Abstract
Selective elimination of mitochondria by autophagy (mitophagy) is a crucial developmental process to dispose of disintegrated or superflous organelles. However, little is known about underlying regulatory mechanisms. We have investigated mitophagy in response to conditional overexpression of the a2 mating-type locus gene lga2, which encodes a small mitochondrial protein critically involved in uniparental mitochondrial DNA inheritance during sexual development of Ustilago maydis. In this study, we show that conditional overexpression of lga2 efficiently triggers mitophagy that is dependent on atg8 and atg11, consistent with selective autophagy. lga2-triggered mitophagy is preceded by mitochondrial dysfunction, including depletion of mitochondrial RNA transcripts, and is mechanistically distinct from starvation-induced mitophagy despite a common requirement for atg11. In particular, lga2-triggered mitophagy strongly depends on the mitochondrial fission factor Dnm1, but it is only slightly affected by N-acetylcysteine, which is an inhibitor of starvation-induced mitophagy. To further delineate the role of mitochondrial fission, we analyzed lga2 effects in Δfis1 mutants. This revealed that mitochondrial fragmentation was only attenuated and mitophagy was largely unaffected. In further support of a Fis1-independent role for Dnm1, mitochondrial association of green fluorescent protein-tagged Dnm1 as well as Dnm1-opposed mitochondrial fusion during sexual development were fis1 independent. In conclusion, our results specify the role of the mitochondrial fission factor Dnm1 in mitophagy and uncover differences between mitophagy pathways in the same cellular system.
INTRODUCTION
Autophagy is a catabolic process that typically involves de novo formation of double-membrane bilayers, which engulf cellular targets to become autophagosomes. Subsequent fusion with lytic compartments leads to formation of autophagic bodies, which disintegrate and deliver their cargo for hydrolysis by enzymes resident within lysosomes or vacuoles (14, 28). Nonselective or bulk autophagy is induced under nutrient starvation conditions and involves degradation of many intracellular components to supply cells with essential metabolic building blocks and energy. On the other side, selective autophagy removes superfluous or damaged organelles as well as protein aggregates (19, 28, 43). Autophagy is governed by the so-called Atg (autophagy-related) proteins, which are classified into core proteins and proteins for selective autophagy. An essential core member is Atg8, a ubiquitin-like protein that is conjugated to the membrane phospholipid phosphatidylethanolamine and participates in the formation and expansion of autophagosomal vesicles. In contrast, Atg11 is dispensable for bulk autophagy of cytosolic proteins but specifically required for selective transport of organelles to lytic compartments (14, 19, 28). Autophagy of mitochondria (termed mitophagy) provides a means to dispose of damaged organelles and thus to contribute to the maintenance of mitochondrial integrity (14, 21, 22, 30, 40, 42, 43). Investigation of mitophagy has gained much attention since the recent discoveries of specific underlying signaling components in both yeast and mammals. A prominent example is the PINK1/Parkin protein couple, which operates in dopaminergic neurons to mediate disposal of damaged mitochondria (references 19 and 43 and references therein). Recently, it was shown that prevention of mitophagy in nitrogen-starved Saccharomyces cerevisiae cells caused accumulation of reactive oxygen species followed by formation of severe mitochondrial DNA (mtDNA) lesions (22). Currently, little is known about the regulatory circuits that underlie induction of mitophagy in response to either starvation conditions or mitochondrial dysfunction (references 11, 14, 19, and 43 and references therein).
There is accumulating evidence that mitochondrial dynamics participates in the regulation of mitophagy. Mitochondrial dynamics is largely controlled by opposing fusion and fission events involving highly conserved GTPase-containing proteins (42). The dynamin-like protein Dnm1 (termed DRP1 in mammals) is considered a principal player in mitochondrial fission. The tail-anchored outer membrane protein Fis1, which is evenly distributed on the mitochondrial surface, is another conserved fission factor. Yeast Fis1 functions as a membrane receptor that helps recruit Dnm1 to the sites of mitochondrial fission and contributes to the ordered formation of Dnm1 assemblies (26, 33, 34, 39, 42). Recently, it was shown that human Fis1 is dispensable for mitochondria fission and Drp1 recruitment to mitochondria due to the existence of a different Drp1 receptor, which mediates Fis1-independent fission (31). While there is evidence that mitochondrial fission and mitophagy are coordinated in mammalian cells (10, 38, 40), an influence of fission on mitophagy in yeast has been debated for starvation conditions (reference 25 and references therein). Beyond this, a requirement of the mitochondrial fission factor Dnm1 for mitophagy in yeast is only known from a mutant with disturbed cation homeostasis due to depletion of the mitochondrial carrier protein Mdm38 (29).
The smut fungus Ustilago maydis represents a highly attractive model organism for the study of cellular processes (35). Recently, we identified the mitochondrial protein Lga2 as a central component of uniparental mtDNA inheritance in the maize smut fungus U. maydis (2, 8). The lga2 gene resides on the a2 mating-type locus and is highly upregulated during sexual development in response to formation of an active b transcription factor complex in dikaryotic cells (3, 6, 41). In particular, we have shown that lga2 mediates selective mtDNA loss of the a1 mating partner within dikaryotic cells (8). In addition, evidence exists that Lga2 is associated with the mitochondrial surface; however, its mode of action remains to be elucidated (24). To study lga2 effects in U. maydis under more tractable conditions than sexual development, which is intimately coupled to biotrophic growth within the host plant, we previously generated U. maydis strains providing for conditional lga2 overexpression under the arabinose-inducible crg1 promoter. Transfer of these strains from glucose- to arabinose-containing medium led to extensive mitochondrial fragmentation, selective loss of mtDNA, and a severe proliferation defect (6). This raised the question of whether lga2 overexpression additionally triggered mitophagy to dispose of disintegrated organelles, and furthermore, whether this depended on mitochondrial fragmentation.
Here, we demonstrate that conditional overexpression of lga2 in U. maydis efficiently triggers mitophagy in dependence on atg8 and atg11. Our results specify the underlying role of the mitochondrial fission factor Dnm1 and provide evidence for Dnm1-dependent as well as Dnm1-independent mitophagy pathways.
MATERIALS AND METHODS
Strains, growth conditions, and chemicals.
U. maydis strains used in this study are described in Table 1. Strain MC2#4 (6) (designated MC2 if not indicated otherwise), providing for conditional overexpression of lga2 under the arabinose-inducible crg1 promoter, was used as progenitor for production of the MC2Δatg8, MC2Δatg11, MC2Δmca1, and MC2Δfis1 strains. Growth conditions and media (YEPSl [yeast extract-peptone-sucrose] and PD [potato dextrose]) were as described previously (6, 8). For medium-dependent induction, cells were cultured in CMG (complete medium with 1% [wt/vol] glucose) (12) and washed twice in water prior to inoculation. For induction of lga2 expression under the arabinose-inducible crg1 promoter, cells were transferred to CMA (complete medium with 1% [wt/vol] arabinose) at starting optical densities at 600 nm (OD600) between 0.1 and 0.15 if not otherwise specified. The wild-type control strain FB2/pMB2-2 was inoculated at a starting OD600 of 0.06.
Table 1.
U. maydis strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| 521 | a1 b1 | 16 |
| FB1 | a1 b1 | 1 |
| FB2 | a2 b2 | 1 |
| FB2/pMB2-2 | a2 b2 Potef-mtGFP-cbxra | 6 |
| FB2Δmrb1/pMB2-2 | a2 b2 mrb1::hygr Potef-mtGFP-cbxr | 6 |
| FB2Δmca1/pMB2-2 | a2 b2 mca1::hygr Potef-mtGFP-cbxr | This study |
| FB1Δatg8/pMB2-2 | a1 b1 atg8::hygr Potef-mtGFP-cbxr | This study |
| FB2Δatg8/pMB2-2 | a2 b2 atg8::hygr Potef-mtGFP-cbxr | This study |
| FB1Δatg11/pMB2-2 | a1 b1 atg11::hygr Potef-mtGFP-cbxr | This study |
| FB2Δatg11/pMB2-2 | a2 b2 atg11::hygr Potef-mtGFP-cbxr | This study |
| MC2 (FB2/pMB2-2/pCLN1) | a2 b2 Potef-mtGFP-cbxr Pcrg1-lga2-natrb | 6 |
| MC2Δmrb1 | a2 b2 mrb1::hygr Potef-mtGFP-cbxr Pcrg1-lga2-natr | 6 |
| MC2Δdnm1 | a2 b2 dnm1::hygr Potef-mtGFP-cbxr Pcrg1-lga2-natr | 24 |
| MC2Δmca1 | a2 b2 mca1::hygr Potef-mtGFP-cbxr Pcrg1-lga2-natr | This study |
| MC2Δatg8 | a2 b2 atg8::hygr Potef-mtGFP-cbxr Pcrg1-lga2-natr | This study |
| MC2Δatg11 | a2 b2 atg11::hygr Potef-mtGFP-cbxr Pcrg1-lga2-natr | This study |
| MC2Δfis1 | a2 b2 fis1::hygr Potef-mtGFP-cbxr Pcrg1-lga2-natr | This study |
| MC2Δatg8/pMF1p-atg8 | a2 b2 atg8::hygr Potef-mtGFP-cbxr Pcrg1-lga2-natr pMF1p-atg8 | This study |
| FB1/pKS2c | a1 b1 Pcrg1-mtRFP-cbxr | 24 |
| FB2/pKS1c | a2 b2 Pcrg1-mtGFP-cbxr | 24 |
| FB1Δdnm1/pKS2c | a1 b1 dnm1::hygr Pcrg1-mtRFP-cbxr | 24 |
| FB2Δdnm1/pKS1c | a2 b2 dnm1::hygr Pcrg1-mtGFP-cbxr | 24 |
| MF34/pKS2c | a1 b14 Pcrg1-mtRFP-cbxr | 8 |
| GF5/pKS1c | a2 b13 Pcrg1-mtGFP-cbxr | 8 |
| FB2Δlga2/pKS1c | a2 b2 Δlga2::hygr Pcrg1-mtGFP-cbxr | 24 |
| FB1Δfis1/pKS2c | a1 b1 Δfis1::hygr Pcrg1-mtRFP-cbxr | This study |
| FB2Δfis1/pKS1c | a2 b2 Δfis1::hygr Pcrg1-mtGFP-cbxr | This study |
| MF34Δfis1/pKS2c | a1 b14 Δfis1::hygr Pcrg1-mtRFP-cbxr | This study |
| GF5Δfis1/pKS1c | a2 b13 Δfis1::hygr Pcrg1-mtGFP-cbxr | This study |
| BUB7/pKS2/pCudg1 | a1 b3 Pcrg1-mtRFP-cbxr Pcrg1-dnm1-eGFP-natr | 24 |
| BUB7Δfis1/pKS2/pCudg1 | a1 b3 Δfis1::hygr Pcrg1-mtRFP-cbxr Pcrg1-dnm1-eGFP-natr | This study |
For the time course study (see Fig. 2C and D, below), the starting OD600 values were 0.5, 0.5, 0.4, 0.3, and 0.15 (0.075 in case of the wild-type strain) for the 1.5-, 3-, 4.5-, 6-, and 12-h time points, respectively, to yield log-phase cultures at the indicated times. For the 0-h time point, samples were taken immediately before transfer to CMA. For all transfers to starvation conditions, starting OD600 values were between 0.15 and 0.2 if not otherwise specified. Starvation-induced mitophagy was analyzed either in 1:10-diluted YEPSl or in minimal medium without ammonium. Minimal medium contained 0.3% (wt/vol) (NH4)2SO4 (pH 7.0), 6.25% (wt/vol) salt solution (12), 2.0% (wt/vol) glucose (MMG) or 1.0% (wt/vol) arabinose (MMA). For analysis of plating efficiencies, appropriate dilutions were plated on solid PD and incubated at 28°C for 2 days. Strains carrying the pKS1, pKS2, and pCudg1 constructs (Table 1) were analyzed in CMA to enable expression of mitochondrial matrix-targeted green fluorescent protein (mtGFP) or red fluorescent protein (mtRFP) and of the Dnm1-GFP fusion, respectively. The antibiotics carboxin (CBX), hygromycin B (HYG), nourseothricin (NAT), and phleomycin (BLE) used for U. maydis selection were from Riedel-de Haën (Hannover, Germany), Roche (Mannheim, Germany), Werner BioAgents (Jena-Cospeda, Germany), and InvivoGen (Toulouse, France), respectively. All additional chemicals were of analytical grade and were obtained from Sigma (Taufkirchen, Germany) or Roth (Karlsruhe, Germany).
