Fig. 4D). However, BDNF protein was not observed in myoepithelial cells. In absorption control sections of submandibular glands (Fig. 4E), no expression of BDNF was detected with the pre-absorbed antibody. In accordance with the expression of BDNF mRNA, distribution of BDNF protein was predominantly detected in serous cells and weakly detected in ductal cells in all cases.
Fig. 4.
Distribution of BDNF protein in HSG. Immunohistochemical staining of normal submandibular gland with anti-BDNF (A–E). (A) Representative anti-BDNF stained HSG sections at low magnification. (B) BDNF-positive cells were predominantly serous rather than mucous cells. (C, D) BDNF protein was localized in the intercalated, striated (C), and interlobular, intralobular (D) regions. BDNF staining was also observed in the saliva present in the ductal cavity. (E) In negative control sections comprising submandibular gland tissue, expression of BDNF staining is completely absent.
Salivary BDNF concentrations
A comparison of salivary BDNF concentrations in males and females, as determined by ELISA, is presented in Table 1. Although salivary BDNF concentrations in females tended to be higher than in males, the concentrations were not significantly different.
Table 1.
ELISA determination of salivary BDNF concentrations in adult humans
| n | BDNF (pg/ml) | |
|---|---|---|
| Men | 26 | 40.76±4.83 |
| Women | 24 | 52.64±8.42 |
| Total | 50 | 46.58±4.80 |
Concentrations of salivary BDNF are presented as geometric means±SD. There were no significant differences between men and women, p>0.05, Student’s t-test.
IV. Discussion
We previously reported that BDNF expression in the rat submandibular gland is up-regulated by chronic stress [39], and elevated expression of BDNF mRNA and protein was observed in salivary duct cells as a result of immobilization stress and biting behavior [40]. However, BDNF expression is poorly understood in humans, despite the localization of BDNF in rat salivary glands having been demonstrated [51]. While previous studies indicate that the salivary glands are a major source of NGF, and high concentrations of NGF in the submandibular gland have been reported in animals [26, 53], there are currently very few reference values for measuring the expression and localization of NGF in human salivary glands [10]. However, there are many reference values for NGF levels in human saliva [31, 37]. In a previous study, De Vincente et al. examined the expression and localization of neurotrophin proteins in 14 human (4 parotid, 6 submandibular, and 4 sublingual glands) and 5 mouse salivary glands using IHC [10], and found that neurotrophins were not detected in human salivary glands. In the mouse, NGF was the only neurotrophin found in salivary glands (submandibular gland) [10]. The aim of the present study was to clarify the expression and localization of BDNF protein and mRNA in the HSG. We demonstrated that BDNF mRNA and protein were primarily localized in two cell types in the HSG, serous and ductal cells, with the former exhibiting higher expression levels. In addition, BDNF expression and distribution patterns were highly consistent when observed with IHC and ISH. These results provide the first evidence that BDNF is produced in HSG serous and ductal cells. However, fewer positive cells were detected using ISH as compared to IHC. Since we previously reported that expression of BDNF increases in salivary glands under stress conditions [39, 51], we interpret the present results as an indication that individual responses are specific to the stress conditions utilized. Furthermore, we were not able to use the specimens in this study to examine the effects of differences in tissue fixation conditions.
In previous studies, we examined the expression of the BDNF-receptor tyrosine receptor kinase B (TrkB) in rat salivary glands under immobilization stress conditions [51, 52]. Since expression of TrkB was not observed in rat salivary glands, we surmised that BDNF originating from salivary glands does not function in an autocrine fashion. Additionally, since TrkB expression was not observed in the surrounding oral tissues of the rat, including oral mucosa and esophageal mucosa, we concluded that BDNF of salivary gland origin would function in a paracrine manner on other remote organs. However, as we did not examine the expression and localization of TrkB in human salivary glands in the present study, we do not know whether BDNF would function in an autocrine or paracrine manner.
