Abstract
Interleukin (IL)-2 and interferon (IFN)-α combination therapy for metastatic renal cell carcinoma (RCC) improves the prognosis for a subset of patients, while some patients suffer from severe adverse drug reactions with little benefit. To establish a method to predict responses to this combination therapy (approximately 30% response rate), the gene expression profiles of primary RCCs were analyzed using an oligoDNA microarray consisting of 38,500 genes or ESTs, after enrichment of the cancer cell population by laser micro-beam microdissection. The analysis of 10 responders and 18 non-responders identified 24 genes that exhibited significant differential expression between the two groups. In addition, the patients whose tumors did not express HLA-DQA1 or HLA-DQB1 molecules demonstrated poor clinical response. Exclusion of patients with tumors lacking either of these two genes is likely to improve the response rate to IL-2 and IFN-α combination therapy from 30 to 67%, indicating that a simple pretreatment test provides useful information with which to subselect patients with renal cancer in order to improve the efficacy of this treatment and reduce unnecessary medical costs.
Keywords: interleukin-2 and interferon-α combination therapy, HLA-DQA1, HLA-DQB1
Introduction
Renal cell carcinomas (RCCs) account for 2–3% of all malignancies (1,2). Surgical resection is the first choice of treatment for RCC at an early stage. However, when patients present at an advanced stage or have local recurrence or distant metastasis to other organs, immunotherapy, chemotherapy and radiation therapy are applied, although the response rates are poor. Recently, new molecular targeted agents, such as sunitinib and sorafenib, have been developed and are widely used (3–7). Although these drugs have demonstrated a better clinical response than previously used treatments, serious adverse reactions, such as fatigue, mucositis, hand-foot syndrome, diarrhea and hypertension, are often observed and become the cause of discontinuation of the drug administration.
Monotherapy or combination therapy of interleukin (IL)-2 and interferon (IFN)-α has been relatively widely applied for the treatment of advanced kidney cancers. In Japan, to reduce the risk of adverse reactions, the dose of IL-2 is lower than that used in other countries (8–11). While the response rate of monotherapy is as low as 10–20%, that of combination therapy is slightly better, 20–25% (12–17). However, the rate of adverse reactions associated with this combination therapy appears to be lower than recently developed molecular targeting drugs (18,19). Hence, we aimed to define a subset of patients who expect to show a favorable response to this therapy through gene expression profiles of metastatic RCCs, after enrichment of the cancer cells with laser microbeam microdissection technology. In the present study, two antigen-presentation-associated molecules were identified that may predict response to IL-2 and IFN-α combination therapy for metastatic RCC. In addition, this finding may be useful for improving the drug response rate, for contributing to the improvement of the quality of life and prognosis of patients, and reducing unnecessary medical costs to non-responders.
Materials and methods
Patients and tissue samples
Tissue samples from surgically resected RCC and corresponding clinical information were obtained from 21 hospitals (Tokyo University, Okayama University, Sapporo Medical University, Kobe University, Nihon University, Kanazawa University, Isezaki City Hospital, Shinshu University, Kyushu University, Kyoto Prefectural University of Medicine, Osaka Medical Center for Cancer and Cardiovascular Diseases, Hamamatsu Medical University, Sendai Social Insurance Hospital, Iwate Medical University, Okayama Medical Center, Nagoya City University, Tokushima University, Gifu Prefectural General Medical Center, Tokyo Medical University, Tokyo Medical and Dental University, and Tokyo Women’s Medical University Medical Center East, Japan) after each patient provided written informed consent. A total of 42 cancer samples (11 women and 31 men; median age 62.5 years; range 25–75) (Table I) that had been histologically confirmed as RCC, were selected for this study. The clinical stage of each patient was assessed according to the Union International Centre Cancer tumor node metastasis classification. Corresponding normal tissue was also obtained from the distant region of the cancer lesion in the resected kidney tissue. These samples were immediately embedded in TissueTek OCT compound (Sakura, Tokyo, Japan), frozen and stored at −80°C. The frozen tissues were sliced into 8-μm sections using a cryostat (Sakura) and then stained with H&E for histological examination.
Table I.
Clinicopathological features of patients with renal cell carcinoma.