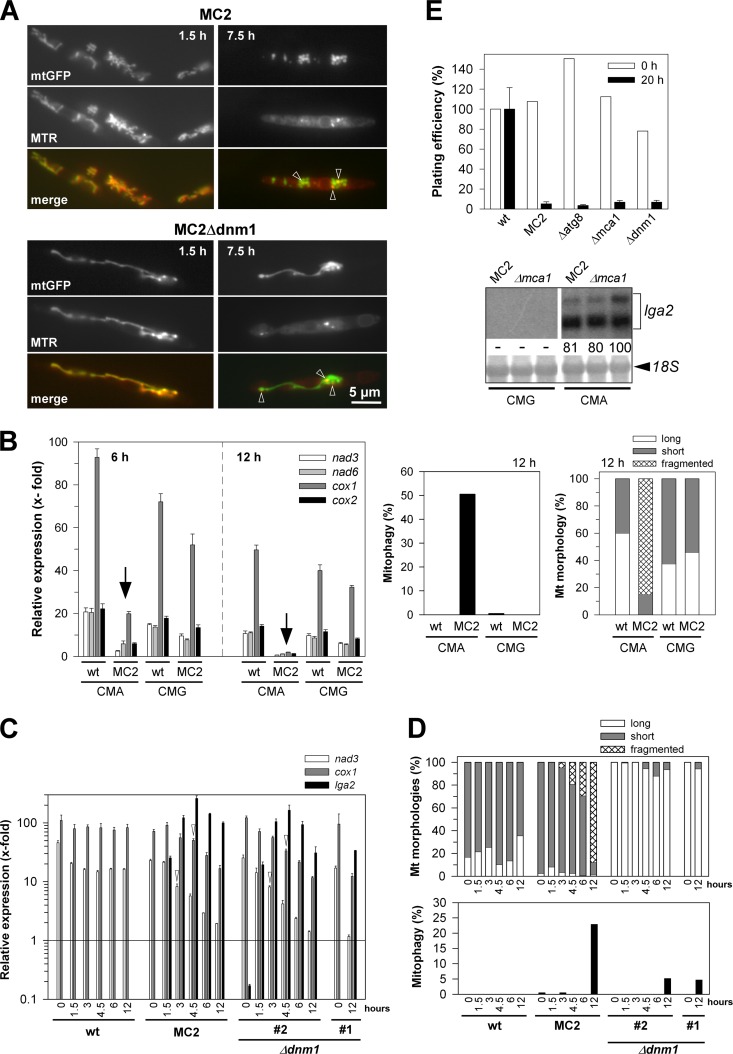
Fig 2.
Analysis of mitochondrial and cellular integrity in response to lga2 overexpression. (A) Reduction of membrane potentials. Cells were removed from cultures of strains MC2 and MC2Δdnm1 at 1.5 and 7.5 h after lga2 induction in CMA. Cells were analyzed by fluorescence microscopy to assess the colocalization between MTR and mtGFP fluorescence (see also Fig. S3 in the supplemental material). (B) Analysis of mtRNA levels in lga2-induced cells (left panel). The U. maydis strains FB2/pMB2-2 (wild type [wt]) and MC2 were transferred from CMG to either CMG or CMA to determine transcript levels of mitochondrial nad3, nad6, cox1, and cox2 genes by qRT-PCR analysis. For the 6-h time point, the starting OD600 values were 0.2 (CMG) or 0.3 (CMA), and for the 12-h time point, the OD600 values were 0.03 (CMG) or 0.1 to 0.15 (CMA). Results were normalized against the expression of the tbp gene (analyzed in each sample and set to 1.0). Given are the means ± standard deviations from four technical replicates. (Right panel) Samples were taken from the same cultures for analysis of mitophagy and mitochondrial morphologies at the 12-h time point. Values refer to the means from two independent cultures analyzed for each strain and condition (n > 100 cells/sample). (C and D) Time course analysis of lga2 effects and their dependence on dnm1. (C) The U. maydis strains FB2/pMB2-2 (wild type [wt]), MC2, and two independent MC2Δdnm1 strains (#1 and #2) were transferred from CMG to CMA to determine nad3 and cox1 transcript levels by qRT-PCR analysis. lga2 levels were analyzed for control. Results were normalized against the expression of the tbp gene (analyzed in each sample and set to 1.0). Given are the means ± standard deviations from three technical replicates. The detection threshold was set to 10−1 (relative units). (D) Samples were collected from cultures for analysis of mitochondrial morphologies (top) and mitophagy (bottom) (n > 200 cells/sample). For both panels C and D, samples from strain MC2Δdnm1#1 were collected only from the 0-h and 12-h time points for comparison. (E) Plating efficiencies were determined before (0 h) and at 20 h after lga2 induction. For the 20-h time point, calculations were from at least two experiments with ≥3 replicates per experiment, each including two independent MC2 mutant strains (means ± standard deviations). For the 0-h time point, average values of at least two replicates are indicated. Colony counts were normalized to the individual OD600 values at the time of plating and refer to the results with the wild-type strain FB2/pMB2-2 (set to 100%) for the two time points. (Bottom) Northern analysis to assess lga2 expression in two independent MC2Δmca1 strains (Δmca1) cultivated in either CMG or CMA for growth periods of 6 h. The parental MC2 strain served as a control. Methylene blue staining (18S) reflects the amounts of RNA loaded. Radioactive signals were quantified (both for lga2), with the strongest signal set to 100.
Mitophagy and mitochondrial fusion assays.
Mitophagy was assayed by fluorescence microscopy based on relocalization of mtGFP to vacuolar compartments as described elsewhere (37). For quantitative determination, cells having accumulated mtGFP within vacuoles were counted positive for mitophagy. Mitophagy was normally assayed 20 to 24 h after transfer to either CMA or to starvation conditions if not otherwise specified. To corroborate the mtGFP signal within vacuoles, the marker dye FM4-64 (Life Technologies GmbH, Darmstadt, Germany; dissolved as a 10 mM stock in dimethyl sulfoxide) was used at a final concentration of 8 μM for staining of vacuolar membranes (see Fig. 1, below). Cells were resuspended in fresh medium in the presence of FM4-64 for 10 min at 25°C in the dark (rotary wheel; 10 rpm) and washed twice with fresh medium. N-Acetylcysteine (NAC) was used at a final concentration of 2.5 mM. Mitochondrial fusion assays were performed as described previously (24).
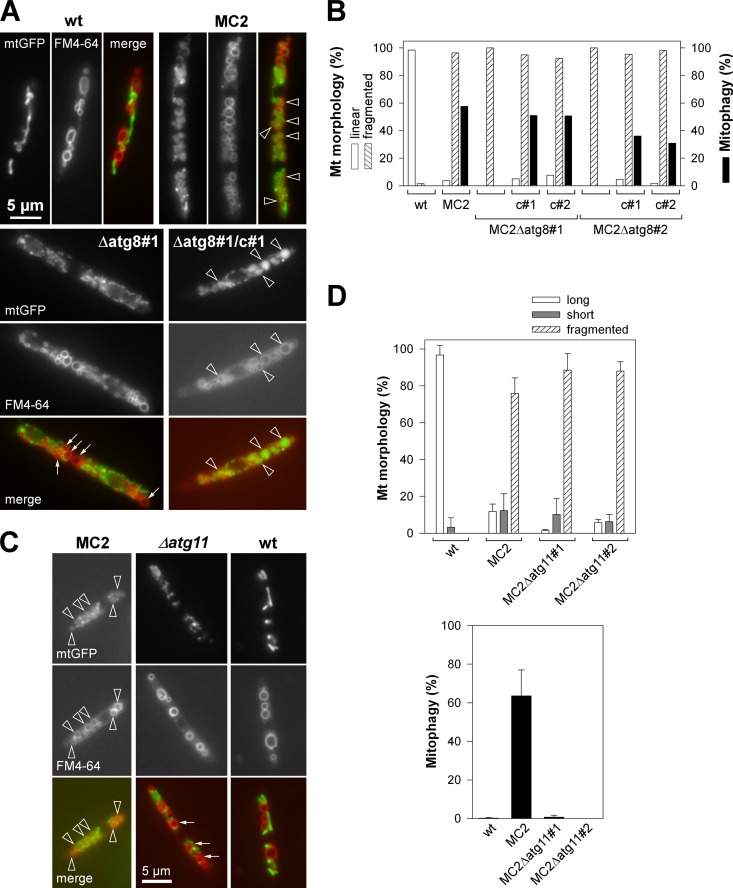
Fig 1.
