Abstract
The filamentous fungus Aspergillus fumigatus is an important opportunistic pathogen that can cause high mortality levels in susceptible patient populations. The increasing dependence on antifungal drugs to control A. fumigatus has led to the inevitable acquisition of drug-resistant forms of this pathogen. In other fungal pathogens, drug resistance is often associated with an increase in transcription of genes such as ATP-binding cassette (ABC) transporters that directly lead to tolerance to commonly employed antifungal drugs. In A. fumigatus, tolerance to azole drugs (the major class of antifungal) is often associated with changes in the sequence of the azole target enzyme as well as changes in the transcription level of this gene. The target gene for azole drugs in A. fumigatus is referred to as cyp51A. In order to dissect transcription of cyp51A transcription and other genes of interest, we constructed a set of firefly luciferase reporter genes designed for use in A. fumigatus. These reporter genes can either replicate autonomously or be targeted to the pyrG locus, generating an easily assayable uracil auxotrophy. We fused eight different A. fumigatus promoters to luciferase. Faithful behaviors of these reporter gene fusions compared to their chromosomal equivalents were evaluated by 5′ rapid amplification of cDNA ends (RACE) and quantitative reverse transcription-PCR (qRT-PCR) analysis. We used this reporter gene system to study stress-regulated transcription of a Hsp70-encoding gene, map an important promoter element in the cyp51A gene, and correct an annotation error in the actin gene. We anticipate that this luciferase reporter gene system will be broadly applicable in analyses of gene expression in A. fumigatus.
INTRODUCTION
Aspergillus fumigatus is an important filamentous fungal pathogen that causes severe invasive infection and pneumonia in immunocompromised hosts, resulting in high mortality rates. The increase in immunocompromised patient population as well as an increase in the incidence of drug resistance among A. fumigatus clinical isolates has aggravated the problem (2, 6, 7, 9, 12, 19, 20, 29, 30, 36, 51). Hence there is an urgent need to identify the molecules and mechanisms that contribute to pathogenesis and discover drug targets that will lead to a better treatment regimen of this important pathogen.
The sequencing and annotation of the A. fumigatus genome (40) have facilitated the process of studying A. fumigatus genes and their role in pathogenesis. However, A. fumigatus is still not genetically very tractable, and limited progress has been made in the understanding of how this saprophytic fungus turns into a prominent opportunistic pathogen. Most of the molecular genetic tools currently in use in A. fumigatus have been borrowed from the A. nidulans system.
The role of promoters in regulating expression of genes at the transcriptional level is important in the mechanism of action of processes across fungal species and beyond. To study this regulatory feature, reporter gene fusions have been employed in many systems as important genetic tools for analyzing the expression of particular genes under various conditions. They have also been used to identify DNA sequences upstream of a gene that function as potential regulatory elements as well as in ultimately determining proteins that bind to these elements. There are no comprehensive gene reporter systems or reagents in place for A. fumigatus. Most of the gene reporter fusions in A. fumigatus so far have been made to characterize individual genes. There has been a study using enhanced green fluorescent protein (eGFP) to analyze differential expression of the A. fumigatus cyclic AMP-dependent protein kinase A gene, pksP, in vitro and in vivo (26). The β-galactosidase-encoding Escherichia coli lacZ gene has been used to characterize individual promoters such as pksP (27) and a putative laccase-encoding gene, abr2 (56). There have also been studies using Gaussia princeps luciferase to study infection models involving the gpdA2 (AFUA_5G01970) promoter (5, 11, 13).
This study describes the generation of autonomous as well as integrating vectors with firefly luciferase as the reporter gene for use in A. fumigatus. The reporter gene system described here was authenticated by comparison with transcript levels measured by quantitative reverse transcription-PCR (qRT-PCR), coupled with 5′ rapid amplification of cDNA ends (RACE) to map the transcription start site of the fusion gene with respect to the native gene. Quantitative analysis of the activity of several promoters fused to the luciferase reporter gene targeted to the pyrG locus led to several insights into the functioning of A. fumigatus promoters. Use of a luciferase reporter gene fused to the promoter of the Hsp70-encoding hspASSA1 uncovered the induction of this gene in the presence of ethanol. Additionally, cyp51A-luciferase fusions provided evidence that the promoter of this gene increased in activity upon introduction of a mutation commonly associated with drug resistance. Finally, analysis of the A. fumigatus actin promoter (from the actA gene) demonstrated that an error was present in the current annotation for this gene and that the ActA promoter drove high levels of luciferase expression. Our hope is that the reporter system and reagents described here will serve as valuable tools for the A. fumigatus research community to study gene function and its regulation in this important opportunistic filamentous fungal pathogen.
MATERIALS AND METHODS
A. fumigatus strains, growth conditions, and transformation.
Two strains were used in this study: the genome-sequenced A. fumigatus Af293 strain (43) and the akuA (AfS35) strain (24), which was used to target A. fumigatus promoter reporter gene fusions to the pyrG locus. These strains were typically grown at 37°C in either rich medium (YG [0.5% yeast extract, 2% glucose]) or in minimal medium (MM) (1% glucose, nitrate salts, trace elements, 2% agar, pH 6.5; trace elements, vitamins, and nitrate salts are as described in the appendix of reference 23). For solid medium, 1.5% agar was added. Media for the growth of uracil auxotrophs were supplemented with 1.2 g/liter each of uracil and uridine. All A. fumigatus strains used in this study were transformed by generating protoplasts as described in reference 42. For regeneration of protoplasts upon transformation, 182 g/liter of sorbitol was added, along with 200 mg/liter of Hygromycin Gold (Invivogen) to select for transformants. Strains with the fusion promoters integrated at the pyrG locus, derived from the AfS35 strain, are listed in Table 1.
Table 1.
A. fumigatus strains used in this study
| Strain | Parent | Description | Source or reference |
|---|---|---|---|
| Af293 | Wild type | 43 | |
| SPF1 | Af293 | hph gpdA-luc::pyrG | This study |
| SPF4 | Af293 | hph luc::pyrG | This study |
| SPF5 | Af293 | hph hspASSA1-luc::pyrG | This study |
| SPF9 | Af293 | hph gpdA2-luc::pyrG | This study |
| AfS35 | loxP::akuA | FGSC (24) | |
| SPF40 | AfS35 | hph luc::pyrG | This study |
| SPF42 | AfS35 | hph hspASSA1-luc::pyrG | This study |
| SPF44 | AfS35 | hph abcA-luc::pyrG | This study |
| SPF46 | AfS35 | hph abcB-luc::pyrG | This study |
| SPF48 | AfS35 | hph gpdA2-luc::pyrG | This study |
| SPF50 | AfS35 | hph F1 cyp51A -luc::pyrG | This study |
| SPF52 | AfS35 | hph cyp51B-luc::pyrG | This study |
| SPF54 | AfS35 | hph F2 cyp51A-luc::pyrG | This study |
| SPF65 | AfS35 | hph C actA-luc::pyrG | This study |
| SPF67 | AfS35 | hph A actA-luc::pyrG | This study |
| SPF69 | AfS35 | hph B actA-luc::pyrG | This study |
| SPF71 | AfS35 | hph T0 cyp51A-luc::pyrG | This study |
| SPF73 | AfS35 | hph T1 cyp51A-luc::pyrG | This study |
| SPF75 | AfS35 | hph F0 cyp51A-luc::pyrG | This study |
| SPF77 | AfS35 | hph T2 cyp51A-luc::pyrG | This study |
Plasmids.
