ABSTRACT
In a previous study of promoters dependent on the Escherichia coli cyclic AMP receptor protein (CRP), carrying tandem DNA sites for CRP, we found that the upstream-bound CRP could either enhance or repress transcription, depending on its location. Here, we have analyzed the interactions between CRP and the C-terminal domains of the RNA polymerase α subunits at some of these promoters. We report that the upstream-bound CRP interacts with these domains irrespective of whether it up- or downregulates promoter activity. Hence, disruption of this interaction can lead to either down- or upregulation, depending on its location.
IMPORTANCE
Many bacterial promoters carry multiple DNA sites for transcription factors. While most factors that downregulate promoter activity bind to targets that overlap or are downstream of the transcription start and −10 element, very few cases of repression from upstream locations have been reported. Since more Escherichia coli promoters are regulated by cyclic AMP receptor protein (CRP) than by any other transcription factor, and since multiple DNA sites for CRP are commonplace at promoters, our results suggest that promoter downregulation by transcription factors may be more prevalent than hitherto thought, and this will have implications for the annotation of promoters from new bacterial genome sequences.
Observation
There are many examples of bacterial promoters that are regulated by the binding of multiple transcription factors. Such regulation provides a way of integrating different environmental cues to modulate gene expression profiles (1). The contribution of each individual transcription factor can be dramatic or subtle, but the overall outcome is transcription regulation that can be fine-tuned to permit adaptation to changing circumstances. At many promoters, two transcription activator proteins can work together by interacting with the C-terminal domains of the two RNA polymerase α subunits (αCTD). This is facilitated by the flexible linker that joins αCTD to the rest of the α subunit, which allows αCTD to extend far upstream to contact activators directly (1).
One activator, for which the contact with αCTD is well documented, is the Escherichia coli cyclic AMP receptor protein (CRP), a global transcription factor that regulates more than 200 transcription units (2, 3). CRP has a surface-exposed patch of amino acid side chains known as activating region 1 (AR1) that interacts with surface side chains of αCTD denoted “the 287 determinant.” Previously, we and others have demonstrated that CRP bound at tandem binding sites at some promoters can function synergistically to activate transcription (4, 5) and that the upstream CRP functions exclusively via contact with αCTD (6). In studies where the promoter-distal DNA site for CRP was systematically moved upstream, locations were found where upstream-bound CRP gave maximum activation, but, surprisingly, at some locations, the distal CRP downregulated promoter activity (7, 8). Hence, in this study, we sought to elucidate the mechanism by which upstream-bound CRP can have a negative effect upon transcription.
Our strategy was to use RNA polymerase containing αCTD derivatized with the Fe-BABE chemical nuclease (9, 10) to compare the juxtaposition of CRP and αCTD at some of the promoters, described by Tebbutt et al. (8), with tandem DNA sites for CRP. These promoters have one DNA site for CRP centered between base pairs 61 and 62, upstream from the transcript start (i.e., position −61.5), and a second DNA site for CRP located further upstream, at positions −93.5, −103.5, −113.5, −117.5, and −122.5. Tebbutt et al. (8) showed that, compared to the activity of the starting promoter CC(−61.5), with a single DNA site for CRP at position −61.5, the second DNA site for CRP at the CC(−93.5)CC(−61.5), CC(−103.5)CC(−61.5), CC(−113.5)CC(−61.5), and CC(−117.5)CC(−61.5) promoters increased CRP-dependent promoter activity. In contrast, the second DNA site for CRP at the CC(−122.5)CC(−61.5) promoter results in reduction of promoter activity, and data from a repeat of Tebbutt’s experiments are listed in Table S1 in the supplemental material. To facilitate our Fe-BABE chemical nuclease experiments, the −10 hexamer element of each promoter was changed from 5′ CATAAT 3′ to the consensus 5′ TATAAT 3′, which improves RNA polymerase binding. The new promoter derivatives were suffixed with “p12T,” and base sequences are shown in Fig. S1 in the supplemental material. Data in Table S1 in the supplemental material show that, for promoters with the p12T substitution, the patterns of activation or repression by upstream-bound CRP are unaffected, though measured promoter activities are higher due to the overall increase in promoter strength.
