Abstract
We report the characterization of a bacterial-type oxygen reductase abundant in the cytoplasm of the anaerobic protozoan parasite Entamoeba histolytica. Upon host infection, E. histolytica is confronted with various oxygen tensions in the host intestine, as well as increased reactive oxygen and nitrogen species at the site of local tissue inflammation. Resistance to oxygen-derived stress thus plays an important role in the pathogenic potential of E. histolytica. The genome of E. histolytica has four genes that encode flavodiiron proteins, which are bacterial-type oxygen or nitric oxide reductases and were likely acquired by lateral gene transfer from prokaryotes. The EhFdp1 gene has higher expression in virulent than in nonvirulent Entamoeba strains and species, hinting that the response to oxidative stress may be one correlate of virulence potential. We demonstrate that EhFdp1 is abundantly expressed in the cytoplasm of E. histolytica and that the protein levels are markedly increased (up to ∼5-fold) upon oxygen exposure. Additionally, we produced fully functional recombinant EhFdp1 and demonstrated that this enzyme is a specific and robust oxygen reductase but has poor nitric oxide reductase activity. This observation represents a new mechanism of oxygen resistance in the anaerobic protozoan pathogen E. histolytica.
INTRODUCTION
The pathogenic protozoan Entamoeba histolytica is the causative agent of amoebiasis and is a leading cause of death by a parasitic disease (29, 34). Upon host infection, E. histolytica is challenged with various oxygen tensions and reactive oxygen species (ROS) in the colon, bloodstream, and liver (34). The susceptibility (14, 15, 26) and transcriptional response (1, 42) of E. histolytica to oxygen and ROS have long been addressed. It has been observed that Entamoeba species and E. histolytica strains with higher virulence are more resistant to oxygen (28) and display higher expression of genes and proteins related to the oxidative stress response (1, 9, 24, 42). Resistance to oxygen-derived stress is thus likely an important component of the E. histolytica virulence framework (28). Oxygen reduction activity has been attributed to different E. histolytica flavoproteins (6, 7, 17), although most produce hydrogen peroxide and thus require further detoxification enzymes. The possible exception is one NADH oxidase, which has been proposed to reduce oxygen to water (6).
Flavodiiron proteins (FDPs) constitute a widespread family of detoxifying enzymes that act as oxygen and/or nitric oxide (NO) reductases (19, 30, 40). However, the substrate preference of FDPs is not understood; some FDPs have been shown to be quite selective toward oxygen, whereas others are more selective toward NO and yet others reduce both substrates with equivalent efficiency (reviewed in reference 38). FDP-encoding genes are found in the genomes of prokaryotes (mostly anaerobes). The few anaerobic protozoan pathogens that contain genes that code for FDPs are Giardia intestinalis (a single gene) and Trichomonas vaginalis (four homologues). These genes were most probably acquired from prokaryotes by lateral gene transfer (3, 4). So far, the protozoan enzymes have been shown to act as oxygen reductases (10, 25, 32).
In the E. histolytica genome (23), four genes that encode FDPs have been identified (3). Two of the E. histolytica FDP-encoding genes have high transcript levels under basal conditions (EhFdp1 or 6.m00467, corresponding to the identical genes EHI_096710 and EHI_152650, and EhFdp2 or 155.m00084, corresponding to EHI_159860 [see Table S1 in the supplemental material]). These genes show no apparent modulation of transcript levels upon exposure to oxidative and nitrosative stresses (42), heat shock (44), the histone deacetylase inhibitor trichostatin A (11, 16), or the DNA methyltransferase inhibitor 5-azacytidine (2) or in a mouse model of intestinal colonization and invasion (13). The gene that encodes E. histolytica FDP1 (EhFdp1; 6.m00467) displays higher transcript levels in E. histolytica than in the nonvirulent species E. dispar (24).
Here we show that bacterial-type EhFdp1 is remarkably abundant in the cytoplasm of E. histolytica and that protein levels increase upon oxygen exposure. Biochemical analyses reveal that recombinant EhFdp1 is a robust oxygen reductase with poor NO reductase activity. These results reveal a previously unknown strategy that the anaerobic pathogen E. histolytica may employ to respond to the variable oxygen tensions encountered in the host during tissue invasion.
Materials and Methods
In silico analyses of E. histolytica FDPs.
Sequences of FDPs were retrieved from NCBI Genomic BLAST by using the sequence of Moorella thermoacetica FDP (accession number Q9FDN7) as the search template. The sequences were aligned with Clustal X for Windows (36). Statistics reports were generated with Genedoc.
Cloning, expression, and purification of EhFdp1.
To determine the cellular localization of EhFdp1 in amoebae, we produced the recombinant protein for antibody production. The gene that encodes full-length EhFdp1 was amplified from E. histolytica genomic DNA (with primers 5′-GCTAGCAAAGCATTGGAAGTAGTAAAAGAC and 5′-GGATCCTTAAGCTTTAAGGGCCTCAGCAAA [where the NheI and BamHI recognition sites are underlined, respectively]), cloned into the Topo TA pCR2.1 vector (Invitrogen), and subcloned into the NheI and BamHI sites of bacterial expression vector pET28b+. The resulting vector (named pET-EhFdp1) was confirmed by sequencing. The same strategy was employed to clone the control gene for E. histolytica rubrerythrin (EhRbr) with primers 5′-GCTAGCGCAACTCTCATTAATCTTTGTAAGG and 5′-GGATCCTTAAATAGAAACTTGAACTCTGAAG.