| ID | Gender | Age | Histological type | Response | Group | Prediction |
|---|---|---|---|---|---|---|
| KIS-020 | M | 63 | Clear cell carcinoma | PR | Responder | Learning |
| KIS-021 | F | 71 | Clear cell carcinoma | PR | Responder | Learning |
| KIS-028 | M | 71 | Clear cell carcinoma | PR | Responder | Learning |
| KIS-034 | M | 55 | Clear cell carcinoma | PR | Responder | Learning |
| KIS-035 | F | 74 | Clear cell carcinoma | PR | Responder | Learning |
| KIS-038 | M | 63 | Clear cell carcinoma | PR | Responder | Learning |
| KIS-044 | M | 66 | Clear cell carcinoma | CR | Responder | Learning |
| KIS-050 | M | 60 | Clear cell carcinoma | PR | Responder | Learning |
| KIS-058 | M | 56 | Clear cell carcinoma | PR | Responder | Learning |
| KIS-061 | M | 65 | Clear cell carcinoma | CR | Responder | Learning |
| KIS-001 | M | 69 | Clear cell carcinoma | NC | Non-responder | Learning |
| KIS-005 | M | 25 | Combined type (cyst-associated and papillary renal cell carcinoma) | PD | Non-responder | Learning |
| KIS-006 | M | 44 | Clear cell carcinoma | PD | Non-responder | Learning |
| KIS-007 | M | 71 | Clear cell carcinoma | PD | Non-responder | Learning |
| KIS-008 | M | 67 | Clear cell carcinoma | PD | Non-responder | Learning |
| KIS-016 | F | 67 | Clear cell carcinoma | NC | Non-responder | Learning |
| KIS-026 | M | 64 | Clear cell carcinoma | NC | Non-responder | Learning |
| KIS-032 | M | 60 | Clear cell carcinoma | NC | Non-responder | Learning |
| KIS-040 | F | 68 | Clear cell carcinoma | PD | Non-responder | Learning |
| KIS-046 | M | 51 | Clear cell carcinoma | PD | Non-responder | Learning |
| KIS-048 | M | 67 | Clear cell carcinoma | PD | Non-responder | Learning |
| KIS-049 | F | 51 | Clear cell carcinoma | NC | Non-responder | Learning |
| KIS-051 | M | 73 | Clear cell carcinoma | NC | Non-responder | Learning |
| KIS-052 | F | 71 | Combined type (clear cell, granular cell and spindle cell carcinoma) | PD | Non-responder | Learning |
| KIS-057 | F | 60 | Clear cell carcinoma | PD | Non-responder | Learning |
| KIS-059 | M | 60 | Clear cell carcinoma | NC | Non-responder | Learning |
| KIS-060 | M | 56 | Clear cell carcinoma | NC | Non-responder | Learning |
| KIS-066 | M | 61 | Clear cell carcinoma | NC | Non-responder | Learning |
| KIS-012 | M | 62 | Clear cell carcinoma | PR | Responder | Test |
| KIS-029 | F | 57 | Clear cell carcinoma | PR | Responder | Test |
| KIS-043 | M | 64 | Clear cell carcinoma | PR | Responder | Test |
| KIS-045 | M | 62 | Papillary renal cell carcinoma | PR | Responder | Test |
| KIS-065 | F | 39 | Combined type (clear cell carcinoma and spindle cell carcinoma) | PR | Responder | Test |
| KIS-003 | F | 56 | Clear cell carcinoma | NC | Non-responder | Test |
| KIS-004 | M | 58 | Clear cell carcinoma | MR | Non-responder | Test |
| KIS-015 | M | 40 | Clear cell carcinoma | NC | Non-responder | Test |
| KIS-023 | M | 71 | Clear cell carcinoma | MR | Non-responder | Test |
| KIS-030 | F | 51 | Clear cell carcinoma | NC | Non-responder | Test |
| KIS-033 | M | 72 | Clear cell carcinoma | PD | Non-responder | Test |
| KIS-047 | M | 76 | Clear cell carcinoma | MR | Non-responder | Test |
| KIS-056 | M | 69 | Clear cell carcinoma | MR | Non-responder | Test |
| KIS-064 | M | 61 | Clear cell carcinoma | PD | Non-responder | Test |
Response, response to IL-2 and IFN-α combination therapy for RCC with lung metastasis; Responder, CR or PR; Non-responder, PD, NC or MR. CR, complete response (shrinkage rate 100%); PR, partial response (shrinkage rate ≥50 to <100%); MR, minor response (shrinkage rate ≥25 to <50%); NC, no change (shrinkage rate ≥−25 to <25%); PD, progressive disease (shrinkage rate ≤−25%). Shrinkage rate, tumor shrinkage rate in pulmonary metastasis.
Preparation and analysis of RNA
Total RNA purified using the RNeasy Micro kit (Qiagen, Germany) was quantified in a Nano Drop spectrophotometer (ND 1000). The RNAs, whose ratios of OD 260/280 nm were between 1.7 and 2.0, were used for further analysis.
Microarray analysis
According to the protocol of the T7-Oligo(dT) Promoter Primer kit and IVT Labeling kit (Affymetrix), RNAs were reversely transcribed using oligo(dT) promoter primers with a T7 recognition site in the first-strand cDNA synthesis. Following purification of the product of the double-strand cDNA after second-strand cDNA synthesis, in vitro transcription (IVT) reaction of biotinylated complementary RNA was carried out using T7 RNA polymerase. Labeled RNA was fragmented and hybridized to the array named Affymetrix Human Genome U133 Plus 2.0. The arrays were washed, stained and scanned using the GeneChip 3000 7G scanner (Affymetrix). Signal intensities and the absolute call dataset were generated with Affymetrix Gene Chip Operating Software (AGCC) using the MAS5.0 algorithm.
