Abstract
To identify ways to improve the efficiency of generating chimeric mice via microinjection of blastocysts with ES cells, we compared production and performance of ES-cell derived chimeric mice using blastocysts from two closely related and commonly used sub-strains of C57BL/6. Chimeras were produced by injection of the same JM8.N4 (C57BL/6NTac) derived ES cell line into blastocysts of mixed sex from either C57BL/6J (B6J) or C57BL/6NTac (B6NTac) mice. Similar efficiency of production and sex-conversion of chimeric animals was observed with each strain of blastocyst. However, B6J chimeric males had fewer developmental abnormalities involving urogenital and reproductive tissues (1/12, 8%) compared with B6NTac chimeric males (7/9, 78%). The low sample size did not permit determination of statistical significance for many parameters. However, in each category analyzed the B6J-derived chimeric males performed as well, or better, than their B6NTac counterparts. Twelve of 14 (86%) B6J male chimeras were fertile compared with 6 of 11 (55%) B6NTac male chimeras. Ten of 12 (83%) B6J chimeric males sired more than 1 litter compared with only 3 of 6 (50%) B6NTac chimeras. B6J male chimeras produced more litters per productive mating (3.42 ± 1.73, n=12) compared to B6NTac chimeras (2.17 ± 1.33, n=6). Finally, a greater ratio of germline transmitting chimeric males was obtained using B6J blastocysts (9/14; 64%) compared with chimeras produced using B6NTac blastocysts (4/11; 36%). Use of B6J host blastocysts for microinjection of ES cells may offer improvements over blastocysts from B6NTac and possibly other sub-strains of C57BL/6 mice.
Keywords: ES cell chimeric mice, C57BL/6J, JM8.N4 ES cells
INTRODUCTION
The use of mouse embryonic stem cells and homologous recombination has revolutionized the field of functional mammalian genomics (van der Weyden et al. 2002; Glaser et al. 2005). An international consortium is applying this powerful technology to generate ES cell lines with targeted cre-recombinase conditional mutant alleles of each mouse gene (Skarnes et al. 2011). These potentially valuable ES cell lines are being distributed to the scientific community, but their conversion into a whole animal frequently requires participation of a core facility. The increasing demand for ES- cell injection services makes it important to identify new methods, or refine established methods, to convert ES cell based resources into mice using fewer animals and with improved efficiency.
Relatively few studies have investigated the effect of the host embryo strain in production of germ-line transmitting chimeric mice. Genetic analysis of chimeric mice made by aggregating pre-compaction stage mouse embryos indicated that the relative contribution of two male embryos to the somatic and germline tissues of such animals could vary dramatically (Mintz 1968; Krzanowska et al. 1991; MacGregor 2002). Studies involving mixing of mouse ES cells with blastocysts in different strain combinations revealed similar effects. For example, Papaioannou and Johnson reported that 129/Sv-derived ES cells produced good chimerism with germline transmission following injection of C57BL/6J blastocysts, while use of random-bred CD-1 embryos produced poor chimeras with minimal contribution to the germline (Papaioannou and Johnson 1993). Ledermann and Bürki described that C57BL/6 – derived ES cells colonized the germline of chimeras formed using BALB/c but not 129/Sv host blastocysts (Ledermann and Burki 1991). Similarly, Schuster-Gossler et al showed that C57BL/6J-derived ES cells could contribute to the germ line following injection into C57BL/6J-Tyrc-2J/J but not FVB/N blastocysts, (Schuster-Gossler et al. 2001). The reasons why different combinations of ES cells and host embryos vary in their ability to produce germline-transmitting chimeras remain unclear.
More recently, investigators have used novel methods such as laser drilling of a hole in the zona pellucida of 8-cell stage embryos to improve the efficiency of production of chimeras from injected ES cells using different combinations of inbred or hybrid B6 ES cells into Swiss Webster embryos, or BALB/c ES cells into C57BL/6NTac embryos (Poueymirou et al. 2007). This novel approach is a major advance, but requires relatively sophisticated and expensive laser drilling equipment and compatible optics. The international knockout mouse consortium is using ES cells derived from C57BL/6NTac mice for its project. In looking for ways to modify established methods to improve both production and performance of ES cell derived chimeric mice, we compared use of two closely related and commonly used strains of C57BL/6 mice as sources of host blastocysts for production of ES-cell derived chimeric mice.
Inbred male mice vary in their reproductive performance (Krzanowska 1971; Chubb 1992; Handel 2011). Amongst commonly used inbred strains, C57BL/6J mice have larger testis weight, increased number of Sertoli cells per gram testis weight and increased sperm output compared to many other inbred strains, including C57BL/6JByJ and C57BL/10J (Chubb 1992; Handel 2011). The genetic basis for this difference is not published although multiple loci are known to affect testis weight in mice (Le Roy et al. 2001). C57BL/6J (B6J) mice have a mutant allele of the Nnt locus that encodes nicotinamide nucleotide transhydrogenase (NNT) (Toye et al. 2005). This mutation, which arose spontaneously in the production facility at Jackson Laboratory (Bar Harbor, ME) between 1976 and 1984, involves an in-frame deletion of exons 7–11 and a missense (M35T) mutation in the mitochondrial leader peptide sequence that results in reduced expression of Nnt mRNA and no functional protein (Toye et al. 2005; Huang et al. 2006). The NntC57BL/6J mutant allele appears to be restricted to the C57BL/6J strain and sub-strains developed from it after the mutation arose (Mekada et al. 2009; Huang et al. 2006). Strains with the NntC57BL/6J deletion allele include C57BL/6J, C57BL/6JJcl and C57BL/6JmsSlc. Strains not carrying this mutation include C57BL/6NCrl, C57BL/6JEi, C57BL/6JByJ, C57BL/10J, C57L/J, C58/J, FVB/N, C3H/HeJ, DBA/2J, BALB/cJ, CAST/EiJ, SJL/J, SPRET/EiJ, MOLF/EiJ and AKR/J, NOD and 129Sv/J. Based on correspondence from The Jackson Laboratory, the commercial stock of the albino-B6 strain (B6(Cg)-Tyrc-2J/J, Jax Lab stock number 000058, formerly known as C57BL/6J-Tyrc-2j/J) that is frequently used to produce host blastocysts for injection of C57BL/6NTac ES cells is homozygous for the Nnt C57BL/6J deletion allele. Whether the mutation in Nnt in C57BL/6J mice contributes to the increased testis weight, number of Sertoli cells and sperm output in this strain compared to C57BL/6JByJ and C57BL/10J is currently unknown.