EhFdp1 expression was accomplished in Escherichia coli BL21(DE3)Gold cells, and protein purification was done as previously described for bacterial FDPs (40). Briefly, pET-EhFdp1 was transformed into E. coli BL21(DE3)Gold cells and plated onto LB plates containing kanamycin. Bacteria were grown at 30°C in minimal medium (M9) supplemented with 100 μM FeSO4 · 7H2O, induced at an optical density at 600 nm of 0.5 with 100 μM isopropyl-β-d-thiogalactopyranoside (IPTG), and harvested 7 h after induction (cells overexpressing the control protein EhRbr were grown under identical conditions). Cells were disrupted at 900 lb/in2 in a French press; the soluble fraction was separated from the membranes by ultracentrifugation (2 h at ∼100,000 × g, 4°C) and dialyzed overnight against 10 mM Tris-HCl–9% glycerol (pH 7.5). Pure EhFdp1 was obtained in two anion-exchange chromatographic steps. Protein purity was assessed by SDS-PAGE.
Immunofluorescence microscopy.
Antibodies were obtained by injecting 1 mg of pure EhFdp1 into two rabbits over a period of 11 weeks (Open Biosystems). Immunofluorescence assays (IFAs) were carried out as previously described (45). Briefly, mid-log-phase E. histolytica HM-1:IMSS trophozoites were chilled (10 min, 4°C) and transferred to chamber slides to adhere (1 h, 37°C), rinsed with phosphate-buffered saline (PBS), fixed and permeabilized in a mixture of acetone-methanol (50:50), blocked (3% bovine serum albumin in PBS, 30 min), and then incubated for 1 h with anti-EhFdp1 antibody (1:500; Open Biosystems) or preimmune serum as a control. Parasites were rinsed with PBS-Tween and incubated with Alexa 488-conjugated anti-rabbit antibody (1:1,000; Molecular Probes). To probe for membrane localization, trophozoites were coincubated with antibody for the membrane protein Gal/GalNAc lectin (anti-HgL antibody at 1:50; kind gift of W. A. Petri, Jr.) and Alexa 594-conjugated anti-mouse antibody (1:500; Molecular Probes) as described in reference 5. To assess the effect of oxygen exposure on EhFdp1 localization, mid-log-phase E. histolytica HM-1:IMSS trophozoites were seeded into 2-ml chamber slides and left to adhere to the slides for 1 h at 37°C, after which 1.5 ml of medium was removed, leaving a large headspace (1.5 ml); the chamber was kept unsealed to allow gas exchange. After 1 h of oxygen exposure, amoebae were fixed, permeabilized, and incubated with anti-EhFdp1 and anti-HgL antibodies as described above. Images were collected with a Zeiss LSM700 confocal microscope and filtered using Volocity software (Improvision).
Western blot analysis.
We sought to confirm the effect of oxygen exposure on EhFdp1 expression in E. histolytica HM-1:IMSS by Western blot analysis. Mid-log-phase parasites were seeded into 8-ml culture tubes and incubated overnight. The next morning, medium of all of the cultures was replaced. Six milliliters of medium was removed from the oxygen-challenged samples, and the tubes were placed horizontally. At different time points, parasites were harvested by centrifugation for 5 min at 1,000 × g and washed once with PBS. Trophozoites were lysed in lysis buffer (50 mM HEPES-KOH, 50 mM KCl, 5 mM MgCl2, 0.5% NP-40) containing protease inhibitors (20 mM dithiothreitol, 18.75 μM E-64, 0.8 μg/ml leupeptin, 4× Halt* phosphatase inhibitor cocktail) (Thermo Scientific), and lysates were subjected to 12.5% SDS-PAGE and blotted onto polyvinylidene difluoride membranes. Anti-EhFdp1 antibody (1:500) and HRP-conjugated anti-rabbit antibody (1:1,000) (Cell Signaling) were used (1 h of incubation each), and the signal was detected with ECL+ (GE Healthcare). Primary (anti-actin, 1:1,000; MP Biomedicals) and secondary (HRP-conjugated anti-mouse antibody at 1:1,000; Cell Signaling) antibodies were used for the actin control.
Biochemical characterization of recombinant EhFdp1.
The quaternary structure of recombinant EhFdp1 was assessed by analytical size exclusion chromatography (120-ml Superdex S-200 column connected to an Åkta Prime fast protein liquid chromatography system [GE Healthcare]). The column was previously equilibrated and run at 0.5 ml/min with 20 mM Tris-HCl buffer–18% glycerol–150 mM NaCl (pH 7.5). Dextran blue (molecular mass, 1,000 kDa) was used as an internal standard. The column was calibrated with the molecular size markers ferritin (440 kDa), aldolase (158 kDa), albumin (67 kDa), ovalbumin (43 kDa), and RNase (13.7 kDa). The iron content of recombinant EhFdp1 was determined by the 2,4,6-tripyridyl-1,3,5-triazine (TPTZ) method (12), whereas flavin content measurement and identification were done by an adaptation of the method described in reference 35. Briefly, after flavin extraction with trichloroacetic acid and centrifugation to remove the precipitated protein, the flavin solution was neutralized with ammonium acetate and its visible spectrum was recorded, allowing flavin identification and quantitation. Whereas flavin mononucleotide (FMN) has its absorption maximum at 445 nm (ε = 11,300 M−1 cm−1), flavin adenine dinucleotide absorbs maximally at 450 nm (ε = 12,500 M−1 cm−1).
Spectroscopic analysis of EhFdp1.
The UV-visible spectra of purified EhFdp1 and the NADH:oxygen oxidoreductase assay results reported below were recorded with a Shimadzu UV-1603 spectrophotometer. Electron paramagnetic resonance (EPR) spectra were collected with a Bruker EMX spectrometer equipped with an Oxford Instruments ESR-900 continuous-flow helium cryostat. Since nonheme diiron sites have EPR signals only in a mixed-valence state, we reduced the protein under anaerobic conditions with sodium dithionite in the presence of redox mediators covering a wide range of redox potentials and recorded EPR spectra to detect the signal corresponding to a mixed-valence (FeIII-FeII) diiron site with g values below 2.0 (37).