Identification of genes associated with the clinical response to the treatment
Treatment responses, in detail, evaluated according to the Response Criteria for Urological Cancer Treatment (20), which are nearly identical to the WHO criteria (21), are documented in Tables I and II. We applied a random permutation test to identify genes whose expression levels were significantly different between the responder (clinical response: CR, complete response or PR, partial response) and non-responder groups (MR, minor response; NC, no change or PD, progressive disease); favorable response or poor response was defined by the tumor shrinkage rate of pulmonary metastasis.
Table II.
Clinical response and classification of the two groups.
| Group | Clinical response | No. of patients |
|---|---|---|
| Responder | ||
| CR | 2 | |
| PR | 13 | |
| Non-responder | ||
| MR | 4 | |
| NC | 12 | |
| PD | 11 | |
| Total | 42 |
Responder group, CR or PR; non-responder group, PD, NC or MR. CR, complete response (shrinkage rate 100%); PR, partial response (shrinkage rate ≥50 to <100%); MR, minor response (shrinkage rate ≥25 to <50%); NC, no change (shrinkage rate ≥−25 to <25%); PD, progressive disease (shrinkage rate ≤−25%). Shrinkage rate, tumor shrinkage rate in pulmonary metastasis. Efficacy rate 35.7%.
The mean (μ) and standard deviation (σ) were calculated from the log-transformed relative expression ratios of each gene in responder (r) and non-responder (n) cases. A discrimination score (DS) for each gene was defined as follows: DS = (μr-μn)/(σr+σn).
We performed permutation tests to evaluate the ability of individual genes to distinguish between 10 samples in the responders and 18 samples in the non-responders; samples were randomly permutated between the two groups at 1,000,000 times. Since the DS dataset of each gene showed a normal distribution, we calculated a P-value for the user-defined grouping (22).
Quantitative RT-PCR
We identified 24 genes (Table III) that showed significantly different levels of expression between the responder and non-responder groups, based on microarray analysis, and subsequently focused on two immunologically important genes, HLA-DQA1 and HLA-DQB1. To examine the possibility of adapting our prediction system for clinical use, we performed quantitative real-time RT-PCR of the two genes and evaluated their expression levels in all 42 samples by calculating relative expression ratios of each sample. Extracted RNAs were reversely transcribed using the SuperScript VILO cDNA Synthesis kit (Invitrogen) following the supplier’s protocol. For the quantification of mRNA levels, real-time quantitative PCR was performed with LightCycler 480 (Roche). The sequences of each primer and probe were as follows: internal control (β2M), forward primer 5′-TAGGAGGGCTGGCAACTTAG-3′; reverse primer 5′-CCAAGATGTTGATGTTGGATAAGA-3′; and TaqMan Probe 5′-GGGAGCAG-3′; predictive gene1 (HLA-DQA1), forward primer 5′-ACTATTCTCTGGCCCGGTTT-3′; reverse primer 5′-TACCCCAGGCATGTCTTTGT-3′; and TaqMan probe 5′-CTCCTCCA-3′; predictive gene2 (HLA-DQB1), forward primer 5′-AGCATTTTGGGGTGTCAAGT-3′; reverse primer 5′-ACACAGCACTCACCAAACCA-3′; and TaqMan probe 5′-CAGAGGAG-3′.
Table III.
List of 24 discriminating genes.
