Abstract
The Hat1 histone acetyltransferase has been implicated in the acetylation of histone H4 during chromatin assembly. In this study, we have characterized the Hat1 complex from the fission yeast Schizosaccharomyces pombe and have examined its role in telomeric silencing. Hat1 is found associated with the RbAp46 homologue Mis16, an essential protein. The Hat1 complex acetylates lysines 5 and 12 of histone H4, the sites that are acetylated in newly synthesized H4 in a wide range of eukaryotes. Deletion of hat1 in S. pombe is itself sufficient to cause the loss of silencing at telomeres. This is in contrast to results obtained with an S. cerevisiae hat1Δ strain, which must also carry mutations of specific acetylatable lysines in the H3 tail domain for loss of telomeric silencing to occur. Notably, deletion of hat1 from S. pombe resulted in an increase of acetylation of histone H4 in subtelomeric chromatin, concomitant with derepression of this region. A similar loss of telomeric silencing was also observed after growing cells in the presence of the deacetylase inhibitor trichostatin A. However, deleting hat1 did not cause loss of silencing at centromeres or the silent mating type locus. These results point to a direct link between Hat1, H4 acetylation, and the establishment of repressed telomeric chromatin in fission yeast.
INTRODUCTION
During nucleosome assembly, newly synthesized H4 is acetylated prior to its deposition onto DNA (2, 40, 71, 83). In humans, Drosophila, and Tetrahymena, the acetylation of new H4 takes place in a conserved pattern, at lysines 5 and 12 (the sites are K4 and K11 in Tetrahymena, due to a deletion of the usual arginine residue at position 3) (24, 76). Deacetylation of new H4 occurs over the next 30 to 60 min (40, 73) and is required for proper chromatin maturation (6). The acetylation of new H4 may facilitate the import of H3/H4 dimers into the nucleus (5, 15, 21, 26, 31, 87). Moreover, recent studies using Physarum as a model system have provided evidence that the K5/K12 diacetylation of H4 is required for efficient nucleosome assembly in that system (26). It therefore seems likely that the rigorous conservation of the diacetylation of nascent H4 reflects an important role in the import/assembly process.
The most likely candidate for the enzyme that acetylates newly synthesized H4 is Hat1 (Kat1), a type B histone acetyltransferase (HAT) (7, 60). Hat1 acetylates free H4 at lysines 5 and 12 in vitro, consistent with the acetylation pattern of new H4 (22, 24, 46, 64, 68, 75, 82). In many organisms Hat1 is associated with p46 (termed Hat2p in Saccharomyces cerevisiae), which stimulates its enzymatic activity (59, 72, 82). Although it was long thought of as predominantly a cytoplasmic enzyme, it is now clear that Hat1 is also present in nuclei (1, 51, 63, 68, 72, 82). In S. cerevisiae, the nuclear Hat1 complex also contains the protein Hif1p (1, 63).
Several lines of evidence have indicated that Hat1 is involved in DNA damage repair (12, 14, 66) and can be recruited to the sites of DNA double-strand breaks (67). Moreover, in combination with mutations of the acetylatable lysines of histone H3, deletion of HAT1 causes loss of telomeric silencing in budding yeast (44, 63). However, deletion of HAT1 without mutating H3 has no effect on either silencing or DNA repair in that system (44, 66). This suggests that the acetylation of H3 and H4 can act redundantly during silencing and repair, possibly during chromatin assembly.
Although they are both unicellular organisms classified as fungi, S. cerevisiae and Schizosaccharomyces pombe are estimated to be separated by approximately one billion years of evolution (39). Significant differences between them include the manner of cell division (budding as opposed to fission), the structure of centromeres (in S. cerevisiae the kinetochore consists of a single nucleosome; in S. pombe, centromeres are much more mammalian-like and are up to 100 kb long), and the presence of introns in ∼45% of S. pombe genes (<5% of the genes in S. cerevisiae have introns [85]). In addition, the yeasts differ dramatically in silencing mechanisms. Unlike S. pombe, S. cerevisiae does not exhibit the methylation of histone H3 at K9 (or use RNA interference [RNAi] or an Swi6/HP1 homologue) for silencing at telomeres, centromeres, or the mating type loci. S. pombe uses all these methods at all these sites (19, 20). In light of these differences and of their evolutionary separation, it is not necessarily expected that budding and fission yeasts share a requirement for Hat1 to effect telomeric silencing.