Oxygen and NO consumption assays.
Amperometric measurements were carried out with oxygen- and NO-specific Clark-type electrodes. Oxygen consumption was measured with a Clark-type O2 electrode plugged into a biological oxygen monitor (model 5300; Yellow Springs). NO consumption was measured under anaerobic conditions with a NO-selective Clark-type electrode (ISO-NO Mark II; World Precision Instruments). To maintain anaerobic conditions, the reaction chamber was flushed with argon and filled with degassed buffer and contaminant O2 was further scavenged by glucose oxidase (17 U/ml), catalase (130 U/ml), and glucose (2 mM). The NO electrode was calibrated by recording the signal produced by sequential additions of aliquots of a 2 mM NO stock solution prepared by flushing pure NO gas through a previously degassed ultrapure water solution (with vacuum-argon cycles). The NO gas was purified by flowing through a 1 M NaOH trap to remove higher N-oxides and a water trap to remove aerosols prior to flushing of the degassed ultrapure water. The O2 and NO consumption rates of E. coli lysates overexpressing EhFdp1 and the control protein EhRbr were measured with 1 mM NADH as a source of electron equivalents. Cell lysates were obtained by three freeze-thaw cycles, and the soluble fraction was separated by centrifugation (15 min, 12,000 × g, 5°C). The oxygen and NO reductase activities of pure recombinant EhFdp1 were determined by assembling an in vitro electron transfer (ET) chain comprising E. coli NADH:flavorubredoxin oxidoreductase and the truncated rubredoxin domain of flavorubredoxin with NADH as the primary electron source (10, 41). This ET chain was analyzed for the ability to transfer electrons to EhFdp1 by visible spectrophotometric assays following (i) the reduction of EhFdp1 under anaerobic conditions and (ii) the oxidation of NADH to NAD+ monitored at 340 nm at the expense of oxygen reduction by EhFdp1 mediated by the ET chain.
RESULTS AND DISCUSSION
FDPs in E. histolytica.
In the E. histolytica genome (23), four genes that encode FDPs have been identified (3). These have 53 to 66% amino acid sequence identity and 72 to 81% similarity among themselves (see Fig. S1 in the supplemental material). Amoebic FDPs have relatively poor sequence similarity to the single FDP-encoding gene from G. intestinalis (23 to 25% identity, 42 to 44% similarity) and to the four homologues from T. vaginalis (22 to 39% identity, 42 to 58% similarity). This suggests that although protozoan FDPs were all proposed to have been acquired by lateral gene transfer from prokaryotes (3, 4), they are likely to have originated from phylogenetically distant prokaryotic sources. Amoebic FDPs display similar levels of sequence identity and similarity to FDPs from selected prokaryotes that have been previously characterized (23 to 37% identity, 42 to 57% similarity). This sequence alignment clearly shows that the amoebic FDP sequences have the two structural domains that constitute the structural prototype of this protein family: a C-terminal flavodoxin-like domain containing an FMN and an N-terminal β-lactamase-like domain harboring a diiron site (39). A structural feature common to the FDPs studied to date is that the minimal functional quaternary arrangement consists of a head-to-tail homodimer that places the flavin from one monomer in close contact with the diiron active site from the opposing monomer (39). Moreover, the amoebic sequences also contain the strictly conserved ligands for the diiron active site, as well as the residues that compose the flavin binding site (38).
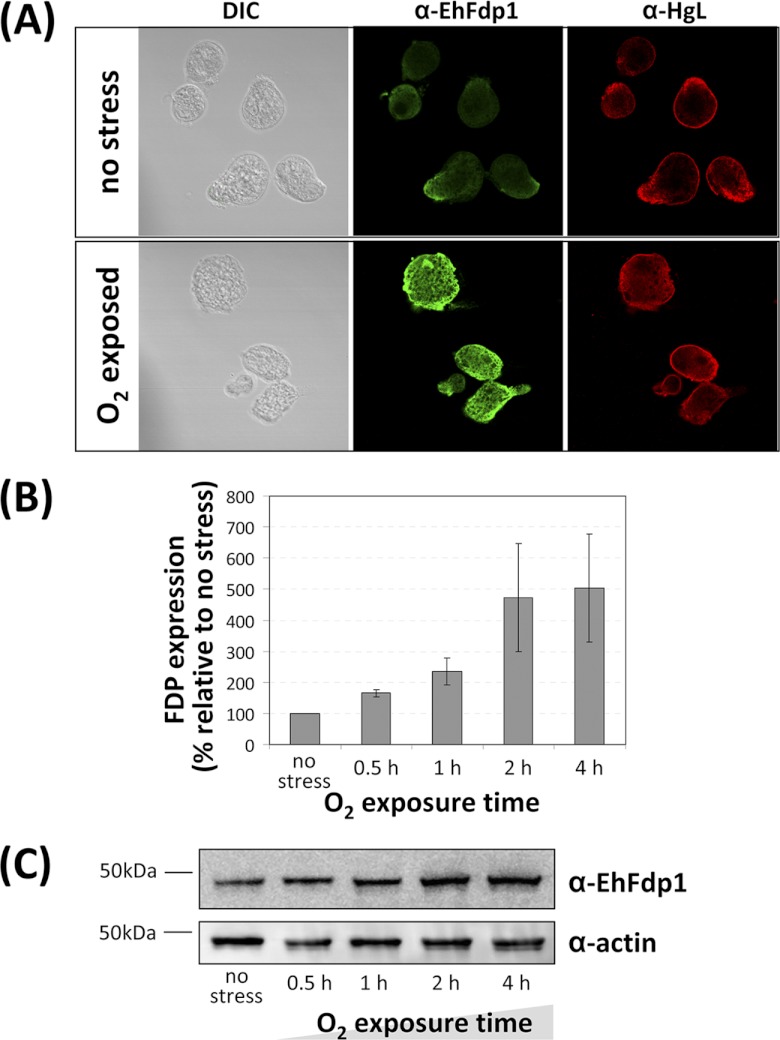
EhFdp1 is abundant in the cytoplasm of anaerobically grown amoebae and increases in abundance upon oxygen exposure.