We also demonstrated here that a 14 kDa protein, corresponding to BDNF, was detected as a single band in western blot analysis of HSG tissue. Similarly, BDNF was detected in saliva samples as a single band of the expected size. At the mRNA level, RT-PCR analysis of these samples yielded a specific band corresponding to BDNF. Based on these results, mature BDNF protein is present in HSG and whole saliva. Although BDNF mRNA and protein expression has been observed in rodent submandibular glands [48, 51], we did not investigate the presence of BDNF in rodent saliva. In addition, a recent proteomic analysis of human saliva failed to identify growth factors such as NGF and BDNF [9]. Recently, Mandel et al. performed deglycosylation and plasmin treatment of saliva, and observed that saliva samples contained many bands corresponding to BDNF (24, 32 and 34 kDa) [24]. There was considerable variation in the expression and relative concentrations of each form of the protein, and not all participants expressed every form. In their study, the specificity of the anti-BDNF antibody was confirmed by peptide neutralization, as well as by a lack of cross reactivity with the other neurotrophins, thereby verifying that the antibody specifically binds BDNF. The existence of multiple higher molecular weight forms of BDNF (24, 32 and 34 kDa) has also been reported in experiments using cultured neuronal and non-neuronal cells [28, 47]. It has been suggested that the various bands represent differentially glycosylated and glycosulphated forms of pro- and mature BDNF, and that salivary pro-BDNF is cleaved to mature BDNF by plasmin. To our knowledge, this is the first report of BDNF in salivary secretions, whether in humans or other species [24]. However, the observation of single bands in this study is consistent with the fact that we did not perform deglycosylation or plasmin treatment of saliva and HSG tissues.
In the present study, we used ELISA to measure salivary BDNF concentrations in a total of 50 adult men and women. Our results indicate that although salivary BDNF concentrations tended to be higher in females than in males, there were no significant differences between men and women. It is important to note that all samples were collected between 9 and 10 a.m. in this study. In contrast to the present observations, Mandel et al. reported that women had significantly higher levels of salivary BDNF than men [25], with all samples being collected between 12 and 1 p.m. There are numerous factors known to affect salivary protein levels, including circadian rhythm, salivary flow-rate, stress, and infection [36]. Furthermore, since we have found that salivary BDNF concentrations exhibit diurnal variation, the observation of significant gender differences in saliva BDNF concentrations may be subject to diurnal variation (data not shown). In the future, we plan to further investigate the effect of various parameters on salivary BDNF concentrations, including diurnal variation, gender, daily variation, salivary flow-rate, stress, and infection.
With regard to the physiological roles of the various growth factors present in saliva, it has been reported that wound licking accelerates wound healing in rodents [4, 16]. We also propose that BDNF may interact with and complement other salivary growth factors in maintaining the balance between proliferation, survival, and death of cells. We measured the concentration of HGF, using an ELISA system, in saliva and blood before and after an operation for salivary gland tumor and found that HGF levels were significantly increased after surgery [49]. Thus, it is highly possible that the presence of growth factors in saliva serves to accelerate wound healing. Conversely, blood NGF concentrations decrease in mice that have undergone submandibular gland resection [2]. Although the details of the route taken by the growth factors produced in the salivary glands to the blood are unknown, reabsorption from the sublingual area is considered a likely explanation. Indeed, nitroglycerin tablets are administered at the sublingual area because this location has a thin mucous membrane and an abundance of blood vessels. The openings of the submandibular and sublingual glands are located in this area and it is reasonable to hypothesize that growth factors in saliva can be reabsorbed from the sublingual area. Although NGF is not detected in human salivary glands, other unknown neurotrophic factors could be produced in the salivary glands and reabsorbed from the sublingual area, thereby affecting the central nervous system. We propose that saliva could be assessed as a useful indicator of growth factor production, assuming that growth factors are beneficial to the living body.
In summary, the present study represents the first description of the expression of BDNF in serous and ductal cells of the HSG, indicating that salivary BDNF originates in the submandibular gland. In subsequent studies, we are planning to focus on further analysis of BDNF function in saliva.
V. Acknowledgments
This research was supported in part by Kakenhi Grants-in-Aid for Young Scientists (Start-up, #1989023, B, #23792157) for J.S. as well as Scientific Research grants (B, #20390467, B, #23390420) for K.T. from the Japan Society for the Promotion of Science.