In previous work we have shown that, in contrast to results obtained for S. cerevisiae, deletion of hat1 in the fission yeast S. pombe causes heightened sensitivity to DNA damage in the absence of concurrent mutations of histone H3 (14). To further explore possible differences in Hat1 function between these two evolutionarily distant fungal systems, we have purified and analyzed the Hat1 complex from fission yeast. We find that Hat1 is associated with Mis16 (an orthologue of RbAp46 and Hat2p), and we confirm that, unlike HAT2 in budding yeast, mis16 is an essential gene (38). As with most eukaryotes, the S. pombe Hat1 complex acetylates lysines 5 and 12 of histone H4. However, deletion of hat1 in and of itself causes loss of telomeric silencing in S. pombe, without concurrent mutations of the H3 N-terminal domain. Loss of Hat1 did not reduce silencing at centromeres or the mating type locus. Surprisingly, deletion of hat1 caused an increase in the acetylation of H4 at telomeres, rather than the reverse. Our results demonstrate that Hat1 is essential for the establishment of telomeric silencing and organization in fission yeast, and they suggest that the proper diacetylation of H4 during chromatin assembly is required to foster the generation of heterochromatin at telomeres in S. pombe.
MATERIALS AND METHODS
S. pombe strain construction.
S. pombe was cultured and maintained in YEA medium (37). The genotypes of the strains are listed in Table 1. The construction of KTP1 was performed as follows. The S. pombe Hat1 protein (SPAC139.06; Uniprot accession number Q9UTM7) was tandem affinity purification (TAP) tagged using a modified PCR-based method (9). PCR primers hat1-tagfor (5′-CTACCCAAGCTTAAGGAAGATTCGCCTCGAAAACGCCAAAAACTTGCTCAATCTTCTTCCCGGATCCCCGGGTTAATTAA-3′) and hat1-tagrev (5′-AAGCTTTCAAAAGCAAATTATATAAAAAGTAATTGCGTCCAATAGTGTAATTTAGTCGATGAATTCGAGCTCGTTTAAAC-3′) were used to amplify the TAP tag from plasmid pFA6A-CBP 4.5X protein A (TEV)-kanMX6. This amplicon, containing sequence homology to the COOH terminus of Hat1, was integrated into a wild-type strain (975) by transformation as previously described (43). Transformed cells were plated onto YEA and G418 (100 μg/ml) plates. Candidate cells were selected for their resistance to G418; integration was confirmed by PCR and DNA sequencing.
Table 1.
S. pombe strains
| Strain | Genotype | Source or reference |
|---|---|---|
| 975 | h+, wild type | 50 |
| 972 | h−, wild type | 50 |
| FY336 | h− ade6-210 leu1-32 ura4-DS/E TM1::ura4 | 3 |
| FY496 | h+ ade6-210 leu 1-32 ura4-DS/E imr1L(dg-glu) NcoI::ura4 oriL | 3 |
| FY648 | h+ ade6-210 leu 1-32 ura4-DS/E otr1R(dg-gluBamHI-SpeI fragment) SphI::ura4 | 3 |
| FY1872 | h90 ade6-210 leu1-32 ura4-DS/E otrRSph1::ade6 TEL2L-ura4 | 54 |
| mis16-myc | h− leu1 ura4 mis16-myc [ura4+] | 38 |
| FWP93 | h− ura4::fbp1-lacZ leu1-32 ade6-210 | C. Hoffman |
| LBP6 | h+ hat1D::kan | 14 |
| KTP7 | h−/h+ mis16+/mis16Δ::kan ade6-M216/ade6-M210 leu1+/leu1-32 | This study |
| KTP22 | h+ ura4 hat1-4xPACTAP | This study |
| KTP24 | h− hat1Δ::kan ade6-210 leu1-32 ura4::fbp1-lacZ | This study |
| KTP25 | h+ hat1Δ::kan ade6-210 leu1-32 ura4::fbp1-lacZ | This study |
| KTP29 | h− hat1Δ::kan ade6-210 leu1-32 ura4-DS/E imr 1L(dg-glu) NcoI::ura4 oriI | This study |
| KTP30 | h− hat1Δ::kan ade6-210 leu1-32 ura4-DS/E otr1R(dg-gluBamHI-SpeI fragment) sphI::ura4 | This study |
| KTP33 | h− hat1Δ::kan ade6-210 leu1-32 ura4-DS/E TM1::ura4 | This study |
| KTP35 | h+ hat1Δ::kan ade6-210 leu1-32 ura4-DS/E tRNA Phe-otr 1L(XhoI-BamHI fragment) HpaI::ura4 | This study |
| KTP36 | h90? hat1Δ::kan ade6-210 leu1-32 ura4-DS/E otr1 Rsph1::ade6 TEL2L-ura4 12C | This study |
| KTP40 | h+ hat1-4xPACTAP mis16-myc | This study |
| CHP1608 | h90 ura4-DS/E leu1-32 ade6-M210 kinte::ura4+ hatΔ::kan | This study |
| CHP1609 | h90 ura4-DS/E leu1-32 ade6-M210 kinte::ura4+ | This study |
A Hat1-TAP Mis16-myc-tagged strain (KTP40) was constructed by tetrad dissection of a cross between KTP1 and mis16-myc strains. Tetrad dissections on YEA plates were replica plated onto −ura, 0.04% 5-fluoroorotic acid (5-FOA), and G418 plates to select for ura+, G418-resistant cells.