| Accession no. | Symbol | P-value | Sign | Gene title |
|---|---|---|---|---|
| NM_004193 | GBF1 | 8.87E-07 | - | Golgi-specific brefeldin A resistance factor 1 |
| NM_005575 | LNPEP | 1.49E-06 | + | Leucyl/cystinyl aminopeptidase |
| NM_005923 | MAP3K5 | 3.47E-06 | + | Mitogen-activated protein kinase kinase kinase 5 |
| AW452656 | ----- | 6.95E-06 | - | cDNA FLJ37989 fis, clone CTONG2011676 |
| NM_007081 | RABL2B | 1.36E-05 | - | RAB, member of RAS oncogene family-like 2B |
| X63381 | MEF2A | 2.26E-05 | + | Myocyte enhancer factor 2A |
| AK093779 | LOC399900 | 2.45E-05 | - | Hypothetical gene supported by AK093779 |
| X00452 | HLA-DQA1 | 2.83E-05 | + | Major histocompatibility complex, class II, DQ α1 |
| AK023514 | TDP1 | 3.76E-05 | + | Tyrosyl-DNA phosphodiesterase 1 |
| NM_018835 | RC3H2 | 3.99E-05 | + | Ring finger and CCCH-type zinc finger domain 2 |
| AL096842 | MTUS1 | 4.05E-05 | + | Mitochondrial tumor suppressor 1 |
| NM_007121 | NR1H2 | 5.80E-05 | - | Nuclear receptor subfamily 1, group H, member 2 |
| AA131302 | ----- | 8.88E-05 | - | Transcribed locus, weakly similar to NP_001039959.1 dynamin 1-like (Bos taurus) |
| NM_003170 | SUPT6H | 1.16E-04 | - | Suppressor of Ty 6 homolog (S. cerevisiae) |
| NM_001190 | BCAT2 | 1.17E-04 | - | Branched chain aminotransferase 2, mitochondrial |
| BF060747 | LOC130576 | 1.20E-04 | + | Hypothetical protein LOC130576 |
| BC000580 | PH-4 | 1.57E-04 | - | Hypoxia-inducible factor prolyl 4-hydroxylase |
| NM_024605 | ARHGAP10 | 1.91E-04 | + | Rho GTPase activating protein 10 |
| BG231758 | ----- | 1.92E-04 | + | Transcribed locus, strongly similar to NP_001025836.1 tuftelin interacting protein 11 (Gallus gallus) |
| AB029026 | TACC1 | 2.56E-04 | + | Transforming, acidic coiled-coil containing protein 1 |
| M16276 | HLA-DQB1 | 2.66E-04 | + | Major histocompatibility complex, class II, DQ β1 |
| NM_012463 | ATP6V0A2 | 3.51E-04 | + | ATPase, H+ transporting, lysosomal V0 subunit a2 |
| J03225 | TFPI | 3.61E-04 | + | Tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| AJ011597 | BDNFOS | 4.45E-04 | - | Brain-derived neurotrophic factor opposite strand |
P-values were calculated by random permutation tests. Information was retrieved from AGCC annotation database (Affymetrix).
+, gene-increased expression in the responder group;
-, gene-increased expression in the non-responder group.
PCR reactions were optimized for the number of cycles to ensure product intensity within the logarithmic phase of amplification.
Results
Laser microbeam microdissection (LMM) was carried out to enrich cancer cell populations from each of 42 tumor tissues from patients that had met the inclusion criteria of this clinical trial as reported previously (21). To attempt to establish a prediction method for clinical responses to IL-2 and IFN-α combination therapy, we analyzed gene expression profiles of microdissected renal cancer cells using an oligoDNA microarray consisting of 38,500 genes or ESTs. Analyzing 10 responders (CR or PR) and 18 non-responders (PD or NC), we identified 24 genes that showed significantly different levels of expression between the responder and non-responder groups according to the two following definitions: i) expression information based on signal intensities higher than the cutoff level in >60% of samples of at least one group; ii) random permutation test P-values <0.0005 (Table III). Among the 24 genes selected, the expression levels of 14 genes were higher in the responder group, including two immunologically important genes, HLA-DQA1 (P=2.83E-05) and HLA-DQB1 (P=2.66E-04), that are known to be HLA class II molecules having critical roles in antigen presentation. Of the remaining 22 genes, three genes have also been implicated in several roles in immunological responses: MAP3K5 (ASK1) is suggested to be related to the natural immunity to the stress response. TFPI is suggested to function as a negative regulator of cytokine expression. In addition, NR4A2 has been identified as a candidate target molecule for the treatment of multiple sclerosis, one of the autoimmune diseases of the central nervous system.
Since IL-2 and IFN-α treatment is expected to enhance patient immunity with the subsequent attack by immune cells on cancer cells, these two HLA class II molecules were considered to likely play key roles in clinical response. Hence, we focused on these two molecules measuring the expression levels quantitatively and comparing them to clinical response. We confirmed the relativity of expression level and clinical response using the receiver operating characteristic (ROC) curve, and the best cutoff value for the expression level of each gene was determined (Fig. 1). The results demonstrated that IL-2 and IFN-α combination therapy showed poor response rates of 85.7 and 83.3%, respectively, when the expression level of HLA-DQA1 or HLA-DQB1 was lower than the cutoff value (Fig. 2). If patients having a lower expression of these genes were excluded, than the rate of effectiveness of this therapy is expected to be 57.1% in the case of HLA-DQA1 and 61.1% in the case of HLA-DQB1, respectively. However, since HLA-DQA1 and HLA-DQB1 are known to form a heterodimer in the antigen presentation process, we hypothesized that if either one was expressed lower than the cutoff level, the tumor cells were unlikely to produce an HLA-class II molecule sufficiently and were then unable to present the antigen(s) effectively. Considering this hypothesis, if this treatment was withheld to the patients whose tumors expressed a lower level of either HLA-DQA1 or HLA-DQB1 than each cutoff value, than the response rate could be expected to reach as high as 66.7% (Fig. 2, Table IV).
Figure 1.