LTM dependence on atg8 and on atg11. (A) Cultures of MC2Δatg8 (Δatg8#1) and the complemented strain (Δatg8#1/c#1) were transferred from CMG to CMA for 22 h of growth, followed by FM4-64 staining. Fluorescence microscopy was used to detect mitochondrial structures (mtGFP) and vacuoles stained with FM4-64. Representative images are shown. Bar, 5 μm (all panels). (B) Quantification of mitochondrial morphology states and mitophagy of two independent MC2Δatg8 mutants (#1 and #2) and complemented strains (c#1 and c#2) at 20 h after lga2 induction in CMA. Data show the percentages of cells with linear (tubular morphology) or fragmented mitochondria (punctuate pattern) and percentage of cells displaying mitophagy (black). n ≥ 200 cells/sample. (C) The same approach as described for panel A was applied to MC2Δatg11 mutants. (A and C) Note the enclosure of mtGFP within vacuolar compartments in strain MC2 and the complemented Δatg8 mutant (arrowheads), in contrast with the cytoplasmic, dotted mtGFP pattern in the Δatg8 and Δatg11 mutants (arrows). (D) Quantification of mitochondrial morphologies and mitophagy in lga2-induced MC2 strains in dependence on atg11, by fluorescence microscopy. Means ± standard deviations were derived from four replicate cultures using two independent Δatg11 mutants. Long, fluorescent structures spanning >1/3 of the cell length; short, tubules <1/3 of the cell length. n ≥ 100 cells per strain and sample. For all panels, strains FB2/pMB2-2 (wild type [wt]) and MC2 served as controls.
DNA and RNA procedures.
Escherichia coli K-12 strain TOP10 (Life Technologies) was used as host for plasmid amplifications. U. maydis DNA isolation and manipulation were performed as described previously (24). For Northern analysis, 32P-labeled fragments of the indicated genes were used. Radioactive labeling of DNA was performed with the NEBlot kit (New England BioLabs [NEB], Frankfurt a.M., Germany). Detection and quantification of the signals were performed using a Storm PhosphorImager (Molecular Dynamics, CA) and ImageQuant software. Restriction enzymes were from NEB, and oligonucleotides were obtained from MWG (Ebersberg, Germany). The correctness of all plasmid constructs was verified by sequencing (automatic DNA isolation and sequencing [ADIS]; Max-Planck-Institute, Cologne, Germany, or LMU Munich Sequencing Service [http://www.gi.bio.lmu.de/sequencing/index_html]). For Northern blot analysis, the lga2 fragment was isolated from pCLN1 by using StuI/NdeI (6). The atg8 fragment spanned the entire open reading frame (ORF).
qRT-PCR analysis.
For analysis of mtDNA expression in response to lga2 overexpression, total RNA was isolated from cells cultivated for 6 and 12 h, respectively, in either CMA (lga2 induction) or in CMG (control). For analysis of mtDNA expression under starvation conditions, total RNA was isolated from cells cultivated for 20 h in either MMG-N (nitrogen starvation) or in MMG (control). RNA was treated using the Ambion Turbo DNase kit (Life Technologies). For the production of cDNA, the SuperScript III first-strand cDNA synthesis kit and random hexamer primers were used (Life Technologies). Quantitative real-time PCR (qRT-PCR) analysis was performed using the Platinum SYBR qPCR Supermix-UDG kit (Life Technologies) in the presence of 10 nM fluorescein (Bio-Rad, Munich, Germany). Gene specific primers were the following: 5′-CGGTCAAACACGAAACCCTTTCTC-3′/5′-AGCTCCAGTACCAAACTCGTAGAC-3′ (nad3), 5′-CACTCGGTGCTGTTCTTTCTGG-3′/5′-CAGGAAAAGTACTGTTACTGCTCC-3′ (nad6), 5′-GCTCCTGATATGGCATTCCCTCG-3′/5′-CCTGTTCCTGCCCCTTGTTCTAC-3′ (cox1), 5′-TCCGGTTCATCGTAACTGGTGC-3′/5′-ACTCCTTCACGCTCGATAAGAACG-3′ (cox2). Results were normalized against the expression of the probable TATA box binding factor gene tbp (um10143) as described previously (44).
Production of Δmca1, Δatg8, Δatg11, and Δfis1 deletion strains.
Deletion strains were produced according to a PCR-based protocol (15). Sequences flanking the mca1 ORF (um01408) were amplified (Taq polymerase; Roche) from chromosomal DNA of strain FB2, using the primer combinations (SfiI sites underlined) 5′-CTTCATAGCTGCACGTCGACCCTCG-3′/5′-CACGGCCTGAGTGGCCGATTCAGTGGATGAGTGTGCCAG-3′ (left flank) and 5′-GTGGGCCATCTAGGCCGCGCTCCGCTTTTCATGTCTG-3′/5′-GACGAGTTGCAAAAACCCCGAGTAG-3′ (right flank). The deletion covered the entire ORF. Sequences flanking the atg8 ORF (um05567) were amplified (Taq polymerase; Roche) from chromosomal DNA of strain FB2, using the primer combinations (SfiI sites underlined) 5′-GTCGACGTAACAGCCAAGATG-3′/5′-CACGGCCTGAGTGGCCCATCGTGGCGCTATAAGACTC-3′ (left flank) and 5′-GTGGGCCATCTAGGCCGTACCAGTGTGCTGAAC-3′/5′-CGTGAATGAGCAGCACTCACAGAC-3′ (right flank). The deletion covered the entire ORF extended by 18 bp of untranslated 3′ region. Sequences flanking the fis1 ORF (um03919) were amplified (Phusion high-fidelity DNA polymerase; NEB) from chromosomal DNA of strain 521, using the primer combinations (SfiI sites underlined) 5′-GAGGCGCAACGACGCAAGCAG-3′/5′-CACGGCCTGAGTGGCCTGCGTGCGAGCACCAAGGTTG-3′ (left flank) and 5′-GTGGGCCATCTAGGCCCGTCGCTAAATTACTCCGTTCTC-3′/5′-GAGGTATCCCGACAACTGGTTC-3′ (right flank). The deletion covered the entire ORF except for the 3′-terminal 9 bp. Sequences flanking the atg11 ORF (um03341) were amplified (Phusion high-fidelity DNA polymerase; NEB) from chromosomal DNA of strain 521, using the primer combinations (SfiI sites underlined) 5′-CGCATCTCACATGCAATGTG-3′/5′-CACGGCCTGAGTGGCCCATTGGCCGACGTACAGCTG-3′ (left flank) and 5′-GTGGGCCATCTAGGCCTCGCTGCGGTGATCGGAACG-3′/5′-ACGTCGAAGCTACACCATCG-3′ (right flank). The deletion covered the entire ORF except for the 5′-terminal 3 bp and 3′-terminal 12 bp. PCR products were cleaved with SfiI for ligation with the Hyg resistance cassette isolated with SfiI from pBShhn (15). Gene replacement constructs were cloned into pCR4-TOPO or pCR2.1 (Life Technologies) and were reamplified for transformation (Δmca1, Δfis1, and Δatg11 [in case of transformation of strain FB1/pMB2-2]). For transformation of the Δatg8 construct, the plasmid was cut at the BglII site. For transformation of the Δatg11 construct (strains FB2/pMB2-2 and MC2), the 4,225-bp BamHI/NaeI fragment was isolated. Homologous recombination in U. maydis transformants (Table 1) was confirmed by diagnostic PCR to demonstrate the absence of the endogenous gene as well as integration of left and right borders (15).
Complementation of Δatg8 mutants.
For complementation of Δatg8 mutants, genomic DNA of strain FB2 was amplified (Phusion high-fidelity DNA polymerase; NEB), using the primer combination (EcoRI sites underlined) 5′-CGGAATTCCTCTGTGCTGGATGTGTCTG-3′/5′-CGGAATTCCCATGAGATCGATCTTGTGG-3′ spanning the atg8 ORF extended by 1,317 bp of the 5′ region and 97 bp of the 3′ region. The PCR product was cleaved with EcoRI and inserted into the MfeI site of plasmid pMF1-p (BLE resistance) (4). The resulting construct, pMF1p-atg8, was cleaved with ScaI prior to transformation into MC2Δatg8 strains. Ectopic insertion was verified by PCR using the primer combination 5′-GAGAGGATCCGTCAGAAGTAC-3′/5′-CTGTCCGAAAGTGTTCTCGC-3′ spanning the atg8 ORF.
Microscopy.
Microscopy was performed as described previously (6). For microscopic analysis, cells were immobilized in agarose and first analyzed for RFP fluorescence to avoid photoconversion of GFP. Samples were observed with differential interference contrast (DIC) optics or under fluorescence microscopy (excitation/emission for enhanced GFP [eGFP], 450 to 490/515 to 565 nm; for RFP or FM4-64, 546/>590 nm). All microscopic imaging was performed at ×100 magnification. Images were processed using AxioVision software (www.zeiss.de/axiovision) and Adobe Photoshop image software, using only those processing functions that could be applied equally to all pixels of the image.
