Abstract
Growth-hormone-secreting pituitary adenomas (GHomas) account for approximately 20% of all pituitary neoplasms. However, the pathogenesis of GHomas remains to be elucidated. To explore the possible pathogenesis of GHomas, we used bead-based fiber-optic arrays to examine the gene expression in five GHomas and compared them to three healthy pituitaries. Four differentially expressed genes were chosen randomly for validation by quantitative real-time reverse transcription-polymerase chain reaction. We then performed pathway analysis on the identified differentially expressed genes using the Kyoto Encyclopedia of Genes and Genomes. Array analysis showed significant increases in the expression of 353 genes and 206 expressed sequence tags (ESTs) and decreases in 565 genes and 29 ESTs. Bioinformatic analysis showed that the genes HIGD1B, HOXB2, ANGPT2, HPGD and BTG2 may play an important role in the tumorigenesis and progression of GHomas. Pathway analysis showed that the wingless-type signaling pathway and extracellular-matrix receptor interactions may play a key role in the tumorigenesis and progression of GHomas. Our data suggested that there are numerous aberrantly expressed genes and pathways involved in the pathogenesis of GHomas. Bead-based fiber-optic arrays combined with pathway analysis of differentially expressed genes appear to be a valid method for investigating the pathogenesis of tumors.
Keywords: pituitary adenomas, growth-hormone-secreting pituitary adenomas, bead-based fiber-optic array, gene expression profiling, pathway analysis
Introduction
Growth-hormone-secreting pituitary adenomas (GHomas) account for approximately 20% of all pituitary neoplasms (1). Acromegaly results from supraphysiological GH release from pituitary somatotroph adenomas in 99% of cases, resulting in persistently high circulating levels of GH, which may affect the cardiovascular system. Cardiovascular effects, such as hypertension, cardiomyopathy and valvular heart disease, are associated with increased morbidity and premature mortality, and significant increases in both respiratory disorders and malignancies have been reported (2). Retrospective cohort studies have suggested that mortality in patients with acromegaly is at least twice that present in the general population (3).
However, the pathogenetic mechanisms that underlie pituitary adenomas are complex and remain enigmatic. Several studies examining X-chromosome inactivation have shown that primary pituitary adenomas are monoclonal (4). Heterozygous activating somatic point mutations in the α-subunit of the stimulatory G protein have been identified in up to 40% of human GHomas (5). The presence of these mutations appears to correlate with a densely granulated ultrastructural morphology of somatotroph tumors and possibly with greater GH responsiveness to inhibition by the somatostatin analog octreotide (6). However, GHomas very rarely exhibit activating ras mutations in invasive or metastatic lesions (7). Uniquely, pituitary mitotic activity is relatively low, even in invasive adenomas. Several growth factors, including dysregulated receptors for fibroblast growth factors, dopamine, estrogen and nerve growth factor (8), have been implicated, predominantly in prolactinoma pathogenesis, but not uniformly in acromegaly. The pathogenesis of GHoma remains to be elucidated.
It is increasingly clear that improved diagnostic methods and a better understanding of disease mechanisms may be derived from a careful analysis of microarray measurements (9). Some reports have used microarrays to study differential gene expression in GHomas (10,11). However, with the rapid development of human genetics and genetic research, more and more genes will be identified and their functions will be elucidated. Therefore, further research will likely identify new genes involved in the pathogenesis of GHomas. To explore the possible pathogenesis of GHomas, we used a human genome-wide bead-based fiber-optic array (Illumina Human WG-6 v3.0), which has the characteristics of high density, high repeatability and high sensitivity, to identify the differentially expressed genes between GHomas and healthy pituitary tissues. We then analyzed the differential gene expression profiles using bioinformatic and pathway analyses to search for new candidate genes that may be implicated in the pathogenesis of GHomas.
Materials and methods
Tissue specimens
Five sporadic GHomas were obtained from patients at Tiantan Hospital (Beijing, China), following transsphenoidal surgery. The clinical and pathological characteristics of these adenomas are listed in Table I. Portions of the surgical specimens were immediately frozen in liquid nitrogen and stored at −80°C. Three healthy pituitary glands were used as controls, obtained from three adult males within 12 h of their being involved in fatal accidents. Informed consent was obtained from the patients and the study was approved by the local ethics committee.
Table I.