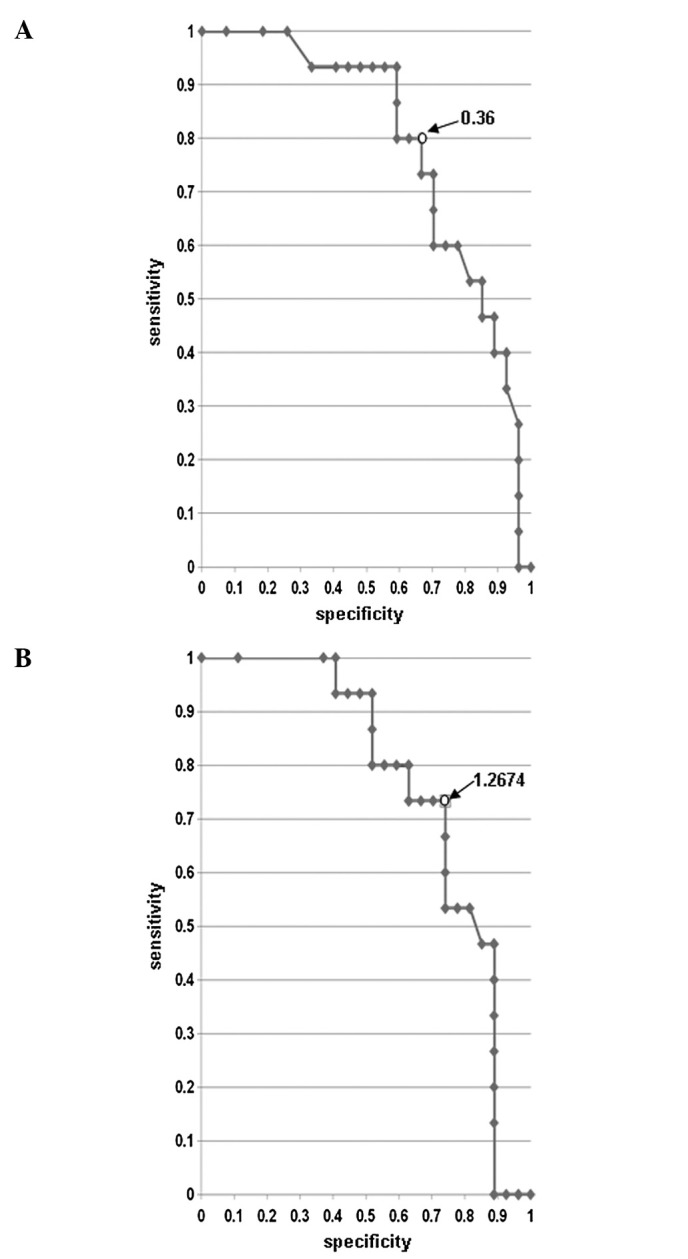
Receiver operating characteristic (ROC) curve. Result of microarray data. Sensitivity shows the ratio of the number of samples for which a value is higher than each cutoff value in the responder group. Specificity shows the ratio of the number of samples for which a value is higher than each cutoff value in the non-responder group. Specificity indicates the number for which the ‘ratio of the number of samples for which a value is higher than each cutoff value in the non-responder group’ is subtracted from ‘1’. The point where ‘sensitivity’ and ‘specificity’ were both near ‘1’ [in short where ‘(1 - sensitivity)2 + (1 - specificity)2’ was the lowest] was determined to be the optimal cutoff value for the expression level of each gene based on the ROC curve. (A) ROC curve of HLA-DQA1. The optimum cutoff value (arrow) was 0.36; at this point, sensitivity was 0.80 and specificity was 0.67. (B) ROC curve of HLA-DQB1. The optimum cutoff value (arrow) was 1.2674; at this point, sensitivity was 0.73 and specificity was 0.74.
Figure 2.