Immunoblot analysis.
Mitochondrial and cytosolic protein fractions were prepared as described previously (6). Immunoblot analysis using anti-GFP and anti-cytochrome c heme lyase (CCHL) antibodies was performed as described previously (24). The anti-α-tubulin antibody (clone DM1A) was from Sigma. Purified GFP (Roche) was used as a positive control.
Databases.
Protein sequences were compared using the NCBI BLAST database (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Genomic sequences of U. maydis were retrieved from the Broad Institute (http://www.broadinstitute.org/annotation/genome/ustilago_maydis/Home.html). U. maydis protein sequences (um numbers) were retrieved from the MIPS (Munich Information Center for Protein Sequences) Ustilago maydis Genome Database (MUMDB; http://mips.helmholtz-muenchen.de/genre/proj/ustilago). Protein subdomains were predicted with PFAM (http://pfam.sanger.ac.uk/search). Transmembrane regions were predicted by using TMHMM (http://www.expasy.ch/; http://www.cbs.dtu.dk/services/TMHMM-2.0/). ClustalX was used for comparative sequence alignments (http://www.clustal.org/).
RESULTS
lga2-triggered mitophagy.
Previously, we reported that conditional overexpression of lga2 (MC2 strains) interferes with respiratory activity and causes selective loss of mtDNA, indicative of mitochondrial dysfunction (6). Mitochondrial dysfunction is known to trigger mitophagy (19, 21). To assess whether mitophagy was induced in response to lga2 overexpression, we used a reporter assay based on relocalization of mitochondrial matrix-targeted GFP (mtGFP) as previously reported for yeast (37). This revealed accumulation of diffuse mtGFP within vacuolar compartments (Fig. 1A). lga2-triggered mitophagy (here termed LTM) was normally attained in 30 to 50% of the cells in individual experiments. Counterstaining with FM4-64 verified colocalization of vacuolar mtGFP fluorescence and vacuolar compartments in the majority of cells analyzed (Fig. 1A; see also Fig. S1A in the supplemental material). This solidified our assay system based on detection of diffuse mtGFP fluorescence within rounded vacuolar compartments. The Atg8 protein is essential for autophagy (28), and a requirement of U. maydis Atg8 for autophagy in response to carbon deprivation has recently been demonstrated (27). To further validate LTM, MC2Δatg8 mutants (Table 1) were analyzed for vacuolar accumulation of mtGFP. This revealed complete absence of vacuolar mtGFP fluorescence under conditions of lga2 overexpression (Fig. 1A and B). Counterstaining with FM4-64 further showed that the lack of mitophagy was not due to impaired integrity of vacuoles in Δatg8 mutants under conditions of lga2 overexpression (see Fig. S1A in the supplemental material). To verify that the lack of mitophagy resulted from the Δatg8 mutation, we performed a complementation experiment. The results showed that LTM was restored after introduction of an ectopic atg8 gene, expressed under its own regulatory elements (Fig. 1A and 1B; see also Fig. S1A in the supplemental material). In addition, Northern analysis confirmed atg8 expression in analyzed wild-type strains, the absence of a signal in the Δatg8 mutant, as expected, and signals of different strengths in complemented strains (see Fig. S1B in the supplemental material). The stronger signal might have resulted from either multiple copy numbers or position effects of the ectopically integrated complementation construct (pMF1p-atg8).
Mitophagy represents a selective autophagic event (17, 19, 21, 43). Atg11 is required for all types of selective autophagy in yeast (19). The U. maydis genome predicts an atg11 homolog (MIPS number um03341; NCBI accession number XP_759488.1) whose deduced 1,828-amino-acid sequence displays an atg11 motif (positions 1052 to 1191) with 36 to 46% sequence identity to the homologous regions of Aspergillus nidulans and Laccaria bicolor, respectively, but only 20% identity to the corresponding region of S. cerevisiae Atg11 (see Fig. S2A in the supplemental material). Consistent with the absence of Atg11 homologs in mammals (19), BLASTP revealed no mammalian homologs of the predicted U. maydis Atg11 domain (data not shown). Next, we analyzed LTM in MC2Δatg11 mutant strains. Interestingly, vacuolar mtGFP fluorescence was almost abolished under conditions of lga2 overexpression, despite maintenance of extensive mitochondrial fragmentation (Fig. 1C and D). In addition, counterstaining with FM4-64 confirmed integrity of vacuolar compartments in Δatg11 mutants (see Fig. S1A in the supplemental material). This indicates that LTM represents a selective type of autophagy.
lga2-induced mitochondrial dysfunction and mtRNA depletion.
The strong induction of mitophagy in response to lga2 overexpression was consistent with lga2-triggered mitochondrial dysfunction. Previously, we showed that lga2 overexpression causes a reduction in the membrane potential, as determined from staining with the marker dye CM-H2Xros (here termed MTR), whose fluorescence decreases upon low membrane potential (6). To get clues on the conditions underlying LTM, we analyzed whether this loss preceded mitophagy and required mitochondrial fragmentation. We therefore examined MTR fluorescence in MC2 and MC2Δdnm1 strains. As previously shown, the MC2 strains we used displayed comparable levels of lga2 overexpression under inductive conditions in CMA (24) (see below). In the absence of lga2 expression (wild-type control) or up to 3 h after induced lga2 expression, MTR fluorescence predominantly appeared throughout mitochondrial structures. In contrast, MTR fluorescence significantly decreased after 4.5 h of lga2 induction (see Fig. S3 in the supplemental material). In particular, after 6 and 7.5 h, MTR staining appeared faded and was primarily confined to small foci in a high percentage of lga2-expressing cells. Interestingly, this reduction was also detected in MC2Δdnm1 cells despite the absence of mitochondrial fragmentation (Fig. 2A [arrowheads]; see also Fig. S3 in the supplemental material). In contrast, mitophagy was detected earliest between 6 and 8 h after lga2 induction in arabinose-containing medium (see Fig. S1C in the supplemental material). Taken together, the loss in membrane potential did not depend on mitochondrial fragmentation and preceded mitophagy induction.
To assess whether lga2-induced dysfunction affected mitochondrial transcription, we performed qRT-PCR analysis using four mitochondrial genes (nad3, nad6, cox1, and cox2) as markers. These genes are distributed over the 57-kb mitochondrial genome of U. maydis (NCBI accession number DQ157700) and are encoded on either strand. Samples were taken at 6 and 12 h after transfer to lga2-inducing conditions. At the 12-h time point, we detected a high degree of mitochondrial fragmentation as well as mitophagy under conditions of lga2 overexpression (strain MC2), while mitophagy was not detected at the 6-h time point (Fig. 2B, right panels, and data not shown). Strikingly, mtRNA transcript levels dropped 3.5- to 8.5-fold at the 6-h time point and 10- to 26-fold at the 12-h time point, in comparison with the wild-type control strain (FB2/pMB2-2) incubated under identical conditions (Fig. 2B, arrows). In contrast, under noninducing conditions in CMG, mtRNA transcript levels of the two strains were similar. The increased transcript levels of cox1 relative to the other tested genes likely reflect its larger size, since random hexamer primers were used for the reverse transcription (see Materials and Methods).
To gain further insight into underlying relationships, we monitored the time course of mtRNA transcript levels, along with the occurrence of mitochondrial fragmentation and mitophagy. In addition, based on recent findings suggesting that mitochondrial fission supports selective removal of damaged organelles (43), Δdnm1 mutants were included to assess mitophagy in dependence on mitochondrial fragmentation. As shown for nad3 and cox1, transcript levels gradually decreased from 3 to 4.5 h after lga2 induction in the wild type and Δdnm1 mutant strain analyzed (Fig. 2C, arrowheads). Overexpression of lga2 was explicitly verified during the time course (Fig. 2C). Mitochondrial fragmentation was already detected 3 h after lga2 induction, and as expected from a previous investigation (24), was absent from Δdnm1 mutants (Fig. 2D). In contrast, mitophagy was not detected at the 6-h time point and, interestingly, was markedly reduced in the Δdnm1 mutants at the 12-h time point (Fig. 2D). The complete time course analysis was independently performed with the additional mutant strain (MC2Δdnm1#1), and this confirmed strongly reduced mitophagy (2.5% versus 25.7% in the parental MC2 strain at the 12-h time point; n > 200 cells), despite similar reductions of nad3 and cox1 levels (data not shown). In summary, lga2-triggered mitochondrial fragmentation, loss of mtRNA transcript levels, and membrane potential changes precede mitophagy induction. Importantly, the effects on respiratory activity were maintained in Δdnm1 mutants despite impaired mitophagy induction (see below).
Is lga2-induced damage linked to caspase-dependent cell death?
Our previous analysis showed that lga2 overexpression leads to a severe growth defect (6). Because induction of mitophagy may be linked to cell death (5), we asked whether lga2 overexpression interfered with cell viability in a caspase-dependent manner. The U. maydis genome predicts only a single metacaspase gene (MIPS number um01408; NCBI accession number XP_757555.1), here termed mca1. The deduced 402-amino-acid sequence displayed 51% identity to its 432-amino-acid homolog in S. cerevisiae (NCBI accession number NP_014840), which is involved in cell death in response to external stimuli or aging (23). To assess an influence on cell viability, we measured the percentage of colony growth in response to lga2 overexpression in the Δmca1 mutant. However, as in the parental control strain (MC2), the plating efficiency of the Δmca1 mutant remained reduced by more than 90% (Fig. 2E). Furthermore, plating efficiencies were similarly affected in the Δatg8 mutant, excluding an underlying role of autophagy, and also in the Δdnm1 mutant, consistent with our previous investigation (24) and the above results. Together, these findings substantiated that Lga2 affected the respiratory activity, suggesting that this was the reason for the growth defect, whereas evidence for lga2-triggered programmed cell death was lacking.