Clinical and pathological characteristics of the patients.
| Patient | Gender | Age | Clinical features | Tumor size (cm) | Immunohistochemistry |
|---|---|---|---|---|---|
| 246 | F | 36 | Acromegaly visual defect | 2.5×1.8×3.0 | GH2+ |
| 178 | M | 50 | Acromegaly | 2.8×1.7×3.2 | GH2+ |
| 193 | M | 20 | Acromegaly visual loss | 2.4×2.6×3.7 | GH3+ |
| 187 | M | 56 | Acromegaly visual defect | 1.2×2.5×0.9 | GH3+ |
| 235 | F | 27 | Acromegaly | 2.6×1.5×2.8 | GH3+ |
| 1(1) | M | 34 | Normal | - | Normal |
| 1(2) | M | 36 | Normal | - | Normal |
| 3(1) | M | 31 | Normal | - | Normal |
The samples were analyzed using bead-based fiber-optic array and qRT-PCR.
RNA extraction
Total RNA was extracted from healthy pituitaries (50–100 mg) and GHomas (50–100 mg) using the Unizol reagent protocol (Shanghai Biostar, Shanghai, China). The integrity of the RNA in each sample was determined by denaturing agarose gel electrophoresis, and the mRNA was quantified by spectrophotometry.
RNA amplification
RNA was amplified using the TotalPrep RNA amplification kit protocol (Ambion, Austin, TX, USA). In brief, total RNA was used as a template, and the T7 oligonucleotide was used for priming prior to the reverse transcription synthesis of the first strand cDNA. DNA polymerase and RNase H were used to simultaneously degrade the RNA and to synthesize the second strand cDNA. cDNA purification removed the remaining RNA, primers, enzymes and salts that would inhibit in vitro transcription. In vitro transcription generated multiple copies of biotinylated cRNA from the double-stranded cDNA templates, in the amplification and labeling step. cRNA purification removed the unincorporated NTPs, salts, enzymes and inorganic phosphates. Following purification, the cRNA was used with direct hybridization array kits (Illumina, San Diego, CA, USA).
Hybridization
A total of 1.5 μg of cRNA was hybridized to each array. Sample labeling and hybridization on Illumina WG-6 v3.0 Sentrix Bead Chips (Illumina) were performed at a single Illumina BeadStation facility according to the manufacturer’s instructions. These chips contained over 48,000 transcripts for a total of 37,804 genes, which covered 25,158 well-characterized genes from the human genome and 12,646 expressed sequence tags (ESTs).
Image scanning and data analysis
Illumina BeadArray Reader image analysis software (Illumina) was used to acquire the original signal value for each probe. In order to reduce random errors generated during the experiment and to improve the comparability between the different samples, the quantile normalization method was used to correct the data. The data were then exported into Significance Analysis of Microarrays (SAM) software to calculate the differential expression and false discovery rate (FDR) between tumor and healthy tissues. Filtering was performed to identify genes overexpressed or underexpressed at least 2.0 fold and to determine q-values (similar to the p-value, the q-value calculated the probability that the expression differences and FDR between the two sets of data were due to chance) of <5% in tumors compared to healthy pituitary tissues. Further analysis as to the potential relevance of the differentially expressed genes was performed using the Genbank database (http://www.ncbi.nlm.nih.gov/Genbank/). The differentially expressed genes were analyzed using pathway analysis methods utilizing the R programming language and Bioconductor software package. Hypergeometric tests were used to classify the enrichment of genes in a particular pathway, and the FDR was calculated to correct the p-values.
qRT-PCR
To verify the results of the microarray analysis, four differentially expressed genes were randomly selected, and their expression levels were measured in a blind manner. qRT-PCR was performed as previously described (12) using a SYBR Green I dye detection kit (Applied Biosystems, Foster City, CA, USA), and β-actin was used as an internal control. The fold change of each gene was calculated using the 2−ΔΔCt method, as previously described (13). The expression levels of these four genes were measured in three healthy pituitary glands and five GHomas. The four genes analyzed were growth hormone secretagogue receptor (GHSR; GenBank accession no. NM_198407.1), gonadotropin-releasing hormone receptor (GNRHR; GenBank accession no. NM_001012763.1), homeobox B2 (HOXB2; GenBank accession no. NM_002145.3) and glucagon receptor (GCGR; GenBank accession no. NM_000160.2). The primers for these genes were selected using Primer5.0 and synthesized by Generay Biotech Co. (Shanghai, China), and are listed in Table II.
Table II.
