Abstract
One of the most critical factors in gene expression studies using quantitative real-time PCR is the choice of reference gene. Many of the commonly used reference genes have been shown to vary during a number of biological processes as well as between tissues. It is therefore important to always verify the stability of the gene of choice for all new tissues and experimental conditions. Here, we used two publicly available computer software packages (GeNorm and NormFinder) to investigate the stability of eight potential reference genes in formalin-fixed paraffin-embedded (FFPE) samples from normal oral tissue of different origin as well as from oral squamous cell carcinomas. Both programs found the tubulin α-6 chain (TUBA6) and ribosomal protein S13 (RPS13) to have the most stable expression between malignant and non-malignant tissue. NormFinder also found TUBA6 to be the most stable gene when samples were grouped according to tissue origin. FFPE samples constitute a large research resource, which considerably increases the number of samples available for analysis, leading to more reliable conclusions. Verification of a proper reference gene in oral FFPE tissue is therefore of great importance for future studies.
Keywords: oral squamous cell carcinoma, reference gene, tubulin α-6 chain, quantitative real-time PCR
Introduction
Squamous cell carcinoma of the head and neck (SCCHN) is the sixth most common malignancy worldwide. Approximately 500,000 new cases and 320,000 deaths are reported annually (1). Despite major technical improvements in both surgery and radiotherapy, the 5-year survival has not changed during the past three decades and remains at approximately 50% (2–4). The head and neck region includes several different sites such as the oral cavity, pharynx, larynx, nasal cavity, paranasal sinuses and salivary glands, and the clinical behaviour, prognosis and response to treatment varies between tumours from different sites. Major contributing factors to the non-improved 5-year survival are the lack of good biomarkers for predicting treatment response and prognosis and the fact that tumours are often detected at a late stage (4,5).
One commonly used strategy for the identification of novel therapeutic targets and biomarkers is the investigation of levels of gene expression in tumours compared to normal tissue, and a number of possible targets for treatment that are overexpressed at the mRNA level, for example EGFR and VEGF, are currently used in clinical trials (6,7).
Quantitative real-time PCR (q-PCR) is the most commonly used method in small scale gene expression studies due to its rapidity, high sensitivity and good reliability (8,9). One of the most debated and critical factors for the detection of differences in gene expression is normalization, and errors here may result in misinterpretation of the results (10). Even though a variety of strategies have been employed, the normalization of results using a single internal constitutively expressed reference gene is generally accepted as the standard approach (9,10). The most basic requirement on an internal reference gene is that it is stably expressed between cells or tissues under the conditions being investigated. In recent years, it has become clear that no single such gene exists; instead, many of the commonly used reference genes have been shown to vary, for example during differentiation, cell growth and different disease states, illustrating the need to verify the stability of a reference gene in all tissues and under all experimental conditions used (9,11).
Archival formalin-fixed paraffin-embedded (FFPE) patient material is a large source for research. However, due to the fragmented nature of RNA in these samples, they have been of limited use (12,13). Methods for the extraction and analysis of RNA from these samples have been developed during the last few years, and studies utilizing FFPE samples are becoming more common (14,15). RNA from these samples is not only fragmented but also highly modified, rendering it resistant to reverse transcriptase reactions. Verification of proper reference genes is therefore of great importance in these samples.
Here, we investigated the stability of eight potential reference genes in FFPE samples from normal oral tissue and oral squamous cell carcinomas (OSCCs). Two publicly available software packages (GeNorm and NormFinder) were used to analyse data, and tubulin α-6 chain (TUBA6) and ribosomal protein S13 (RPS13) were identified as stable reference genes in both normal and malignant oral tissue.
Materials and methods
Samples and RNA extraction
FFPE samples from 10 patients with OSCC and 10 patients with benign oral conditions, such as hyperplasia, were available for analysis. The material comprised tissue from the gingiva (tumour, n=5; control, n=2), hard palate (control, n=2), tongue (tumour, n=4; control, n=4) and buccal mucosa (tumour, n=1; control, n=2). The study was approved by the local Ethics Committee at the Umeå University (dnr 01-057 and dnr 08-003M). FFPE samples were cut into 5-μm sections and, depending on the size of the sample, 3–20 sections were pooled giving an approximate total area of 1 cm2. RNA was isolated using the High Pure RNA Paraffin kit (Roche Diagnostics GmbH, Mannheim, Germany) according to the manufacturer’s instructions. RNA from the pharyngeal squamous cell carcinoma cell line FaDu was extracted by the use of TRIzol (Invitrogen, Carlsbad, CA, USA) and used when creating standard curves for all primers in the q-PCR reaction. The purity of the RNA samples was evaluated by the 260/280-and the 260/230-nm absorbance ratio.