DNA manipulations were done according to the method of Sambrook et al. (48) or according to reagent manufacturer instructions. The autonomous plasmid containing the AMA1 origin of replication to generate promoter-reporter gene fusions for propagation in A. fumigatus was derived from pPRTII (25). To generate a gpdA2-luciferase fusion construct in pPTRII, the following fragments were PCR amplified: CEN6-ARSH and ScURA3 genes from pRS316 (49), the gpdA2 promoter from A. fumigatus strain Af293, either Renilla reniformis luciferase from pH12.7 (55) or firefly luciferase from pLG3 (Promega), and the gpdA2 transcription terminator region from the Af293 genome. These PCR fragments were targeted to SmaI/KpnI-digested, gapped pPTRII, using recombinational cloning in Saccharomyces cerevisiae (41), with ScURA3 as a selection marker. The primers were designed such that each PCR fragment had an overlapping 40-bp sequence with the adjoining DNA fragment while the terminal DNA fragments had a 40-bp overlapping sequence with the termini of gapped pPTRII to enable simultaneous cloning of the DNA pieces using homologous recombination in S. cerevisiae. The vector harboring firefly luciferase as a reporter gene was named pSP5, and the plasmid having Renilla luciferase was named pSP7. The pyrithiamine resistance marker from pSP5 and pSP7 was swapped with the hygromycin resistance marker cassette (Hph) using recombinational cloning in S. cerevisiae to form plasmids pSP15 and pSP17, respectively.
The integrating vector for targeting the A. fumigatus promoter-firefly luciferase fusion constructs to pyrG was generated in the pDHT/sk vector (38) amenable to Agrobacterium tumefaciens-mediated transformation (57). Initially, ScCEN6-ARSH and ScURA3 genes were PCR amplified from pRS316 (49) and cloned in pDHT gapped by PmlI and SphI digestion, by homologous recombination in S. cerevisiae, to form pSP19. To generate a gpdA2-luciferase fusion construct in pSP19, the following fragments were PCR amplified: 1 kb DNA upstream of pyrG from the Af293 genome, the Hph selection cassette from pHPH3, the gpdA2 promoter from A. fumigatus strain Af293, either Renilla luciferase from pH12.7 or firefly luciferase from pLG3, and 1 kb DNA downstream of pyrG from the Af293 genome that also served as the transcription terminator. These PCR fragments were targeted to pSP19 termini gapped with SacI and KpnI digestion, using recombinational cloning in S. cerevisiae, with ScURA3 as a selection marker. The primers were designed such that each PCR fragment had an overlapping 40-bp sequence with the adjoining DNA fragment while the terminal DNA fragments had a 40-bp overlapping sequence with the termini of SacI- and KpnI-digested, gapped pSP19, to enable simultaneous cloning of the DNA pieces using homologous recombination. The vector harboring Renilla luciferase as a reporter gene was named pSP21, and the plasmid having firefly luciferase was named pSP23. The gpdA2 promoter was removed from pSP23 using SmaI/SnaB1 digestion, followed by blunt-end ligation using T4 DNA ligase. The resulting firefly luciferase integrating vector without any A. fumigatus promoter was named pSP25. Different A. fumigatus promoters were cloned in pSP25 by targeting 1 kb DNA upstream of annotated genes, PCR amplified from the Af293 genome, to PmeI/NotI-digested, gapped pSP25. The primers for the above PCR was designed such that it contained 40 bp overlapping with the termini of the gapped plasmid such that the A. fumigatus promoter was placed directly upstream of the ATG start codon of the firefly luciferase gene. The list of the plasmids derived from pSP25, with different Afu promoters upstream of the reporter gene, used in this study is given in Table 2.
Table 2.
Plasmids used in this study
| Plasmid | Description | Source or reference |
|---|---|---|
| pPRTII | ptrA AMA1 lacZ bla | 25 |
| pSP5 | pPTRII ScCEN6 ARSH4 ScURA3 gpdA2-Fluc | This study |
| pSP7 | pPTRII ScCEN6 ARSH4 ScURA3 gpdA2-Rluc | This study |
| pSP15 | pSP5 hph | This study |
| pSP17 | pSP7 hph | This study |
| pDHT | A. tumifaciens vector kan | 38 |
| pSP19 | pDHT ScCEN6 ARSH4 ScURA3 | This study |
| pSP21 | pSP19 pyrG::hph gpdA2-Rluc::pyrG | This study |
| pSP23 | pSP19 pyrG hph gpdA2-luc::pyrG | This study |
| pSP25 | pSP23 pyrG hph luc::pyrG | This study |
| pSP27 | pSP25 pyrG hph A actA-luc::pyrG | This study |
| pSP28 | pSP25 pyrG hph hspASSA1-luc::pyrG | This study |
| pSP29 | pSP25 pyrG hph abcA-luc::pyrG | This study |
| pSP30 | pSP25 pyrG hph abcB-luc::pyrG | This study |
| pSP31 | pSP25 pyrG hph gpdA2-luc::pyrG | This study |
| pSP33 | pSP25 pyrG hph F1 cyp51A -luc::pyrG | This study |
| pSP34 | pSP25 pyrG hph cyp51B-luc::pyrG | This study |
| pSP35 | pSP25 pyrG hph B actA -luc::pyrG | This study |
| pSP51 | pSP25 pyrG hph C actA -luc::pyrG | This study |
| pSP52 | pSP25 pyrG hph T0 cyp51A -luc::pyrG | This study |
| pSP53 | pSP25 pyrG hph T1 cyp51A -luc::pyrG | This study |
| pSP56 | pSP25 pyrG hph F0 cyp51A -luc::pyrG | This study |
| pSP57 | pSP25 pyrG hph T2 cyp51A -luc::pyrG | This study |
Luciferase assays.
Renilla and firefly luciferase assays were done using the Renilla and firefly luciferase assay kits, respectively (Biotium, Inc.). To prepare cell lysates for luciferase assays, A. fumigatus spores were grown in 5 ml YG medium at 37°C for 12 to 14 h, and the mycelia were collected with an ashless Whatman 40 filter paper using a vacuum pump and washed in deionized water. This mycelium was then transferred to a 1.5-ml microcentrifuge tube containing 100 μl of 0.5-mm glass beads (Biospec, Inc.). Then, 50 μl of the cell lysis buffer (Biotium) was added to wet the mycelium. Using a battery-operated pestle grinder (catalog no. 03-392-106; Fisherbrand), the mycelium was ground for 1 min. Then, 200 μl of cell lysis buffer was added to the mycelium-glass bead slurry and vortexed at 4°C for 5 min. The mycelium was then centrifuged at 16,000 × g for 5 min, and the supernatant was used as the cell lysate for the luciferase assay. A 2-μl sample of the cell lysate was added to half-area flat-bottom 96-well plates in triplicate. For the Renilla luciferase assay, 5 μl of Renilla luciferase assay enhancer (Biotium) and 5 μl of Renilla luciferase assay solution (2× coelenterazine solution; Biotium) were added to the cell lysate. For the firefly luciferase assay, 10 μl of firefly luciferase assay solution (containing 0.2 mg/ml luciferin; Biotium) was added to the cell lysate. The 96-well plate was then mixed on a Titramax platform for 15 s. Luminescence was immediately measured using the IVIS 100 imaging system (Caliper Life Sciences). The number of photons emitted per second (luminescence) was then normalized to the amount of total protein present in the cell lysate per ml using a Bradford assay. The luciferase assay data depicted in this paper is quantitative, and the values shown are the average of luminescence from at least two independent experiments done in triplicates, with standard deviations between experiments shown in the form of error bars.
Real-time PCR.
Quantitative reverse transcription (qRT)-PCR was performed as described in reference 44 with the following modifications. Mycelium (100 mg) collected by filtration was ground into fine powder in liquid nitrogen using a mortar and pestle. Total RNA was then isolated from the ground mycelium using the RNeasy plant minikit (Qiagen). Synthesis of cDNA was performed using an iScript cDNA synthesis kit (Bio-Rad) with 1 μg of RNA as the template. For the quantitative PCR, primers were designed using the Primer Select program from the DNAStar software package. Primer concentrations were optimized for each gene, and annealing profiles were analyzed to evaluate nonspecific amplification by primer dimers. Control reaction mixtures including RNA instead of cDNA were made for each gene. The threshold cycle (CT) values were determined in the logarithmic phase of amplification for all genes, and the average CT value for each sample was calculated from three replicates. The CT value of the gene coding for actin (actA) was used for normalization of variable cDNA levels, and the fold difference in transcript levels was determined with respect to abcA. Amplification was carried out in the iCycler apparatus from Bio-Rad in a two-step process as follows: a denaturation step of 3 min at 95°C and 40 cycles of 95°C for 10 s each, with annealing and extension at 60°C for 45 s each. The reporter signals were analyzed using the iCycler iQ software (Bio-Rad).