As Nnt is expressed in many tissues (Hoek and Rydstrom 1988; Arkblad et al. 2001) and loss of NNT is associated with increased cellular oxidative stress (Arkblad et al. 2005), we reasoned that Nnt wildtype ES cells might contribute differently to tissues of chimeric embryos following their injection into a Nnt-null host blastocyst, compared to injection into a Nnt wildtype blastocyst and that this might be manifest in chimeric animals with differences in developmental abnormalities and breeding performance. Here, we show improvement of multiple parameters of chimeric male mice generated by microinjection of a sub-line of C57BL/6NTac (B6NTac)-derived JM8.N4 ES cells (Pettitt et al. 2009) into B6J (Nnt-null) blastocysts compared to B6NTac (Nnt wild type) blastocysts. Use of B6J blastocysts as a host for production of chimeric mice with B6NTac-derived ES cells may offer improved efficiency for production of germ line transmitting ES cell-derived chimeric mice.
MATERIALS AND METHODS
Mice
Sources, history and diet of mouse strains at vendors - C57BL/6J (Nnt −/−; Cat # 000664, F226 (Jan 2010), maintained on diet 5K52 (PMI); Jackson Laboratory, Sacramento, CA). C57BL/6NTac (Nnt +/+; Cat # B6-F, B6-M, Taconic, Oxnard, CA. Maintained on NIH#31M diet (Taconic); Hall to Jax in 1948; Jax at F32 to NIH in 1951; NIH at F151 to Taconic in 1991). After arrival at UCI, mice were housed in individually ventilated cages (Techniplast, Philadelphia, PA) provided with Teklad 2920X irradiated diet (Harlan, Indianapolis, IN) and non-acidified tap water ad libitum. Mice were maintained on a light cycle of 14 hours on, 10 hours off, with lights on at 06.30 and off at 20.30. Studies involving animals were approved by the UCI Institutional Animal Care and Use Committee.
Preparation of blastocysts
Female B6J mice (age 24–25 days at PMSG injection) were mated to B6J stud males after superovulation by IP injection of 5 IU of pregnant mare serum gonadotropin (PMSG, obtained from the National Institutes of Health National Hormone & Peptide Program) between 14.00 and 14.30, followed 46–48 hours later by 5 IU of human chorionic gonadotropin (hCG, catalog number C1063, Sigma-Aldrich, St. Louis, MO). Female B6NTac mice were superovulated and mated with B6NTac males in the Taconic production facility in Oxnard, CA, and arrived at 1.5 or 2.5 days post-coitus (dpc). Uteri were removed on the morning of e3.5 and blastocysts flushed using FHM media buffered with 10mM HEPES, pH 7.3 (Millipore, Billerica, MA). Blastocysts were maintained in FHM at 37°C before and after injection.
Culture of ES cells, microinjection and embryo transfer
Feeder-independent JM8.N4 (B6NTac-derived) ES cells were electroporated with a vector targeting the ROSA26 locus on chromosome 6 to introduce a cDNA under transcriptional control of the endogenous ROSA26 locus. The cDNA is preceded by three copies of an SV40 polyadenylation signal (3xpA), which is flanked by loxP sites. In the absence of cre recombinase the cDNA is not expressed (data not shown). One of the resulting clones, JM8_2H5, shown to be correctly targeted using Southern blots hybridized with probes specific for the 5’ and 3’ ends (data not shown), was cultured in KnockOut DMEM (Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum (ES cell characterized, HyClone Laboratories, Logan, UT), supplemented with 2mM L-glutamine, 1µM β-mercaptoethanol, 1000 U/ml LIF (ESGRO, Millipore), and G418 at 150µg/ml, without antibiotics.
JM8_2H5 ES cells were trypsinized immediately before microinjection and maintained in ES cell medium on ice until use. Approximately 15–20 ES cells with small, round and smooth morphology were injected into the cavity of each blastocyst using a microinjection pipette while visualizing embryos using an inverted microscope equipped with differential interference contrast optics and a cooling stage (Brook Industries, Lake Villa, IL) set to 10°C. The number of cells injected per blastocyst was recorded during each injection session using a video camera. Surviving embryos were transferred to the uterus of pseudo-pregnant HSD:ICR (CD-1) female mice via insertion of the transfer pipette through the utero-tubal junction as described (Chin and Wang 2001). To minimize experimental variation, all ES cell culture was performed by S.W. and all embryo manipulation including microinjection was performed by K.-X. S.