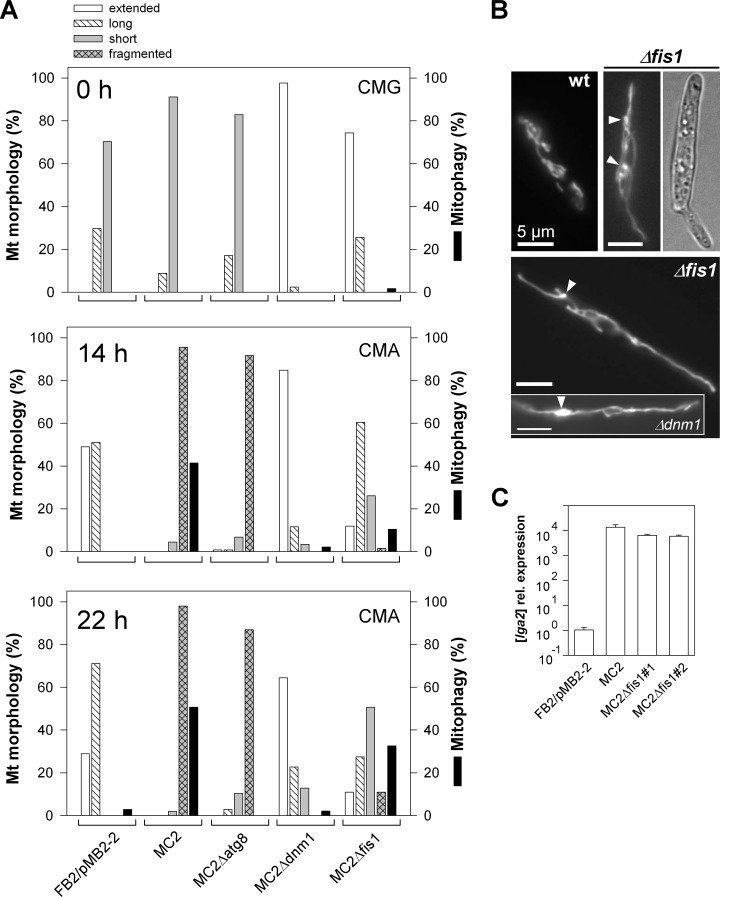
The role of mitochondrial fission in lga2-triggered mitophagy.
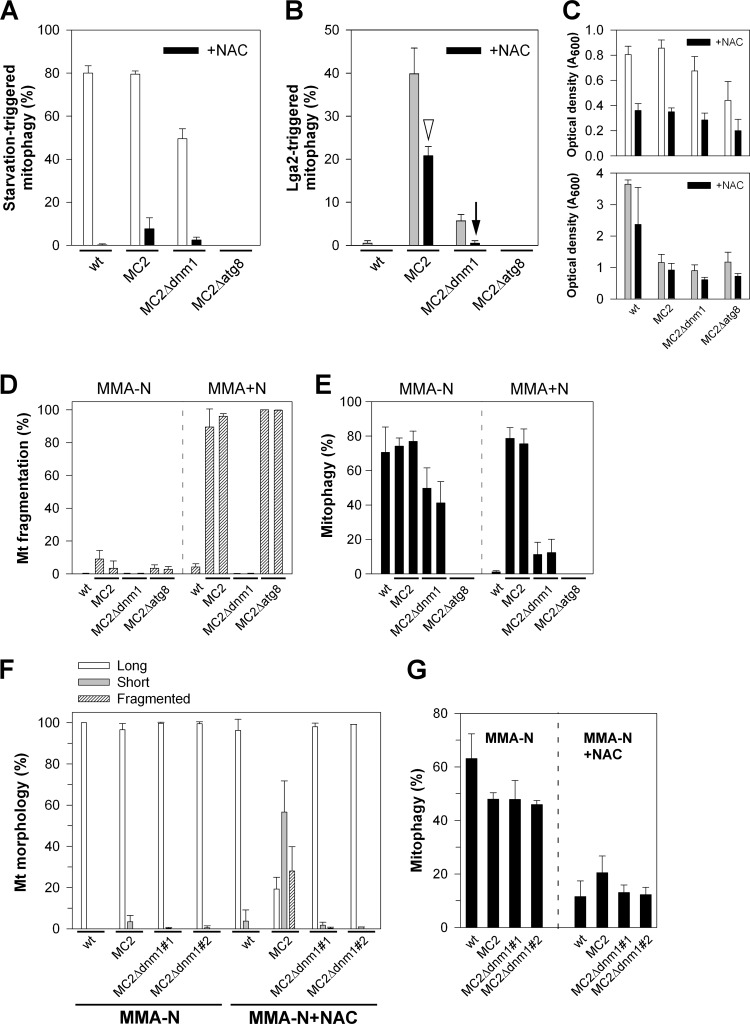
Prompted by the finding of a requirement of dnm1 for LTM (see above), we further examined the role of mitochondrial fission, using U. maydis Δdnm1 as well as Δfis1 mutant strains. First, these experiments corroborated the strongly reduced LTM in Δdnm1 mutants (≤3% mitophagy in two independent Δdnm1 mutants analyzed) (Fig. 3A). Counterstaining with FM4-64 further revealed that the integrity of vacuoles was not compromised in MC2Δdnm1 cells (see Fig. S4 in the supplemental material). The Fis1 protein is highly conserved in eukaryotic cells and may serve to anchor Dnm1 on the mitochondrial surface (26, 33, 34, 39). The U. maydis fis1 gene (MIPS number um03919; NCBI accession number XP_760066.1) predicts a protein of 152 amino acids, which exhibits 40% identity to the Fis1 sequence of S. cerevisiae and 70% identity to the predicted homolog of L. bicolor (NCBI accession number XP_001874659.1). The deduced U. maydis sequence further predicts a single transmembrane domain which, as in yeast, maps to the C-terminal region (see Fig. S2B in the supplemental material). As expected for the absence of a fission factor, mitochondria of Δfis1 mutants were predominantly extended or displayed interconnected networks (Fig. 3A, 0 h, and B). In addition, bulbous structures were detected, similar to those of Δdnm1 mutants (Fig. 3B, arrowheads). Under conditions of lga2 overexpression, as verified by qRT-PCR analysis (Fig. 3C), mitochondrial fragmentation was attenuated in MC2Δfis1 mutants, but it was less severely affected than in MC2Δdnm1 mutants (Fig. 3A). In addition, compared with the frequency of mitophagy in parental MC2 cells (41% at 14 h and 51% at 22 h), mitophagy was only partially reduced (10 to 11% at 14 h and 24 to 33% at 22 h) in two independent Δfis1 mutants analyzed (Fig. 3A). This excluded a critical role of Fis1 for the induction of LTM.
Fig 3.
Mitochondrial morphology states and mitophagy in various mutant backgrounds. (A) Cells were transferred from CMG (0 h) to CMA for growth periods of 14 and 22 h. (See Fig. 1D for classification of mitochondrial morphologies.) Extended, a net-like or single linear tubule throughout the cell. Percentages of cells displaying mitophagy are also shown. n > 100 cells/sample. One representative example of each of two independent mutants analyzed, which gave very similar results, is shown. (B) Log-phase cultures of strains FB2/pKS1 (wild type [wt]) and FB2Δfis1/pKS1 (Δfis1) in CMA were analyzed by fluorescence and DIC light microscopy. Note the formation of interconnected and bulbous structures (arrowheads) in the Δfis1 mutant, in contrast to interrupted tubules in the wild-type control. The inset shows the mitochondrial morphology of strain FB2Δdnm1/pKS1. Bars, 5 μm. (C) Verification of lga2 overexpression in Δfis1 mutants by qRT-PCR analysis. RNA was extracted from samples at 6 h after lga2 induction. Differences in transcript levels were calculated relative to those in strain FB2/pMB2-2 (set to 1.0). Data shown are the means ± standard deviations from four technical replicates.
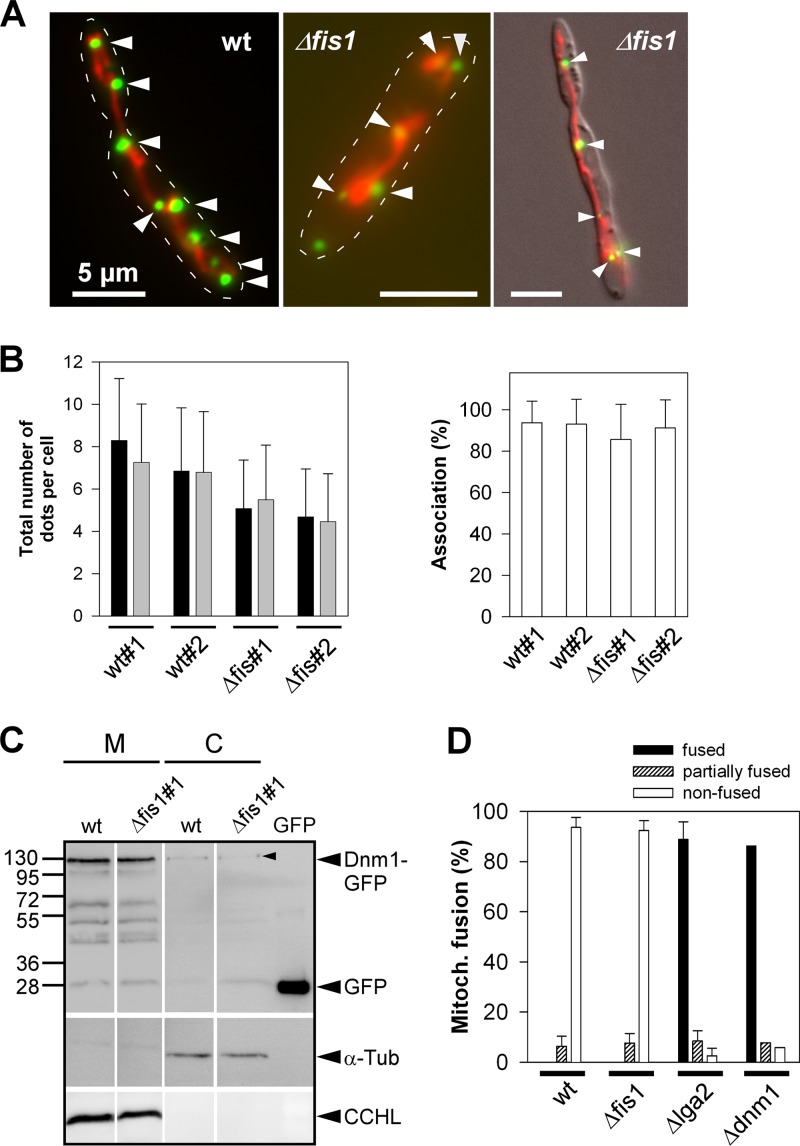
Fis1 is not required for recruitment of Dnm1 to mitochondria.