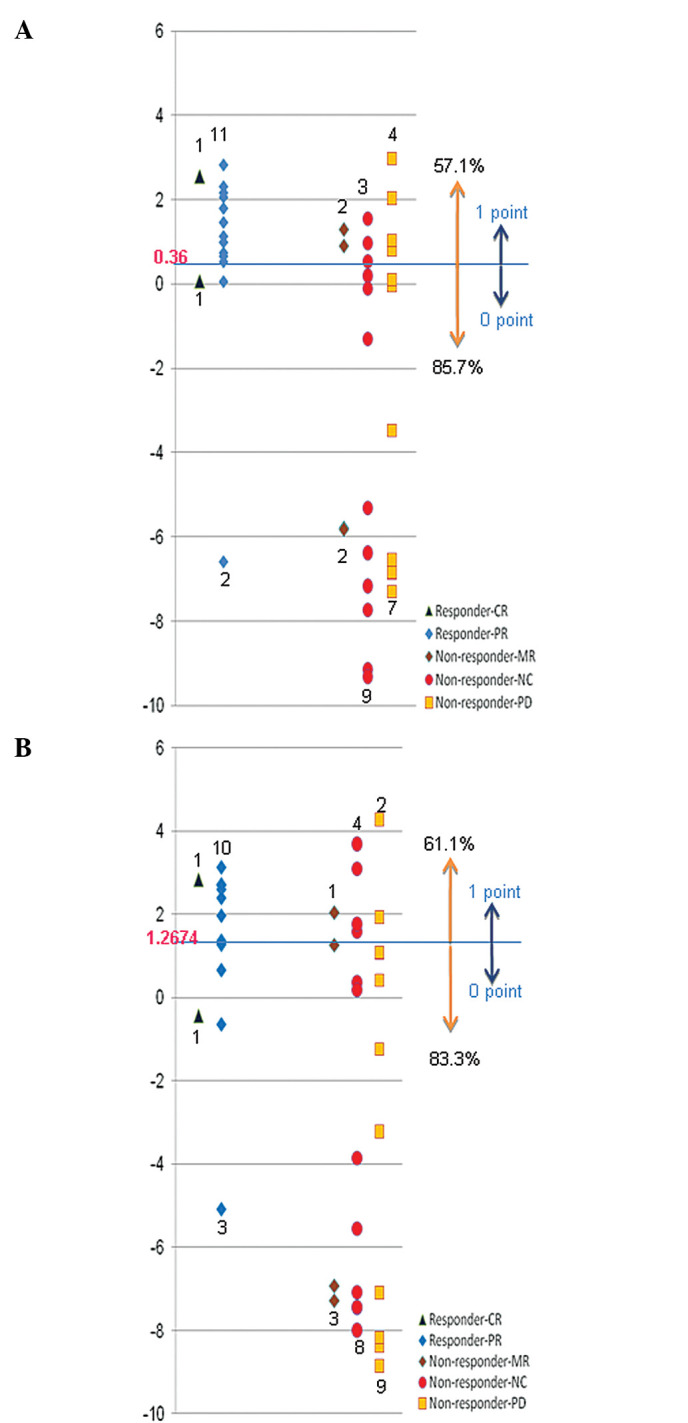
Expression pattern. The figures of the spindle show the value, which was divided by the expression level of the normal tissue and transformed to a logarithm. Dark blue squares, CR; blue diamonds, PR; brown diamonds, MR; red circles, NC; orange squares, PD. Samples whose expression levels were higher than these cutoff values were assigned a score of ‘1’ and samples whose expression levels were lower than these cutoff values were assigned a score of ‘0’. (A) Expression pattern of HLA-DQA1. (B) Expression of HLA-DQB1.
Table IV.
Scoring-based expression pattern 1.
| Group | Score
|
||
|---|---|---|---|
| 0 | 1 | 2 | |
| Non-responder (n=27) (MR + PD + NC) | 16 | 6 | 5 |
| Responder (n=15) (CR + PR) | 2 | 3 | 10 |
Summary table of the scoring-based expression pattern of Fig. 2. Score: 0, HLA-DQA1 and HLA-DQB1 expression values both less than the cutoff value; 1, HLA-DQA1 (or HLA-DQB1) expression value less than the cutoff value and HLA-DQB1 (or HLA-DQA1) expression value greater than the cutoff value; 2, HLA-DQA1 and HLA-DQB1 expression values both greater than the cutoff value. CR, complete response; PR, partial response; MR, minor response; NC, no change; PD, progressive disease.
Discussion
To screen genes that may be associated with the clinical response of advanced RCCs to IL-2 and IFN-α combination therapy, oligo DNA microarray analysis was applied in combination with LMM to obtain precise expression profile data of thousands of genes simultaneously. The following random permutation test of expression data identified 24 candidate genes that exhibited significant differential expression between the ‘responder’ and ‘non-responder’ groups (Table III). Since two immunologically important genes, HLA-DQA1 and HLA-DQB1, were included in the set of 24 genes, the relative expression levels were measured and then compared to clinical responses by applying the ROC curve for these two genes (Figs. 1 and 2; Table IV). As HLA-DQA1 and HLA-DQB1 are known to form a heterodimer, we hypothesized that when either gene exhibited a low level of expression in tumors, the antigen presentation from the tumor cells for immunotherapy may be insufficient. When such patients were excluded from this particular treatment, the rate of effectiveness of this therapy was expected to improve to 65–70% (Table IV).
To further apply our prediction system in clinical use, we attempted to establish the quantitative RT-PCR method. As a result, the mRNA levels measured by the quantitative RT-PCR method were found to be quite consistent to those obtained from the microarray analysis with Pearson’s correlation coefficient and Spearman’s rank-correlation coefficient of 0.89 and 0.83 for HLA-DQA1, respectively, and 0.78 and 0.65 for HLA-DQB1, respectively (Table V, Fig. 3).
Table V.