Quantitative real-time PCR (q-PCR)
Primers were designed using Primer 3 software, and their specificity was evaluated using the primer search in NCBI BLAST. All amplicons were kept under 100 bp in order to optimize the conditions for the amplification of degraded RNA and were also designed to span an intron, to avoid the amplification of genomic DNA (Table I) (16). First-strand cDNA was synthesised using the Cloned AMV First-Strand cDNA Synthesis kit (Invitrogen) using 200 ng of RNA and random primers in a 20-μl reaction mixture. The cDNA reaction (1 μl) was used as template for the amplification of TUBA6, S100A6, ACTB, OAZ1, GAPDH, RPS13, RPL27 and HPRT1, respectively. The q-PCR reaction was carried out in a 20-μl reaction using IQ SYBR Green Supermix (BioRad, Hercules, CA, USA) according to the manufacturer’s recommendations. Reactions were carried out in triplicate. For a selection of genes (RPS13, OAZ1 and RPL27) all samples having a standard deviation (σ) > 0.3 between the three replicates were re-run at a later occasion for the evaluation of the reproducibility of the assay.
Table I.
Gene and primer information.
| Gene | Accession no. | Full name | Location | Primer | Size (bp) | Efficiencya |
|---|---|---|---|---|---|---|
| TUBA6 | NM_032704 | Tubulin α-6 chain | 12q12-q14 | F: CCGGGCAGTGTTTGTAGACT | 99 | 2.04 |
| R: TTGCCTGTGATGAGTTGCTC | ||||||
| S100A6 | NM_014624 | S100 calcium binding protein A6 | 1q21 | F: ACAAGCACACCCTGAGCAAGA | 99 | 1.92 |
| R: CCATCAGCCTTGCAATTTCA | ||||||
| ACTB | NM_001101 | β-actin | 7p15-p12 | F: CCAACCGCGAGAAGATGAC | 96 | 1.88 |
| R: CAGAGGCGTACAGGGATAGC | ||||||
| OAZ1 | NM_004152 | Ornithine decarboxylase antizyme | 19p13.3 | F: GAGCCGACCATGTCTTCATT | 84 | 1.99 |
| R: CAAAGCCCAAAAAGCTGAAG | ||||||
| GAPDH | NM_002046 | Glyceraldehyde-3-phosphate dehydrogenase | 12p13 | F: CTCTGCTCCTCCTGTTCGAC | 99 | 2.00 |
| R: TTGACTCCGACCTTCACCTT | ||||||
| RPS13 | NM_001017 | Ribosomal protein S13 | 11p15 | F: CAGTCGGCTTTACCCTATCG | 95 | 1.99 |
| R: CCCTTCTTGGCCAGTTTGTA | ||||||
| RPL27 | NM_000988 | Ribosomal protein L27 | 17q21.1-21.2 | F: ATGAAACCTGGGAAGGTGGT | 90 | 2.00 |
| R: TGAGGTGCCATCATCAATGT | ||||||
| HPRT1 | NM_000194 | Hypoxanthine phosphoribosyltransferase 1 | Xq21.6 | F: TCCTTGGTCAGGCAGTATAATCC | 90 | 1.99 |
| R: GTCTGGCTTATATCCAACACTTCG |
An efficiency of 2 represents a doubling with every PCR cycle.
Statistical analysis
Ct values from the PCR reaction were transformed to relative quantities using the relative standard curve method (17). Identical PCR reactions carried out at two different time points generate different relative quantities, and a dimensionless dispersion parameter like the coefficient of variation (CV) is therefore needed to evaluate the reproducibility of the assay. CV is calculated by dividing the σ-value by the mean. Ratios, necessary for calculating the CV-values, were obtained by dividing samples to one arbitrarily chosen sample analysed with the same primer and from the same run. The σ-value and the mean for each ratio was then calculated between the two runs and divided to calculate the CV-values.
For comparing the stabilities of the candidate genes, two publicly available software packages, GeNorm, a Visual Basic application tool for Microsoft Excel, and NormFinder, a Microsoft Excel add-in, were used (18,19) according to the developer’s instructions. In NormFinder, samples were grouped according to the origin of tissue or the presence or absence of malignancy. A Mann-Whitney U test was used to pre-evaluate the stability of the genes.