Biopsy of potential chimeric animals and breeding analysis
To identify animals with contribution from injected ES cells a tail biopsy was collected from each weaned pup and solubilized at 55°C overnight in 0.2ml of 0.1M Tris, pH 8.5, 5mM EDTA, 0.2% SDS, 0.2M NaCl and 3.4µg/µl of Proteinase K (Sigma-Aldrich, St. Louis, MO), followed by centrifugation at 10,000 × g for 5 min. The supernatant was used for PCR without further purification. To analyze breeding efficiency each male chimera was housed with two B6NTac female mice. Matings were established simultaneously in the same ventilated cage rack and continued for 4 months. Offspring were removed following birth and a biopsy taken for genotyping analysis. Females were kept in the cage throughout the mating period to permit mating during post-partum estrus. To provide the best opportunity to reproduce and to rule out the possibility that any females might be sterile, males who failed to sire offspring after 4 months of being housed with the 2 females, were caged for an additional 3 months with proven breeder B6NTac female mice.
Extraction of total DNA from tissues
Total genomic DNA was isolated from one testis, heart (mesoderm), lung (endoderm) and brain (ectoderm) from each male chimera. If an animal was a hermaphrodite and had only one testis, the testis was used for histology without DNA extraction. Each tissue sample was homogenized in 5ml of 50mM Tris-HCl (pH 8.8 at 20°C), 20mM EDTA. After adding SDS to 0.5% and Proteinase K to 250µg/ml, the sample was mixed by inverting the tube three times and incubated at 55°C overnight. The solution was extracted three times with an equal volume of equilibrated phenol, then chloroform : isoamyl alcohol (25:1). After ethanol precipitation, total DNA was washed in 70% ethanol, briefly air dried, and solubilized in 10mM Tris pH 7.4, 1mM EDTA.
Genotyping by semi-quantitative PCR analysis of Nnt locus or neo
B6NTac and B6J strains were genotyped for their respective Nnt alleles using a three primer, two allele PCR assay that discriminates between the wild-type allele of Nnt (B6NTac) and the mutant allele lacking exons 7–11 in B6J mice (Nicholson et al. 2010). The coding sequence for aminoglycoside transferase (neo) was detected by PCR using Neo-For (5’-GGG CGCCCGGTTCTTTTTGTCA-3’) and Neo-Rev (5’- CACACCCAGCCGGCCACAGTCG -3’) primers that produces a 516 bp product. Amplification was performed in 30µl, using 1x Qiagen Taq polymerase buffer (Qiagen, Valencia, CA) with a final concentration of 0.83mM MgCl2, 0.33µM Neo-For, 0.33µM Neo-Rev, 0.33mM dNTP’s, 1% DMSO, and 1.25 units of Qiagen Taq polymerase. Amplification conditions used were initial melt 94°C, 5 min; then 35 cycles of 94°C, 45 sec, 57°C, 15 sec, 72°C, 45 sec; followed by a final extension of 5 min at 72°C.
Quantitative PCR analysis of neo in total DNA extracted from chimeric animals
To enable reproducible pipetting of DNA, an aliquot of each genomic DNA was sheared by passing ten times through a 22g needle attached to a 1ml syringe barrel. To analyze the relative contribution of ES cells to different tissues from each chimera, we quantified the amount of neo target, derived from the JM8_2H5 ES cell line, relative to an autosomal locus (Slc25a4, referred to here as Ant1, which encodes adenine nucleotide translocator 1). The assay was validated using total genomic DNA from heterozygous ROSA22 mice (Campbell et al. 2002), which also have a single copy of a neo coding sequence, serially diluted with wild type B6NTac genomic DNA. This assay could reproducibly detect as low as a 1% contribution of neo positive ES cell DNA (data not shown).
Quantitative PCR (qPCR) was performed with a real-time PCR instrument (Chromo4, BioRad, Hercules, CA) equipped with Opticon Monitor 3 software (BioRad). Assays were performed using FastStart Taq DNA Polymerase (Roche) and PrimeTime qPCR Assays (IDT) utilizing hydrolysis probes containing FAM or HEX 5' reporter dyes paired with 3' Iowa Black FQ (BkFQ) quenchers. Names of primers, their sequences and modifications were as follows (all 5’ – 3’). Ant1_Probe_1.1.pt (FAM-CCCGCTGGACTTTGCTAGGACC-BkFQ); Ant1_For_1.1.pt (GCTACTTTGCTGGTAACCTGG); Ant1_Rev_1.1.pt (TGAATTCTCGCTGGGAAGATC); neo.1.pt_probe (HEX-CACTGAAGCGGGAAGGGACTGG-BkFQ); neo.1.pt_For (AGGTGAGATGACAGGAGATCC); neo.1.pt_Rev (AATGAACTGCAGGACGAGG). The linear dynamic range and sensitivity of the assay was determined using samples containing DNA from a ROSA22 (neo positive) heterozygous animal mixed with wild type B6NTac DNA. The assay reliably detected at least a 1% contribution of ES cell DNA and was linear over at least two orders of magnitude. The quantity of cells originating from hemizygous neo+ ES cells in adult chimeric mouse tissues was calculated with the ΔΔCt method using Ant1 as a reference gene. Assays were performed in triplicate for each sample to determine technical error, represented as the standard deviation from the mean. Reaction sizes of 25µl contained 1x buffer (Roche FastStart Taq DNA polymerase), 400µM dNTPs, 1mM MgCl2, 500nM Ant1_For_1.1.pt, 500nM Ant1_Rev_1.1.pt, 250 nM Ant1_Probe_1.1.pt, 500nM neo.1.pt_For, 500nM neo.1.pt_Rev, 250nM neo.1.pt_probe, 1 unit of FastStart Taq polymerase (Roche) and 2µl of sheared genomic DNA at 50ng/µl. Cycling conditions were 95°C, 5 min; followed by 35 cycles of 95°C for 30 sec and 60°C for 10 sec with the relative amount of FAM and HEX recorded at the end of each cycle.