Southern blot analysis.
Genomic DNA was isolated from A. fumigatus as described in reference 39. Genomic DNA (1 mg) was subjected to overnight digestion with BclI. Southern blotting was performed as described in reference 48. The probe used corresponded to the firefly luciferase gene and was labeled using [α-32P]dCTP (Perkin Elmer) by random primer labeling using the Klenow fragment of DNA polymerase I.
5′ RACE.
5′ rapid amplification of cDNA ends (RACE) was carried out using the ExactSTART eukaryotic mRNA 5′-RACE kit (Epicentre) according to the manufacturer's instructions. Briefly, total RNA was extracted from Afu mycelium ground with a mortar and pestle in the presence of liquid nitrogen using an RNeasy plant minikit (Qiagen). Total RNA (10 mg) was subjected to APex heat-labile alkaline phosphatase to convert uncapped RNA into a nonligatable 5′-hydroxyl RNA. The reaction product was cleaned up using an RNeasy Minelute kit (Qiagen). The 5′ cap was then removed from 5′ intact mRNAs using tobacco acid pyrophosphatase. The 5′-monophosphate poly(A) RNA thus generated was ligated to 5′-RACE acceptor oligonucleotide (with PCR priming site) using T4 RNA ligase. First-strand cDNA was synthesized from the above template using Moloney murine leukemia virus (MMLV)-reverse transcriptase using a primer that bound to the 5′-RACE acceptor oligonucleotide. RNA from this reaction was removed using RNase. Using the above-described reaction product as the template, second-strand cDNA was synthesized and amplified by PCR using a forward primer that bound to the 5′-RACE acceptor oligonucleotide and a gene-specific reverse primer (corresponding to either the firefly luciferase or the gene of A. fumigatus promoter under study). This PCR product was purified using a DNA Minelute kit (Qiagen) and used as the template for the sequencing reaction using a nested gene-specific primer.
RESULTS
Development of episomal/integrating vectors for promoter-luciferase reporter gene assays in A. fumigatus.
One of the key regulatory steps in any organism is control of gene expression at the level of promoter function. In the case of the filamentous fungal pathogen Aspergillus fumigatus, very little information exists concerning transcriptional control regions and the factors that exert regulatory effects there. Our goal was to develop reporter plasmids that would facilitate analysis of promoter function in A. fumigatus.
To accomplish this goal, we tested the utility of two different genes encoding luciferase enzymes, from either Renilla reniformis or Photinus pyralis (firefly). These enzymes use different substrates to generate light but are both very sensitive indicators of gene expression when used as reporter cassettes. Each of these two different luciferase genes was inserted into two different plasmid constructs to allow introduction into A. fumigatus strains. We generated both an episomally replicating plasmid and an integrative vector for delivery of reporter genes. The episomal vector used was derived from pPTRII (25) with the AMA1 origin of replication and was modified to contain a hygromycin resistance cassette for selection in A. fumigatus (Fig. 1A). Similarly, a pDHT-based Ti plasmid was constructed to deliver luciferase gene fusions to the pyrG locus by Agrobacterium tumefaciens-mediated transformation (ATMT) of A. fumigatus (57). Both of these plasmids were modified to contain a ScCEN6-ARSH4 fragment and ScURA3 selection marker to enable facile recombinational cloning in S. cerevisiae. The episomal plasmid was introduced into A. fumigatus by protoplast transformation (42). To target the fusion promoter constructs to the pyrG chromosomal locus by ATMT, the promoter-reporter gene fusion constructs flanked by 1 kb DNA upstream and downstream of the pyrG open reading frame were introduced between the T-DNA repeats of the pDHT integrative plasmid (Fig. 1B). Alternatively, the promoter-reporter gene fusion constructs were targeted to the pyrG locus by releasing it from the integrative vector by restriction enzyme digestion and transforming it into protoplasts of A. fumigatus strain AfS35 (akuAΔ) (24).
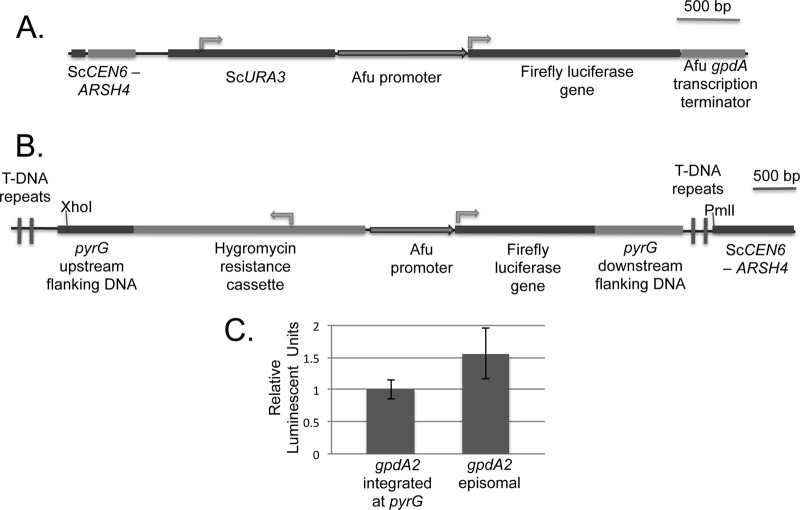
Fig 1.
Map of promoter-reporter fusion gene constructs. (A) Diagram of a region of the pPTRII (25)-based episomal plasmid with the A. fumigatus AMA1 origin of replication and the hygromycin resistance cassette as a selection marker in A. fumigatus shown. S. cerevisiae CEN6-ARSH4 and URA3 were incorporated to propagate the plasmid in S. cerevisiae. The episomal promoter-reporter gene construct consisted of the A. fumigatus promoter of choice, the luciferase reporter gene, followed by the A. fumigatus gpdA2 3′ untranslated region (3′ UTR) containing the likely transcription terminator. Arrows indicate the direction of gene transcription. (B) A partial map of the pDHT-based (38) plasmid for integrating promoter-reporter gene fusion constructs at the pyrG locus is presented. The integrative promoter-reporter gene construct, comprised of the A. fumigatus promoter of choice, the luciferase reporter gene, followed by the pyrG 3′ UTR, was cloned along with the hygromycin cassette and pyrG 5′ UTR flanking DNA within the T-DNA repeats (required by the Agrobacterium tumefaciens transformation system) using recombinational cloning in S. cerevisiae. Alternately, the plasmid can be digested by XhoI and PmlI and transformed into A. fumigatus protoplasts. (C) Comparison of a gpdA2-firefly luciferase fusion gene expressed in A. fumigatus from the AMA1-containing episomal plasmid versus one integrated into the chromosome at the pyrG locus, as measured by in vitro luminescent assay. Enzyme activities were normalized to those produced from the integrated luciferase fusion gene.
To test the efficacy of these reporter genes in A. fumigatus, the gpdA2 (AFUA_5G01030) promoter was used to provide presumably strong and constitutive transcription. A PCR fragment corresponding to DNA immediately 5′ of the annotated ATG for this locus to 1,000 bp upstream was prepared and inserted adjacent to the luciferase coding sequences. The resulting plasmids were then introduced into episomal or integrated contexts as appropriate. Hygromycin-resistant transformants were selected, and properly integrated reporter genes were detected by their simultaneous acquisition of uracil auxotrophy. Representative transformants were grown in liquid media and assayed for the level of luciferase activity produced (Fig. 1C).