Scoring-based expression pattern 2.
| Group | Score
|
||
|---|---|---|---|
| 0 | 1 | 2 | |
| Non-responder (n=27) (MR + PD + NC) | 16 | 6 | 5 |
| Responder (n=15) (CR + PR) | 1 | 4 | 10 |
Summary table of scoring-based expression pattern of quantitative RT-PCR. Score: 0, HLA-DQA1 and HLA-DQB1 expression values both less than the cutoff value; 1, HLA-DQA1 (or HLA-DQB1) expression value less than the cutoff value and HLA-DQB1 (or HLADQA1) expression value greater than the cutoff value; 2, HLA-DQA1 and HLA-DQB1 expression values both greater than the cutoff value. CR, complete response; PR, partial response; MR, minor response; NC, no change; PD, progressive disease.
Figure 3.
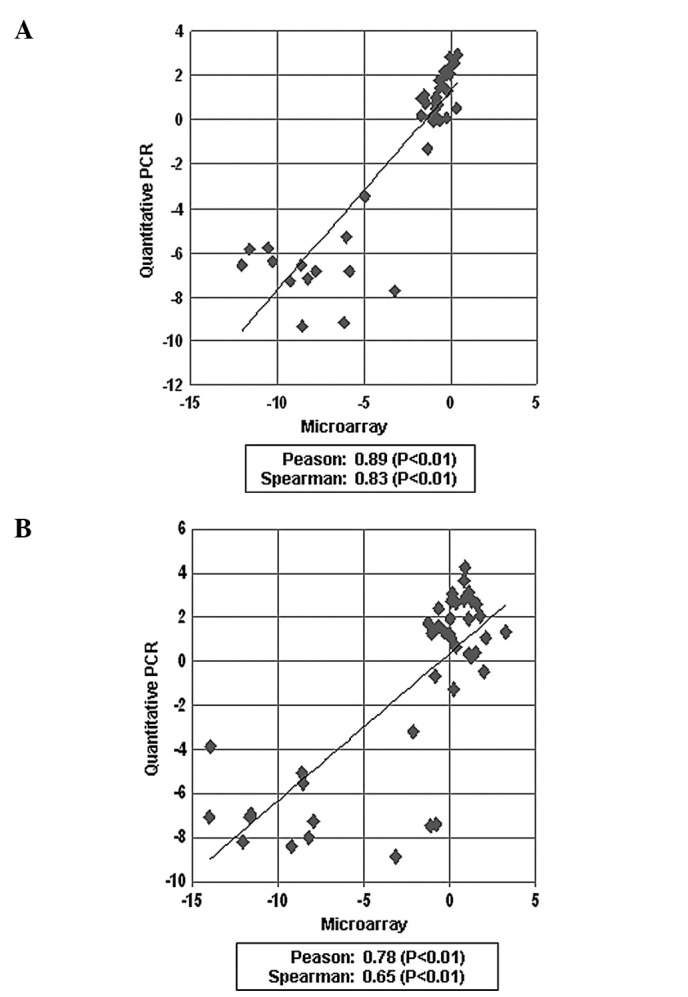
Comparison of microarray data for HLA-DQA1 and HLA-DQB1 with quantitative RT-PCR data. (A) Correlation of microarray data and real-time PCR for HLA-DQA1. (B) Correlation of microarray data and real-time PCR for HLA-DQB1.
In conclusion, the expression levels of HLA-DQA1 and HLA-DQB1 are reliable candidate markers for predicting the response to IL-2 and IFN-α combination therapy for RCC and provide useful information for the establishment of personalized treatment.
Acknowledgments
We thank Ms. Noriko Sudo for the data and sample management, Ms. Noriko Ikawa for the preparation of the samples by cryostat, Ms. Kyoko Kijima for the preparation of materials and Drs Yataro Daigo, Koichi Matsuda, Ryuji Hamamoto, Ryo Takata, Chikako Fukukawa, Yosuke Harada, Jae-Hyun Park, Masahiro Ajiro, Suyoun Chung, Akira Togashi, Yasuo Mochiduki, Shinya Hayami, Cho Hyuu-Soo, Goji Toyokawa, Masashi Takawa, Tadashi Kizaki, Mitsuko Nakashima and Kazuma Kiyotani for the helpful discussion and comments. This study was supported by Shionogi & Co., Ltd.