Statistical analysis
Statistical methods used and the P values obtained are reported in Table 1. Significance was defined as P ≤ 0.05.
TABLE 1.
Comparison of parameters of chimeric animals generated by injection of JM8_2H5 ES cells into C57BL/6J or C57BL/6NTac blastocysts.
| B6NTac | B6J | Notes | |
|---|---|---|---|
| # Pups produced from injected blastocysts | 31 | 31 | |
| # Chimeras (% of live born pups) a | 19 (67%) | 25 (81%) | P = 0.161, Fisher’s exact test, two tailed. |
| # Chimeras Male : Female (%M : %F) | 15M : 4F (79% M : 21% F) |
20M : 5F (80% M : 20% F) |
|
| # Male chimeras surviving to adult (%) | 11 / 15 (73 %) b | 18 / 20 (90%) c | |
| # Chimeras mated | 11 | 14 | |
| # Fecund (%) | 6 (55%) | 12 (86%) | P = 0.178, Fisher’s exact test, two tailed. |
| Total litters | 13 | 41 | |
| Litters per productive mating |
2.17 ± 1.33; 0.54 (n=6) |
3.42 ± 1.73; 0.5 (n=12) |
P = 0.141; two-tailed unpaired t-test. |
| Litter size (Mean ± SD; SEM (n)) | 6.3 ± 2.7; 0.7 (n=13) |
7.3 ± 2.8; 0.4 (n=41) |
P = 0.283; two-tailed unpaired t-test. |
| # Fertile chimeras with >1 litter | 3 / 6 (50%) | 10 / 12 (83%) | |
| # Neo positive offspring – germ-line transmitting (GLT) chimeras only (%) |
27/51 (53%) | 87/221 (39%) | |
| # GLT per all male chimeras bred (%) | 4/11 (36 %) | 9/14 (64 %) | P = 0.238, Fisher’s exact test, two tailed. |
| # GLT per fecund male chimeras bred (%) | 4/6 (67%) | 9/12 (75%) | |
| Mean body mass of male chimeras (g) | 55.1 ± 8.3: 2.8 | 47.7 ± 5.5; 1.6 | P = 0.038; two-tailed |
| (Mean ± SD; SEM (n)) | (n=9) | (n=12) | Welch’s unpaired t-test. |
| # Chimeras with abnormal urogenital tract or mammary gland. |
7 / 9 (78%) | 1 / 12 (8%) | P = 0.002, Fisher’s exact test, two tailed. |
| Testis wt (mg) d | 53.9 ± 30.6; 7.7 | 80.6 ± 23.2; 4.9 | P = 0.007; two-tailed |
| (Mean ± SD; SEM (n)) | (n=16) | (n=23) | Welch’s unpaired t-test. |
| Percentage abnormal or atrophic seminiferous tubules. |
25.3 ± 20.4; 6.8 (n=9) |
18.7 ± 23.2; 6.7 (n=12) |
P = 0.494; two-tailed Welch’s unpaired t-test. |
| (Mean ± SD; SEM (n)) | |||
| Seminal vesicle wt (mg) (Mean ± SD; SEM (n)) |
575 ± 320; 107 (n=9) |
701 ± 282; 81 (n=12) |
P = 0.355; two-tailed unpaired t-test. |
Footnotes
Chimerism defined by positive PCR signal using neo or Nnt polymorphism assay.
Four B6/NTac chimeric males died before reaching breeding age; one each was euthanized due to hydrocephaly or cleft jaw, while two others died of unknown causes.
Two B6/J-host chimeric males died of unknown causes before reaching breeding age.
In comparison, the testis weight in an adult B6J or B6NTac animal is similar, ranging between 112 – 122 mg.
RESULTS
Identification and initial characterization of chimeric animals
Microinjection of ES cells into C57BL/6J (B6J) or C57BL/6NTac (B6NTac) blastocysts produced a total of 31 offspring from each type of host blastocyst (Table 1). To identify animals with contribution from ES cells we performed semi-quantitative PCR analysis of DNA from tail biopsy of each animal. Animals generated using B6J (Nnt mutant) embryos were genotyped for the presence of both neo and the wild type allele of Nnt present in JM8_2H5 (B6NTac) ES cells. Genotypes of chimeras generated using B6NTac embryos were informative for neo alone. By this assay, 19 of 31 animals produced using B6NTac blastocysts, and 25 of 31 animals produced using B6J blastocysts were chimeric (Table 1). Hereafter, we refer to these animals as “B6NTac chimeras” and “B6J chimeras”. When analyzed at weaning, 15 (79%) of the 19 B6NTac chimeras and 20 (80%) of the 25 B6J chimeras had male external genitalia. Female chimeras were not analyzed further. Two male chimeras from each host strain blastocyst background died due to unknown causes after weaning but before being tested for fertility. Two additional B6NTac chimeras were euthanized due to developmental abnormalities (one each; hydrocephalus and deformed lower jaw). Eleven B6NTac and fourteen B6J male chimeras remained for further analysis (Table 1).
Breeding analysis of male chimeras
To analyze fecundity and to determine whether the injected ES cells had contributed to the germ line, we bred surviving chimeric males with two B6NTac females in trios for at least four months. Six of 11 (55%) B6NTac chimeric males were fertile while 12 of 14 (86%) of B6J chimeric males produced offspring (Table 1). Each fecund chimeric male sired a first litter of pups within 21 – 25 days after establishing matings, with the exception of one B6NTac chimeric male who first sired pups 11 weeks after being mated. During the 4-month mating period a greater proportion (10 of 12; 83%) of fertile B6J chimeras sired more than a single litter compared with B6NTac chimeras (3 of 6; 50%). Five of 11 (45%) B6NTac chimeric males but only 2 of 14 (14%) B6J chimeric males failed to produce offspring, despite substituting their females with proven breeder females for an additional 3 months of breeding. During the entire breeding period, fecund B6NTac chimeric males sired fewer litters per productive mating (2.17 ± 1.33, n=6) than did B6J chimeric males (3.42 ± 1.73, n=12). To identify germ line transmission of DNA derived from ES cells, offspring of chimeras were genotyped for the presence of neo. Four of 11 (36%) B6NTac male chimeras produced offspring arising from ES cell-derived gametes while a larger proportion of B6J chimeras, 9 of 14 (64%), produced offspring from ES cell-lineage gametes (Table 1).