Several important findings arose from these experiments. First, the R. reniformis luciferase-expressing transformants failed to produce any detectable luciferase activity. We also tested these same constructs in the heterologous S. cerevisiae host and found readily assayable luciferase activity (data not shown). Second, the firefly luciferase-expressing plasmids produced readily detectable luciferase enzyme and focused our experiments on the use of these constructs. Finally, we compared the firefly luciferase levels in transformants corresponding to the episomally maintained or the integrated gpdA2-luciferase fusion genes. There was a modest but reproducible increase in the luciferase levels driven by the episomally carried gpdA2-luciferase fusion gene (Fig. 1C). However, owing to the uncertain replication status of the AMA1-based episomal plasmid, we restricted our studies to integrated firefly luciferase fusions targeted to the pyrG locus.
Analysis of different A. fumigatus promoters by in vitro luciferase assay.
To validate the ability of this reporter gene system to faithfully reproduce A. fumigatus promoter function, we compared the ability of a number of different promoters to drive luciferase activity. As described above for gpdA2, DNA fragments corresponding to approximately 1,000 bp immediately upstream of the annotated ATG were prepared from each gene and inserted into the integrating firefly luciferase reporter gene vector. The AfS35 strain of A. fumigatus was employed, as this mutant strain lacks the akuA gene and exhibits enhanced homologous recombination (24). Transformants containing integrated reporter gene fusions were selected on hygromycin-containing medium and counterscreened for their uracil auxotrophy. Representative transformants were grown to mid-log phase, cell-free protein extracts prepared, and luciferase activities measured. All activities were normalized to a fusion gene lacking any inserted A. fumigatus promoter fragment (Fig. 2A). Correct genomic integration was confirmed by Southern blotting (Fig. 2C) and PCR analyses (data not shown).
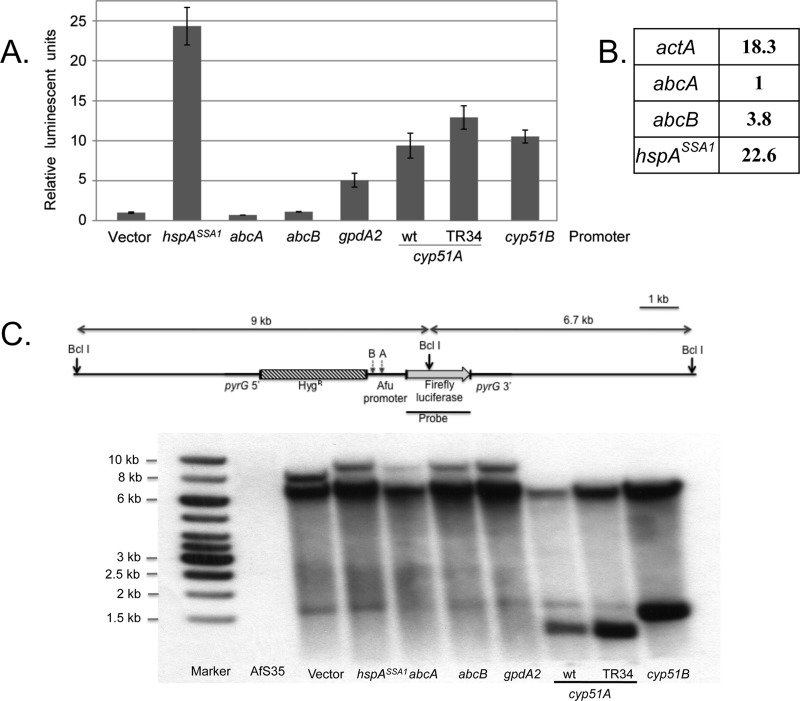
Fig 2.
Characterization and validation of luciferase fusion genes. (A) All luciferase fusion genes were integrated at the pyrG locus in the AfS35 strain (akuAΔ). Transformants were grown to mid-log phase, and protein extracts were prepared and assayed for levels of luciferase activity as described above. Enzyme activities were normalized to the level produced in the absence of an inserted A. fumigatus promoter (Vector). Inserted promoters included hspASSA1, corresponding to the A. fumigatus homologue of the ScSSA1 Hsp70-encoding gene; abcA and abcB are two A. fumigatus homologues of ABC transporter ScPDR5; gpdA2 encodes an A. fumigatus glyceraldehyde-3-dehydrogenase isoform, cyp51A and cyp51B are the genes encoding the lanosterol 14α demethylase, while TR34 indicates the presence of a 34-bp duplication in the cyp51A promoter associated with azole resistance. (B) Quantitative RT-PCR analysis of transcripts from the Af293 strain was carried out using total RNA and the indicated gene-specific primers. Transcript levels were normalized to those from the abcA gene. (C) Southern blot analysis to confirm single-copy targeted integration of the fusion promoter constructs at the pyrG locus. Genomic DNA was digested with BclI, which cuts once within the firefly luciferase gene, and also within the cyp51A and cyp51B promoters. The relative positions of the BclI restriction sites present in either cyp51A (A) or cyp51B (B) are indicated. The probe was prepared from the indicated segment of the luciferase gene. The expected DNA fragment sizes upon single integration at pyrG are indicated along the lanes. A nonspecific, cross-hybridizing DNA fragment is also seen around 1.5 kb.
Along with gpdA2, we also selected two other promoters from genes that could be expected to exhibit strong transcription of the luciferase reporter gene. These promoters corresponded to the actin-encoding gene (actA) and a homologue of the S. cerevisiae SSA1 gene called hspASSA1 (AFUA_1G07440). ScSSA1 is robustly expressed and regulated (10), attributes we anticipated might be conserved with its A. fumigatus counterpart. The hspASSA1 promoter placed upstream of the luciferase gene produced the highest levels of enzyme activity among the initial promoter fusions, while, to our surprise, the actA-luciferase fusion gene produced no detectable luciferase activity. The reason for the unexpected behavior of the actA-luciferase fusion is presented below.
Prompted by our interest in drug resistance in A. fumigatus, we prepared five other fusion gene constructs. Two promoters were taken from A. fumigatus genes encoding ATP-binding cassette transporter proteins that exhibited the highest degree of sequence conservation with a major ABC transporter from S. cerevisiae called PDR5 (reviewed in references 17, 22, and 45). These genes were designated abcA (AFUA_2G15130) and abcB (AFUA_1G14330), respectively. Additionally, fusion genes were constructed between luciferase and the two genes encoding the lanosterol α14 demethylase enzyme: cyp51A and cyp51B. A fusion gene containing an allele of cyp51A commonly associated with elevated azole resistance was also prepared. This form of cyp51A contains a duplication of a 34-bp element in the promoter that is believed to lead to enhanced expression of the relevant transcript and is referred to as cyp51ATR34.
Both cyp51A and cyp51B produced readily detectable and similar levels of luciferase activity. Introduction of the cyp51ATR34 fusion promoter led to increased expression of luciferase enzyme, consistent with enhanced promoter function in this mutant. Surprisingly, levels of expression from either the abcA or abcB promoter fusions were very low: not significantly greater than enzyme activity produced by a construct lacking an inserted Afu promoter. We attribute the latter to the presence of the divergently placed but strong constitutive Aspergillus nidulans promoter (46) that drives expression of the adjacent hygromycin resistance gene.
Confirmation that the luciferase reporter gene system was faithfully reflecting transcription of the endogenous Afu genes came from a comparison of the various luciferase activities and mRNA levels measured by quantitative reverse transcription-PCR (qRT-PCR). Total RNA was prepared from wild-type cells, and levels of four different transcripts were compared by qRT-PCR. Transcripts were normalized to actA (AFUA_6G04740), and fold differences in mRNA transcript levels were compared with respect to AbcA (Fig. 2B).