VI. References
- 1.Adlard P. A., Cotman C. W. Voluntary exercise protects against stress-induced decreases in brain-derived neurotrophic factor protein expression. Neuroscience. 2004;124:985–992. doi: 10.1016/j.neuroscience.2003.12.039. [DOI] [PubMed] [Google Scholar]
- 2.Alleva E., Francia N. Psychiatric vulnerability: suggestions from animal models and role of neurotrophins. Neurosci. Biobehav. Rev. 2009;33:525–536. doi: 10.1016/j.neubiorev.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 3.Aloe L., Alleva E., Bohm A., Levi-Montalcini R. Aggressive behavior induces release of nerve growth factor from mouse salivary gland into the bloodstream. Proc. Natl. Acad. Sci. U S A. 1986;83:6184–6187. doi: 10.1073/pnas.83.16.6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bodner L. Effect of parotid submandibular and sublingual saliva on wound healing in rats. Comp. Biochem. Physiol. A. Comp. Physiol. 1991;100:887–890. doi: 10.1016/0300-9629(91)90309-z. [DOI] [PubMed] [Google Scholar]
- 5.Bodner L., Dayan D., Rothchild D., Hammel I. Extraction wound healing in desalivated rats. J. Oral. Pathol. Med. 1991;20:176–178. doi: 10.1111/j.1600-0714.1991.tb00916.x. [DOI] [PubMed] [Google Scholar]
- 6.Chohen S. Purification of a nerve-growth promoting protein from the mouse salivary gland and its neuro-cytotoxic antiserum. Proc. Natl. Acad. Sci. U S A. 1960;46:302–311. doi: 10.1073/pnas.46.3.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chohen S. Isolation of a mouse submaxillary gland protein accelerating incisor eruption and eyelid opening in the newly born animal. J. Biol. Chem. 1962;237:1555–1562. [PubMed] [Google Scholar]
- 8.Dagogo-Jack S., Atkinson S., Kendall-Taylor P. Homologous radioimmunoassay for epidermal growth factor in human saliva. J. Immunoassay. 1985;6:125–136. doi: 10.1080/01971528508063025. [DOI] [PubMed] [Google Scholar]
- 9.Denny P., Hagen F. K., Hardt M., Liao L., Yan W., Arellanno M., Bassilian S., Bedi G. S., Boontheung P., Cociorva D., Delahunty C. M., Denny T., Dunsmore J., Faull K. F., Gilligan J., Gonzalez-Begne M., Halgand F., Hall S. C., Han X., Henson B., Hewel J., Hu S., Jeffrey S., Jiang J., Loo J. A., Ogorzalek Loo R. R., Malamud D., Melvin J. E., Miroshnychenko O., Navazesh M., Niles R., Park S. K., Prakobphol A., Ramachandran P., Richert M., Robinson S., Sondej M., Souda P., Sullivan M. A., Takashima J., Than S., Wang J., Whitelegge J. P., Witkowska H. E., Wolinsky L., Xie Y., Xu T., Yu W., Ytterberg J., Wong D. T., Yates J. R., 3rd, Fisher S. J. The proteomes of human parotid and submandibular/sublingual gland salivas collected as the ductal secretions. J. Proteome. Res. 2008;7:1994–2006. doi: 10.1021/pr700764j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Vincente J. C., Garcia-Suárez O., Esteban I., Santamaria J., Vega J. A. Immunohistochemical localization of neurotrophins and neurotrophin receptors in human and mouse salivary glands. Ann. Anat. 1998;180:157–163. doi: 10.1016/S0940-9602(98)80016-2. [DOI] [PubMed] [Google Scholar]
- 11.Egan M. F., Kojima M., Callicott J. H., Goldberg T. E., Kolachana B. S., Bertolino A., Zaitsev E., Gold B., Goldman D., Dean M., Lu B., Weinberger D. R. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112:257–269. doi: 10.1016/s0092-8674(03)00035-7. [DOI] [PubMed] [Google Scholar]
- 12.Ghinelli E., Johansson J., Rios J. D., Chen L. L., Zoukhri D., Hodges R. R., Dartt D. A. Presence and localization of neurotrophins and neurotrophin receptors in rat lacrimal gland. Invest. Opthalmol. Vis. Sci. 2003;44:3352–3357. doi: 10.1167/iovs.03-0037. [DOI] [PubMed] [Google Scholar]