Analysis of internal organs and testis of chimeric males
After breeding, chimeras were necropsied and the gross morphology of internal organs analyzed with particular attention to the reproductive system. The mean body mass of B6NTac chimeric males (55.1 ± 8.3 g, n=9) was significantly greater than that of B6J chimeras (47.7 ± 5.5 g, n=12). This difference is similar to that observed in non-chimeric animals from the different strain backgrounds of mice used to produce blastocysts. Analysis of 142 day old stud male B6J (29.88 ±1.42 g, n=12) and B6NTac (32.24 ± 3.71 g, n=18) mice in our colony housed under identical conditions showed a similar difference in body weight. The mean testicular weight of B6NTac-host chimeras (53.9 ± 30.6 mg, n=16) was significantly less than that of B6J-host chimeras (80.6 ± 23.2 mg; n=23). Again, a similar difference is observed in different strain backgrounds of mice used to produce blastocysts, with average paired testis weight of adult (8–10 wk) C57BL/6J mice being 182 ± 8 mg (n=5) (Handel 2011), while the values for C57BL/6NTac mice are 113 ± 6 mg (n=10) (B6 physiological data, Feb 2004, Taconic.com). The mean weight of seminal vesicles was also lower in B6NTac chimeras (575 ± 320 mg; n=9) compared with B6J chimeras (701 ± 282 mg; n=12). Seven of 9 B6NTac chimeric males had abnormal internal organs. The most common abnormality, found in 4 out of 9 (44%) animals, was the presence of mammary glands on one side of the body. The mass and internal structure of these glands were similar to that found in adult virgin females (data not shown). Three of 9 B6NTac chimeras had markedly enlarged kidneys or adrenal glands and two animals were hermaphrodites. In contrast, only 1 of 12 (8 %) of B6J chimeras analyzed showed any abnormality of internal organs, this animal being a hermaphrodite (Table 1).
The status of spermatogenesis in chimeras was examined by quantifying the number of atrophic seminiferous epithelia in testis histology (data not shown). The mean percentage of atrophic tubule cross-sections was similar in B6NTac chimeras (25.3 ± 20.4 %, n=9) and B6J chimeras (18.7 ± 23.2, n=12). As anticipated, there was a general inverse correlation between percentage of atrophic tubules and testis weight. However, chimeras produced from either strain background with a relatively large proportion of atrophic tubules and reduced testis weight could be fertile and fecund. Conversely, chimeras on either strain background could have testis weights approaching that of non-chimeric control animals, and have few atrophic seminiferous tubules, but fail to sire offspring.
Quantification of relative contribution of ES cells to testis, heart, lung and brain in chimeric males
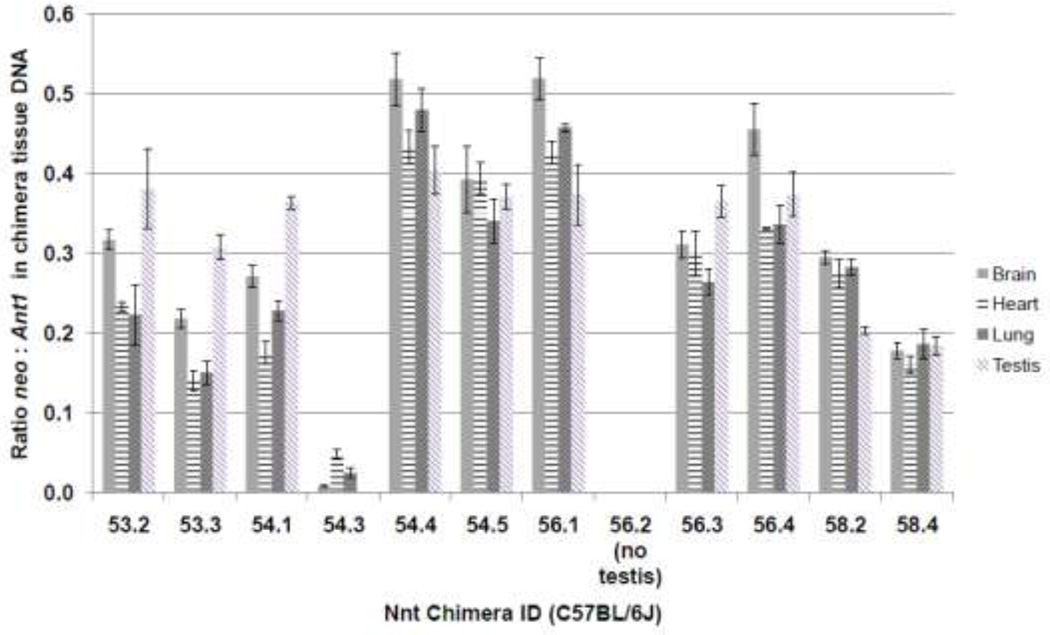
To determine if there was a correlation between contribution of ES cells to the testis, and ability to transmit ES-derived gametes (i.e. GLT-chimera), total genomic DNA was isolated from one testis of chimeras and analyzed by qPCR for the relative ratio of neo (JM8_2H5 ES cell derived) to a locus (Ant1; official name Slc25a4) present in both ES cells and the host blastocyst. The PCR assay used could detect at least 1% contribution of ES cells (data not shown) and linear through over a hundred fold difference in DNA concentration (data not shown). We analyzed total genomic DNA from testes from eleven B6J chimeras and four B6NTac chimeras (Fig. 1). Each GLT-chimera had at least 30% contribution of ES cell-derived DNA to the testis, although 1 animal (#56.4) out of 11 having at least 30% contribution did not transduce the ES cell derived allele. Two B6J chimeras (#58.2 and 58.4), each with approximately 20% contribution of ES cell DNA to the testis, were fertile, although no ES cell-derived gamete was transmitted during the breeding analysis.