The construction of an S. pombe diploid strain heterozygous for deletion of mis16 (SPCC1672.10, Uniprot accession number O94244) (KTP7) was performed as follows. PCR primers hat2delfor2 (5′-ATGTCAGAGGAAGTAGTCCAGGATGCACCTCTCGAGAATAATGAACTCAATGCCGAGATACGGATCCCCGGGTTAATTAA-3′) and hat2rev2 (5′-TGGTGTTATAGAAATGTAGTCTGATTTATAACAGTAGTTTTGATGTATTTACAAGGCGACGAATTCGAGCTCGTTTAAAC-3′) were used to amplify the kanMX6 selectable marker from plasmid pFA6a-3HA-kanMX6. The amplicon, which contains sequence homology to regions flanking mis16, was integrated into diploid strains by transformation. Deletion of mis16 was confirmed by colony PCR using primers hat2test2 (5′-TTCAGACTTAAGAGTGCGCTAG-3′) and hat2test (5′-TAGTACGGAGAGAGCCCTGG-3′). To determine if mis16 is essential, tetrad dissections of diploid transformants were performed on YEA plates, grown at 30°C, and replica plated to G418 (400 μg/ml) plates. Other yeast strains used in this study were constructed by mating and tetrad dissection.
Tandem affinity purification.
Tandem affinity purification of the Hat1 complex was performed using published protocols (65, 69).
Mass spectrometry.
Protein samples were subjected to electrophoresis through approximately 2 cm of a 10% SDS-polyacrylamide gel. Lanes were excised above the dye front and fixed for 30 min with a solution containing 50% methanol and 5% acetic acid. Gel fragments were washed with distilled water and analyzed using an LCQ Deca ion trap mass spectrometer (Tapalin Biological Mass Spectrometry Facility at Harvard University).
MMS assays.
The analysis of the sensitivities of wild-type and mutant S. pombe strains was performed as previously described (66). Five-fold serial dilutions were made and spotted on Edinburgh minimal medium (EMM) plates (57) containing 0.01% methyl methanesulfonate (MMS) (Sigma-Aldrich). Plates were incubated for 3 to 4 days at 30°C.
Gel electrophoresis and immunoblotting.
To separate the subunits of the Hat1 complex, purified extracts were subjected to SDS-PAGE in 10% to 12.5% polyacrylamide gels. Immunoblotting was performed according to the Western-Star system (Applied Biosystems). Anti-c-myc (sc-40; Santa Cruz Biotechnology) was diluted 1:5,000 in blocking buffer; secondary antibodies conjugated to alkaline phosphatase were diluted 1:5,000.
HAT assays.
In vitro histone acetyltransferase (HAT) filter binding assays using H4 peptides were performed as previously described (14). For the acetylation of H4 peptides and recombinant H4, Hat1p was affinity purified (tobacco etch virus [TEV] protease eluate) from KTP1. For 100-μl reaction mixtures, the following were combined: 100 mM sodium butyrate (pH 7.2) to a 5 mM concentration, 10-mg/ml acetylated bovine serum albumin (GE Healthcare) to a 1-mg/ml concentration, 0.5 μg recombinant H4 (14-697; Millipore), 1 mM unlabeled acetyl coenzyme A (acetyl-CoA) (Sigma-Aldrich) to a 10 μM concentration, and 79 μl of purified Hat1p. The reaction mixture components were mixed, incubated at 30°C for 1 h, and cooled on ice. Recombinant H4 was precipitated with 25% trichloroacetic acid (TCA) and washed with acidified acetone (0.05 N HCl) and then acetone. Protein pellets were vacuum dried, resuspended in sample buffer, and subjected to SDS-PAGE in a 13% polyacrylamide gel. The resolved proteins were transferred to an Immobilon-P transfer membrane and immunoblotted as described above. The acetylated (ac) antibodies used and their dilutions are as follows: anti-acH4 K5/K12 (13) was diluted 1:5,000, anti-acH4 K5 (07-290; Millipore) was diluted 1:600, anti-acH4 K12 (07-595; Millipore) was diluted 1:800, anti-acH4 K8 (07-378; Millipore) was diluted 1:2000, and anti-total H4 (05-858; Millipore) was diluted 1:30,000.
Silencing assays.
Telomeric (KTP36) and centromeric (KTP29, KTP33, and KTP35) marker strains along with the indicated controls were used to inoculate overnight cultures in YEL (37). Cultures were resuspended at 2 × 106 cells/ml. Five-fold serial dilutions of the suspensions were made and spotted onto one of the following: YEA, YEA containing 0.04% 5-FOA, EMMG, or EMMG-ura plates (57). Plates were incubated at 30°C for 2 to 3 days. To test the effect of histone hyperacetylation on telomeric silencing, 3-ml cultures were seeded with 6.2 × 105 cells treated with 50 μg/ml trichostatin A (TSA) (Wako) or methanol (vehicle) in YEL at 30°C (27). Cells were grown for ∼44 h (∼3 doublings in TSA; methanol-treated cells were reseeded if overgrown). Cells were then counted and resuspended at 2 × 106 cells/ml in the absence of TSA, and then 2.5- to 5-fold serial dilutions of the suspensions were made and 5 μl of each dilution was spotted on EMMG (minimal glutamate), EMMG-ura, and 5-FOA (1 g/liter) plates, all in the absence of TSA. Plates were incubated for 2 to 4 days at 30°C.