Results
Selection of reference gene candidates and design of primers
Eight genes previously described to have housekeeping properties were selected for the investigation of stability in FFPE samples of normal oral tissue and SCCHN. This included a number of commonly used reference genes, e.g., ACTB and GAPDH, but also more recently suggested reference genes such as OAZ1 and TUBA6 (18,20). Genes of different functions and location in the genome were selected in order to reduce the risk of co-regulation between genes, which would influence particularly results from GeNorm calculations. However, the two ribosomal genes RPL27 and RPS13, which are likely to show correlated expression, were included, as both were reported to have stable expression over a large number of tissues and conditions in a recent study covering 13,629 gene expression arrays (20). Primers optimal for the amplification of degraded RNA were designed for each gene (21). Data on selected genes and primers are summarized in Table I.
Expression levels of the eight candidate genes
All samples were analyzed for one gene at a time on a single PCR plate. RNA extracted from the cell line FaDu, which highly expresses all eight selected reference genes, was used in parallel for obtaining standard curves. All primers showed high efficiency (1.9-2.1) except ACTB, which had a slightly lower efficiency of 1.88 (Table I). The PCR reaction first included two replicates of each sample, but this was increased to three as standard deviations >0.3 were noted between some of the replicates. All samples run with two replicates were re-analyzed using three replicates. To investigate the reproducibility of the assay, all samples showing a σ > 0.3 for three of the genes were also re-analyzed a second time, and CV-values were calculated between the runs. The mean CV was 0.05, showing that the use of three replicates was sufficient to provide highly reproducible means. A bar graph over the means and σ-values of ratios between the two runs is shown in Fig. 1.
Figure 1.
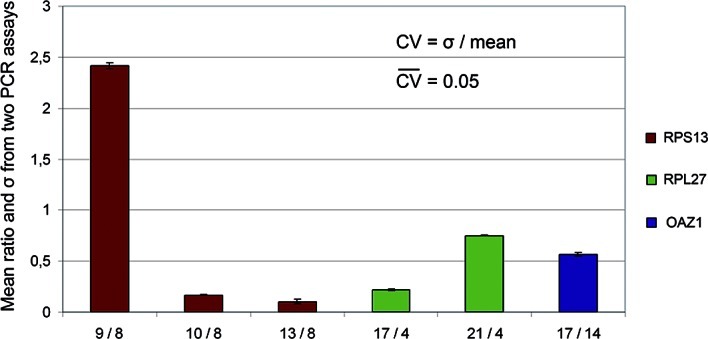
Chart of the mean and σ between two PCR runs. Four samples (8, 9, 10 and 13) amplified with RPS13 primers (red) showed a σ > 0.3 in the first run and were therefore re-run a second time. Ratios were calculated by dividing all samples by one arbitrary sample from the same PCR run, and means between the two runs were calculated for each ratio. The CV was then calculated by dividing the σ-value by the mean. The same was performed for RPL27 (three samples, 4, 17 and 21) and OAZ1 (two samples, 17 and 14). All Ct-values were transformed to relative quantities before calculations.
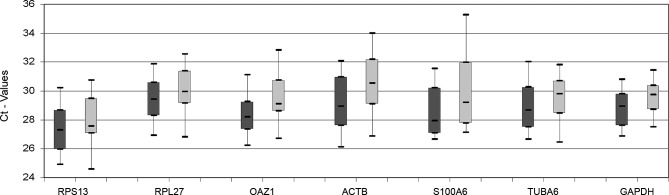
HPRT1, one of the candidate reference genes generally showed high Ct-values and could not be detected in some of the samples. This gene was therefore excluded from further analysis. The other seven genes were all detected between 25–35 cycles. Data for malignant and non-malignant samples are shown as separate box plots in Fig. 2. The expression ranges span from 6.2 cycles (GAPDH) to 10.7 cycles (ACTB). None of the genes showed a significantly different expression between tumours and controls using the Mann-Whitney U test. Although a significance test could indicate a gene’s unfitness as a reference, it is important to keep in mind that these calculations were based on non-normalized data relying on equal input material, which is difficult to control; hence, the reason there is a need for good reference genes. Due to this circular problem, a number of programs (GenNorm and NormFinder) using alternative strategies to simple significance testing have been developed for the evaluation of reference gene stability (10).
Figure 2.