To provide further evidence that Dnm1 mediates lga2 effects independent of Fis1, we examined whether the Δfis1 mutation affected the intracellular distribution of Dnm1-GFP fusion proteins (expressed under the inducible crg1 promoter contained in plasmid pCudg1). These fusions have previously been shown to reside in multiple spots of various sizes along mitochondrial tubules (Fig. 4A) (24). However, cytoplasmic dispersal of focal GFP fluorescence was not detected in Δfis1 mutants (Fig. 4A). Furthermore, the total number of GFP spots was only weakly diminished, and the percentages of mitochondria-associated GFP spots were similar between mutant and wild-type strains (Fig. 4B), indicating that Dnm1-GFP can efficiently aggregate along mitochondria in the absence of Fis1. These results were corroborated by cell fractionation and immunoblot analysis, in which Dnm1-GFP signals of similar strength were preferentially detected in the mitochondrial fraction of both the parental wild-type strain and the Δfis1 mutant derivative (Fig. 4C). Additional bands detected in both mitochondrial fractions likely represented degradation products, as previously observed (24). Taken together, these findings suggested that mitochondrial recruitment of Dnm1-GFP largely occurred in a Fis1-independent manner in U. maydis.
Fig 4.
Analysis of Dnm1-GFP localization and mitochondrial fusion in dependence on fis1. (A) Fluorescence microscopy of log-phase cultures of pCudg1 strains (wild type [wt] and Δfis1; see Table 1) in CMA. Green fluorescence, Dnm1-GFP spots; red fluorescence, mtRFP. Dnm1-GFP spots associated along mitochondrial tubules (arrowheads). Stippled lines mark cell boundaries added using Adobe Photoshop. (Right panel) Overlay with the DIC image. Bars, 5 μm. (B) Log-phase cultures of pCudg1 strains in CMA (two independent wt and Δfis1 strains) analyzed by fluorescence microscopy to determine the total number of Dnm1-GFP spots per cell (left panel; black and gray bars refer to replicate cultures; data are means ± standard deviations; n ≥ 50 cells/culture) and the relative percentage of mitochondria-associated Dnm1-GFP spots (right panel; means ± standard deviations; n ≥ 30 cells/culture). (C) Intracellular distribution of Dnm1-GFP. Strains BUB7/pKS2/pCudg1#1 (wt) and the Δfis1 mutant derivative (#1) were cultivated in CMG, then shifted to CMA (starting OD600 value of 0.25) for a period of 3 h to provide for Dnm1-GFP expression. Proteins from mitochondrial (M) and cytosolic (C) fractions (15 to 20 μg loaded for each) were subjected to SDS-PAGE and immunostained using either anti-GFP, anti-α-tubulin (α-Tub), or anti-CCHL antibody. GFP (12 ng) was loaded for size comparison. All lanes for one immunodetection are from the same blot. Note the only faint signals of cytoplasmic Dnm1-GFP (arrowhead). Expected sizes for Dnm1-GFP and GFP were 119.1 and 26.9 kDa, respectively. (D) Determination of mitochondrial fusion rates. Mitochondrial fusion was assayed in matings of compatible cells as described previously (24). Fused, nonfused, and partially fused divisions correspond to colocalization of GFP and RFP fluorescence in parts of mitochondrial tubules. n ≥ 50 hyphae/strain combination and experiment. Counting results were represented as means ± standard deviations from three to four experiments (a1Δdnm1/a2Δdnm1 [Δdnm1] and a1/a2Δlga2 [Δlga2] served as controls [Table 1]). For the Δdnm1 control, only a single experiment was performed.
To substantiate a Fis1-independent role of Dnm1, we compared mitochondrial fusion rates in dikaryotic cells between wild-type and Δfis1 mutant strains. Previous investigations showed that mitochondrial fusion among mating partners is efficiently blocked in dikaryotic wild-type cells but strongly promoted in the absence of either lga2 or dnm1 (24). If Dnm1-opposed fusion requires Fis1, mitochondrial fusion rates should be increased in Δfis1 mutants, accordingly. However, mitochondrial fusion rates were not stimulated in the absence of fis1 (Fig. 4D), providing additional evidence for a Fis1-independent role of Dnm1 acting downstream of Lga2.
LTM is distinct from starvation-induced mitophagy.
Mitophagy in yeast is known to be induced under amino acid or nitrogen starvation conditions (17). For U. maydis, atg8-dependent autophagy has been reported for conditions of carbon deprivation (27). We detected mitophagy after transfer of log-phase cells to either water-diluted YEPSl or nitrogen-deprived minimal medium (MMG-N). Diluted YEPSl containing 0.1% (wt/vol) yeast extract provides for amino acid starvation conditions (11). As expected, starvation-induced mitophagy was abolished in Δatg8 mutants, and this was accompanied by reduced lengths or the appearance of clumpy structures of otherwise extended mitochondrial tubules (see Fig. S5A to C in the supplemental material). Next, we analyzed Δatg11 mutants for mitophagy under starvation conditions. S. cerevisiae atg11 is required for mitophagy under starvation-mimicking conditions but dispensable for bulk autophagy (20, 25). Consistently, vacuolar accumulation of mtGFP fluorescence was abolished in U. maydis Δatg11 strains under N starvation conditions. Furthermore, the appearance of long, mitochondrial tubules was significantly reduced, as observed with Δatg8 mutants (see Fig. S5D and E in the supplemental material), suggesting that failure of mitophagy impacts mitochondrial dynamics. The same experimental approach was applied to FB1 (a1b1) cells, and the results confirmed the strict atg11 requirement for mitophagy irrespective of the mating type (data not shown).
Based on the requirement of atg11 for both LTM and starvation-triggered mitophagy, we explored underlying differences in their regulation. First, we analyzed a requirement of dnm1 for starvation-triggered mitophagy. Intriguingly, mitophagy was only slightly affected in Δdnm1 mutants under N starvation conditions (Fig. 5A, white bars) compared with the pronounced requirement of dnm1 for LTM (Fig. 5B, gray bars). To provide additional evidence for different regulation, cells were treated with NAC, which is known to inhibit starvation-induced mitophagy by increasing the cellular reduced glutathione pool (reference 19 and references therein). Despite a negative influence on cell growth (Fig. 5C), the effect of NAC was clearly selective in that starvation-induced mitophagy was severely reduced (Fig. 5A, black bars), whereas LTM of strain MC2 was approximately halved (Fig. 5B, arrowhead). Interestingly, residual LTM in the MC2Δdnm1 strain was abolished in the presence of NAC, suggesting that dnm1-independent mitophagy is susceptible to NAC (Fig. 5B, arrow). Next, we analyzed whether lga2 effects are maintained under starvation conditions. For this purpose, cells were inoculated in arabinose-containing N starvation medium (MMA-N). Intriguingly, lga2-triggered mitochondrial fragmentation was efficiently prevented under these conditions, and this also included the Δatg8 mutant, excluding that autophagy itself was responsible for this effect. In contrast, under equivalent conditions in the presence of a nitrogen source (MMA+N), lga2-triggered dnm1-dependent mitochondrial fragmentation was seen as expected, and furthermore, these conditions provided for dnm1-dependent mitophagy (Fig. 5D and E). Together, this suggested that lga2 effects are blocked under N starvation conditions. To dissect whether this was due to the starvation medium or resulted from starvation-induced signaling, lga2 effects were analyzed in MMA-N in the presence of NAC. Despite low cell proliferation under the applied starvation conditions, lga2 expression under the crg1 promoter remained inducible, as verified by qRT-PCR analysis (see Fig. S6 in the supplemental material). Interestingly, in the presence of NAC, the percentage of long mitochondrial tubules was significantly reduced in strain MC2 in favor of short or fragmented mitochondria (Fig. 5F). Furthermore, the MC2 strain displayed the highest levels of mitophagy in the presence of NAC (Fig. 5G). The residual levels of mitophagy in the remaining strains may be explained by incomplete inhibition of mitophagy by NAC. Together, these results indicated that lga2 effects were at least in part prevented by starvation-mediated signaling in support of different regulation underlying starvation-induced mitophagy versus LTM.
Fig 5.
Comparison of mitophagy-inducing conditions. (A and B) Influence of NAC on starvation-induced mitophagy (A) and LTM (B). Cells were transferred from CMG to MMG-N with or without NAC (A) or to CMA with or without NAC (B) and assayed for mitophagy. Means ± standard deviations are for data from analysis in duplicate of two independent MC2Δdnm1 and MC2Δatg8 strains (or four replicates of the control strains FB2/pMB2-2 [wt] and MC2). n ≥ 100 cells/sample. (C) Optical densities of the analyzed strains. (D and E) Analysis of lga2 effects under starvation conditions. Strains FB2/pMB2-2 (wt) and two independent of each of MC2 (MC2#4 and #5 [6]), MC2Δdnm1 (derived from MC2#4 and #5), and MC2Δatg8 strains with comparable levels of lga2 overexpression were used. Cells were transferred from CMG to either MMA-N or MMA+N for analysis of mitochondrial fragmentation (D) and mitophagy (E). Means ± standard deviations from three replicates (n ≥ 100 cells/sample). (F and G) Partial restoration of lga2 effects under starvation conditions in the presence of NAC. Strains were transferred from CMG to MMA-N with or without NAC for analysis of mitochondrial morphologies (F) and mitophagy (G). Means ± standard deviations are from three (Δdnm1 strains) to four (wt and MC2) replicate cultures per strain analyzed (n ≥ 100 cells/sample).