Chromatin immunoprecipitation (ChIP).
Cells were grown to 1 × 107 cells/ml in 50 ml of YEL. The culture was fixed with 3% paraformaldehyde for 30 min at 30°C with gentle shaking. Glycine was added to a 0.125 M concentration. After centrifugation for 5 min at 3,000 rpm, cells were washed three times with 1 ml ice-cold phosphate-buffered saline (PBS) containing 50 mM sodium butyrate. Pelleted cells were resuspended in 400 μl of ice-cold lysis buffer (50 mM HEPES-KOH [pH 7.5], 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% deoxycholate, 1 mM phenylmethylsulfonyl fluoride [PMSF], Complete tablet [EDTA free; Roche]) containing 23.5 μM depudecin, 10 μg/ml TSA, and 50 mM sodium butyrate. The cells were combined with 0.6 g of ice-cold glass beads and lysed three times for 5 min each in a mini-bead beater (Biospec Products). The base of the tube was punctured, placed on top of a 5-ml round-bottom glass tube, and centrifuged for 1 min at 1,500 rpm. Lysis buffer was added to bring the volume of the lysate up to 750 μl. The lysate was sonicated 12 times on ice to shear the chromatin to ∼600 bp.
Chromatin immunoprecipitations were performed with either anti-acH4 K12 (Millipore), anti-acH4 K5/12, or anti-acH4 K8/16 antibodies (13), using protein A-Sepharose beads. Rabbit IgG (Bethyl Laboratories) and rabbit nonimmune serum were used for control immunoprecipitations. Input fractions were removed from the lysate prior to immunoprecipitation, adjusted to 50 mM Tris (pH 8.0), 10 mM EDTA, and 1% SDS, incubated overnight at 65°C, cooled to room temperature, and then precipitated with ethanol and vacuum dried. The pellet was resuspended in 50 mM Tris (pH. 8.0)–1% SDS and treated with proteinase K for 2 h at 37°C. The input fraction was extracted twice with phenol-chloroform, precipitated, and resuspended in 20 μl Tris-EDTA (TE). Immunoprecipitated chromatin beads (bound fraction) were washed and DNA extracted as described above.
Real-time PCR.
To amplify immunoprecipitated ura4 DNA sequence, real-time PCR was performed using primers Ura4Chip-F (5′-CAAGGCCTCAAAGAAGTTGG-3′) and Ura4Chip-R (5′-GATGATATCGCTACCGCAG-3′). To amplify immunoprecipitated fus1 DNA sequence, real-time PCR was performed using primers fus1F (5′-AGAGCACAACCCCGTCC-3′) and fus1R (5′-TTTGCTATTGGTAGTACCGTAGCC-3′).
Analysis of real-time PCR data was performed according to published methods (58). Triplicate threshold cycle (CT) values for input and bound fractions were averaged from chromatin immunoprecipitations (ChIPs) using antiserum or nonimmune serum. Average net CT values for ura4 and fus1 primers were calculated by subtracting the average input CT from the average bound CT for the two primers. For each individual primer, the average net CT for FY1872 was subtracted from the average net CT of KTP36 to give the average net CT difference between the hat1Δ and wild-type strains. The negative of this value was used as an exponent for the base 1.9 to calculate the relative level of immunoprecipitated acetylated histones at the ura4 or fus1 DNA sequence. The calculations are summarized by the following equations: average net CT ura4 = average bound CT ura4 − average input CT, average net CT fus1 = average bound CT fus1 − average input CT, average net CT ura4 difference = average net KTP36 CT ura4 − average net FY1872 CT ura4, average net CT fus1 difference = average net KTP36 CT fus1 − average net FY1872 CT fus1, relative level of IP histones at ura4 in hat1Δ mutant over wild type = 1.9−average netCT ura4 difference, and relative level of IP histones at fus1 in hat1Δ mutant over WT = 1.9−average netCT fus1 difference.
F-tests were performed to determine if the variances from the ChIPs using antiserum were significantly different (<0.05) from those from the ChIPs using nonimmune serum. t tests (two-sample unequal variances or two-sample equal variances) were performed to determine if the difference between the ChIPs using antiserum was significantly different (<0.05) from that between the ChIPs using nonimmune serum.