The relative expression of these different mRNA species is reflected by the luciferase activities produced by each reporter gene. The best-expressed mRNA under these conditions corresponded to the hspASSA1 gene that was 23-fold higher than the abcA transcript; a difference entirely consistent with that predicted by comparison of the luciferase activities produced from the two different reporter genes. Expression of actA was 18-fold elevated compared to that of abcA. This differential exhibited good agreement once appropriate actA reporter constructs were prepared (see below). To ensure that authentic A. fumigatus transcription start sites were being used when these relatively small promoter fragments were transferred to the heterologous pyrG locus, we compared 5′ endpoints of several genes that were produced both from the endogenous locus and when present in the fusion context by 5′ RACE (5′ rapid analysis of cDNA ends).
5′ RACE analysis to map transcription start sites of the fusion promoters.
To confirm that the fusion gene promoters were still utilizing the authentic A. fumigatus 5′ start sites of transcription, even when moved to a different chromosomal location, we carried out 5′ rapid analysis of cDNA ends, or RACE, analysis. Reverse transcription primers specific for cyp51A, hspASSA1, abcA, abcB, gpdA2, and actA were used along with a primer specific for luciferase sequences. Use of these two types of primers allowed us to directly compare transcription start sites used by the native mRNA and the chimeric luciferase fusion gene. In every case, the primary PCR product was sequenced, in order to avoid any artifacts that could be introduced by cloning a rare cDNA product. Total RNA was annealed to either the native primers or luciferase-specific primers, and cDNAs were prepared and amplified using 5′ RACE (Fig. 3).
Fig 3.
5′ RACE analysis to map transcription start sites of A. fumigatus promoters. Total RNA derived from relevant A. fumigatus promoter-reporter gene fusion strains targeted to the pyrG locus was used to generate cDNA. These cDNAs were sequenced using gene-specific primers directing sequencing toward the 5′ end of the transcript. Primers were generated that corresponded to either the native A. fumigatus gene or the firefly luciferase coding sequence. The ATG codons present in the 5′ UTRs from abcA and abcB are shown in bold. The length of each 5′ UTR is indicated at the right. The 5′ untranslated sequences from both the authentic and the fusion genes were identical in all cases tested, except for the gpdA2 promoter. In the case of gpdA2, there were seven additional nucleotides (shown in lower case) found in the transcript produced from the native gene compared to the fusion gene.
This analysis demonstrated that for every gene examined, except gpdA2, a unique transcription start site was identified. Importantly this same transcription start site was used by both the mRNA produced from the normal chromosomal locus as well as from the luciferase fusion promoter integrated at pyrG. Only the gpdA2 transcript exhibited two different 5′ endpoints, although these were separated by only 7 bp. Comparison of the wild-type cyp51A and cyp51ATR34 transcripts determined that the presence of the duplicated region in the TR34 allele did not alter the start point of transcription. In the cases of both abcA and abcB, out-of-frame ATG sequences were detected in the relatively long 5′ untranslated regions (UTRs) of these two genes. The significance of these ATG codons is unclear, but their position in the transcript has been associated with translational control elements in other, better-characterized mRNAs (reviewed in reference 53).
The Afu hspASSA1 promoter is induced by ethanol.
Having established that good general agreement existed between mRNA and luciferase levels driven by the reporter fusions, we wanted to confirm that this reporter gene system would be responsive to acute changes in environmental conditions. Since Hsp70-encoding genes have often been observed to respond to changes in the environment, we tested the hspASSA1-luciferase fusion gene for its response to several different stress agents. Transformants were grown to mid-log phase and challenged with several different stress conditions for 2 h, and then luciferase activities were determined as described previously (Fig. 4A).
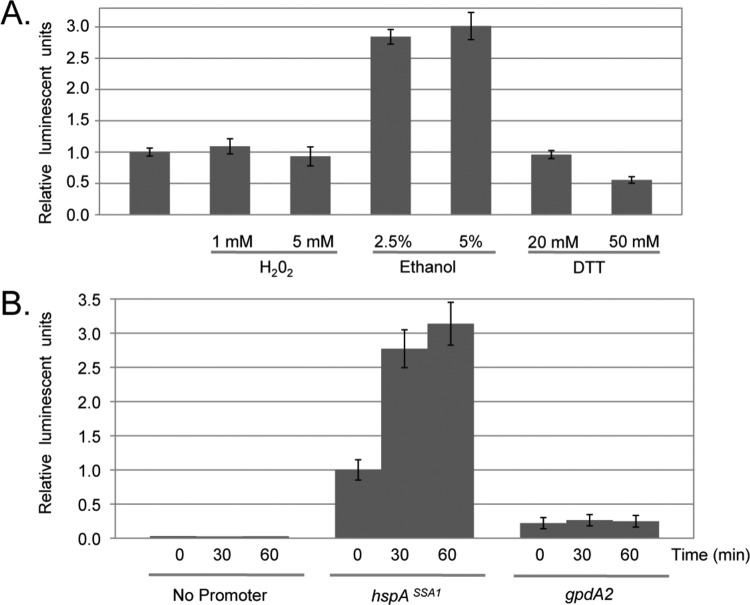
Fig 4.
Analysis of the hspASSA1 promoter. (A) A representative transformant containing the hspASSA1-luciferase fusion gene promoter targeted to the pyrG locus in AfS35 was grown in the presence of different stress conditions. In each case, the stress was given for 2 h before mycelia were harvested for luciferase assay. Luciferase activities were normalized to those produced by the hspASSA1-luciferase fusion gene in the absence of stress. (B) Mid-log-phase transformants containing the indicated luciferase fusions were stressed by the addition of 5% ethanol. Aliquots were removed at the indicated time points to assess the kinetics of hspASSA1 promoter induction. As controls, the luciferase levels driven in the absence of an inserted promoter (No Promoter) or by the gpdA2 promoter were also determined.
Treatment of hspASSA1-luciferase-containing transformants with 2.5 or 5% ethanol led to a roughly 3-fold increase in luciferase activity. Using this induction regimen, we did not observe significant elevation in luciferase expression when transformants were subjected to heat-, H2O2- (not shown), or dithiothreitol (DTT)-induced stress.
Next, the kinetics of ethanol induction were evaluated. Transformants containing luciferase fusions to either hspASSA1 or gpdA2, along with the base luciferase fusion plasmid lacking any inserted Afu promoter, were grown to mid-log phase and challenged with 5% ethanol for 0, 30, or 60 min. At each time point, luciferase activity assayed was normalized to the total protein extracted from the cell lysate (Fig. 4B).
Rapid induction of hspASSA1-luciferase was detected as full activation of the fusion gene and was observed within 30 min of ethanol exposure. Neither the gpdA2 nor the promoter-less luciferase fusion genes showed any response to the ethanol challenge. These data indicate that this Hsp70-encoding gene is selectively responsive to stress regimens likely to be experienced by A. fumigatus.
Deletion analysis of the cyp51A promoter.
Use of a reporter gene system facilitates the functional analysis of promoters (8). The cyp51A promoter plays an important role in azole tolerance in A. fumigatus, as most resistant isolates contain both a change in the coding sequence for the lanosterol 14α demethylase enzyme and a duplication of a 34-bp element present in the promoter region (31, 33–35, 37, 52). The presence of this duplication in the cyp51A promoter is referred to as the TR34 promoter. To evaluate the contribution of this 34-bp region in cyp51A to expression of the gene, several different constructs were prepared containing different segments of the cyp51A promoter fused to luciferase (Fig. 5A). The 34-bp region in cyp51A was duplicated to form cyp51ATR34 in two constructs (F2 and T2) as well as omitted from two others. All these fusion genes were integrated at the pyrG locus. Representative transformants were grown to mid-log phase, and their luciferase activities were determined. Relative luciferase values were obtained by normalizing to the activity produced by the wild-type cyp51A promoter containing 1.2 kb of 5′ noncoding DNA.