PCR primers used for qRT-PCR.
| Forward primer | Reverse primer | |
|---|---|---|
| GHSR | 5′ CACGGTCCTCTACAGTCTCATCG 3′ | 5′ CACGGTTTGCTTGTGGTTCTG 3′ |
| GNRHR | 5′ TCTACCAGGGAACATAGCATAAAT 3′ | 5′ TGCCAGAGTCCAGCCCTA 3′ |
| HOXB2 | 5′ CACAGAAAAATTCCGTTTGGTAG 3′ | 5′ AAACATAAAAAAGAGGAAAATAGGTC 3′ |
| GCGR | 5′ CGGAACACCAGCAACCAC 3′ | 5′ CCACAGGTAAAATAGATGGAGTTC 3′ |
| β-actin | 5′ GACTTAGTTGCGTTACACCCTTTC 3′ | 5′ TGCTGTCACCTTCACCGTTC 3′ |
Results
Expression profile of GHomas
In the GHoma array, 353 genes and 206 ESTs were overexpressed ≥2-fold and 565 genes and 29 ESTs were underexpressed ≥2-fold. The highest overexpressed and underexpressed genes are listed in Table III. A total of 23 genes and 41 ESTs were exclusively expressed in the GHomas and 120 genes, and 13 ESTs were only expressed in the healthy pituitary tissues.
Table III.
The highest overexpressed and underexpressed genes in GHomas.
| Gene | Accession | Definition | Fold change |
|---|---|---|---|
| GCGR | NM_000160.2 | Glucagon receptor | 158.1 |
| EST | BX089019 | BX089019 Soares_testis_NHT Homo sapiens cDNA clone IMAGp998K243513; IMAGE:1391375, mRNA sequence | 110.3 |
| HS3ST2 | NM_006043.1 | Heparan sulfate (glucosamine) 3-O-sulfotransferase 2 | 99.8 |
| HIGD1B | NM_016438.2 | HIG1 domain family, member 1B | 78.5 |
| GNLY | NM_006433.2 | Granulysin | 42.7 |
| HPGD | NM_000860.3 | Hydroxyprostaglandin dehydrogenase 15-(NAD) | −66.6 |
| GPHA2 | NM_130769.2 | Glycoprotein hormone α2 | −66.0 |
| VASH2 | NM_024749.2 | Vasohibin 2 | −63.5 |
| GJB7 | NM_198568.1 | Gap junction protein, β7, 25 kDa | −56.6 |
| UGT2B7 | XM_943434.1 | Predicted: Homo sapiens UDP glucuronosyltransferase 2 family, polypeptide B7 | −49.8 |
Validation of the differentially expressed genes by qRT-PCR
The microarray analysis showed that GCGR, GHSR and HOXB2 mRNA were increased by 158.1-, 7.2- and 19.3-fold, respectively, whereas the expression of GNRHR mRNA was decreased by 31.6-fold in all GHomas compared to the healthy pituitary tissues. qRT-PCR analysis of these four genes confirmed the changes in mRNA levels that were observed in the microarray analysis. The mean expression level of the four genes is shown in Fig. 1A. Fig. 1B shows the relative expression level of the four genes in the microarray compared to the qRT-PCR analysis.
Figure 1.
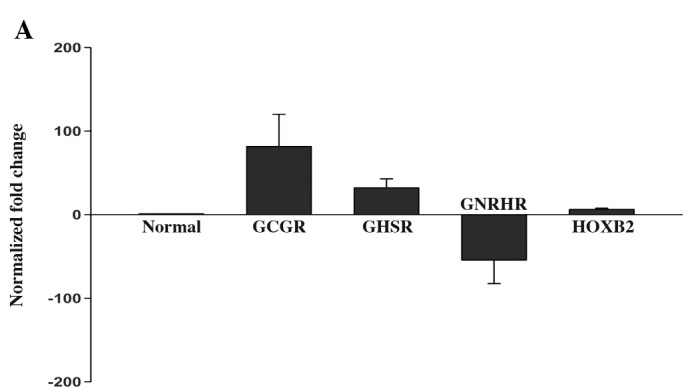
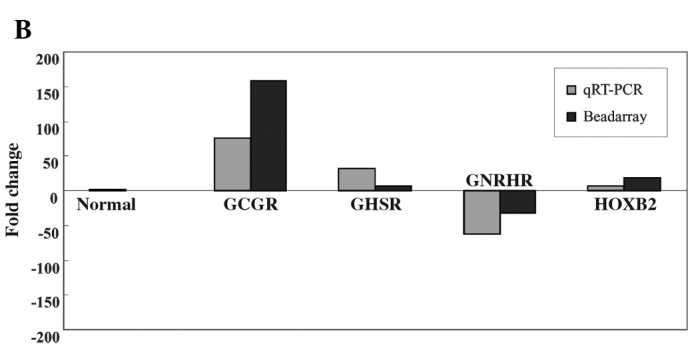
(A) The relative mean expression of GCGR (75-fold), GHSR (31.6-fold), HOXB2 (6.4-fold) and GNRHR (-62.3-fold) mRNA in GHomas compared to human pituitaries based on qRT-PCR analysis. Columns represent the mRNA expression of each gene in GHomas or healthy pituitary glands; bars represent the standard deviation (SD). (B) Relative expression level of these four genes in the microarray compared to the qRT-PCR analysis. The dark bars (right) represent the expression levels as determined by bead-based fiber-optic array (Illumina Human WG-6 v3.0) and the light bars (left) represent the qRT-PCR results.