Box plot of absolute Ct-values for seven reference genes. Dark grey boxes represent controls and light grey boxes represent tumours.
Expression stability of candidate genes in control and tumour samples
Ct-values were transformed to relative quantities using the standard curve method and submitted to two different software packages, GeNorm and NormFinder, for the identification of the most stable genes. GeNorm calculates the stability of genes by pairwise comparisons, generating M-values, and is based on the assumption that the ratio of two proper reference genes will be approximately similar in all samples. According to GeNorm, RPL27 and RPL13 had the most stable expression (Table IIA). Since GeNorm is very sensitive to co-regulation, this could be an artefact of the two ribosomal genes showing highly concordant expression. Data was therefore re-analysed, sequentially excluding one of the two ribosomal genes. Excluding RPL27 resulted in RPS13 being the most stable gene in combination with TUBA6, and the other way around (Table IIB).
Table II.
GeNorm.
| A, All seven genes.
| |
|---|---|
| Gene | M-value |
| RPS13 and RPL27 | 0.286 |
| TUBA6 | 0.385 |
| ACTB | 0.471 |
| GAPDH | 0.543 |
| OAZ1 | 0.598 |
| S100A6 | 0.763 |
| B, RPL27 excluded.
| |
|---|---|
| Gene | M-value |
| RPS13 and TUBA6 | 0.457 |
| ACTB | 0.525 |
| GAPDH | 0.606 |
| OAZ1 | 0.648 |
| S100A6 | 0.817 |
NormFinder, which is a model-based approach that is less sensitive to co-regulation and takes both the overall expression variability as well as the variation between subgroups into consideration, also suggested TUBA6 to be the most stable reference gene, while RPS13 and RPL27 were placed second and fifth, respectively (Table IIIA).
Table III.
NormFinder.
| A, Grouped as malignant or non-malignant.
| |
|---|---|
| Gene | Stability value |
| TUBA6 | 0.161 |
| RPS13 | 0.182 |
| GAPDH | 0.187 |
| ACTB | 0.188 |
| RPL27 | 0.195 |
| OAZ1 | 0.200 |
| S100A6 | 0.314 |
| B, Grouped as gingival and palate or tongue tissue.
| |
|---|---|
| Gene | Stability value |
| TUBA6 | 0.114 |
| RPL27 | 0.138 |
| RPS13 | 0.139 |
| OAZ1 | 0.175 |
| ACTB | 0.194 |
| GAPDH | 0.234 |
| S100A6 | 0.289 |
Expression stability of candidate genes in oral tissue in different sites
As NormFinder takes grouping into account, it was also of interest to determine whether the same genes were stably expressed between tissues at different sites in the oral cavity. Comparing tissue from the gingival and palate (n=9) to tongue tissue (n=8), or tissue from the gingival, palate and buccla mucosa (n=12) to tongue tissue (n=8), TUBA6, RPL27 and RPS13 were the three most stable genes (Table IIIB). The samples from buccal mucosa were too few to be considered a separate group in this comparison, as there should preferably be a minimum of eight samples in each group.
Discussion
Employment of quantitative real-time PCR has exploded in the last two decades, making it the golden standard in studies of gene expression. In recent years, it has become clear that verification of proper reference genes for each tissue and experimental condition is essential for correct interpretation of q-PCR expression data (9,11). This is even more important when using FFPE tissue, as separate mRNAs in these tissues have been shown to degrade to different extents (22). A poor overlap was also observed when examining the stability of genes in FFPE and fresh frozen (FF) tissue, emphasizing the importance of verifying reference genes in FFPE tissue separately (21).
Here, we looked at eight previously identified reference genes and investigated their stability in malignant and non-malignant tissue from different sites in the oral cavity. TUBA6 and RPLS13 had the most stable expression across all samples and conditions. NormFinder and GeNorm both placed TUBA6 and RPS13 as the two most stable reference genes when comparing malignant and non-malignant tissue, and NormFinder also found TUBA6 to be stably expressed across tissues from different sites in the oral cavity. OSCCs comprise a diverse family of tumours, where the site of the tumour has been suggested to effect responsiveness to treatment and clinical parameters (3). Nevertheless, OSCC is in many studies considered one disease where tumours from different sites in the oral cavity are grouped together, and a reference gene verified to be stable across both tissue and disease state is therefore essential. The commonly used reference genes ACTB and GAPDH are generally found in the middle of the stability list. However, when comparing tissue of different sites they were ranked lower, indicating that these genes may be fairly similar between cancer and controls, but less similar between tissues. S100A6 has in a few studies been described to significantly vary between malignant and non-malignant tissues; it was therefore not surprising that we also found it to be the least stable gene under all conditions (23,24). A smaller study of four candidate genes in three cell lines (two SCCHN cell lines and one non-malignant cell line) interestingly found another tubulin gene (TUBB) to be stably expressed in SCCHN cell lines (25).