Fig. 1. Quantification of relative amount of neo DNA in somatic tissues and testis in chimeric animals.
Each bar chart represents the amount of neo target relative to Ant1 in total DNA isolated from a testis, brain, heart or lung from the indicated chimeras. The bar indicates the mean value of three experimental replicates and each error bar represents two standard deviations. A value of 0.4 corresponds to 40% of the DNA originating from ES cell derivatives.
To determine whether there was consistent contribution of ES cells to tissues from different germ layers, as well as whether this was similar to the ES cell contribution to the testis, we also performed qPCR for neo and Ant1 target sequences using total genomic DNA from lung (endoderm), brain (ectoderm) and heart (mesoderm) of B6J chimeras (Fig. 1). In six chimeras (#’s 54.4, 54.5, 56.1, 56.4, 58.2, 58.4) the relative concentration of neo (normalized to Ant1) in testis was similar to, or slightly less than, that found in somatic tissues of the same chimera (Fig. 1). However, in four chimeras (#’s 53.2, 53.3, 54.1 and 56.3) the amount of neo target in testis DNA was greater than that in somatic tissues from the same animal. A plausible reason for the latter finding is that the host blastocyst in each of these four animals was 40,XX. In this scenario any 40,XX host blastocyst-derived germ cells (neo negative) present in the neonatal testis would not progress beyond T1-prospermatogonia (Burgoyne 1987), allowing expansion of the (40,XY) ES cell-derived (neo positive) prospermatogonia to populate the neonatal testis. Regarding the relative amount of neo target in tissues representing different germ layers, with the exception of one animal (# 54.3), brain consistently had an equal or greater contribution of ES cell-derived DNA compared with either lung or heart (Fig. 1).
Discussion
To identify ways to improve the efficiency of production and subsequent reproductive performance of male ES-cell derived chimeric mice in which the ES cells contribute to functional gametes (i.e. germ line transmission (GLT) - chimeras), we compared use of blastocysts from C57BL/6J (B6J) and the closely related strain C57BL/6NTac (B6NTac) as hosts for injected B6NTac-derived ES cells. As the B6J strain is functionally null for Nnt, we reasoned that during development of the chimera, the injected JM8.N4 (B6NTac) derived Nnt wild type (40,XY) ES cells might contribute differently to tissues of the chimera than when they are injected into Nnt wild type C57BL/6NTac hosts. In addition, C57BL/6J mice have superior reproductive characteristics including testis weight and sperm count (Chubb 1992; Handel 2011) that could also influence breeding performance of chimeric animals.
While the numbers of animals analyzed does not permit determination of statistical significance at P<0.05 for all parameters analyzed, of the adult male chimeras bred, a greater ratio of GLT-chimeric males was obtained using B6J blastocysts (9/14; 64%) compared with chimeras produced using B6NTac blastocysts (4/11; 36%). Comparison of several other parameters supports a conclusion that B6J blastocysts may be better hosts for production of chimeric mice compared to B6NTac blastocysts. With the ES cell line injected, the B6J chimeric animals had significantly fewer abnormalities in their urogenital system and mammary glands compared with B6NTac-host chimeras (1/12; 8%, compared with 7/9; 78%). Abnormal development of the reproductive system in chimeric mice can occur following injection of 40,XY ES cells into a 40,XX blastocyst. The abnormalities are usually a consequence of insufficient contribution of 40,XY ES cells to the gonadal rudiments prior to the time of sexual differentiation (i.e. around e10.5) in a chimeric embryo originating from a 40,XX host blastocyst. This can result in a chimera whose reproductive organs and tract can vary from female to inter-sex (hermaphrodite). Defects in normal development of the urogenital system could also account for the unilateral cystic kidney and fluid-filled Müllerian duct derivatives (uterus, oviduct) observed in some B6NTac chimeras. One possibility is that the injected ES cells were better able to contribute to development of the urogenital system in the B6J host-blastocyst derived embryos. It would be of interest to investigate if the low incidence of such defects in B6J chimeras is associated with the injected Nnt wild type ES cells having a competitive advantage in colonizing specific embryonic tissues in B6J blastocyst-derived chimeras compared with B6NTac chimeras.
For all fertile chimeras, B6J chimeras produced more litters than their B6NTac counterparts. The improved breeding performance of B6J chimeras might be due to their reduced adult body mass compared to B6NTac chimeras (47.7 ± 5.5 g for B6J chimeras (n=12), versus 55.1 ± 8.1g for B6NTac chimeras (n=9), both at 38 weeks old). Non-chimeric 20 week-old stud C57BL/6J male mice in our colony also have reduced body mass (29.88 ±1.42 g, n=12) compared to C57BL/6NTac mice (32.24 ± 3.71 g, n=18) housed under identical conditions. Thus, the difference in weight observed in chimeric animals may be independent of the injected ES cell line and due to the strain background of the host blastocyst. Regardless, the reduced weight of the adult male B6J chimeras may contribute to improved breeding performance of these animals as they age relative to their B6NTac counterparts.