Upon host invasion, amoebae are faced with oxygen that rapidly diffuses across their membranes, implying that its potentially toxic effects should be exerted in all cellular compartments. To assess the cellular localization of E. histolytica FDP EhFdp1, we expressed and purified recombinant protein and raised antibodies. Preimmune serum gave no detectable signal in IFAs (data not shown), and we observed that EhFdp1 localizes to the cytoplasm of anaerobically grown amoebae (Fig. 1A), with no overlap with the signal from the membrane protein Gal/GalNAc lectin heavy chain (anti-HgL antibody used as described in reference 5). These results are in concordance with data from FDPs in other systems, which are almost exclusively cytoplasmic. In the cyanobacterium Synechocystis sp. strain PCC 6803, the FDP is proposed to be membrane bound (40, 46) and one of the four T. vaginalis FDPs is localized in the hydrogenosome (32).
Fig 1.
EhFdp1 localizes to the cytoplasm and protein abundance increases upon oxygen exposure. (A) Immunofluorescence microscopy analysis of EhFdp1. Phase-contrast (differential interference contrast [DIC]) transmission images are shown. Antibodies: anti-EhFdp1 (1:500; Open Biosystems), anti-rabbit–Alexa 488 (1:1,000; Molecular Probes), anti-HgL (1:50), and anti-mouse–Alexa 594 (1:500; Molecular Probes). Images were collected with a Zeiss LSM700 confocal microscope and filtered using Volocity software (Improvision). (B) Western blot analysis of the effect of oxygen exposure on EhFdp1 expression in E. histolytica HM-1:IMSS. Anti-EhFdp1 antibody (1:500) and HRP-conjugated anti-rabbit antibody (1:1,000; Cell Signaling) were used (1 h of incubation each), and the signal was detected with ECL+ (GE Healthcare). Primary (anti-actin, 1:1,000; MP Biomedicals) and secondary (HRP-conjugated anti-mouse antibody at 1:1,000; Cell Signaling) antibodies were used for the actin control. Densitometric analysis with ImageJ reveals an increase in EhFdp1 expression with increasing oxygen exposure time from ∼2-fold after 1 h (n = 5; P = 7.0 × 10−3) to ∼5-fold after 4 h (n = 5; P =: 0.029). (C) Representative Western blot assay displaying EhFdp1 levels increasing with oxygen exposure time.
Taking into account the possible involvement of EhFdp1 in oxygen detoxification, we tested the effect of oxygen exposure on protein localization (Fig. 1A). Oxygen exposure was achieved by seeding amoebae into IFA chamber slides and leaving a large headspace (1.5 ml gas from a 2-ml chamber) and leaving the chamber unsealed to allow gas exchange. After 1 h of oxygen exposure, the EhFdp1 localization in oxygen-exposed trophozoites was the same as that in those that did not receive oxygen stress. However, we noted a significant increase in signal intensity in amoebae exposed to oxygen with respect to that in unchallenged ones.
This increased EhFdp1 signal intensity pointed to a modulation of EhFdp1 protein levels upon oxygen exposure, which we further quantitated by Western blot analysis. To this end, amoebae were grown in 8-ml tubes from which 6 ml of medium was removed and which were placed horizontally. EhFdp1 protein levels at different oxygen exposure times were determined by Western blot analysis with antibody to EhFdp1. The protein band detected in the Western blot assays corresponds to monomeric EhFdp1 (47 kDa). Notably, we confirmed that there was a clear increase in EhFdp1 expression with oxygen exposure (Fig. 1B), i.e., ∼2-fold after 1 h (n = 5; P = 7.0 × 10−3) and up to ∼5-fold after 4 h (n = 5; P = 0.029). A representative Western blot assay showing increasing EhFDP1 protein levels with oxygen stress is shown in Fig. 1C. The abundance of EhFdp1 under basal conditions is consistent with the high transcriptional levels observed. However, there were no significant changes in EhFdp1 transcript abundance under a variety of conditions, including response to NO-derived stress (1, 2, 11, 13, 42). FDPs have been shown to be highly and constitutively expressed both in prokaryotes (in some cases incorporating an oxidative stress response operon [8, 18]) and in protozoa (10, 25, 32). However, an increase in expression upon oxygen exposure has been observed for only one other FDP, that from the hyperthermophilic bacterium Thermotoga maritima, which shows an increase in protein levels with unmatched transcriptional levels and is proposed to sustain the growth of this “strict” anaerobe under microaerophilic conditions (20–22). The cytoplasmic abundance and increased protein levels in response to oxygen exposure suggest a potential similar function for EhFdp1 in E. histolytica.
Biochemical and spectroscopic properties of recombinant EhFdp1.
To further understand the functional properties of EhFdp1, we overexpressed the recombinant protein in bacterial cells and purified it to homogeneity in two chromatographic steps (protein purity was confirmed by SDS-PAGE) (Fig. 2A). We assessed the quaternary structure of EhFdp1 by size exclusion chromatography (see Fig. S2 in the supplemental material). EhFdp1 migrated as a dimer, which is in line with structural data for other FDPs (10, 27, 31, 39). Although the most common quaternary structure is tetrameric (27, 39), the “head-to-tail” dimeric arrangement constitutes the minimal functional unit in this protein family as it places the FMN and the diiron centers from opposing monomers in close proximity (for efficient ET).
Fig 2.