RESULTS
S. pombe Hat1is associated with Mis16.
The native Hat1 complex was affinity purified by means of TAP tagging. We have previously shown that the loss of Hat1 in S. pombe causes sensitivity to DNA-damaging agents (14). To verify that TAP-tagged Hat1 remains functional, cells in which Hat1-TAP replaced native Hat1 were tested for normal MMS resistance (see Fig. S1 in the supplemental material).
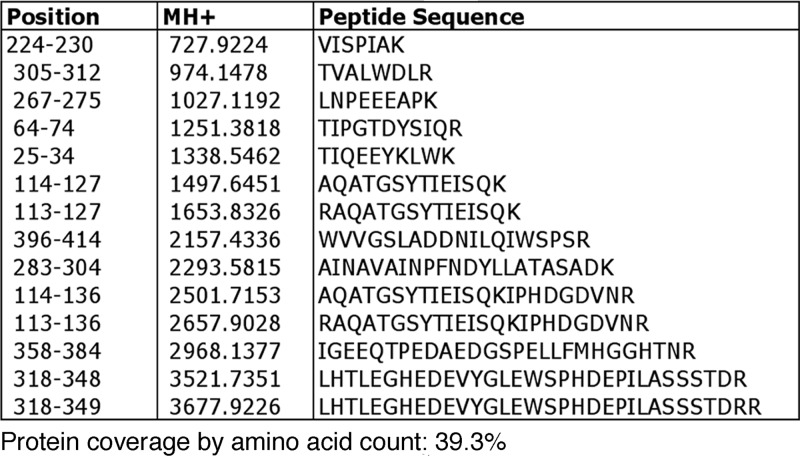
There are several genes in S. pombe that code for homologues of RbAp46/48, including mis16, prw1, and pcf3 (SPAC25H1.06) (25). Analysis of the affinity-purified Hat1 complex by mass spectrometry identified only one other major protein in addition to Hat1 itself: Mis16 (15 peptides, with 39% coverage) (Fig. 1). Mis16 is an RbAp46/48 orthologue that is involved in CENP-A (Cnp1) loading in fission yeast (38, 79). It is required for kinetochore function and histone deacetylation at S. pombe centromeres (38). The association of Hat1 with Mis16 was confirmed by isolating Hat1 from a Hat1-TAP yeast strain that also contained myc-tagged Mis16. Western blotting verified the presence of Mis16-myc in the purified Hat1 complex (see Fig. S2 in the supplemental material). As first demonstrated by Hayashi et al. (38), deletion analysis confirmed that mis16 is an essential gene (see Fig. S3 in the supplemental material). This is in contrast to the case for the HAT2 gene in S. cerevisiae (59, 70).
Fig 1.
Mis16 copurifies with Hat1 during tandem affinity purification. Hat1 was affinity purified from KTP1 (Hat1-TAP) S. pombe cells. Isolated proteins were subjected to SDS-PAGE and analyzed using a LCQ Deca ion trap mass spectrometer. Apart from typical S. pombe contaminants also observed using a non-TAP strain (GAPDH [glyceraldehyde-3-phosphate dehydrogenase] and translation elongation factor 1 alpha 1), the only protein found in addition to Hat1 was Mis16 (14 peptides detected). Mis16 is an S. pombe orthologue of p46/Hat2.
The Hat1 complex acetylates lysines 5 and 12 of histone H4.
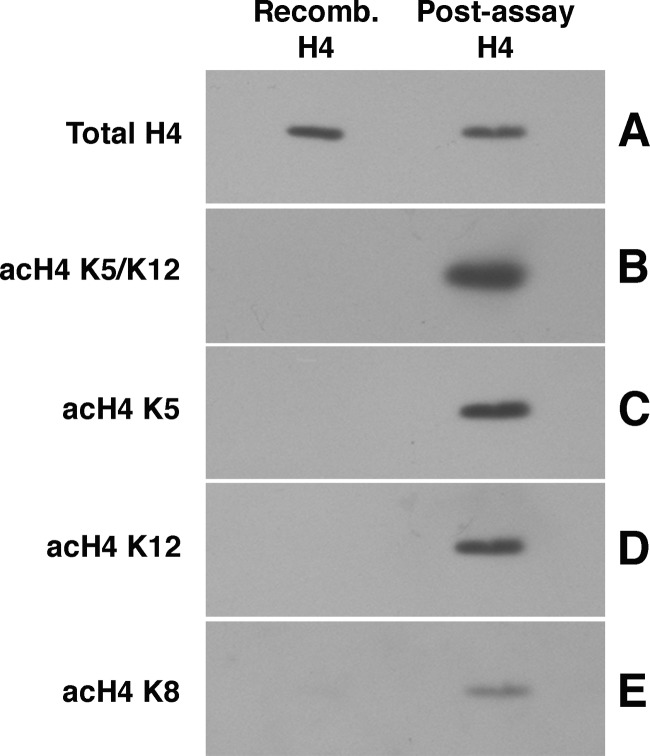
The acetylation activity of the Hat1 complex was then examined. As a first approach, in vitro HAT assays were performed using H4 N-terminal peptides as the substrates. As we have previously shown for Hat1 from budding yeast and human (HeLa) cells (14, 52), Hat1 from S. pombe was able to readily acetylate an unacetylated H4 tail peptide but not a peptide previously acetylated at lysines 5 and 12 (the predicted Hat1 substrate lysines) (Fig. 2). The acetylation of lysines 5 and 12 was then directly tested by means of HAT assays using recombinant H4 as a substrate. Western blotting confirmed that lysines 5 and 12 were robustly acetylated by S. pombe Hat1 (Fig. 3). A weak activity at lysine 8 was also detected; a similar weak activity at this site has also been observed for Hat1 from Drosophila embryos (75) (see Discussion).