Pathway analysis
The KEGG pathways are compiled from multiple literature sources and integrate individual components into a unified pathway. As such, the KEGG pathway database was used to characterize the enrichment of specific pathway components into functionally regulated groups. The differentially expressed genes from the microarray analysis were analyzed utilizing the pathway analysis methods in the R programming language and Bioconductor software package. A total of six KEGG pathways were significantly enriched among the genes associated with GHomas (p<0.05), as shown in Table IV. Of the significant signaling pathways, two were selected for further analysis: the wingless-type (Wnt) signaling pathway and extracellular matrix (ECM)-receptor interactions.
Table IV.
Six highly significant (p<0.05) pathways in GHomas.
| Pathway ID | Pathway title | Gene | p-value |
|---|---|---|---|
| hsa04310 | Wnt signaling pathway | CCND3, LEF1, WNT5B, WNT5A, WIF1, FZD9, FZD2, FZD3, SFRP2, TBL1XR1, NFAT5, VANGL2, FRAT1, FZD7, PRICKLE1, SMAD3 | 0.004 |
| hsa04115 | p53 signaling pathway | TNFRSF10B, GADD45G, CCNE1, CCND3, CCNB2, PMAP1, IGFBP3, RPRM, CDKN1A (p21, CIP1) | 0.012 |
| hsa05217 | Basal cell carcinoma | LEF1, WNT5B, WNT5A, FZD9, FZD2, FZD3, FZD7 | 0.014 |
| hsa05218 | Melanoma | PIK3CD, FGF23, E2F3, PDGFRA, FGF17, PDGFC, FGF2, PIK3R3, CDKN1A | 0.016 |
| hsa05210 | Colorectal cancer | PIK3CD, LEF1, PDGFRA, FZD9, FZD2, FOS, FZD3, PIK3R3, FZD7 | 0.016 |
| hsa04512 | ECM-receptor interaction | LAMA2, ITGA2, COL11A1, COL6A1, COL6A2, COL4A6, SDC4, COL11A2 | 0.018 |
Discussion
These experiments had two goals: First, we sought to identify the differentially expressed genes in GHomas as compared to healthy pituitary tissues, with the aim of identifing novel differentially expressed genes that may be important candidates in the pathogenesis of this poorly understood tumor. Second, we used pathway analysis to explore the possible relationship between the pathogenesis of GHomas and groups of genes functioning together in related pathways.
Analysis of differentially expressed genes
The NCBI database Genbank (http://www.ncbi.nlm.nih.gov/Genbank/) was used to analyze the differentially expressed genes identified in GHomas compared to healthy pituitaries, as well as the genes only expressed in the healthy pituitary tissues or the tumor tissues. We determined that the new genes HIGD1B, HOXB2, ANGPT2, HPGD and BTG2 may be important candidates in the pathogenesis of GHomas. These are discussed below.
HIGD1B (HIG1 domain family, member 1B)
In our study, the expression level of the HIGD1B mRNA was significantly up-regulated and only expressed in GHomas. However, the function of this protein is currently unknown. Denko et al (14) reported that in cultured cells, HIG1 and HIG2 expression is induced by hypoxia and by glucose deprivation. In addition, tumor xenografts derived from human cervical cancer cells display increased expression of HIG1 and HIG2 when they are deprived of oxygen. The role of HIGD1B in the pathogenesis of GHomas is not clear, and the specific mechanisms of action of this protein require further study.
HOXB2 (homeobox B2)
Homeobox genes are a large superfamily whose members function in establishing and maintaining cell fate and cell identity throughout embryonic development (15). Regarding the relationship between Homeobox genes and cancer, numerous studies have been undertaken to examine the differences in Homeobox expression between healthy and neoplastic tissues. Several investigators have explored the hypothesis that Homeobox genes expressed during embryogenesis, but down-regulated during adulthood, are re-expressed in neoplasias – the so-called ‘oncology recapitulates ontology’ hypothesis (16). In our study, the expression level of the HOXB2 mRNA was significantly up-regulated (19.3-fold), and this was validated by qRT-PCR analysis. However, its role in pituitary tumors is unknown and requires further study.