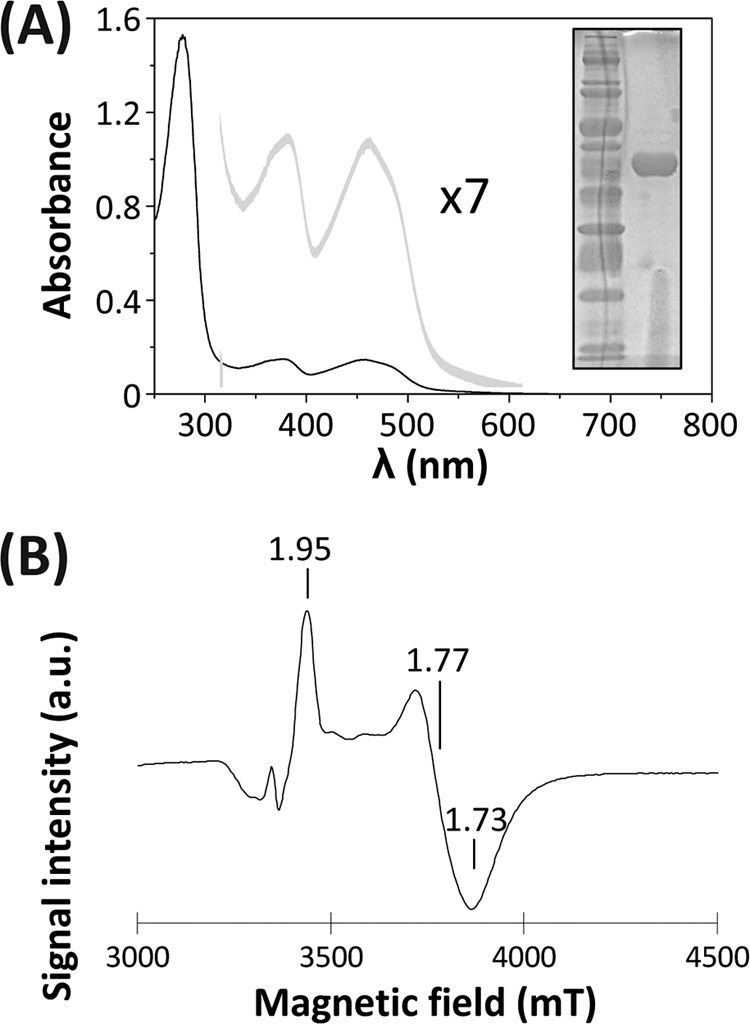
Production of functional recombinant EhFdp1. (A) UV-visible spectrum of pure recombinant EhFdp1 (15 μM in 20 mM Tris-HCl–18% glycerol [pH 7.5]; A280/A457 ratio, 9.1). Gray line, magnified visible region (multiplied by 7 for visualization). Inset, SDS-PAGE (12.5% acrylamide) analysis of purified EhFdp1. (B) EPR spectrum of partially reduced EhFdp1 (190 μM in 50 mM Tris-HCl–18% glycerol [pH 7.5]) showing a rhombic signal attributed to the diiron active site in a mixed-valence (FeIII-FeII) state. Reduction was obtained by sodium dithionite addition in the presence of the redox mediators listed in Table S2 in the supplemental material. The spectrum was recorded with a Bruker EMX spectrometer equipped with an ESR 900 continuous-flow helium cryostat (Oxford Instruments).
Pure recombinant EhFdp1 has a visible spectrum (Fig. 2A) similar to that of bacterial FDPs (40), i.e., one dominated by the flavin moiety spectral features (peaks at 457 and 380 nm) but with a slightly less rounded shape that could hint at subtle differences in the flavin binding site. Flavin determination after acid extraction (35) revealed that recombinant EhFdp1 binds 0.86 mol of flavin per mol of EhFdp1 monomer and identified the flavin as being FMN. The iron content determined by the TPTZ method revealed that recombinant EhFdp1 binds 1.58 mol of Fe per mol of EhFdp1 monomer. These results are consistent with the recombinant EhFdp1 binding one FMN molecule and harboring a nonheme binuclear site (both with ∼80% occupancy). This has been observed for other FDPs either isolated from their natural sources or produced in heterologous systems (reviewed in references 38 and 40).
Since nonheme diiron centers have no detectable spectrum in the visible range or one that is masked by the intense flavin bands in FDPs, we used EPR spectroscopy to probe the nonheme diiron site of EhFdp1. Nonheme diiron centers display characteristic EPR spectra in their mixed-valence state (FeIII-FeII). Since we did not observe any detectable spectrum in the isolated EhFdp1, we reduced the protein under anaerobic conditions with sodium dithionite in the presence of redox mediators (see Table S2 in the supplemental material) covering a wide range of redox potentials and recorded the EPR spectra. The EPR spectrum in Fig. 2B shows a rhombic signal with g values of <2.0 (gx, 1.95; gy, 1.77; gz, 1.73), as observed for other FDPs (from both prokaryotes and protozoa) (32, 37, 43) and in general for nonheme diiron center proteins (reviewed in reference 33). Taken together, the data show that recombinant EhFdp1 is produced as a homodimeric protein and is loaded with cofactors required for a fully functional FDP to be employed in the determination of its in vitro oxygen and NO reductase activities.
EhFdp1 is a robust oxygen reductase.
It has been shown that E. histolytica trophozoites display an oxygen consumption activity that was attributed primarily to an NADH oxidase (6). However, the E. histolytica genome has four genes that encode FDPs (23), strongly suggesting that other potential oxygen-detoxifying enzymes contribute to its oxygen consumption. Despite the number of studies on FDPs from various microbes, no rationale has been established for their substrate preference (oxygen versus NO). To assess the role of EhFdp1 in oxygen and/or NO detoxification by E. histolytica, the O2 and NO reductase activities of recombinant EhFdp1 were determined by amperometric measurements with Clark-type electrodes specific for either O2 or NO. To this end, we started by measuring the consumption of both substrates by bacterial lysates overexpressing EhFdp1 (bacteria expressing another E. histolytica protein, EhRbr, were used as a control). With NADH as the source of electron equivalents in the assays, the soluble extract of EhFdp1-expressing bacteria had a considerable oxygen reductase specific activity of 780 μM O2 min−1 mg total protein−1, ∼5-fold higher than that of the control cells expressing EhRbr. Neither lysate exhibited measurable NO consumption activity under the same conditions. We thus concluded that the 5-fold higher oxygen reductase activity of bacterial lysates expressing EhFdp1 was attributable to oxygen consumption by EhFdp1 and that a nonspecific E. coli component must be mediating ET from NADH to EhFdp1. Moreover, these results already suggested a substrate preference for oxygen that was further confirmed with purified recombinant EhFdp1.