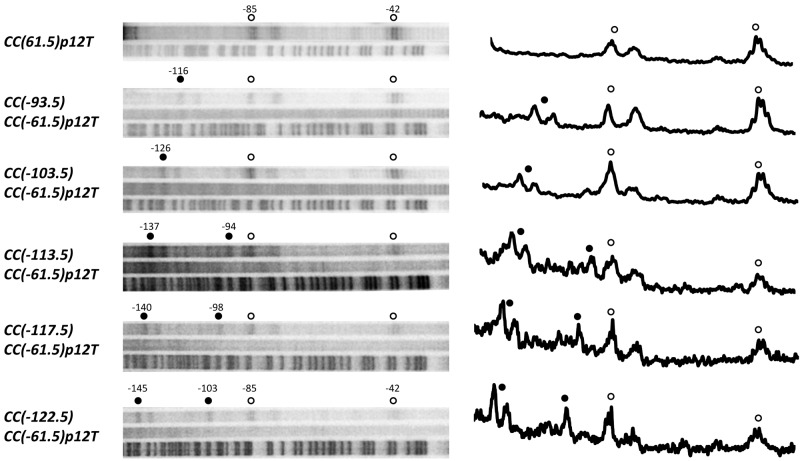
Using purified CRP and purified RNA polymerase holoenzyme, reconstituted with α subunits that had been conjugated with Fe-BABE at residue 302, transcriptionally competent open complexes were formed on 32P end-labeled DNA fragments carrying each of the promoters. Fig. 1 shows the Fe-BABE-mediated DNA cleavage patterns at the different promoters, as revealed by gel and phosphorimager analysis, with a trace along each lane. At each of the promoters, DNA cleavage occurs adjacent to CRP bound at the site centered at position −61.5, and this cleavage is not affected by the upstream-bound CRP. At promoters with tandem CRP sites, additional cleavage occurs adjacent to the upstream CRP, and as the upstream-bound CRP moves from position −93.5 to position −122.5, the cleavage sites move in register. In previous work with RNA polymerase containing α subunits conjugated to Fe-BABE at residue 302, we found that the distance between the center of a CRP site and the positions of upstream and downstream Fe-BABE-mediated DNA cleavage on each strand were 23/24 bp and 18/19 bp, respectively (10). Analysis of our data here reveals identical cleavage patterns, indicating that when bound adjacent to either promoter, proximal or distal CRP, the αCTDs are oriented so as to interact with AR1 of CRP. Taken together, these data indicate that, during the course of the experiment, the αCTDs are able to visit each available DNA position adjacent to CRP and interact with AR1, including at the CC(−122.5)CC(−61.5) promoter, where upstream-bound CRP downregulates promoter activity.
FIG 1 .
Fe-BABE-mediated DNA cleavage patterns at the different promoters, as revealed by gel and phosphoimager analysis, with a trace along each lane.
To further analyze the downregulation of the CC(−122.5)CC(-61.5) promoter by upstream-bound CRP, we exploited the HL159 substitution that renders the CRP AR1 surface defective (2). This was combined with a second substitution, EV181, that changes the DNA binding specificity of CRP so that it recognizes an altered DNA binding sequence (11, 12). Here, we denote the altered DNA site for CRP as QQ, and we constructed QQ(−93.5)CC(−61.5), QQ(−117.5)CC(−61.5), and QQ(−122.5)CC(−61.5) (see Fig. S1 in the supplemental material). Activity from the promoters was analyzed in E. coli cells that express wild-type CRP from the chromosome and mutant CRP derivatives supplied in trans from a plasmid. The data, shown in Table 1, indicate that the presence of a defective AR1 in the upstream CRP results in a marked decrease in promoter activity at the CC(−93.5)CC(−61.5) and CC(−117.5)CC(−61.5) promoters, thus confirming that the distal CRP at these promoters enhances transcription via the AR1-287 interface. However, the presence of a defective AR1 on the upstream CRP at the CC(−122.5)CC(−61.5) promoter resulted in an increase in overall activity, indicating that disruption of the AR1-287 interface at this promoter relieves the repressive effect exerted by the upstream CRP. We conclude that this repressive effect involves a direct contact between the upstream DNA-bound transcription factor and RNA polymerase, and we suggest that, by interacting with αCTD, CRP is restraining RNA polymerase from escaping from the initiation complex.
TABLE 1 .
Expression from the QQ(−n)CC(−61.5) promoter derivatives in the presence of CRP mutantsa
| Promoter | Expression |
|
|
|---|---|---|---|
| CRP EV181 (A) | CRP EV181HL159 (B) | ||
| QQ(−93.5)CC(−61.5) | 3,910 | 1,024 | 381 |
| QQ(−117.5)CC(−61.5) | 1,256 | 943 | 133 |
| QQ(−122.5)CC(−61.5) | 3,77 | 541 | 77 |
The table shows the β-galactosidase activities measured in Escherichia coli strain M182 crp+ cells, containing pRW50 carrying different promoter derivatives and different pDCRP plasmids encoding derivatives of CRP. Each data point is the average from at least 3 independent experiments that differed by less than 5%.
Implications.
There are few documented cases of transcription factors that repress transcription when bound upstream of promoters. For example, at the E. coli guaB promoter, CRP bound to a DNA site at position −117.5 decreases promoter activity, although the precise mode of action is still unknown (13), and at the E. coli yfiD promoter, activation by FNR, a transcription factor related to CRP, is suppressed by a second upstream-bound FNR molecule, though αCTD is not involved (14). Here, we have uncovered a mechanism that is similar to that of the p4 protein from the Bacillus subtilis phage, φ29, where, at the phage A2c promoter, p4 binds to a DNA site at position −71.5 and interacts with αCTD to mediate repression (15). The significance of all these findings is that they point to a layer of complexity within bacterial transcription regulatory regions that will have consequences for the annotation of both published and newly sequenced bacterial genomes. Hence, annotation based simply on DNA binding site identification may provide false information about how different signals are integrated to regulate genes. The most sophisticated bioinformatic analysis will be unable to accurately predict regulation of any promoter without equally sophisticated knowledge of the mechanisms by which transcription factors interact at promoters.