ANGPT2 (angiopoietin 2)
Angiopoietins are members of a novel family of angiogenic factors and participate in the formation of blood vessels. Of the four currently known family members, the best characterized are angiopoietin (ANG)-1 and -2, both of which function as ligands for the endotheliumspecific tyrosine kinase receptor TIE-2. ANG-1 plays an important role in maintaining vessel integrity. ANG-2, which is thought to be an endogenous antagonist of the action of ANG-1, competes for binding to the TIE-2 receptor. This blocks ANG-1-induced TIE-2 autophosphorylation during vasculogenesis, and subsequently leads to the loosening of cell-matrix and cell-cell contacts, allowing access to angiogenic inducers (17).
Nag et al (18) reported that the localization of both ANG-1 and ANG-2 occurs in scattered periodic acid-Schiff (PAS)-positive adenohypophysial cells rather than in endothelial cells. Accumulated evidence also indicates that the production of ANG-2 is implicated in tumor progression (19). Our study showed that ANG-2 mRNA was overexpressed by up to 5.6-fold in GHomas. We therefore believe that ANG-2 may play an important role in promoting the angiogenesis, progression and invasion of these tumors.
HPGD (15-hydroxyprostaglandin dehydrogenase; 15-PGDH)
15-PGDH is a prostaglandin degrading enzyme that catalyses the oxidization of the 15(S)-hydroxyl group of PGE2 to yield an inactive 15-keto PGE2. PGE2 is related to carcinogenesis through immunosuppression, the inhibition of apoptosis, an increase in the metastatic potential of epithelial cells and the promotion of angiogenesis (20). Genetic deletion of 15-PGDH in mice leads to increased tissue levels of PGE2 (21).
Previous studies identified tumor suppressor activity for 15-PGDH in colon, lung, bladder and breast carcinomas, and suggested that epigenetic silencing of this enzyme occurs by DNA methylation and histone modification (22). The expression of 5-PGDH in benign tumors has yet to be reported. Our study showed that the expression of the 15-PGDH mRNA in GHomas was significantly down-regulated (-66.6-fold). We therefore speculate that this gene also functions as a tumor suppressor gene in GHomas, and plays a key role in the tumorigenesis and progression of these tumors. However, the specific mechanisms affected by 5-PGDH in these tumors require further study.
BTG2 (BTG family, member 2)
BTG2 is an antiproliferative tumor suppressor protein, as its overexpression leads to the blockade of cells at the G1 phase of the cell cycle (23). It has also been demonstrated that BTG2 expression is induced by cytotoxic and genotoxic stress through a p53-dependent mechanism (24). Farioli-Vecchioli et al (25) reported that the down-regulation of BTG2 is observed in pre-neoplastic lesions as well as in human and murine medulloblastomas. This previous report provides the earliest evidence that BTG2 acts as a tumor suppressor in the central nervous system. Our study showed that the expression of BTG2 mRNA in GHomas was down-regulated. We therefore speculate that this gene plays a key role in the tumorigenesis and progression of these tumors.
Pathway analysis
Single-gene analysis may overlook important effects that occur on pathways, as cellular processes often affect sets of genes acting in concert. An increased expression of 20% of all gene members in a metabolic pathway may dramatically alter the flux through the pathway, and may therefore be more important than a 20-fold increase in any single gene (26). Therefore, we used the KEGG pathway database (http://www.genome.jp/kegg/pathway.html) to analyze pathways with a p-value <0.05 (Table IV). The study suggested that the Wnt signaling pathway and ECM-receptor interactions play key roles in the pathogenesis of GHomas.
The Wnt signaling pathway
The signaling transduction pathway mediated by the Wnt proteins is highly conserved between species. It influences embryonic development, cell polarity and adhesion, apoptosis and tumorigenesis, and it interacts with other cell signaling pathways (27). There are at least three different Wnt pathways: the canonical, the planar cell polarity and the Wnt/Ca2+ pathway. In the canonical Wnt pathway, the major effect of Wnt ligand binding to its receptor is the stabilization of cytoplasmic β-catenin through inhibition of the β-catenin degradation complex. Subsequently, β-catenin is then free to enter the nucleus and activate Wnt-regulated genes through its interaction with the T-cell factor (TCF) family of transcription factors and concomitant recruitment of coactivators.