Fig 2.
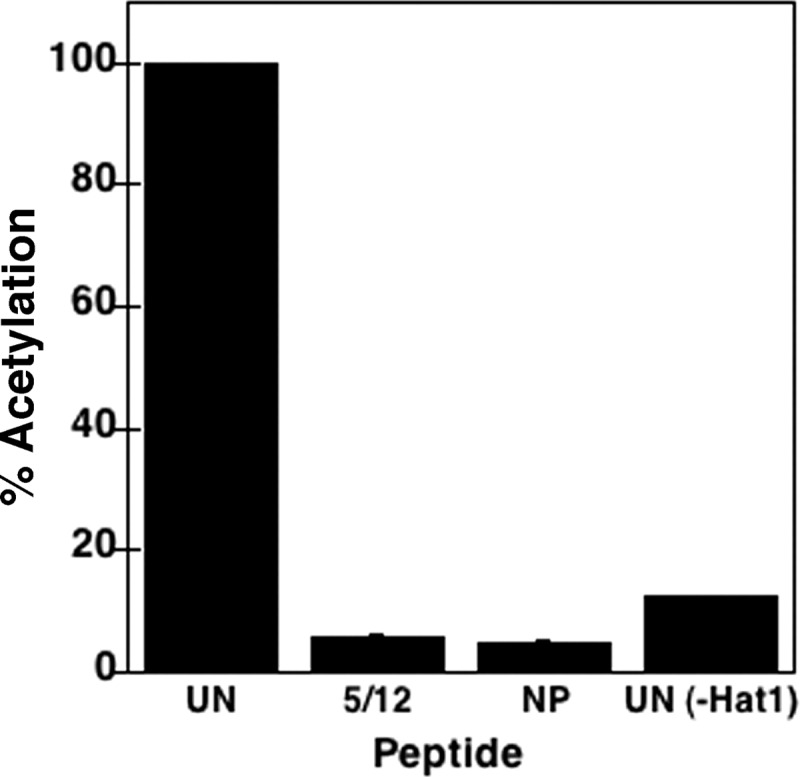
Acetylation of H4 N-terminal peptides by S. pombe Hat1 in vitro. Unacetylated (UN) and K5/K12-diacetylated (5/12) H4 N-terminal peptides were incubated in vitro for 30 min at 37°C with S. pombe Hat1 and [3H]acetyl-CoA. Reactions were also performed without added peptide (NP) and with the unacetylated peptide minus Hat1 (UN-Hat1). Results are expressed as a percentage of radioactivity incorporated into the unacetylated peptide.
Fig 3.
Acetylation of recombinant H4 by S. pombe Hat1. Recombinant H4 was incubated with Hat1 and unlabeled acetyl-CoA. Proteins from the reaction were resolved by electrophoresis and analyzed by Western blotting using antibodies that recognize total H4 (A) or H4 acetylated at K5 and/or K12 (B), K5 (C), K12 (D), or K8 (E). Acetylation at K16 was not detected above background (not shown).
Hat1 is essential for telomeric, but not all, silencing in fission yeast.