SUPPLEMENTAL MATERIAL
Materials and Methods. Download Text S1, DOCX file, 0.1 MB.
Base sequence of promoters used in this work. Download Figure S1, DOCX file, 0.1 MB.
Expression from the CC(−61.5) promoter and the CC(−n)CC(−61.5) promoter derivatives.
Oligonucleotide primers used in this work.
ACKNOWLEDGMENTS
This work was funded by a program grant from the Wellcome Trust.
We are grateful to Christine Webster for technical support.
Footnotes
Citation Lee DJ, Busby SJW. 2012. Repression by cyclic AMP receptor protein at a distance. mBio 3(5):e00289-12. doi:10.1128/mBio.00289-12.
REFERENCES
- 1. Lee DJ, Minchin SD, Busby SJW. Activating transcription in bacteria. Annu. Rev. Microbiol., in press [DOI] [PubMed] [Google Scholar]
- 2. Busby S, Ebright RH. 1999. Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 293:199–213 [DOI] [PubMed] [Google Scholar]
- 3. Lawson CL, et al. 2004. Catabolite activator protein: DNA binding and transcription activation. Curr. Opin. Struct. Biol. 14:10–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Joung JK, Le LU, Hochschild A. 1993. Synergistic activation of transcription by Escherichia coli cAMP receptor protein. Proc. Natl. Acad. Sci. U. S. A. 90:3083–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Busby S, et al. 1994. Transcription activation by the Escherichia coli cyclic AMP receptor protein. Receptors bound in tandem at promoters can interact synergistically. J. Mol. Biol. 241:341–352 [DOI] [PubMed] [Google Scholar]
- 6. Lloyd GS, Niu W, Tebbutt J, Ebright RH, Busby SJ. 2002. Requirement for two copies of RNA polymerase alpha subunit C-terminal domain for synergistic transcription activation at complex bacterial promoters. Genes Dev. 16:2557–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Belyaeva TA, Rhodius VA, Webster CL, Busby SJ. 1998. Transcription activation at promoters carrying tandem DNA sites for the Escherichia coli cyclic AMP receptor protein: organisation of the RNA polymerase alpha subunits. J. Mol. Biol. 277:789–804 [DOI] [PubMed] [Google Scholar]
- 8. Tebbutt J, Rhodius VA, Webster CL, Busby SJ. 2002. Architectural requirements for optimal activation by tandem CRP molecules at a class I CRP-dependent promoter. FEMS Microbiol. Lett. 210:55–60 [DOI] [PubMed] [Google Scholar]
- 9. Murakami K, et al. 1997. Positioning of two alpha subunit carboxy-terminal domains of RNA polymerase at promoters by two transcription factors. Proc. Natl. Acad. Sci. U.S.A. 94:11274–11278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee DJ, Busby SJ, Lloyd GS. 2003. Exploitation of a chemical nuclease to investigate the location and orientation of the Escherichia coli RNA polymerase alpha subunit C-terminal domains at simple promoters that are activated by cyclic AMP receptor protein. J. Biol. Chem. 278:52944–52952 [DOI] [PubMed] [Google Scholar]
- 11. Ebright RH, et al. 1987. Role of glutamic acid-181 in DNA-sequence recognition by the catabolite gene activator protein (CAP) of Escherichia coli: altered DNA-sequence-recognition properties of [Val181]CAP and [Leu181]CAP. Proc. Natl. Acad. Sci. U. S. A. 84:6083–6087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gunasekera A, Ebright YW, Ebright RH. 1992. DNA sequence determinants for binding of the Escherichia coli catabolite gene activator protein. J. Biol. Chem. 267:14713–14720 [PubMed] [Google Scholar]
- 13. Husnain SI, Busby SJ, Thomas MS. 2009. Downregulation of the Escherichia coli guaB promoter by upstream-bound cyclic AMP receptor protein. J. Bacteriol. 191:6094–6104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marshall FA, et al. 2001. A novel promoter architecture for microaerobic activation by the anaerobic transcription factor FNR. Mol. Microbiol. 39:747–753 [DOI] [PubMed] [Google Scholar]
- 15. Monsalve M, Calles B, Mencía M, Rojo F, Salas M. 1998. Binding of phage phi29 protein p4 to the early A2c promoter: recruitment of a repressor by the RNA polymerase. J. Mol. Biol. 283:559–569 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Materials and Methods. Download Text S1, DOCX file, 0.1 MB.
Base sequence of promoters used in this work. Download Figure S1, DOCX file, 0.1 MB.
Expression from the CC(−61.5) promoter and the CC(−n)CC(−61.5) promoter derivatives.
Oligonucleotide primers used in this work.