Receptors for the Wnt proteins are members of the frizzled family of transmembrane proteins encoded by the frizzled genes (Fzs). Currently, 10 or more human Fzs genes have been identified (28). The Fzs proteins have an N-terminal cysteine-rich extracellular domain and seven membrane-spanning α helices linked by hydrophilic loops ending with an intracellular C-terminus (28). The extracellular ligand (a specific Wnt) binds to the cysteine-rich domain (CRD) of a specific Fzs, altering the interaction of the CRD with the transmembrane domain. These changes result in a rearrangement of the transmembrane α helices, a remodelling of the cytosolic face of the protein and an alteration of downstream signalling (27). In our study, the expression of FZD2, FZD3, FZD7 and FZD9 mRNA was down-regulated; however, the specific mechanism by which this occurred is not clear, and requires further study.
In the nucleus, stabilized β-catenin associates with the TCF/lymphoid enhancer factor (LEF) family of transcription factors to activate specific target genes. De-repression by TCF/LEF/β-catenin complexes enables the transcription of more than 30 different genes. These include c-myc, cyclin D1, c-jun, fra-1, matrilysin and members of the Ap-1 family of genes, including fos, fosB and junB (27). Our study showed that the expression of LEF1 and CCND3 mRNA in GHomas was up-regulated. We speculated that this plays a crucial role in the tumorigenesis and progression of these tumors.
Secreted Wnt antagonists are involved in regulating the Wnt pathways. These Wnt inhibitors are divided into two main families containing either secreted frizzled-related proteins (sFRPs) or the Dickkopf (DKK) proteins. The sFRP family is comprised of five sFRPs and Wnt inhibitory factor 1 (WIF1). Elston et al (29) used microarray analysis to compare pituitary tumors to healthy pituitary glands, demonstrating that WIF1 mRNA expression is markedly underexpressed in all pituitary tumor subtypes. In addition, they found significantly reduced mRNA expression of two other Wnt pathway inhibitors, sFRP2 and sFRP4, and suggested that the Wnt pathways are important in pituitary tumorigenesis. Our experimental results are in agreement with their report, as we observed that SFRP2 and WIF1 mRNA were significantly underexpressed in GHomas. Based on the above analysis, we propose that the Wnt pathway may play an important role in promoting the tumorigenesis and progression of GHomas.
ECM-receptor interactions
The ECM is a three-dimensional network of proteins, glycosaminoglycans and other macromolecules. It has a structural support function, as well as a role in cell adhesion, migration, proliferation and survival. ECM components, including laminins, collagen and fibronectin, or synthetic substrates, such as poly-D-lysine, representing a cationic polypeptide, affect the proliferation and function of healthy and neoplastic cells (30). Such effects have been demonstrated in many different cultured tumor cells, including endocrine tumor cells (31). For example, laminin was found to regulate the development and growth of breast, prostate, colon, thyroid and ovarian cancers (32). In our study, LAMA2, COL11A1, COL6A1, COL6A2, COL4A6 and COL11A2 mRNA were observed to be underexpressed in GHomas.
Apart from being growth factors, ECM components are considered to be the main control elements of cellular proliferation, acting though integrin surface receptors. Integrins are a diverse family of glycoproteins that form heterodimeric receptors for ECM molecules. This family of proteins forms at least 25 distinct pairings among its 18 α-subunits and 8 β-subunits, with each pairing being specific for a unique set of ligands (33). Integrin α2β1 is formed by a non-covalent association of the α2 subunit as a monogamous partner to the promiscuous β1 subunit. The expression of integrin α2 has been correlated with metastatic behavior in breast cancer, hepatocarcinoma and rhabdomyosarcoma (34). In our study, the integrin α2 (ITGA2) mRNA was expressed at a lower level in GHomas.
ECM components and integrins constitute one of the few clear examples of factors differentially expressed during pituitary pathogenesis (32). The ECM likely plays a role in the inhibition of invasion and metastasis in pituitary tumors (32). Consistent with this previous report, our study showed that all the differentially expressed genes involved in this pathway were underexpressed in GHomas. The role of ECM-receptor interactions in GHomas may be to inhibit their invasion and malignant transformation.
In conclusion, in this study we identified a number of genes and pathways that may play an important role in the tumorigenesis and progression of GHomas. Expression analysis of tumors using bead-based fiber-optic arrays appears to be a valid method for identifying genes important in tumor pathogenesis. Moreover, further characterization of gene expression profiles based on pathway analysis may play a vital role in elucidating the pathogenesis of tumors.