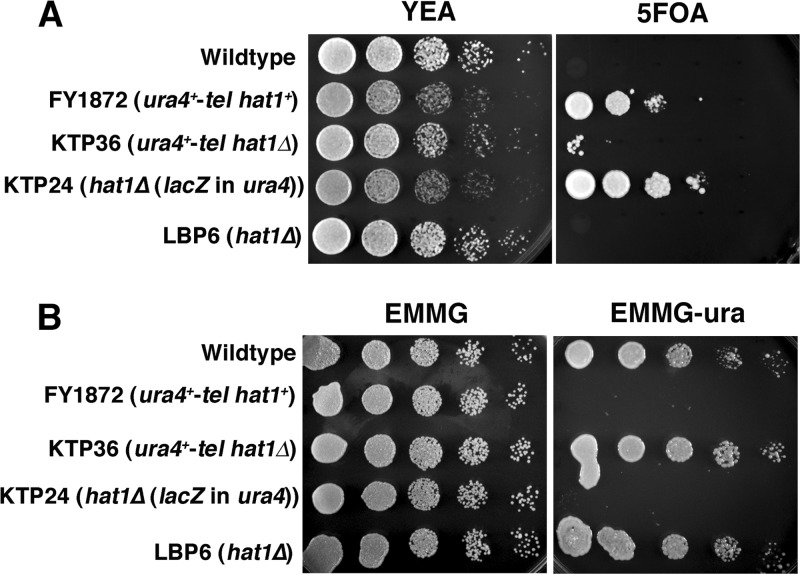
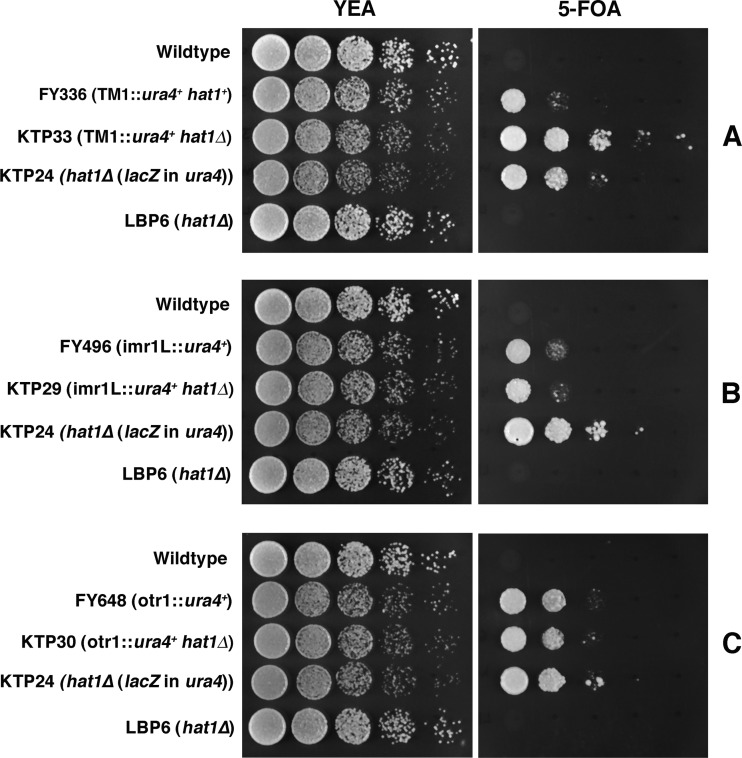
To study the role of Hat1 in telomeric silencing, a hat1Δ yeast strain that possessed as its sole copy of ura4 a gene inserted at a subtelomeric region was generated. When ura4 is silent, growth on the counterselective agent 5-FOA can occur. Conversely, expression of ura4 will cause cell death on 5-FOA but permit growth on medium lacking uracil. As expected, wild-type cells and the original hat1Δ strain LBP6 (both possessing an active ura4 gene in the normal euchromatic location) failed to grow on 5-FOA (Fig. 4A and B). However, strain FY1872 (hat1+, with a silent subtelomeric ura4 marker) did show growth on 5-FOA, as did hat1Δ cells in which ura4 had been disrupted by lacZ. Significantly, deletion of hat1 derepressed subtelomeric ura4, causing cell death on 5-FOA (Fig. 4A) while permitting growth on medium lacking uracil (Fig. 4B). Moreover, transforming cells with a hat1 expression vector restored telomeric silencing (see Fig. S4 in the supplemental material). In contrast, deleting hat1 did not cause the loss of silencing at any centromeric region tested (Fig. 5). In fact, a slight increase in silencing at the central core region was consistently observed (strain KTP33 in Fig. 5). Despite this increase in silencing, loss of Hat1 did not significantly alter the mitotic stability of minichromosome Ch16, as determined by the half-red colony sector assay of Allshire et al. (3) (data not presented). Moreover, deleting hat1 did not reduce silencing at the silent mating type locus (see Fig. S5 in the supplemental material), as judged by comparing 5-FOA resistance in hat1+ and hat1Δ strains bearing the kint2::ura4+ reporter (34).
Fig 4.
hat1 deletion results in the loss of telomeric silencing in S. pombe. (A) Wild-type and experimental yeast strains were cultured on YEA plates in the presence (5FOA) or absence (YEA) of 5-FOA. Spot cultures represent 5-fold dilutions. Cells were grown for two (YEA) or three (5-FOA) days at 30°C. FY1872, ura4-telomeric marker (tel); KTP36, ura4-tel hat1Δ; KTP24, ura4 disrupted, hat1Δ; LBP6, hat1Δ. (B) Wild-type and experimental yeast strains were cultured on EMMG plates in the presence (EMMG) or absence (EMMG−ura) of added uracil. Spot cultures represent 5-fold dilutions. Cells were grown for two (EMMG) or three (EMMG−ura) days at 30°C. FY1872, ura4-tel; KTP36, ura4-tel hat1Δ; KTP24, ura4 disrupted, hat1Δ; LBP6, hat1Δ.