Primers used on RNA from FFPE samples should span an intron, not exceed 100 bp, preferably be located away from the 5′ end, and also be specific to the gene/splice form of interest (21). The primer pair amplifying ACTB was not optimal, showing a reduced efficiency of 1.88. This was most probably caused by one of the primers having a second perfect match to the gene ACTA1, causing competition for primer binding and lowered PCR efficiency. This illustrates the importance and also the difficulties in designing primers for the amplification of RNA from FFPE material, particularly for genes of large conserved families. The verification of the efficiency of amplification is therefore required, though not always performed.
Retrospective clinical studies using FFPE samples have the potential to involve an extensive number of patients and thus increase our understanding of neoplastic progression and response to therapy. Identification of the appropriate reference genes will help to better extract adequate information from these samples. In a recent study of normal and malignant tongue FFPE, we used TUBA6 as a reference gene in the conformational q-PCR reactions of gene expression array data, and observed high concordance between the two methods (26). Confirmation included both highly and poorly expressed genes as well as large and small expression differences, and showed that TUBA6 is a proper normaliser, at least for tongue tissue.
Taken together, a number of studies on reference genes in different tissues have been published lately (27–29). However, to our knowledge this is the first study concerning reference genes in FFPE samples from normal oral tissue and OSCC. According to the results from this study as well as our previous results, TUBA6 is an appropriate reference gene for FFPE samples from the oral cavity (26).
Acknowledgments
This study was supported by grants from the Lion’s Cancer Research Foundation, Umeå University, the Swedish Cancer Society (contract no. 08 0371) and the Kempe foundation.
References
- 1.Shibuya K, Mathers CD, Boschi-Pinto C, Lopez AD, Murray CJ. Global and regional estimates of cancer mortality and incidence by site. II. Results for the global burden of disease 2000. BMC Cancer. 2002;2:37. doi: 10.1186/1471-2407-2-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Geisler SA, Olshan AF. GSTM1, GSTT1 and the risk of squamous cell carcinoma of the head and neck: a mini-HuGE review. Am J Epidemiol. 2001;154:95–105. doi: 10.1093/aje/154.2.95. [DOI] [PubMed] [Google Scholar]
- 3.Rautava J, Luukkaa M, Heikinheimo K, Alin J, Grenman R, Happonen RP. Squamous cell carcinomas arising from different types of oral epithelia differ in their tumor and patient characteristics and survival. Oral Oncol. 2007;43:911–919. doi: 10.1016/j.oraloncology.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 4.Bettendorf O, Piffko J, Bankfalvi A. Prognostic and predictive factors in oral squamous cell cancer: important tools for planning individual therapy? Oral Oncol. 2004;40:110–119. doi: 10.1016/j.oraloncology.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 5.Culliney B, Birhan A, Young AV, Choi W, Shulimovich M, Blum RH. Management of locally advanced or unresectable head and neck cancer. Oncology. 2008;22:1152–1161. [PubMed] [Google Scholar]
- 6.Lothaire P, de Azambuja E, Dequanter D, et al. Molecular markers of head and neck squamous cell carcinoma: promising signs in need of prospective evaluation. Head Neck. 2006;28:256–269. doi: 10.1002/hed.20326. [DOI] [PubMed] [Google Scholar]
- 7.Shirai K, O’Brien PE. Molecular targets in squamous cell carcinoma of the head and neck. Curr Treat Options Oncol. 2007;8:239–251. doi: 10.1007/s11864-007-0030-4. [DOI] [PubMed] [Google Scholar]
- 8.Ginzinger DG. Gene quantification using real-time quantitative PCR: an emerging technology hits the mainstream. Exp Hematol. 2002;30:503–512. doi: 10.1016/s0301-472x(02)00806-8. [DOI] [PubMed] [Google Scholar]
- 9.Huggett J, Dheda K, Bustin S, Zumla A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005;6:279–284. doi: 10.1038/sj.gene.6364190. [DOI] [PubMed] [Google Scholar]