- 13.Givalois L., Arancibia S., Alonso G., Tapia-Arancibia L. Expression of brain-derived neurotrophic factor and its receptors in the median eminence cells with sensitivity to stress. Endocrinology. 2004;145:4737–4747. doi: 10.1210/en.2004-0616. [DOI] [PubMed] [Google Scholar]
- 14.Goto F., Saruta J., Kanzaki S., To M., Tsutsumi T., Tsukinoki K., Ogawa K. Various levels of plasma brain-derived neurotrophic factor in patients with tinnitus. Neurosci. Lett. 2012;510:73–77. doi: 10.1016/j.neulet.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 15.Hand A. R. In “Orban’s Oral Histology and Embryology”, ed. by S. N. Bhaskar. Mosby; St Louis: 1980. Salivary glands; pp. 336–370. [Google Scholar]
- 16.Hutson J. M., Niall M., Evans D., Fowler R. Effect of salivary glands on wound contraction in mice. Nature. 1979;279:793–795. doi: 10.1038/279793a0. [DOI] [PubMed] [Google Scholar]
- 17.Hwang D. L., Wang S., Chen R. C., Lev-Ran A. Trauma, especially of the submandibular glands, causes release of epidermal growth factor into bloodstream in mice. Regul. Pept. 1991;34:133–139. doi: 10.1016/0167-0115(91)90172-d. [DOI] [PubMed] [Google Scholar]
- 18.Kobashi H., Yaoi T., Oda R., Okajima S., Fujiwara H., Kubo T., Fushiki S. Lysophospholipid receptors are differentially expressed in rat terminal Schwann cells, as revealed by a single cell rt-PCR and in situ hybridization. Acta Histochem. Cytochem. 2006;39:55–60. doi: 10.1267/ahc.06002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kondo Y., Saruta J., To M., Shiiki N., Sato C., Tsukinoki K. Expression and role of the BDNF receptor-TrkB in rat adrenal gland under acute immobilization stress. Acta Histochem. Cytochem. 2010;43:139–147. doi: 10.1267/ahc.10027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le A., Saverin M., Hand A. R. Distribution of dendritic cells in normal human salivary glands. Acta Histochem. Cytochem. 2011;44:165–173. doi: 10.1267/ahc.11010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee T., Saruta J., Sasaguri K., Sato S., Tsukinoki K. Allowing animals to bite reverses the effects of immobilization stress on hippocampal neurotrophin expression. Brain Res. 2008;1195:43–49. doi: 10.1016/j.brainres.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 22.Lewin G. R., Barde Y. A. Physiology of the neurotrophins. Annu. Rev. Neurosci. 1996;19:289–317. doi: 10.1146/annurev.ne.19.030196.001445. [DOI] [PubMed] [Google Scholar]
- 23.Lipps B. V. Isolation of nerve growth factor (NGF) from human body fluids; saliva, serum and urine: comparison between cobra venom and cobra serum NGF. J. Nat. Toxins. 2000;9:349–356. [PubMed] [Google Scholar]
- 24.Mandel A. L., Ozdener H., Utermohlen V. Identical of pro- and mature brain-derived neurotrophic factor in human saliva. Arch. Oral Biol. 2009;54:689–695. doi: 10.1016/j.archoralbio.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mandel A. L., Ozdener H., Utermohlen V. Brain-derived neurotrophic factor in human saliva: ELISA optimization and biological correlates. J. Immunoassay Immunochem. 2011;32:18–30. doi: 10.1080/15321819.2011.538625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mathison R., Davison J. S., Befus A. D. Neuroendocrine regulation of inflammation and tissue repair by submandibular gland factors. Immunol. Today. 1994;15:527–532. doi: 10.1016/0167-5699(94)90209-7. [DOI] [PubMed] [Google Scholar]
- 27.Morris-Wiman J., Sego R., Brinkley L., Dolce C. The effects of sialoadenectomy and exogenous EGF on taste bud morphology and maintenance. Chem. Senses. 2000;25:9–19. doi: 10.1093/chemse/25.1.9. [DOI] [PubMed] [Google Scholar]