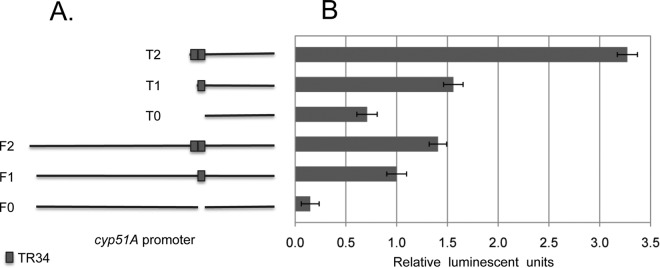
Fig 5.
Deletion mapping the cyp51A promoter. (A) Maps of the various cyp51A promoter fusions constructed and tested for luciferase activity are shown. F0, F1, and F2 represent the presumably full-length cyp51A promoter with 0, 1, and 2 34-bp (TR) elements, respectively. T0, T1, and T2 represent the truncated cyp51A promoter with 0, 1, and 2 34-bp (TR) elements, respectively. (B) The luciferase activities produced by transformants containing integrated copies of the cyp51A promoter derivatives shown to the left are presented. Luciferase activities are normalized to those produced by the wild-type full-length cyp51A fusion gene.
The effect of the TR34 element could be seen in the context of the 1.2-kb cyp51A promoter fragment, but the increased expression represented a modest change (Fig. 5). Interestingly, removal of the wild-type 34-bp region from the cyp51A promoter (F0) led to a 90% reduction in expression compared to the wild-type promoter (F1). The 34-bp repeat plays a major role in function of the wild-type cyp51A promoter.
To examine the contribution of DNA regions upstream of the 34-bp element in cyp51A promoter activity, a series of 5′ truncation mutants were generated. A construct containing only 239 bp upstream of the cyp51A transcription start site (T0) still produced 70% of the luciferase activity driven by the full 1.2-kb promoter fragment. This −239 construct lacked the 34-bp element. Returning 1 copy (T1) or the duplicated copy (T2) of the 34-bp element led to a greater than 2-fold or nearly 5-fold increase in luciferase expression, again illustrating the stimulatory nature of this 34-bp repeat on expression.
Definition of the actA promoter.
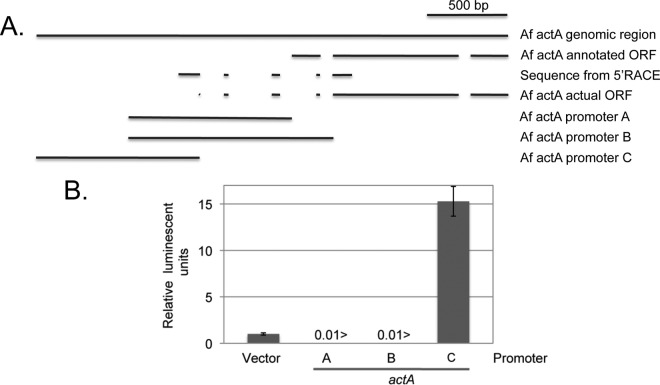
An initially puzzling finding came from attempts to use the actA promoter as a representative of a strong, constitutive promoter, as it often is in other fungi (32). Analysis of the actA mRNA by qRT-PCR indicated that this transcript was quite abundant (Fig. 2B). However, when 1 kb upstream of the presumptive actA ATG was inserted into the luciferase fusion plasmid, no detectable luciferase activity was produced (Fig. 6B). This first actA-luciferase fusion construct is referred to as the promoter A fusion. The failure of the promoter A fusion to drive luciferase prompted us to revisit the annotation of actA that indicated the first 50 amino acids of this fungal actin were unique to A. fumigatus. A second luciferase fusion gene construct (promoter B) was produced, forming a translational fusion between the predicted actA coding sequence and luciferase. The fusion joint within actA corresponded to a position that was highly conserved between actin proteins, although the 5′ end of this clone was identical to the promoter A fusion. This new chimera also failed to produce luciferase activity.
Fig 6.
Analysis of the actA transcript and promoter. (A) Diagram of the theoretical and empirical 5′ region of the actA transcript is shown. Af actA annotated ORF refers to the open reading frame (ORF) expected from the current A. fumigatus annotation. Gaps represent predicted introns. Sequence from 5′ RACE denotes regions of the actA locus found in the cDNA product from this gene. actA actual ORF represents the location of the coding sequences for the actin derived from the cDNA sequence (and by comparison with A. nidulans). The bounds of the DNA fragments used to test for actA promoter activity as fusion genes to luciferase are indicated as A, B, and C. (B) The luciferase activities produced from transformants containing a single copy of the indicated actA-luciferase reporter genes. Vector denotes a transformant with no inserted A. fumigatus promoter. Enzyme activities were normalized to those produced in the absence of any inserted A. fumigatus promoter.
We next carried out 5′ RACE analysis of the 5′ end of the endogenous actA mRNA and found a very different splicing pattern than that predicted from the genomic annotation (Fig. 6A). Based on this new information, a third actA-luciferase fusion gene was constructed (promoter C). This luciferase reporter gene produced readily detectable enzyme activity and represented one of the strongest fusion genes produced here.
DISCUSSION
The work described here provides an important new tool for analysis of gene expression in the filamentous fungal pathogen Aspergillus fumigatus. Development of reporter gene systems was extremely valuable in the progression of the analysis of promoter function in S. cerevisiae (16) as well as Candida albicans (54). Previous studies in A. fumigatus have described the use of lacZ-based reporter systems (4, 27, 28, 50, 56) in addition to expression of firefly luciferase as an indicator of the presence of A. fumigatus in infection models (4, 5, 11, 13). Here we detail the generation of both integrating and autonomously replicating luciferase reporter vectors that will allow analysis and dissection of promoter regions from this fungus. Given the limited understanding of the mechanisms underlying control of gene expression in A. fumigatus, progress in this area is a crucial need. Comparison of the levels of the reporter gene fusions described here with their relative transcription levels assessed recently by RNA-seq (15) also supports the view that these constructs faithfully reflect native expression levels.
By the reporter gene analysis from this study, we provide new insights about the regulation and function of several A. fumigatus promoters. Here we demonstrate that the hspASSA1 promoter responds to exposure to ethanol. These data also argue that the promoter region alone is sufficient to drive this ethanol induction of gene expression. We were surprised to find that this promoter was not activated by the presence of DTT, H2O2, or heat (data not shown). One possible explanation for the relatively limited range of induction of the hspASSA1 promoter is the presence of multiple other hsp70 homologues in the A. fumigatus genome. This is common in eukaryotic organisms and suggests the increased specialization of Hsp70 function in this fungus as seen in other fungal species. DTT, for example, is often associated with an increase in the unfolded protein response and as such may signal directly to the endoplasmic reticulum (ER)-associated Hsp70 protein in A. fumigatus. This organism expresses an Hsp70 with a high level of sequence similarity with an ER-localized Hsp70 from S. cerevisiae: Kar2 (47) (designated AFUA_2G04620). This A. fumigatus gene may be the DTT-responsive Hsp70 in this fungus. The reporter gene system here will allow the comparison of these different Hsp70-encoding gene promoters.
One of the few examples of an A. fumigatus gene in which transcription has been implicated as an integral part of its function is the cyp51A gene. Changes in both the cyp51A promoter and its coding sequence are required to express a lanosterol 14α demethylase protein that confers resistance to azole drugs (34). Experiments from several groups have established that the mRNA levels of cyp51A are elevated in these azole-resistant strains (1). More recent data have implicated the hypoxic regulator SrbA in stimulation of cyp51A transcription upon depletion of oxygen (1, 3). The binding site for SrbA lies in the upstream region of cyp51A but has not been precisely defined. We believe that SrbA binds an element distinct from the 34-bp element, as previous experiments determined that a cyp51A promoter fragment lacking the 34-bp element was bound by recombinant SrbA (3). Further experiments are required to confirm this suggestion.