As with inbred C57BL/6J and C57BL/6NTac males, the mean testis weight of B6J chimeric males was significantly greater than that of B6NTac chimeras. However, no consistent correlation was found between mean testis weight and the proportion of abnormal seminiferous tubules, and fecundity (i.e. numbers of litters sired per male) for chimeras from either host-blastocyst background. For example, two B6NTac chimeras with 31% and 28% atrophic tubules were fertile and transmitted ES cell-derived gametes, although these animals each sired only one litter during the test period. In comparison, two B6J chimeras with 43% and 45% atrophic tubules each sired between 3 and 6 litters from ES-cell derived sperm during the same period. Conversely, a B6NTac chimera with testis weights of 91 and 97 mg, and < 1% abnormal tubules failed to sire offspring, as did B6J chimera # 58.2 which had testis weights of 94 and 102 mg and <1% atrophic tubules.
Since the first report fifty years ago of chimeric mice produced by embryo aggregation, coat color genetics has enabled simple visual assessment of the degree of contribution of genetically distinct cells in a chimeric mouse (Tarkowski 1961). Although simple, this method is semi-quantitative and influenced by investigator bias, which confounds comparison of results from different studies. Quantitative PCR allows accurate quantification of the relative contribution of cells of different genotypes in a tissue, although it cannot provide information on the distribution of cells in that tissue. Methods for quantitation of coding sequence for neo (commonly used while targeting mutations in ES cells) in mouse tissues will enable accurate and reproducible quantification of the percentage of chimerism in different tissues. When used in conjunction with JM8.A3 ES cells, which are derived from C57BL/6NTac ES cells in which function of the Agouti locus has been restored (Pettitt et al. 2009), quantification can be performed using both qPCR and coat color. Based on our data with the combination of JM8.N4 ES cells and C57BL/6J or C57BL/6NTac embryos, we predict that a threshold of 30% neo (or other ES derived) chimerism may be a useful working predictor for determining whether a chimeric male may transmit ES-derived DNA through the germline. This prediction is based on an assumption that the proportion of ES cell derived DNA found in heart, brain, lung and testis is similar to that found in tail, which is comprised of ectoderm and mesoderm derivatives. In conclusion, use of C57BL/6J host blastocysts for microinjection of ES cells may offer improvements over blastocysts from C57BL/6NTac and possibly other sub-strains of C57BL/6N mice.
Acknowledgements
We thank Dr. W. Skarnes for the generous gift of JM8.N4 C57BL/6NTac ES cells and Dr. G. Melkonian and S. Donovan for assistance with genotyping. This work was supported by funds from UCI’s Office of Research, the Chao Family Comprehensive Cancer Center Support Grant (P30CA062203) from the National Cancer Institute, and by grants to D.C.W. and G.R.M. (HD-45913, HL-102862) from the National Institutes of Health. H.-W. C. was supported by a fellowship from the George E. Hewitt Foundation for Medical Research.
Footnotes
Author Contributions – C.S.Y. and H.-W. C. performed experiments and interpreted data; J.M prepared all histology; S.W. performed all ES cell culture, semi-quantitative PCR and analyzed data; K.-X. S. performed all microinjection and embryo manipulation; K.G.W. co-supervised the study, trained J.M, and H.-W. C., performed some of the experiments and analyzed data; D.C.W. provided the JM8_2H5 ES cell line; T.J.F. coordinated the study, performed some of the experiments, analyzed data and edited the manuscript; G.R.M. conceived the study, performed some of the experiments, analyzed data and wrote the manuscript.
Literature cited
- Arkblad EL, Betsholtz C, Mandoli D, Rydstrom J. Characterization of a nicotinamide nucleotide transhydrogenase gene from the green alga Acetabularia acetabulum and comparison of its structure with those of the corresponding genes in mouse and Caenorhabditis elegans. Biochim Biophys Acta. 2001;1520(2):115–123. doi: 10.1016/s0167-4781(01)00257-3. [DOI] [PubMed] [Google Scholar]
- Arkblad EL, Tuck S, Pestov NB, Dmitriev RI, Kostina MB, Stenvall J, Tranberg M, Rydstrom J. A Caenorhabditis elegans mutant lacking functional nicotinamide nucleotide transhydrogenase displays increased sensitivity to oxidative stress. Free Radic Biol Med. 2005;38(11):1518–1525. doi: 10.1016/j.freeradbiomed.2005.02.012. [DOI] [PubMed] [Google Scholar]
- Burgoyne PS. The role of the mammalian Y chromosome in spermatogenesis. Development. 1987;101(Suppl):133–141. doi: 10.1242/dev.101.Supplement.133. [DOI] [PubMed] [Google Scholar]
- Campbell PK, Waymire KG, Heier RL, Sharer C, Day DE, Reimann H, Jaje JM, Friedrich GA, Burmeister M, Bartness TJ, Russell LD, Young LJ, Zimmer M, Jenne DE, MacGregor GR. Mutation of a novel gene results in abnormal development of spermatid flagella, loss of intermale aggression and reduced body fat in mice. Genetics. 2002;162(1):307–320. doi: 10.1093/genetics/162.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin HJ, Wang CK. Utero-tubal transfer of mouse embryos. Genesis. 2001;30(2):77–81. doi: 10.1002/gene.1036. [DOI] [PubMed] [Google Scholar]