So far, no putative redox partners for EhFdp1 have been identified in E. histolytica's genome. Several FDPs have been shown to accept electrons from a hybrid ET chain consisting of E. coli flavorubredoxin oxidoreductase (EcRdR) and the rubredoxin domain (EcRd) of flavorubredoxin (the FDP from E. coli) (see Fig. S3A in the supplemental material) (10, 41). Therefore, we used spectrophotometric assays to analyze the ability of EcRdR and EcRd to mediate ET from NADH to EhFdp1. When EhFdp1 was incubated with subcatalytic amounts of EcRdR and EcRd under anaerobic conditions, the visible spectrum of isolated oxidized EhFdp1 (Fig. 2A) was bleached within seconds upon the addition of NADH (data not shown). This result confirmed that the hybrid ET chain could be used to assess the oxygen and/or NO reductase activity of EhFdp1.
The oxygen reductase activity of EhFdp1 was initially probed by an aerobic spectrophotometric assay monitoring NADH consumption at 340 nm (Fig. 3A). In this assay, the ET chain with EhFdp1 as the terminal oxygen reductase consumes NADH at 45 μmol NADH s−1 μmol EhFdp1−1 with “zero”-order kinetics, indicating high substrate specificity. This observation was further confirmed by amperometric measurements with an oxygen-specific Clark-type electrode. Assembling the same ET chain in the reaction chamber, it was observed that EhFdp1 directly reduces oxygen to water (Fig. 3B) at a considerable rate (∼21 μmol O2 s−1 μmol EhFdp1−1), comparable to that of other oxygen-reducing FDPs. Importantly, this occurs without the apparent formation of ROS, as no effect was seen upon catalase addition. Experiments with different concentrations of the ET partners suggested that the activity determined is limited by electron delivery from the ET chain to EhFdp1, which implies that the oxygen reductase activity may be an underestimation, as it was assessed in the absence of the amoebic true redox partner(s).
Fig 3.

EhFdp1 is an oxygen reductase. Oxygen consumption was assayed by spectrophotometric (NADH consumption) and amperometric (O2 consumption determined with a Clark-type oxygen electrode) measurements. Assays were done at room temperature in 50 mM Tris-HCl–18% glycerol (pH 7.5). Electron delivery to EhFDP1 was achieved by assembling a hybrid ET chain composed of NADH, EcRdR, and EcRd. (A) Spectrophotometric NADH consumption was assayed with a reaction mixture volume of 1 ml and sequential addition of 160 μM NADH, 0.17 μM EcRdR, 2.65 μM EcRd, and 36 nM EhFdp1. The decrease in A340 shows NADH oxidation at the expense of oxygen reduction mediated by the ET chain with EhFdp1 as the terminal oxygen reductase. (B) Oxygen consumption was assayed with a reaction mixture volume of 1.5 ml and the sequential addition of 1 mM NADH, 0.2 μM EcRdR, 2 μM EcRd, and 150 nM EhFdp1. Oxygen depletion upon the addition of substoichiometric EhFdp1 is shown; addition of catalase (Kat) yielded no production of oxygen, suggesting that EhFdp1 reduces O2 directly to water.
Contrary to the efficient oxygen-scavenging ability of EhFdp1, anaerobic NO consumption assays using the same ET chain showed that EhFdp1 has poor NO reductase activity (∼0.1 μmol NO s−1 μmol EhFdp1−1), which was ∼200-fold lower than the rate of oxygen consumption (see Fig. S3C in the supplemental material). Moreover, the NO reductase assays were performed with a large excess of electron donors with respect to the EhFdp1 concentration (unlike the O2 assays), which suggests that the poor NO reductase activity was not caused by a limited electron supply.
Taken together, these experiments show that EhFdp1 efficiently reduces oxygen to water without generating ROS. It thus constitutes a robust line of defense in the parasite. Moreover, EhFdp1 displays a significant specificity for oxygen rather than NO, similar to oxygen-reducing FDPs from other organisms, notably including those from other protozoan pathogens such as G. intestinalis and T. vaginalis (10, 32). EhFdp1 thus represents one further line of defense against oxygen exposure that contributes to sustained amoebic viability in microaerophilic or aerobic environments within the host. The reasons for substrate preference in this protein family remain elusive, since the structural and sequence features of FDPs with opposing substrate preferences are essentially identical, particularly the active-site iron ligands. However, small differences in the protein environment around the diiron site could contribute to substrate selectivity. It remains to be determined whether the abundance of EhFdp1 could account for a partial NO-scavenging ability, despite the sluggish in vitro activity. Overall, EhFdp1 and its protozoan homologues are appealing targets for inhibition studies, since they have roles in important aspects of parasite biology and lack sequence or structural homologues in the host.
Supplementary Material
ACKNOWLEDGMENTS
We are deeply grateful to Anisha Patel and Richard Pearson for help with cloning and to Hussein Alramini for help with immunofluorescence microscopy.
This work was supported by FCT grant SFRH/BPD/26895/2006 and Gulbenkian Foundation short-term grant 99504/2008 to J.B.V., FCT project grants PTDC/QUI-BIQ/111080/2009 and PTDC/SAU-MIC/111447/2009 to J.B.V., FCT project grant PTDC/BIA-PRO/67263/2006 to M.T., and a grant from the NIAID (AI-053724) to U.S.