References
- 1.Ferlay J, Bray F, Pisani P, Parkin DM. GLOBOCAN 2002: Cancer Incidence, Mortality and Prevalence Worldwide. IARC CancerBase No. 5, Version 2.0. IARC Press; Lyon: 2004. [Google Scholar]
- 2.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 3.Motzer RJ, Dror Michaelson M, Redman BG, et al. Activity of SU11248, a multitargeted inhibitor of vascular endothelial growth factor receptor and platelet-derived growth factor receptor, in patients with metastatic renal cell carcinoma. J Clin Oncol. 2006;24:16–24. doi: 10.1200/JCO.2005.02.2574. [DOI] [PubMed] [Google Scholar]
- 4.Motzer RJ, Rini BI, Bukowski RM, et al. Sunitinib in patients with metastatic renal cell carcinoma. JAMA. 2006;295:2516–2524. doi: 10.1001/jama.295.21.2516. [DOI] [PubMed] [Google Scholar]
- 5.Motzer RJ, Hutson TE, Tomczak P, et al. Sunitinib versus inter-feron alpha in metastatic renal-cell carcinoma. N Engl J Med. 2007;356:115–124. doi: 10.1056/NEJMoa065044. [DOI] [PubMed] [Google Scholar]
- 6.Escudier B, Eisen T, Stadler WM, et al. Sorafenib in advanced clear-cell renal-cell carcinoma. N Engl J Med. 2007;356:125–134. doi: 10.1056/NEJMoa060655. [DOI] [PubMed] [Google Scholar]
- 7.Szcylik C, Demkow T, Staehler M, et al. Randomized phase II trial of first-line treatment with sorafenib versus interferon in patients with advanced renal cell carcinoma: final results. J Clin Oncol. 2007;25:5025. doi: 10.1200/JCO.2008.19.3342. [DOI] [PubMed] [Google Scholar]
- 8.Aso Y, Tazaki H, Umeda T, Marumo K. A phase II study of S-6820 (recombinant interleukin-2) on renal cell carcinoma (in Japanese) Biotherapy. 1989;3:999–1007. [Google Scholar]
- 9.Aso Y, Homma Y, Tazaki H, et al. A phase II trial of S-6820 (recombinant interleukin-2) on renal cell carcinoma refractory to interferon (in Japanese) Hinyokigeka. 1995;8:75–86. [Google Scholar]
- 10.Hayashi T, Miyagawa Y, Tsujimura A, Nonomura N, Minami M, Okuyama A. A case of renal cell carcinoma with multiple lung metastases refractory to interferon-α showing complete remission by interleukin-2 monotherapy. Int J Urol. 2006;13:805–808. doi: 10.1111/j.1442-2042.2006.01408.x. [DOI] [PubMed] [Google Scholar]
- 11.Kobayashi M, Ikeda H, Nukui A, et al. Clinical outcome and prognostic survival factors in patients with advanced renal cell carcinoma treated with very low-dose interleukin-2, interferon-α and tegafur-uracil: a single-institution experience. Int J Clin Oncol. 2008;13:257–262. doi: 10.1007/s10147-007-0752-0. [DOI] [PubMed] [Google Scholar]
- 12.Wirth MP. Immunotherapy for metastatic renal cell carcinoma. Urol Clin North Am. 1993;20:283–295. [PubMed] [Google Scholar]
- 13.Horoszewicz JS, Murphy GP. An assessment of the current use of human interferons in therapy of urological cancers. J Urol. 1989;142:1173–1180. doi: 10.1016/s0022-5347(17)39022-5. [DOI] [PubMed] [Google Scholar]
- 14.Bukowski RM. Natural history and therapy of metastatic renal cell carcinoma: the role of interleukin-2. Cancer. 1997;80:1198–1220. doi: 10.1002/(sici)1097-0142(19971001)80:7<1198::aid-cncr3>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 15.Motzer RJ, Russo P. Systemic therapy for renal cell carcinoma. J Urol. 2000;163:408–417. [PubMed] [Google Scholar]
- 16.Klapper JA, Downey SG, Smith FO, et al. High-dose interleukin-2 for the treatment of metastatic renal cell carcinoma: a retrospective analysis of response and survival in patients treated in the Surgery Branch at the National Cancer Institute between 1986 and 2006. Cancer. 2008;113:293–301. doi: 10.1002/cncr.23552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Belldegrun AS, Klatte T, Shuch B, et al. Cancer-specific survival outcomes among patients treated during the cytokine era of kidney cancer (1989–2005): a benchmark for emerging targeted cancer therapies. Cancer. 2008;113:2457–2463. doi: 10.1002/cncr.23851. [DOI] [PubMed] [Google Scholar]
- 18.Akaza H, Tsukamoto T, Onishi T, Miki T, Kinouchi T, Naito S. A low-dose combination therapy of interleukin-2 and interferon-α is effective for lung metastasis of renal cell carcinoma: a multi-center open study. Int J Clin Oncol. 2006;11:434–440. doi: 10.1007/s10147-006-0596-z. [DOI] [PubMed] [Google Scholar]
- 19.Akaza H, Kawai K, Tsukamoto T, et al. Successful outcomes using combination therapy of interleukin-2 and interferon-α for renal cell carcinoma patients with lung metastasis. Jpn J Clin Oncol. 2010;40:684–689. doi: 10.1093/jjco/hyq027. [DOI] [PubMed] [Google Scholar]
- 20.WHO Handbook for Reporting Results of Cancer Treatment . WHO Offset Publication No. 48. World Health Organization; Geneva: 1979. [Google Scholar]
- 21.Japanese Urological Association and Japanese Society of Pathology Response criteria for urological cancer treatment. Jpn J Urol. 1992;83:447–472. [PubMed] [Google Scholar]
- 22.Golub TR, Slonim DK, Tamayo P, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]