References
- 1.Pollock BE, Jacob JT, Brown PD, Nippoldt TB. Radiosurgery of growth hormone-producing pituitary adenomas: factors associated with biochemical remission. J Neurosurg. 2007;106:833–838. doi: 10.3171/jns.2007.106.5.833. [DOI] [PubMed] [Google Scholar]
- 2.Rajasoorya C, Holdaway IM, Wrightson P, Scott DJ. Ibbertson HK determinants of clinical outcome and survival in acromegaly. Clin Endocrinol. 1994;41:95–102. doi: 10.1111/j.1365-2265.1994.tb03789.x. [DOI] [PubMed] [Google Scholar]
- 3.Beauregard C, Truong U, Hardy J, Serri O. Long-term outcome and mortality after transsphenoidal adenomectomy for acromegaly. Clin Endocrinol. 2003;58:86–91. doi: 10.1046/j.1365-2265.2003.01679.x. [DOI] [PubMed] [Google Scholar]
- 4.Herman V, Fagin J, Gonsky R, Kovacs K, Melmed S. Clonal origin of pituitary adenomas. J Clin Endocrinol Metab. 1990;71:1427–1433. doi: 10.1210/jcem-71-6-1427. [DOI] [PubMed] [Google Scholar]
- 5.Farrell WE, Clayton RN. Molecular genetics of pituitary tumours. Trends Endocrinol Metab. 1998;9:20–26. doi: 10.1016/s1043-2760(98)00006-x. [DOI] [PubMed] [Google Scholar]
- 6.Ezzat S, Kontogeorgos G, Redelmeier DA, Horvath E, Harris AG, Kovacs K. In vivo responsiveness of morphological variants of growth hormone-producing pituitary adenomas to octreotide. Eur J Endocrinol. 1995;133:686–690. doi: 10.1530/eje.0.1330686. [DOI] [PubMed] [Google Scholar]
- 7.Karga HJ, Alexander JM, Hedley-Whyte ET, Klibanski A, Jameson JL. Ras mutations in human pituitary tumors. J Clin Endocrinol Metab. 1992;74:914–919. doi: 10.1210/jcem.74.4.1312542. [DOI] [PubMed] [Google Scholar]
- 8.Asa SL, Ezzat S. The pathogenesis of pituitary tumors. Annu Rev Pathol. 2009;4:97–126. doi: 10.1146/annurev.pathol.4.110807.092259. [DOI] [PubMed] [Google Scholar]
- 9.Mao S, Dong G. Discovery of highly differentiative gene groups from microarray gene expression data using the gene club approach. J Bioinform Comput Biol. 2005;3:1263–1280. doi: 10.1142/s0219720005001545. [DOI] [PubMed] [Google Scholar]
- 10.Evans CO, Young AN, Brown MR, Brat DJ, Parks JS, Neish AS, Oyesiku NM. Novel patterns of gene expression in pituitary adenomas identified by complementary deoxyribonucleic acid microarrays and quantitative reverse transcription-polymerase chain reaction. J Clin Endocrinol Metab. 2001;86:3097–3107. doi: 10.1210/jcem.86.7.7616. [DOI] [PubMed] [Google Scholar]
- 11.Morris DG, Musat M, Czirjak S, Hanzely Z, Lillington DM, Korbonits M, Grossman AB. Differential gene expression in pituitary adenomas by oligonucleotide array analysis. Eur J Endocrinol. 2005;153:143–151. doi: 10.1530/eje.1.01937. [DOI] [PubMed] [Google Scholar]
- 12.Ellestad LE, Carre W, Muchow M, Jenkins SA, Wang X, Cogburn LA, Porter TE. Gene expression profiling during cellular differentiation in the embryonic pituitary gland using cDNA microarrays. Physiol Genomics. 2006;25:414–425. doi: 10.1152/physiolgenomics.00248.2005. [DOI] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Denko N, Schindler C, Koong A, Laderoute K, Green C, Giaccia A. Epigenetic regulation of gene expression in cervical cancer cells by the tumor microenvironment. Clin Cancer Res. 2000;6:480–487. [PubMed] [Google Scholar]
- 15.Lewis MT. Homeobox genes in mammary gland development and neoplasia. Breast Cancer Res. 2000;2:158–169. doi: 10.1186/bcr49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lappin TR, Grier DG, Thompson A, Halliday HL. HOX genes: seductive science, mysterious mechanisms. Ulster Med J. 2006;75:23–31. [PMC free article] [PubMed] [Google Scholar]
- 17.Wang HL, Deng CS, Lin J, Pan DY, Zou ZY, Zhou XY. Expression of angiopoietin-2 is correlated with vascularization and tumor size in human colorectal adenocarcinoma. Tohoku J Exp Med. 2007;213:33–40. doi: 10.1620/tjem.213.33. [DOI] [PubMed] [Google Scholar]