Footnotes
Published ahead of print 13 July 2012
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1. Akbar MA, et al. 2004. Genes induced by a high-oxygen environment in Entamoeba histolytica. Mol. Biochem. Parasitol. 133:187–196 [DOI] [PubMed] [Google Scholar]
- 2. Ali IK, Ehrenkaufer GM, Hackney JA, Singh U. 2007. Growth of the protozoan parasite Entamoeba histolytica in 5-azacytidine has limited effects on parasite gene expression. BMC Genomics 8:7 doi:10.1186/1471-2164-8-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Andersson JO, Sjogren AM, Davis LA, Embley TM, Roger AJ. 2003. Phylogenetic analyses of diplomonad genes reveal frequent lateral gene transfers affecting eukaryotes. Curr. Biol. 13:94–104 [DOI] [PubMed] [Google Scholar]
- 4. Andersson JO, Hirt RP, Foster PG, Roger AJ. 2006. Evolution of four gene families with patchy phylogenetic distributions: influx of genes into protist genomes. BMC Evol. Biol. 6:27 doi:10.1186/1471-2148-6-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baxt LA, Baker RP, Singh U, Urban S. 2008. An Entamoeba histolytica rhomboid protease with atypical specificity cleaves a surface lectin involved in phagocytosis and immune evasion. Genes Dev. 22:1636–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brown DM, Upcroft JA, Upcroft P. 1995. Free radical detoxification in Giardia duodenalis. Mol. Biochem. Parasitol. 72:47–56 [DOI] [PubMed] [Google Scholar]
- 7. Bruchhaus I, Richter S, Tannich E. 1998. Recombinant expression and biochemical characterization of an NADPH:flavin oxidoreductase from Entamoeba histolytica. Biochem. J. 330(Pt 3):1217–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Das A, Coulter ED, Kurtz DM, Jr, Ljungdahl LG. 2001. Five-gene cluster in Clostridium thermoaceticum consisting of two divergent operons encoding rubredoxin oxidoreductase–rubredoxin and rubrerythrin-type A flavoprotein–high-molecular-weight rubredoxin. J. Bacteriol. 183:1560–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Davis PH, Zhang X, Guo J, Townsend RR, Stanley SL., Jr 2006. Comparative proteomic analysis of two Entamoeba histolytica strains with different virulence phenotypes identifies peroxiredoxin as an important component of amoebic virulence. Mol. Microbiol. 61:1523–1532 [DOI] [PubMed] [Google Scholar]
- 10. Di Matteo A, et al. 2008. The O2-scavenging flavodiiron protein in the human parasite Giardia intestinalis. J. Biol. Chem. 283:4061–4068 [DOI] [PubMed] [Google Scholar]
- 11. Ehrenkaufer GM, Eichinger DJ, Singh U. 2007. Trichostatin A effects on gene expression in the protozoan parasite Entamoeba histolytica. BMC Genomics 8:216 doi:10.1186/1471-2164-8-216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fischer DS, Price DC. 1964. A simple serum iron method using the new sensitive chromogen tripyridyl-S-triazine. Clin. Chem. 10:21–31 [PubMed] [Google Scholar]
- 13. Gilchrist CA, et al. 2006. Impact of intestinal colonization and invasion on the Entamoeba histolytica transcriptome. Mol. Biochem. Parasitol. 147:163–176 [DOI] [PubMed] [Google Scholar]
- 14. Gillin FD, Diamond LS. 1980. Entamoeba histolytica and Entamoeba invadens: effects of temperature and oxygen tension on growth and survival. Exp. Parasitol. 49:328–338 [DOI] [PubMed] [Google Scholar]
- 15. Gillin FD, Diamond LS. 1981. Entamoeba histolytica and Giardia lamblia: effects of cysteine and oxygen tension on trophozoite attachment to glass and survival in culture media. Exp. Parasitol. 52:9–17 [DOI] [PubMed] [Google Scholar]
- 16. Isakov E, Siman-Tov R, Weber C, Guillen N, Ankri S. 2008. Trichostatin A regulates peroxiredoxin expression and virulence of the parasite Entamoeba histolytica. Mol. Biochem. Parasitol. 158:82–94 [DOI] [PubMed] [Google Scholar]
- 17. Jeelani G, et al. 2010. Two atypical l-cysteine-regulated NADPH-dependent oxidoreductases involved in redox maintenance, l-cystine and iron reduction, and metronidazole activation in the enteric protozoan Entamoeba histolytica. J. Biol. Chem. 285:26889–26899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Johnston S, et al. 2009. A genomic island of the sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough promotes survival under stress conditions while decreasing the efficiency of anaerobic growth. Environ. Microbiol. 11:981–991 [DOI] [PubMed] [Google Scholar]
- 19. Kurtz DM., Jr 2007. Flavo-diiron enzymes: nitric oxide or dioxygen reductases? Dalton Trans. 37:4115–4121 [Google Scholar]
- 20. Lakhal R, et al. 2011. Oxygen uptake rates in the hyperthermophilic anaerobe Thermotoga maritima grown in a bioreactor under controlled oxygen exposure: clues to its defence strategy against oxidative stress. Arch. Microbiol. 193:429–438 [DOI] [PubMed] [Google Scholar]
- 21. Le Fourn C, Fardeau ML, Ollivier B, Lojou E, Dolla A. 2008. The hyperthermophilic anaerobe Thermotoga maritima is able to cope with limited amount of oxygen: insights into its defence strategies. Environ. Microbiol. 10:1877–1887 [DOI] [PubMed] [Google Scholar]