- 28.Mowla S. J., Farhadi H. F., Pareek S., Atwal J. K., Morris S. J., Seidah N. G., Murphy R. A. Biosynthesis and post-translational processing of the precursor to brain-derived neurotrophic factor. J. Biol. Chem. 2001;276:12660–12666. doi: 10.1074/jbc.M008104200. [DOI] [PubMed] [Google Scholar]
- 29.Müller D., Davidoff M. S., Bargheer O., Paust H. J., Pusch W., Koeva Y., Jezek D., Holstein A. F., Middendoriff R. The expression of neurotrophins and their receptors in the prenatal and adult human testis: evidence for functions in Leydig cells. Histochem. Cell Biol. 2006;126:199–211. doi: 10.1007/s00418-006-0155-8. [DOI] [PubMed] [Google Scholar]
- 30.Nakahashi T., Fujimura H., Altar C. A., Li J., Kambayashi J., Tandon N. N., Sun B. Vascular endothelial cells synthesize and secrete brain-derived neurotrophic factor. FEBS Lett. 2000;470:113–117. doi: 10.1016/s0014-5793(00)01302-8. [DOI] [PubMed] [Google Scholar]
- 31.Nam J. W., Chung J. W., Kho H. S., Chung S. C., Kim Y. K. Nerve growth factor concentration in human saliva. Oral Dis. 2007;13:187–192. doi: 10.1111/j.1601-0825.2006.01265.x. [DOI] [PubMed] [Google Scholar]
- 32.Nanda R., Catalanotto F. A. Long-term effects of surgical desalivation upon taste acuity, fluid intake, and taste buds in the rat. J. Dent. Res. 1981;60:69–76. doi: 10.1177/00220345810600011401. [DOI] [PubMed] [Google Scholar]
- 33.Olsen P. S., Poulsen S. S., Kirkegaard P., Nexø E. Role of submandibular saliva and epidermal growth factor in gastric cytoprotection. Gastroenterology. 1984;87:103–108. [PubMed] [Google Scholar]
- 34.Porter S. R., Scully C., Hegarty A. M. An update of the etiology and management of xerostomia. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2004;97:28–46. doi: 10.1016/j.tripleo.2003.07.010. [DOI] [PubMed] [Google Scholar]
- 35.Ricci A., Felici L., Mariotta S., Mannino F., Schmid G., Terzano C., Cardillo G., Amenta F., Bronzetti E. Neurotrophin and neurotrophin receptor protein expression in the human lung. Am. J. Respir. Cell Mol. Biol. 2004;30:12–19. doi: 10.1165/rcmb.2002-0110OC. [DOI] [PubMed] [Google Scholar]
- 36.Rudney J. D. Does variability in salivary protein concentrations influence oral microbial ecology and oral health? Crit. Rev. Oral Biol. Med. 1995;6:343–367. doi: 10.1177/10454411950060040501. [DOI] [PubMed] [Google Scholar]
- 37.Ruhl S., Hamberger S., Betz R., Sukkar T., Schmalz G., Seymour R. A., Hiller K. A., Thomason J. M. Salivary proteins and cytokines in drug-induced gingival overgrowth. J. Dent. Res. 2004;83:322–326. doi: 10.1177/154405910408300410. [DOI] [PubMed] [Google Scholar]
- 38.Saruta J., Tsukinoki K., Sasaguri K., Ishii H., Yasuda M., Osamura Y. R., Watanabe Y., Sato S. Expression and localization of Chromogranin A gene and protein in human submandibular gland. Cells Tissues Organs. 2005;180:237–244. doi: 10.1159/000088939. [DOI] [PubMed] [Google Scholar]
- 39.Saruta J., Lee T., Shirasu M., Takahashi T., Sato C., Sato S., Tsukinoki K. Chronic stress affects the expression of brain-derived neurotrophic factor in rat salivary glands. Stress. 2010;13:53–60. doi: 10.3109/10253890902875167. [DOI] [PubMed] [Google Scholar]
- 40.Saruta J., Kondo Y., Sato C., Shiiki N., Tsukinoki K., Sato S. Salivary glands as the source of plasma brain-derived neurotrophic factor in stressed rats engaged in biting behavior. Stress. 2010;13:238–247. doi: 10.3109/10253890903296728. [DOI] [PubMed] [Google Scholar]
- 41.Saruta J., Sato S., Tsukinoki K. The role of neurotrophins related to stress in saliva and salivary glands. Histol. Histopathol. 2010;25:1317–1330. doi: 10.14670/HH-25.1317. [DOI] [PubMed] [Google Scholar]