- 18.Nag S, Nourhaghighi N, Venugopalan R, Asa SL, Stewart DJ. Angiopoietins are expressed in the normal rat pituitary gland. Endocr Pathol. 2005;16:67–73. doi: 10.1385/ep:16:1:067. [DOI] [PubMed] [Google Scholar]
- 19.Hu B, Guo P, Fang Q, et al. Angiopoietin-2 induces human glioma invasion through the activation of matrix metalloprotease-2. Proc Natl Acad Sci USA. 2003;100:8904–8909. doi: 10.1073/pnas.1533394100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jang TJ, Ji YS, Jung KH. Decreased expression of 15-hydroxyprostaglandin dehydrogenase in gastric carcinomas. Yonsei Med J. 2008;49:917–922. doi: 10.3349/ymj.2008.49.6.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coggins KG, Latour A, Nguyen MS, Audoly L, Coffman TM, Koller BH. Metabolism of PGE2 by prostaglandin dehydrogenase is essential for remodeling the ductus arteriosus. Nat Med. 2002;8:91–92. doi: 10.1038/nm0202-91. [DOI] [PubMed] [Google Scholar]
- 22.Wolf I, O’Kelly J, Rubinek T, et al. 15-hydroxyprostaglandin dehydrogenase is a tumor suppressor of human breast cancer. Cancer Res. 2006;66:7818–7823. doi: 10.1158/0008-5472.CAN-05-4368. [DOI] [PubMed] [Google Scholar]
- 23.Rouault JP, Falette N, Guehenneux F, et al. Identification of BTG2, an antiproliferative p53-dependent component of the DNA damage cellular response pathway. Nat Genet. 1996;14:482–486. doi: 10.1038/ng1296-482. [DOI] [PubMed] [Google Scholar]
- 24.Cortes U, Moyret-Lalle C, Falette N, et al. BTG gene expression in the p53-dependent and -independent cellular response to DNA damage. Mol Carcinog. 2000;27:57–64. [PubMed] [Google Scholar]
- 25.Farioli-Vecchioli S, Tanori M, Micheli L, et al. Inhibition of medulloblastoma tumorigenesis by the antiproliferative and prodifferentiative gene PC3. FASEB J. 2007;21:2215–2225. doi: 10.1096/fj.06-7548com. [DOI] [PubMed] [Google Scholar]
- 26.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karim R, Tse G, Putti T, Scolyer R, Lee S. The significance of the Wnt pathway in the pathology of human cancers. Pathology. 2004;36:120–128. doi: 10.1080/00313020410001671957. [DOI] [PubMed] [Google Scholar]
- 28.Jones SE, Jomary C. Secreted Frizzled-related proteins: searching for relationships and patterns. Bioessays. 2002;24:811–820. doi: 10.1002/bies.10136. [DOI] [PubMed] [Google Scholar]
- 29.Elston MS, Gill AJ, Conaglen JV, et al. Wnt pathway inhibitors are strongly down-regulated in pituitary tumors. Endocrinology. 2008;149:1235–1242. doi: 10.1210/en.2007-0542. [DOI] [PubMed] [Google Scholar]
- 30.Danen EH, Yamada KM. Fibronectin, integrins, and growth control. J Cell Physiol. 2001;189:1–13. doi: 10.1002/jcp.1137. [DOI] [PubMed] [Google Scholar]
- 31.Huet C, Pisselet C, Mandon-Pepin B, Monget P, Monniaux D. Extracellular matrix regulates ovine granulosa cell survival, proliferation and steroidogenesis: relationships between cell shape and function. J Endocrinol. 2001;169:347–360. doi: 10.1677/joe.0.1690347. [DOI] [PubMed] [Google Scholar]
- 32.Paez-Pereda M, Kuchenbauer F, Arzt E, Stalla GK. Regulation of pituitary hormones and cell proliferation by components of the extracellular matrix. Braz J Med Biol Res. 2005;38:1487–1494. doi: 10.1590/s0100-879x2005001000005. [DOI] [PubMed] [Google Scholar]
- 33.Gerger A, Hofmann G, Langsenlehner U, et al. Integrin alpha-2 and beta-3 gene polymorphisms and colorectal cancer risk. Int J Colorectal Dis. 2009;24:159–163. doi: 10.1007/s00384-008-0587-9. [DOI] [PubMed] [Google Scholar]
- 34.Sawhney RS, Cookson MM, Omar Y, Hauser J, Brattain MG. Integrin alpha2-mediated ERK and calpain activation play a critical role in cell adhesion and motility via focal adhesion kinase signaling: identification of a novel signaling pathway. J Biol Chem. 2006;281:8497–8510. doi: 10.1074/jbc.M600787200. [DOI] [PubMed] [Google Scholar]