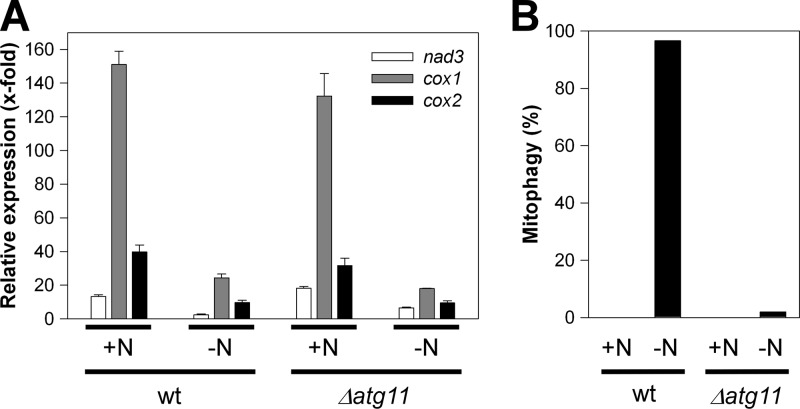
To further compare the conditions underlying the two mitophagy pathways studied, we analyzed nad3, cox1, and cox2 transcript levels 20 h after transfer to MMG-N. In addition, we included the Δatg11 mutant strain to judge an influence of mitophagy on mtRNA transcript levels under starvation conditions. This indicated a 4- to 6-fold reduction relative to MMG+N medium conditions for the wild-type strain, which likely reflects reduced metabolic activity under starvation conditions. The same reduction (3- to 7-fold) was seen for the Δatg11 mutant unable to undergo mitophagy, thus ruling out that mitophagy itself was responsible for reduced mtRNA transcript levels during starvation (Fig. 6). Hence, both lga2 induction and N starvation conditions imposed significantly reduced mtRNA levels, although this was more pronounced under conditions of lga2 overexpression (Fig. 2B).
Fig 6.
Analysis of mtRNA levels under N starvation conditions and dependence on atg11. (A) The U. maydis strains FB2/pMB2-2 (wild type [wt]) and FB2Δatg11/pMB2-2 were transferred from CMG to either MMG or MMG-N to determine transcript levels of mitochondrial nad3, cox1, and cox2 levels after a 20-h incubation period by qRT-PCR analysis. The starting OD600 values were 0.05 (MMG) and 0.15 (MMG-N). Results were normalized against the expression of the tbp gene (analyzed in each sample and set to 1.0). Shown are the means ± standard deviations from four technical replicates. (B) Samples were taken from the same cultures as described above for analysis of mitophagy (n > 50 cells/sample).
DISCUSSION
In this study, we have used inducible lga2 overexpression as a trigger for mitochondrial dysfunction, followed by mitophagy. Based on different requirements for dnm1 and susceptibility to NAC, we have provided evidence that lga2-triggered mitophagy is mechanistically distinct from starvation-induced mitophagy. Moreover, this study has revealed further insight into the requirement of mitochondrial fission for mitophagy. Lga2 is decisive for uniparental mtDNA inheritance in U. maydis (2, 8). Future studies need to address whether LTM plays an underlying role either directly or to dispose of affected organelles having lost their DNA.
Mitophagy-inducing conditions.
Selective autophagy has gained much interest in recent years based on its potential role in mitochondrial quality control and homeostasis from yeast to mammals (21, 22, 30, 40, 42, 43). In contrast to nonselective macroautophagy of cytosol in bulk, selective types of autophagy rely on specific recognition of the cargoes prior to sequestration by isolation membranes. In S. cerevisiae, specific recognition is mediated by the adapter protein Atg11, which subsequently interacts with Atg8 for delivery of the cargo into autophagosomes (19, 20, 43). Studies performed in yeast have shown that atg11 is required for mitophagy under N starvation as well as stationary-phase conditions, while it is not required for autophagy of bulk cytoplasmic cargo (17, 19, 20, 25). Protein sequence alignment revealed that homologs of yeast Atg11 are confined to fungal species, with the Atg11 motif being particularly well conserved among ascomycetes (E value, <107). Thus far, a requirement of Atg11 for selective autophagy is only known for S. cerevisiae. Despite little sequence similarity to the yeast counterpart (see Fig. S2A in the supplemental material), this study has clearly assigned U. maydis Atg11 to mitophagy. In particular, we have shown that atg11 is essential for mitophagy in response to starvation and lga2-imposed mitochondrial dysfunction. Rare mitophagic events detected in U. maydis atg11 mutants may therefore be due to bulk autophagy.
At present, little is known about the conditions that define damage-induced mitophagy. Neither mammalian nor yeast mutant cells lacking mtDNA are prone to mitophagy (11, 36). Hence, selective loss of mtDNA as it occurs during sexual development may not necessarily be a trigger. In yeast, interference with the mitochondrial ATP-generating system is involved in the induction of mitophagy (32). Furthermore, mitophagy has been detected in yeast in response to depletion of the mitochondrial K+/H+ exchanger Mdm38, which leads to a reduced membrane potential (29). Conversely, while mitophagy can be induced in mammalian cells upon depolarization of the mitochondrial membrane potential by an uncoupler, application of either CCCP or the F1F0-ATPase inhibitor oligomycin is not sufficient to do so in yeast, but instead may interfere with the autophagic flux (references 11 and 14 and references therein). Likewise, mitophagy was not induced in U. maydis cells treated with various concentrations of CCCP that were sufficient to severely impair cell proliferation (data not shown). As it appeared from the time course study, lga2-triggered induction of mitochondrial fragmentation, depletion of mtRNA transcripts, and loss of membrane potential are rapid events that clearly precede mitophagy induction. The finding that the latter two events are independent of mitochondrial fragmentation suggests that LTM is sponsored by at least two inputs, namely, reduction of respiratory activity and Dnm1-dependent fission.
The influence of mitochondrial fission on mitophagy.
A reduction in the mitochondrial membrane potential may well explain impaired fusion in response to conditional lga2 overexpression (reference 30 and references therein). Conceivably, mitochondrial fragmentation reflects the number of defective mitochondria destined for mitophagy. In support of this, our finding suggests that Dnm1-mediated fission favors mitophagy caused by mitochondrial dysfunction. We furthermore used U. maydis Δfis1 mutants to obtain additional clues on the underlying role of mitochondrial fission. This revealed that lga2-triggered mitochondrial fragmentation was only attenuated in Δfis1 mutants, but not prevented as in Δdnm1 mutants (24), providing an explanation why LTM was only partially affected in Δfis1 mutants.
Fis1 is a highly conserved mitochondrial fission factor, which in yeast is involved in the recruitment of Dnm1 to the mitochondrial surface (26, 33, 34, 39). In agreement, U. maydis Δfis1 mutants display net-like mitochondrial morphologies reminiscent of U. maydis Δdnm1 and S. cerevisiae fis1Δ mutants (13, 24). In favor of a Fis1-independent mode for Dnm1, mitochondrial association of Dnm1-GFP was maintained in U. maydis Δfis1 mutants. In S. cerevisiae, the Fis1 protein is not essential for, but contributes to, recruitment of Dnm1 to the mitochondrial membrane and to formation of Dnm1 assemblies. Explicitly, yeast fis1Δ mutants display at least 50% fewer assemblies that are associated with mitochondria. In addition, Fis1 contributes to the ordered aggregation and orientation of Dnm1 assemblies on mitochondria (26, 33, 34). Despite this, mitochondrial fission is not blocked in yeast fis1Δ mutants (13). To this end, we cannot exclude an influence of Fis1 on the proper distribution of Dnm1 assemblies in U. maydis. The finding that endogenous lga2 expression under mating conditions strongly interfered with fusion among parental mitochondria in dependence on dnm1 (24) but independent of fis1 (Fig. 4D), supports a Fis1-independent mode of Dnm1. In yeast, the Num1 protein has been shown to act as an additional Fis1-independent Dnm1 docking site on mitochondria (7, 42). Moreover, firm evidence for a Fis1-independent mode of human Drp1 has recently been provided. In particular, the small membrane-spanning protein Mff is an essential factor in mitochondrial recruitment of Drp1 independent of Fis1, suggesting that Mff functions as a Drp1 receptor and Fis1 functions downstream of Mff (31). In S. cerevisiae, mitochondrial fission additionally involves the two adapter proteins Mdv1 and Caf4, which both are yeast specific (42). Conversely, no Mff homologs exist in yeast (31) or other fungal genomes as judged from BLASTP analysis (data not shown). This suggests marked differences in the control of mitochondrial fission throughout taxa. In this regard, it will be exciting to identify yet-hypothetical Dnm1-associated proteins in U. maydis and assess their role in LTM.
Conflicting reports exist about the role of fission in the induction of mitophagy. Kanki et al. (18) reported that mitophagy is reduced by ∼20 to 40% in yeast dnm1Δ mutants under N starvation conditions in the presence of a fermentable carbon source. Furthermore, mitophagy in response to depletion of Mdm38 was inhibited in yeast dnm1Δ mutants (29). A role of the mitochondrial fission factors Drp1 and Fis1 in mitophagy has been reported for mammals (40). A link between mitochondrial fission and induction of mitophagy has further been suggested based on Parkin-induced degradation of mitofusins under mitochondria-damaging conditions in mammalian cells. In support of this, inhibition of Drp1-mediated mitochondrial fission prevented Parkin-induced mitophagy (38). In addition, mitochondrial fusion was found to protect mammalian cells from mitophagy under starvation conditions (10). Conversely, analysis of mitophagy in response to ectopic expression of wild-type and mutant forms of Fis1 in mammalian cells has indicated that mitochondrial dysfunction rather than induced fission promotes mitophagy (9). Moreover, a recent investigation studying rapamycin-induced mitophagy, including S. cerevisiae dnm1Δ and fis1Δ mutants, clearly ruled out a role of mitochondrial fission in mitophagy of yeast cells (25). Our study could bring some light to this debate, since based on the same cellular system, we have shown that marked dnm1 dependency only concerns damage-induced, but not starvation-induced, mitophagy and, consistent with the study of Gomes and Scorrano (9), did not depend on Fis1.