The preliminary analysis of the cyp51A promoter suggests the presence of multiple, interacting regulator sites in this transcriptional control region. Consideration of the effects of varying the 34-bp element from 0 to 2 (TR34) copies in the context of either 1.2 kb of 5′ flanking region from cyp51A or only 288 of upstream DNA demonstrated very different expression patterns. The 34-bp element was critically important in maintaining wild-type expression, while introduction of the TR34 repeat caused a modest increase in expression. Strikingly, the fusion construct containing 288 bp of 5′ flanking DNA drove 7-fold-higher levels of luciferase expression than a much larger construct lacking the 34-bp element. This suggests the possibility that negative regulatory elements upstream of the 34-bp region act to inhibit the function of the promoter. This inhibition is prevented in the presence of either 1 or 2 copies of the 34-bp element.
There also appear to be DNA sequences downstream of the 34-bp element that are significant contributors to cyp51A expression. These positive elements are also responsive to the presence of the 34-bp element, either in single or double copy. Taken together, these data support the view that upstream of the 34-bp element, negative contributors to cyp51A expression reside while downstream of this element, positive effectors will be found. This suggestion can now be readily tested by mutagenesis of the cyp51A promoter using the reporter gene system developed here.
The construction of the actA-luciferase genes also led to correction of the current annotation for this gene in the A. fumigatus genome. All reporter gene fusions were designed based on the annotation in the GenBank database and in every other case, these fusions behaved as expected. We selected actA for this analysis, as this gene is typically well expressed in other fungi. The finding that two different reporter constructs failed to produce any detectable luciferase activity prompted a more detailed consideration of the presumptive N terminus of A. fumigatus actA. Along with our 5′ RACE analysis, we also examined the known splicing pattern of A. nidulans actA (14) and found this to contain a very different transcript structure from that predicted by the A. fumigatus genomic annotation. This analysis also points out the utility of this reporter system to confirm the likely gene structure of A. fumigatus loci, irrespective of the presence of homologues in other fungi.
The 5′ RACE analysis provided several important findings related to A. fumigatus transcription. Most apparent was that the majority of transcripts exhibited a unique transcription start site, at least in this small sample of genes. This is in marked contrast to S. cerevisiae, in which global transcription start site mapping indicates that most gene contain multiple transcription start sites (58). The 5′ leader regions detected in the 5′ RACE analysis were also generally fewer than 70 nucleotides, with no AUG codons in this region. The two notable exceptions were the ABC transporter-encoding genes, in which both long leaders and an AUG codon were found. Preliminary data suggest that this leader is important for expression as deletion mutations removing the 3′ segment of the leader failed to express luciferase activity (data not shown), but more analyses are required to confirm this suggestion.
Increased availability of molecular biological tools for use in A. fumigatus is a necessary step toward accelerating the understanding of the biology of this important fungal pathogen. Even in the more extensively studied filamentous fungus A. nidulans, analysis of the molecular basis of transcriptional regulation remains at an early stage compared to research on organisms like S. cerevisiae (recently reviewed in reference 18). The combination of reagents aimed at the study of single genes, like the reporter system described here, coupled with genomic approaches such as RNA-seq and construction of a library of heterologously regulated genes (21), will help to refine our understanding of transcriptional mechanisms in this filamentous fungus.
ACKNOWLEDGMENTS
This work was supported in part by NIH R21 AI92331 and the Carver Medical Research Foundation.
We thank Ricardo Almeido and Gustavo Goldman for valuable assistance in the early stages of this work.
Footnotes
Published ahead of print 27 July 2012
REFERENCES
- 1. Albarrag AM, et al. 2011. Interrogation of related clinical pan-azole-resistant Aspergillus fumigatus strains: G138C, Y431C, and G434C single nucleotide polymorphisms in cyp51A, upregulation of cyp51A, and integration and activation of transposon Atf1 in the cyp51A promoter. Antimicrob. Agents Chemother. 55:5113–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arendrup MC, et al. 2010. Development of azole resistance in Aspergillus fumigatus during azole therapy associated with change in virulence. PLoS One 5:e10080 doi:10.1371/journal.pone.0010080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blosser SJ, Cramer RA. 2012. SREBP-dependent triazole susceptibility in Aspergillus fumigatus is mediated through direct transcriptional regulation of erg11A (cyp51A). Antimicrob. Agents Chemother. 56:248–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brock M. 2012. Application of bioluminescence imaging for in vivo monitoring of fungal infections. Int. J. Microbiol. 2012:956794 doi:10.1155/2012/956794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brock M, et al. 2008. Bioluminescent Aspergillus fumigatus, a new tool for drug efficiency testing and in vivo monitoring of invasive aspergillosis. Appl. Environ. Microbiol. 74:7023–7035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bueid A, et al. 2010. Azole antifungal resistance in Aspergillus fumigatus: 2008 and 2009. J. Antimicrob. Chemother. 65:2116–2118 [DOI] [PubMed] [Google Scholar]
- 7. Burgel PR, et al. 2012. High prevalence of azole-resistant Aspergillus fumigatus in adults with cystic fibrosis exposed to itraconazole. Antimicrob. Agents Chemother. 56:869–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Casadaban MJ, Martinez-Arias A, Shapira SK, Chou J. 1983. Beta-galactosidase gene fusions for analyzing gene expression in E. coli and yeast. Methods Enzymol. 100:293–308 [DOI] [PubMed] [Google Scholar]
- 9. Chowdhary A, et al. 2012. Isolation of multiple-triazole-resistant Aspergillus fumigatus strains carrying the TR/L98H mutations in the cyp51A gene in India. J. Antimicrob. Chemother. 67:362–366 [DOI] [PubMed] [Google Scholar]
- 10. Craig E, Ziegelhoffer T, Nelson J, Laloraya S, Halladay J. 1995. Complex multigene family of functionally distinct Hsp70s of yeast. Cold Spring Harb. Symp. Quant Biol. 60:441–449 [DOI] [PubMed] [Google Scholar]
- 11. d'Enfert C, Vecchiarelli A, Brown AJ. 2010. Bioluminescent fungi for real-time monitoring of fungal infections. Virulence 1:174–176 [DOI] [PubMed] [Google Scholar]
- 12. Denning DW, et al. 2011. High-frequency triazole resistance found in nonculturable Aspergillus fumigatus from lungs of patients with chronic fungal disease. Clin. Infect. Dis. 52:1123–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Donat S, et al. 2012. Surface display of Gaussia princeps luciferase allows sensitive fungal pathogen detection during cutaneous aspergillosis. Virulence 3:51–61 [DOI] [PubMed] [Google Scholar]
- 14. Fidel S, Doonan JH, Morris NR. 1988. Aspergillus nidulans contains a single actin gene which has unique intron locations and encodes a gamma-actin. Gene 70:283–293 [DOI] [PubMed] [Google Scholar]
- 15. Gibbons JG, et al. 2012. Global transcriptome changes underlying colony growth in the opportunistic human pathogen Aspergillus fumigatus. Eukaryot. Cell 11:68–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guarente L. 1983. Yeast promoter and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 101:181–191 [DOI] [PubMed] [Google Scholar]
- 17. Gulshan K, Moye-Rowley WS. 2007. Multidrug resistance in fungi. Eukaryot. Cell 6:1933–1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hahn S, Young ET. 2011. Transcriptional regulation in Saccharomyces cerevisiae: transcription factor regulation and function, mechanisms of initiation, and roles of activators and coactivators. Genetics 189:705–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Howard SJ, et al. 2009. Frequency and evolution of azole resistance in Aspergillus fumigatus associated with treatment failure. Emerg. Infect. Dis. 15:1068–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Howard SJ, et al. 2006. Multi-azole resistance in Aspergillus fumigatus. Int. J. Antimicrob. Agents 28:450–453 [DOI] [PubMed] [Google Scholar]