- Chubb C. Genes regulating testis size. Biol Reprod. 1992;47(1):29–36. doi: 10.1095/biolreprod47.1.29. [DOI] [PubMed] [Google Scholar]
- Glaser S, Anastassiadis K, Stewart AF. Current issues in mouse genome engineering. Nat Genet. 2005;37(11):1187–1193. doi: 10.1038/ng1668. [DOI] [PubMed] [Google Scholar]
- Handel M-A. Male reproductive parameters in 14 inbred strains of mice. . MPD:Handel1. Mouse Phenome Database web site. Bar Harbor, Maine USA: The Jackson Laboratory; 2011. [Accessed 11/10/2011]. http://phenome.jax.org. 2011. [Google Scholar]
- Hoek JB, Rydstrom J. Physiological roles of nicotinamide nucleotide transhydrogenase. Biochem J. 1988;254(1):1–10. doi: 10.1042/bj2540001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang TT, Naeemuddin M, Elchuri S, Yamaguchi M, Kozy HM, Carlson EJ, Epstein CJ. Genetic modifiers of the phenotype of mice deficient in mitochondrial superoxide dismutase. Hum Mol Genet. 2006;15(7):1187–1194. doi: 10.1093/hmg/ddl034. [DOI] [PubMed] [Google Scholar]
- Krzanowska H. Influence of Y chromosome on fertility in mice. In: Beatly R, Gluecksohn-Welsch S, editors. Edinburgh Symposium on the Genetics of Spermatozoon. Edinburgh: The University Press; 1971. pp. 370–386. [Google Scholar]
- Krzanowska H, Wabik-Sliz B, Rafinski J. Phenotype and fertilizing capacity of spermatozoa of chimaeric mice produced from two strains that differ in sperm quality. J Reprod Fertil. 1991;91(2):667–676. doi: 10.1530/jrf.0.0910667. [DOI] [PubMed] [Google Scholar]
- Le Roy I, Tordjman S, Migliore-Samour D, Degrelle H, Roubertoux PL. Genetic architecture of testis and seminal vesicle weights in mice. Genetics. 2001;158(1):333–340. doi: 10.1093/genetics/158.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledermann B, Burki K. Establishment of a germ-line competent C57BL/6 embryonic stem cell line. Exp Cell Res. 1991;197(2):254–258. doi: 10.1016/0014-4827(91)90430-3. [DOI] [PubMed] [Google Scholar]
- MacGregor GR. An extreme bias in the germ line of XY C57BL/6<->XY FVB/N chimaeric mice. Reproduction. 2002;124(3):377–386. doi: 10.1530/rep.0.1240377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekada K, Abe K, Murakami A, Nakamura S, Nakata H, Moriwaki K, Obata Y, Yoshiki A. Genetic differences among C57BL/6 substrains. Exp Anim. 2009;58(2):141–149. doi: 10.1538/expanim.58.141. [DOI] [PubMed] [Google Scholar]
- Mintz B. Hermaphroditism, sex chromosomal mosaicism and germ cell selection in allophenic mice. J Anim Sci. 1968;27(Suppl 1):51–60. [PubMed] [Google Scholar]
- Nicholson A, Reifsnyder PC, Malcolm RD, Lucas CA, MacGregor GR, Zhang W, Leiter EH. Diet-induced obesity in two C57BL/6 substrains with intact or mutant nicotinamide nucleotide transhydrogenase (Nnt) gene. Obesity (Silver Spring) 2010;18(10):1902–1905. doi: 10.1038/oby.2009.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaioannou V, Johnson R. Production of chimeras and genetically defined offspring from targeted ES cells. In: Joyner A, editor. Gene targeting - a practical approach. Oxford: IRL Press; 1993. pp. 107–146. [Google Scholar]
- Pettitt SJ, Liang Q, Rairdan XY, Moran JL, Prosser HM, Beier DR, Lloyd KC, Bradley A, Skarnes WC. Agouti C57BL/6N embryonic stem cells for mouse genetic resources. Nat Methods. 2009;6(7):493–495. doi: 10.1038/nmeth.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poueymirou WT, Auerbach W, Frendewey D, Hickey JF, Escaravage JM, Esau L, Dore AT, Stevens S, Adams NC, Dominguez MG, Gale NW, Yancopoulos GD, DeChiara TM, Valenzuela DM. F0 generation mice fully derived from gene-targeted embryonic stem cells allowing immediate phenotypic analyses. Nat Biotechnol. 2007;25(1):91–99. doi: 10.1038/nbt1263. [DOI] [PubMed] [Google Scholar]
- Schuster-Gossler K, Lee A, CP L, HJ P, VW D, VE S, Gossler A, Conover J. Use of coisogenic host blastocysts for efficient establishment of germline chimeras with C57BL/6J ES cell lines. Biotechniques. 2001;31(5):1022–1024. doi: 10.2144/01315st01. [DOI] [PubMed] [Google Scholar]
- Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, Jackson D, Severin J, Biggs P, Fu J, Nefedov M, de Jong PJ, Stewart AF, Bradley A. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474(7351):337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarkowski AK. Mouse chimaeras developed from fused eggs. Nature. 1961;190:857–860. doi: 10.1038/190857a0. [DOI] [PubMed] [Google Scholar]
- Toye AA, Lippiat JD, Proks P, Shimomura K, Bentley L, Hugill A, Mijat V, Goldsworthy M, Moir L, Haynes A, Quarterman J, Freeman HC, Ashcroft FM, Cox RD. A genetic and physiological study of impaired glucose homeostasis control in C57BL/6J mice. Diabetologia. 2005;48(4):675–686. doi: 10.1007/s00125-005-1680-z. [DOI] [PubMed] [Google Scholar]
- van der Weyden L, Adams DJ, Bradley A. Tools for targeted manipulation of the mouse genome. Physiol Genomics. 2002;11(3):133–164. doi: 10.1152/physiolgenomics.00074.2002. [DOI] [PubMed] [Google Scholar]