- 22. Le Fourn C, et al. 2011. An oxygen reduction chain in the hyperthermophilic anaerobe Thermotoga maritima highlights horizontal gene transfer between Thermococcales and Thermotogales. Environ. Microbiol. 13:2132–2145 [DOI] [PubMed] [Google Scholar]
- 23. Loftus B, et al. 2005. The genome of the protist parasite Entamoeba histolytica. Nature 433:865–868 [DOI] [PubMed] [Google Scholar]
- 24. MacFarlane RC, Singh U. 2006. Identification of differentially expressed genes in virulent and nonvirulent Entamoeba species: potential implications for amebic pathogenesis. Infect. Immun. 74:340–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mastronicola D, et al. 2011. Giardia intestinalis escapes oxidative stress by colonizing the small intestine: a molecular hypothesis. IUBMB Life 63:21–25 [DOI] [PubMed] [Google Scholar]
- 26. Murray HW, Aley SB, Scott WA. 1981. Susceptibility of Entamoeba histolytica to oxygen intermediates. Mol. Biochem. Parasitol. 3:381–391 [DOI] [PubMed] [Google Scholar]
- 27. Petoukhov MV, et al. 2008. Quaternary structure of flavorubredoxin as revealed by synchrotron radiation small-angle X-ray scattering. Structure 16:1428–1436 [DOI] [PubMed] [Google Scholar]
- 28. Ramos-Martínez E, et al. 2009. Entamoeba histolytica: oxygen resistance and virulence. Int. J. Parasitol. 39:693–702 [DOI] [PubMed] [Google Scholar]
- 29. Salles JM, Salles MJ, Moraes LA, Silva MC. 2007. Invasive amebiasis: an update on diagnosis and management. Expert Rev. Anti Infect. Ther. 5:893–901 [DOI] [PubMed] [Google Scholar]
- 30. Saraiva LM, Vicente JB, Teixeira M. 2004. The role of the flavodiiron proteins in microbial nitric oxide detoxification. Adv. Microb. Physiol. 49:77–129 [DOI] [PubMed] [Google Scholar]
- 31. Silaghi-Dumitrescu R, Kurtz DM, Jr, Ljungdahl LG, Lanzilotta WN. 2005. X-ray crystal structures of Moorella thermoacetica FprA. Novel diiron site structure and mechanistic insights into a scavenging nitric oxide reductase. Biochemistry 44:6492–6501 [DOI] [PubMed] [Google Scholar]
- 32. Smutná T, et al. 2009. Flavodiiron protein from Trichomonas vaginalis hydrogenosomes: the terminal oxygen reductase. Eukaryot. Cell 8:47–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Solomon EI, et al. 2000. Geometric and electronic structure/function correlations in non-heme iron enzymes. Chem. Rev. 100:235–350 [DOI] [PubMed] [Google Scholar]
- 34. Stanley SL., Jr 2003. Amoebiasis. Lancet 361:1025–1034 [DOI] [PubMed] [Google Scholar]
- 35. Susín S, et al. 1993. Riboflavin 3′- and 5′-sulfate, two novel flavins accumulating in the roots of iron-deficient sugar beet (Beta vulgaris). J. Biol. Chem. 268:20958–20965 [PubMed] [Google Scholar]
- 36. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vicente JB, Teixeira M. 2005. Redox and spectroscopic properties of the Escherichia coli nitric oxide-detoxifying system involving flavorubredoxin and its NADH-oxidizing redox partner. J. Biol. Chem. 280:34599–34608 [DOI] [PubMed] [Google Scholar]
- 38. Vicente JB, Carrondo MA, Teixeira M, Frazao C. 2007. Flavodiiron proteins: nitric oxide and/or oxygen reductases, p 1–19 In Messerschmidt A. (ed), Handbook of metalloproteins. John Wiley & Sons, Ltd., Chichester, United Kingdom [Google Scholar]
- 39. Vicente JB, Carrondo MA, Teixeira M, Frazao C. 2008. Structural studies on flavodiiron proteins. Methods Enzymol. 437:3–19 [DOI] [PubMed] [Google Scholar]
- 40. Vicente JB, Justino MC, Goncalves VL, Saraiva LM, Teixeira M. 2008. Biochemical, spectroscopic, and thermodynamic properties of flavodiiron proteins. Methods Enzymol. 437:21–45 [DOI] [PubMed] [Google Scholar]
- 41. Vicente JB, et al. 2008. Kinetic characterization of the Escherichia coli nitric oxide reductase flavorubredoxin. Methods Enzymol. 437:47–62 [DOI] [PubMed] [Google Scholar]
- 42. Vicente JB, Ehrenkaufer GM, Saraiva LM, Teixeira M, Singh U. 2009. Entamoeba histolytica modulates a complex repertoire of novel genes in response to oxidative and nitrosative stresses: implications for amebic pathogenesis. Cell. Microbiol. 11:51–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vicente JB, et al. 2009. Redox properties of the oxygen-detoxifying flavodiiron protein from the human parasite Giardia intestinalis. Arch. Biochem. Biophys. 488:9–13 [DOI] [PubMed] [Google Scholar]
- 44. Weber C, et al. 2006. Stress by heat shock induces massive down regulation of genes and allows differential allelic expression of the Gal/GalNAc lectin in Entamoeba histolytica. Eukaryot. Cell 5:871–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang H, Alramini H, Tran V, Singh U. 2011. Nucleus-localized antisense small RNAs with 5′-polyphosphate termini regulate long term transcriptional gene silencing in Entamoeba histolytica G3 strain. J. Biol. Chem. 286:44467–44479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang P, Allahverdiyeva Y, Eisenhut M, Aro EM. 2009. Flavodiiron proteins in oxygenic photosynthetic organisms: photoprotection of photosystem II by Flv2 and Flv4 in Synechocystis sp. PCC 6803. PLoS One 4:e5331 doi:10.1371/journal.pone.0005331 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.