- 21. Hu W, et al. 2007. Essential gene identification and drug target prioritization in Aspergillus fumigatus. PLoS Pathog. 3:e24 doi:10.1371/journal.ppat.0030024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jungwirth H, Kuchler K. 2006. Yeast ABC transporters—a tale of sex, stress, drugs and aging. FEBS Lett. 580:1131–1138 [DOI] [PubMed] [Google Scholar]
- 23. Kafer E. 1977. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv. Genet. 19:33–131 [DOI] [PubMed] [Google Scholar]
- 24. Krappmann S, Sasse C, Braus GH. 2006. Gene targeting in Aspergillus fumigatus by homologous recombination is facilitated in a nonhomologous end-joining-deficient genetic background. Eukaryot. Cell 5:212–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kubodera T, Yamashita N, Nishimura A. 2002. Transformation of Aspergillus sp. and Trichoderma reesei using the pyrithiamine resistance gene (ptrA) of Aspergillus oryzae. Biosci. Biotechnol. Biochem. 66:404–406 [DOI] [PubMed] [Google Scholar]
- 26. Langfelder K, Philippe B, Jahn B, Latge JP, Brakhage AA. 2001. Differential expression of the Aspergillus fumigatus pksP gene detected in vitro and in vivo with green fluorescent protein. Infect. Immun. 69:6411–6418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liebmann B, Gattung S, Jahn B, Brakhage AA. 2003. cAMP signaling in Aspergillus fumigatus is involved in the regulation of the virulence gene pksP and in defense against killing by macrophages. Mol. Genet. Genomics 269:420–435 [DOI] [PubMed] [Google Scholar]
- 28. Liebmann B, Muller M, Braun A, Brakhage AA. 2004. The cyclic AMP-dependent protein kinase a network regulates development and virulence in Aspergillus fumigatus. Infect. Immun. 72:5193–5203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lionakis MS, et al. 2005. Increased frequency of non-fumigatus Aspergillus species in amphotericin B- or triazole-pre-exposed cancer patients with positive cultures for aspergilli. Diagn. Microbiol. Infect. Dis. 52:15–20 [DOI] [PubMed] [Google Scholar]
- 30. Lockhart SR, et al. 2011. Azole resistance in Aspergillus fumigatus isolates from the ARTEMIS global surveillance study is primarily due to the TR/L98H mutation in the cyp51A gene. Antimicrob. Agents Chemother. 55:4465–4468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mavridou E, Bruggemann RJ, Melchers WJ, Mouton JW, Verweij PE. 2010. Efficacy of posaconazole against three clinical Aspergillus fumigatus isolates with mutations in the cyp51A gene. Antimicrob. Agents Chemother. 54:860–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McLean M, et al. 1995. Organization of the Saccharomyces cerevisiae actin gene UAS: functional significance of reiterated REB1 binding sites and AT-rich elements. Mol. Microbiol. 18:605–614 [DOI] [PubMed] [Google Scholar]
- 33. Meletiadis J, Mavridou E, Melchers WJ, Mouton JW, Verweij PE. 2012. Epidemiological cutoff values for azoles and Aspergillus fumigatus based on a novel mathematical approach incorporating cyp51A sequence analysis. Antimicrob. Agents Chemother. 56:2524–2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mellado E, et al. 2007. A new Aspergillus fumigatus resistance mechanism conferring in vitro cross-resistance to azole antifungals involves a combination of cyp51A alterations. Antimicrob. Agents Chemother. 51:1897–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Morio F, et al. 2012. High prevalence of triazole resistance in Aspergillus fumigatus, especially mediated by TR/L98H, in a French cohort of patients with cystic fibrosis. J. Antimicrob. Chemother. 67:1870–1873 [DOI] [PubMed] [Google Scholar]
- 36. Mortensen KL, et al. 2011. Aspergillus species and other molds in respiratory samples from patients with cystic fibrosis: a laboratory-based study with focus on Aspergillus fumigatus azole resistance. J. Clin. Microbiol. 49:2243–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mortensen KL, et al. 2010. Environmental study of azole-resistant Aspergillus fumigatus and other aspergilli in Austria, Denmark, and Spain. Antimicrob. Agents Chemother. 54:4545–4549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mullins ED, et al. 2001. Agrobacterium-mediated transformation of Fusarium oxysporum: an efficient tool for insertional mutagenesis and gene transfer. Phytopathology 91:173–180 [DOI] [PubMed] [Google Scholar]
- 39. Nascimento AM, et al. 2003. Multiple resistance mechanisms among Aspergillus fumigatus mutants with high-level resistance to itraconazole. Antimicrob. Agents Chemother. 47:1719–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nierman WC, et al. 2005. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438:1151–1156 [DOI] [PubMed] [Google Scholar]
- 41. Orr-Weaver TL, Szostak JW, Rothstein RJ. 1981. Yeast transformation: a model system for the study of recombination. Proc. Natl. Acad. Sci. U. S. A. 78:6354–6358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Osmani SA, May GS, Morris NR. 1987. Regulation of the mRNA levels of nimA, a gene required for the G2-M transition in Aspergillus nidulans. J. Cell Biol. 104:1495–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pain A, et al. 2004. Insight into the genome of Aspergillus fumigatus: analysis of a 922 kb region encompassing the nitrate assimilation gene cluster. Fungal Genet. Biol. 41:443–453 [DOI] [PubMed] [Google Scholar]
- 44. Paul S, Schmidt JA, Moye-Rowley WS. 2011. Regulation of the CgPdr1 transcription factor from the pathogen Candida glabrata. Eukaryot. Cell 10:187–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Prasad R, Goffeau A. 2012. Yeast ATP-binding cassette transporters conferring multidrug resistance. Annu. Rev. Microbiol. [Epub ahead of print.] doi:10.1146/annurev-micro-092611-150111 [DOI] [PubMed] [Google Scholar]
- 46. Punt PJ, et al. 1990. Functional elements in the promoter region of the Aspergillus nidulans gpdA gene encoding glyceraldehyde-3-phosphate dehydrogenase. Gene 93:101–109 [DOI] [PubMed] [Google Scholar]
- 47. Rose MD, Misra LM, Vogel JP. 1989. KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell 57:1211–1221 [DOI] [PubMed] [Google Scholar]
- 48. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 49. Sikorski RS, Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Smith JM, Tang CM, Van Noorden S, Holden DW. 1994. Virulence of Aspergillus fumigatus double mutants lacking restriction and an alkaline protease in a low-dose model of invasive pulmonary aspergillosis. Infect. Immun. 62:5247–5254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Snelders E, Melchers WJ, Verweij PE. 2011. Azole resistance in Aspergillus fumigatus: a new challenge in the management of invasive aspergillosis? Future Microbiol. 6:335–347 [DOI] [PubMed] [Google Scholar]
- 52. Snelders E, et al. 2008. Emergence of azole resistance in Aspergillus fumigatus and spread of a single resistance mechanism. PLoS Med. 5:e219 doi:10.1371/journal.pmed.0050219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sonenberg N, Hinnebusch AG. 2009. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136:731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Srikantha T, et al. 1996. The sea pansy Renilla reniformis luciferase serves as a sensitive bioluminescent reporter for differential gene expression in Candida albicans. J. Bacteriol. 178:121–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Srikantha T, Zhao R, Daniels K, Radke J, Soll DR. 2005. Phenotypic switching in Candida glabrata accompanied by changes in expression of genes with deduced functions in copper detoxification and stress. Eukaryot. Cell 4:1434–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sugareva V, et al. 2006. Characterisation of the laccase-encoding gene abr2 of the dihydroxynaphthalene-like melanin gene cluster of Aspergillus fumigatus. Arch. Microbiol. 186:345–355 [DOI] [PubMed] [Google Scholar]
- 57. Sugui JA, Chang YC, Kwon-Chung KJ. 2005. Agrobacterium tumefaciens-mediated transformation of Aspergillus fumigatus: an efficient tool for insertional mutagenesis and targeted gene disruption. Appl. Environ. Microbiol. 71:1798–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang Z, Dietrich FS. 2005. Mapping of transcription start sites in Saccharomyces cerevisiae using 5′ SAGE. Nucleic Acids Res. 33:2838–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]