In support of different mitophagy pathways, we found that NAC, known to impact the cellular redox balance (reference 19 and references therein), only targets starvation-induced mitophagy efficiently. The finding that residual LTM in Δdnm1 mutants is abolished in the presence of NAC (Fig. 5B) suggests that the cellular redox balance directs dnm1-independent mitophagy. Moreover, lga2-induced mitochondrial fragmentation was strongly reduced under N starvation conditions, but it was partially restored in the presence of NAC, implying that starvation-induced signaling opposes lga2 effects. In mammalian cells, mitochondria become elongated under nutrient starvation conditions due to protein kinase A-dependent phosphorylation of Drp1, leading to reduced fission activity (references 10 and 30 and references therein). Inactivation of Dnm1 activity under starvation conditions would readily explain the lack of mitochondrial fragmentation in response to lga2 overexpression. In conclusion, our results provide a framework for mechanistic studies to gain further insight into regulatory circuits underlying mitophagy induction under different stress conditions.
Supplementary Material
ACKNOWLEDGMENTS
We thank our students M. Rath for supporting cloning procedures and P. Schneider for supporting mitophagy studies.
This work was performed in the Department of Organismic Interactions of R. Kahmann (Max-Planck-Institute for Terrestrial Microbiology Marburg, Germany) and the Department of Genetics of J. Kämper (Karlsruhe Institute of Technology, Germany).
We are grateful to the Microbiology Department of R. Fischer (Karlsruhe Institute of Technology, Germany) for providing fluorescence microscopy equipment and to J. Kämper for general support.
In addition, we thank two anonymous reviewers for helpful suggestions on the manuscript.
Footnotes
Published ahead of print 27 July 2012
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1. Banuett F, Herskowitz I. 1989. Different a alleles of Ustilago maydis are necessary for maintenance of filamentous growth but not for meiosis. Proc. Natl. Acad. Sci. U. S. A. 86:5878–5882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Basse CW. 2010. Mitochondrial inheritance in fungi. Curr. Opin. Microbiol. 13:712–719 [DOI] [PubMed] [Google Scholar]
- 3. Brachmann A, Weinzierl G, Kämper J, Kahmann R. 2001. Identification of genes in the bW/bE regulatory cascade in Ustilago maydis. Mol. Microbiol. 42:1047–1063 [DOI] [PubMed] [Google Scholar]
- 4. Brachmann A, König J, Julius C, Feldbrügge M. 2004. A reverse genetic approach for generating gene replacement mutants in Ustilago maydis. Mol. Genet. Genomics 272:216–226 [DOI] [PubMed] [Google Scholar]
- 5. Brenner D, Mak TW. 2009. Mitochondrial cell death effectors. Curr. Opin. Cell Biol. 21:871–877 [DOI] [PubMed] [Google Scholar]
- 6. Bortfeld M, Auffarth K, Kahmann R, Basse CW. 2004. The Ustilago maydis a2 mating-type locus genes lga2 and rga2 compromise pathogenicity in the absence of the mitochondrial p32 family protein Mrb1. Plant Cell 16:2233–2248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cerveny KL, Studer SL, Jensen RE, Sesaki H. 2007. Yeast mitochondrial division and distribution require the cortical Num1 protein. Dev. Cell 12:363–375 [DOI] [PubMed] [Google Scholar]
- 8. Fedler M, Luh KS, Stelter K, Nieto-Jacobo F, Basse CW. 2009. The a2 mating-type locus genes lga2 and rga2 direct uniparental mitochondrial DNA (mtDNA) inheritance and constrain mtDNA recombination during sexual development of Ustilago maydis. Genetics 181:847–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gomes LC, Scorrano L. 2008. High levels of Fis1, a pro-fission mitochondrial protein, trigger autophagy. Biochim. Biophys. Acta 1777:860–866 [DOI] [PubMed] [Google Scholar]
- 10. Gomes LC, Di Benedetto G, Scorrano L. 2011. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat. Cell Biol. 13:589–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Graef M, Nunnari J. 2011. Mitochondria regulate autophagy by conserved signalling pathways. EMBO J. 30:2101–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Holliday R. 1974. Ustilago maydis, p 575–595 In King RC. (ed), Handbook of genetics, vol 1 Plenum Press, New York, NY [Google Scholar]
- 13. Jakobs S, et al. 2003. Spatial and temporal dynamics of budding yeast mitochondria lacking the division component Fis1p. J. Cell Sci. 116:2005–2014 [DOI] [PubMed] [Google Scholar]
- 14. Johansen T, Lamark T. 2011. Selective autophagy mediated by autophagic adapter proteins. Autophagy 7:279–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kämper J. 2004. A PCR-based system for highly efficient generation of gene replacement mutants in Ustilago maydis. Mol. Genet. Genomics 271:103–110 [DOI] [PubMed] [Google Scholar]
- 16. Kämper J, et al. 2006. Insights form the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature 444:97–101 [DOI] [PubMed] [Google Scholar]
- 17. Kanki T, Klionsky DJ. 2008. Mitophagy in yeast occurs through a selective mechanism. J. Biol. Chem. 283:32386–32393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kanki T, et al. 2009. A genomic screen for yeast mutants defective in selective mitochondria autophagy. Mol. Biol. Cell 20:4730–4738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kanki T, Klionsky DJ. 2010. The molecular mechanism of mitochondria autophagy in yeast. Mol. Microbiol. 75:795–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim J, et al. 2001. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J. Cell Biol. 153:381–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim I, Rodriguez-Enriquez S, Lemasters JJ. 2007. Selective degradation of mitochondria by mitophagy. Arch. Biochem. Biophys. 462:245–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kurihara Y, et al. 2012. Mitophagy plays an essential role in reducing mitochondrial production of reactive oxygen species and mutation of mitochondrial DNA by maintaining mitochondrial quantity and quality in yeast. J. Biol. Chem. 287:3265–3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Madeo F, et al. 2002. A caspase-related protease regulates apoptosis in yeast. Mol. Cell 9:911–917 [DOI] [PubMed] [Google Scholar]
- 24. Mahlert M, Vogler C, Stelter K, Hause G, Basse CW. 2009. The a2 mating-type-locus gene lga2 of Ustilago maydis interferes with mitochondrial dynamics and fusion, partially in dependence on a Dnm1-like fission component. J. Cell Sci. 122:2402–2412 [DOI] [PubMed] [Google Scholar]
- 25. Mendl N, et al. 2011. Mitophagy in yeast is independent of mitochondrial fission and requires the stress response gene WHI2. J. Cell Sci. 124:1339–1350 [DOI] [PubMed] [Google Scholar]
- 26. Mozdy AD, McCaffery JM, Shaw JM. 2000. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J. Cell Biol. 151:367–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nadal M, Gold SE. 2010. The autophagy genes atg8 and atg1 affect morphogenesis and pathogenicity in Ustilago maydis. Mol. Plant Pathol. 11:463–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. 2009. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 10:458–467 [DOI] [PubMed] [Google Scholar]
- 29. Nowikovsky K, Reipert S, Devenish RJ, Schweyen RJ. 2007. Mdm38 protein depletion causes loss of mitochondrial K+/H+ exchange activity, osmotic swelling and mitophagy. Cell Death Differ. 14:1647–1656 [DOI] [PubMed] [Google Scholar]
- 30. Okamoto K, Kondo-Okamoto N. 2012. Mitochondria and autophagy: critical interplay between the two homeostats. Biochim. Biophys. Acta 1820:595–600 [DOI] [PubMed] [Google Scholar]
- 31. Otera H, et al. 2010. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J. Cell Biol. 191:1141–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Priault M, et al. 2005. Impairing the bioenergetic status and the biogenesis of mitochondria triggers mitophagy in yeast. Cell Death Differ. 12:1613–1621 [DOI] [PubMed] [Google Scholar]
- 33. Schauss AC, Bewersdorf J, Jakobs S. 2006. Fis1p and Caf4p, but not Mdv1p, determine the polar localization of Dnm1p clusters on the mitochondrial surface. J. Cell Sci. 119:3098–3106 [DOI] [PubMed] [Google Scholar]
- 34. Shaw JM, Nunnari J. 2002. Mitochondrial dynamics and division in budding yeast. Trends Cell Biol. 12:178–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Steinberg G, Perez-Martin J. 2008. Ustilago maydis, a new fungal model system for cell biology. Trends Cell Biol. 18:61–67 [DOI] [PubMed] [Google Scholar]
- 36. Suen DF, Narendra DP, Tanaka A, Manfredi G, Youle RJ. 2010. Parkin overexpression selects against a deleterious mtDNA mutation in heteroplasmic cybrid cells. Proc. Natl. Acad. Sci. U. S. A. 107:11835–11840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tal R, Winter G, Ecker N, Klionsky DJ, Abeliovich H. 2007. Aup1p, a yeast mitochondrial protein phosphatase homolog, is required for efficient stationary phase mitophagy and cell survival. J. Biol. Chem. 282:5617–5624 [DOI] [PubMed] [Google Scholar]
- 38. Tanaka A, et al. 2010. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J. Cell Biol. 191:1367–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tieu Q, Nunnari J. 2000. Mdv1p is a WD repeat protein that interacts with the dynamin-related GTPase, Dnm1p, to trigger mitochondrial division. J. Cell Biol. 151:353–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Twig G, et al. 2008. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 27:433–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Urban M, Kahmann R, Bölker M. 1996. Identification of the pheromone response element in Ustilago maydis. Mol. Gen. Genet. 251:31–37 [DOI] [PubMed] [Google Scholar]
- 42. Westermann B. 2010. Mitochondrial fusion and fission in cell life and death. Nat. Rev. Mol. Cell Biol. 11:872–884 [DOI] [PubMed] [Google Scholar]
- 43. Youle RJ, Narendra DP. 2011. Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 12:9–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zheng Y, et al. 2008. The Ustilago maydis Cys2His2-type zinc finger transcription factor Mzr1 regulates fungal gene expression during the biotrophic growth stage. Mol. Microbiol. 68:1450–1470 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.