- 10.Vandesompele J, Kubista M, Pfaffl M. Reference gene validation software for improved normalization. In: Logan J, Edwards K, Saunders N, editors. Real-Time PCR: Current Technology and Applications. Caister. 1st edition. Academic Press; London: 2009. pp. 47–64. [Google Scholar]
- 11.Ruan W, Lai M. Actin, a reliable marker of internal control? Clin Chim Acta. 2007;385:1–5. doi: 10.1016/j.cca.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 12.Masuda N, Ohnishi T, Kawamoto S, Monden M, Okubo K. Analysis of chemical modification of RNA from formalin-fixed samples and optimization of molecular biology applications for such samples. Nucleic Acids Res. 1999;27:4436–4443. doi: 10.1093/nar/27.22.4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Von Ahlfen S, Missel A, Bendrat K, Schlumpberger M. Determinants of RNA quality from FFPE samples. PLoS ONE. 2007;2:e1261. doi: 10.1371/journal.pone.0001261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark-Langone KM, Wu JY, Sangli C, et al. Biomarker discovery for colon cancer using a 761 gene RT-PCR assay. BMC Genomics. 2007;8:279. doi: 10.1186/1471-2164-8-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Penland SK, Keku TO, Torrice C, et al. RNA expression analysis of formalin-fixed paraffin-embedded tumors. Lab Invest. 2007;87:383–391. doi: 10.1038/labinvest.3700529. [DOI] [PubMed] [Google Scholar]
- 16.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 17.Larionov A, Krause A, Miller W. A standard curve based method for relative real time PCR data processing. BMC Bioinformatics. 2005;6:62. doi: 10.1186/1471-2105-6-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 19.Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Jonge HJ, Fehrmann RS, de Bont ES, et al. Evidence based selection of housekeeping genes. PLoS ONE. 2007;2:e898. doi: 10.1371/journal.pone.0000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drury S, Anderson H, Dowsett M. Selection of reference genes for normalization of qRT-PCR data derived from FFPE breast tumors. Diagn Mol Pathol. 2009;18:103–107. doi: 10.1097/PDM.0b013e31817c1ae2. [DOI] [PubMed] [Google Scholar]
- 22.Perez-Novo CA, Claeys C, Speleman F, van Cauwenberge P, Bachert C, Vandesompele J. Impact of RNA quality on reference gene expression stability. Biotechniques. 2005;39:52, 54, 56. doi: 10.2144/05391BM05. [DOI] [PubMed] [Google Scholar]
- 23.Hibbs K, Skubitz KM, Pambuccian SE, et al. Differential gene expression in ovarian carcinoma: identification of potential biomarkers. Am J Pathol. 2004;165:397–414. doi: 10.1016/S0002-9440(10)63306-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yao R, Lopez-Beltran A, Maclennan GT, Montironi R, Eble JN, Cheng L. Expression of S100 protein family members in the pathogenesis of bladder tumors. Anticancer Res. 2007;27:3051–3058. [PubMed] [Google Scholar]
- 25.Campos MS, Rodini CO, Pinto-Junior DS, Nunes FD. GAPD and tubulin are suitable internal controls for qPCR analysis of oral squamous cell carcinoma cell lines. Oral Oncol. 2009;45:121–126. doi: 10.1016/j.oraloncology.2008.03.019. [DOI] [PubMed] [Google Scholar]
- 26.Rentoft M, Laurell G, Coates P, Sjöström B, Nylander K. Gene expression profiling of archival tongue squamous cell carcinomas provides sub-classification based on DNA repair genes. Int J Oncol. 2009;35:1321–1330. doi: 10.3892/ijo_00000450. [DOI] [PubMed] [Google Scholar]
- 27.Cicinnati VR, Shen Q, Sotiropoulos GC, Radtke A, Gerken G, Beckebaum S. Validation of putative reference genes for gene expression studies in human hepatocellular carcinoma using real-time quantitative RT-PCR. BMC Cancer. 2008;8:350. doi: 10.1186/1471-2407-8-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lallemant B, Evrard A, Combescure C, et al. Reference gene selection for head and neck squamous cell carcinoma gene expression studies. BMC Mol Biol. 2009;10:78. doi: 10.1186/1471-2199-10-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lyng MB, Laenkholm AV, Pallisgaard N, Ditzel HJ. Identification of genes for normalization of real-time RT-PCR data in breast carcinomas. BMC Cancer. 2008;8:20. doi: 10.1186/1471-2407-8-20. [DOI] [PMC free article] [PubMed] [Google Scholar]