- 42.Saruta J., Iida M., Kondo Y., To M., Hayashi T., Hori M., Sato S., Tsukinoki K. Chronic stress induces neurotrophin-3 (NT-3) in the rat submandibular gland. Yonsei Med. J. 2012 doi: 10.3349/ymj.2012.53.6.1085. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scarisbrick I. A., Jones E. G., Isackson P. J. Coexpression of mRNAs for NGF, BDNF, and NT-3 in the cardiovascular system of the pre- and postnatal rat. J. Neurosci. 1993;13:875–893. doi: 10.1523/JNEUROSCI.13-03-00875.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seki M., Nawa H., Fukuchi T., Abe H., Takei N. BDNF is upregulated by postnatal development and visual experience: quantitative and immunohistochemical analyses of BDNF in the rat retina. Invest. Ophthalmol. Vis. Sci. 2003;44:3211–3218. doi: 10.1167/iovs.02-1089. [DOI] [PubMed] [Google Scholar]
- 45.Shiiki N., Tokuyama S., Sato C., Kondo Y., Saruta J., Mori Y., Shiiki K., Miyoshi Y., Tsukinoki K. Association between saliva PSA and serum PSA in conditions with prostate adenocarcinoma. Biomarkers. 2011;16:498–503. doi: 10.3109/1354750X.2011.598566. [DOI] [PubMed] [Google Scholar]
- 46.Sugimoto M., Wong D. T., Hirayama A., Soga T., Tomita M. Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics. 2010;6:78–95. doi: 10.1007/s11306-009-0178-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teng H. K., Teng K. K., Lee R., Wright S., Tevar S., Almeida R. D., Kermani P., Torkin R., Chen Z. Y., Lee F. S., Kraemer R. T., Nykjaer A., Hempstead B. L. ProBDNF induces neuronal apoptosis via activation of a receptor complex of p75NTR and sortilin. J. Neurosci. 2005;25:5455–5463. doi: 10.1523/JNEUROSCI.5123-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tirassa P., Manni L., Stenfors C., Lundeberg T., Aloe L. RT-PCR ELISA method for the analysis of neurotrophin mRNA expression in brain and peripheral tissues. J. Biotechnol. 2000;84:259–272. doi: 10.1016/s0168-1656(00)00370-9. [DOI] [PubMed] [Google Scholar]
- 49.Tsukinoki K., Yasuda M., Asano S., Karakida K., Ota Y., Osamura R. Y., Watanabe Y. Association of hepatocyte growth factor expression with salivary gland tumor differentiation. Pathol. Int. 2003;53:815–822. doi: 10.1046/j.1440-1827.2003.01563.x. [DOI] [PubMed] [Google Scholar]
- 50.Tsukinoki K., Yasuda M., Miyoshi Y., Mori Y., Ootsuru M., Saruta J., Sato S., Kaneko A., Watanabe Y., Osamura Y. Role of hepatocyte growth factor and c-Met receptor in neoplastic conditions of salivary glands. Acta Histochem. Cytochem. 2005;38:25–30. [Google Scholar]
- 51.Tsukinoki K., Saruta J., Sasaguri K., Miyoshi Y., Jinbu Y., Kusama M., Sato S., Watanabe Y. Immobilization stress induces BDNF in rat submandibular glands. J. Dent. Res. 2006;85:844–848. doi: 10.1177/154405910608500913. [DOI] [PubMed] [Google Scholar]
- 52.Tsukinoki K., Saruta J., Muto N., Sasaguri K., Sato S., Tan-Ishii N., Watanabe Y. Submandibular glands contribute to increases in plasma BDNF levels. J. Dent. Res. 2007;86:260–264. doi: 10.1177/154405910708600312. [DOI] [PubMed] [Google Scholar]
- 53.Watson A. Y., Anderson J. K., Siminoski K., Mole J. E., Murphy R. A. Cellular and subcellular colocalization of nerve growth factor and epidermal growth factor in mouse submandibular glands. Anat. Rec. 1985;213:365–376. doi: 10.1002/ar.1092130302. [DOI] [PubMed] [Google Scholar]
- 54.Weiffenbach J. M., Schwartz L. K., Atkinson J. C., Fox P. C. Taste performance in Sjogren’s syndrome. Physiol. Behav. 1995;57:89–96. doi: 10.1016/0031-9384(94)00211-m. [DOI] [PubMed] [Google Scholar]
- 55.Yamakoshi T., Park S. B., Jang W. C., Kim K., Yamakoshi Y., Hirose H. Relationship between salivary Chromogranin-A and stress induced by simulated monotonous diving. Med. Biol. Eng. Comput. 2009;47:449–456. doi: 10.1007/s11517-009-0447-y. [DOI] [PubMed] [Google Scholar]