Fig 5.
hat1 deletion does not result in the loss of centromeric silencing. Wild-type and experimental yeast strains were cultured on YEA plates in the presence (5FOA) or absence (YEA) of 5-FOA. Spot cultures represent 5-fold dilutions. Cells were grown for two (YEA) or three (5-FOA) days at 30°C. FY336, ura4-centromeric central core marker; KTP33, ura4-centromeric central core marker, hat1Δ; KTP24, ura4 disrupted, hat1Δ; LBP6, hat1Δ; FY496, ura4-centromeric inner most repeat marker; KTP29, ura4-centromeric inner most repeat marker, hat1Δ; FY648, ura4-centromeric outer repeat marker; KTP30, ura4-centromeric outer repeat marker, hat1Δ.
Deleting hat1 causes increased acetylation of subtelomeric chromatin.
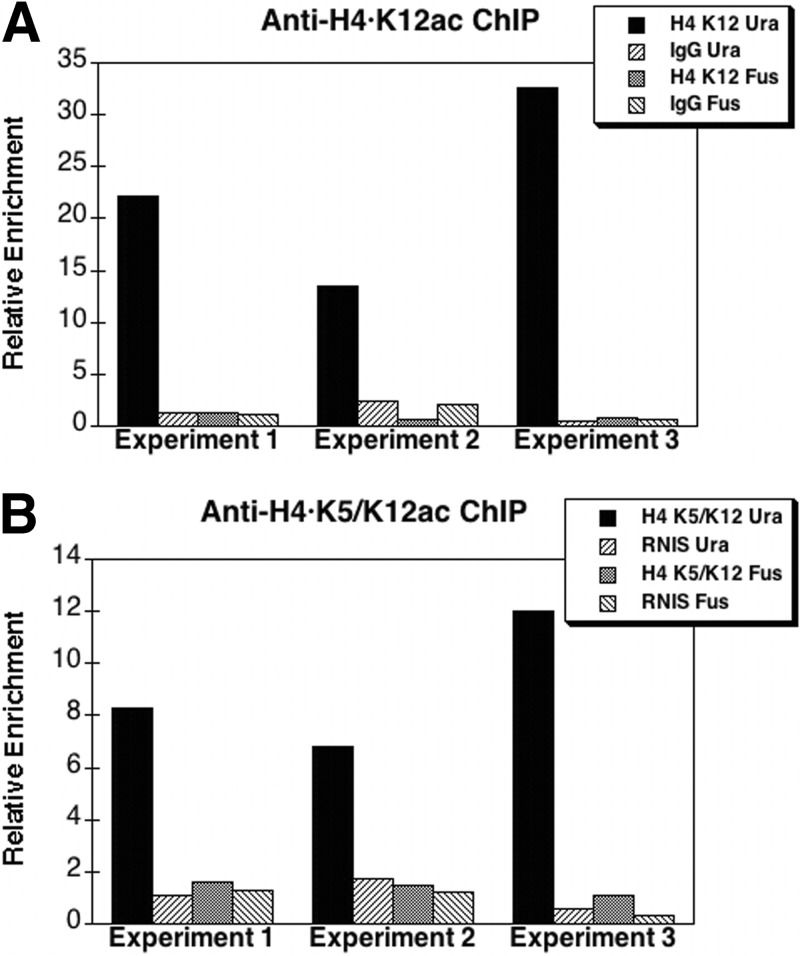
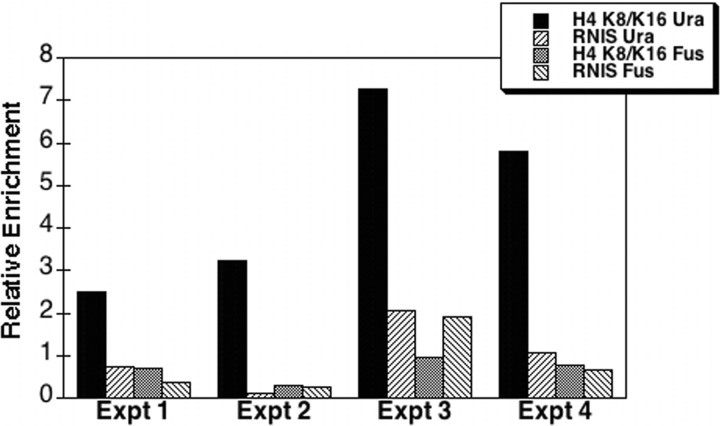
We then asked whether the loss of Hat1 affects histone acetylation at the subtelomeric ura4 locus. ChIP analyses were performed on hat1 wild-type and hat1Δ cells containing the subtelomeric ura4 marker, using antibodies specific for various acetylated states of histone H4. Quantitative real-time PCR was then carried out to measure the immunoprecipitation efficiency of ura4 in comparison to that of the fus1 gene (which is located more than 750 kb from the end of chromosome 1). Because fus1 is transcribed only during nitrogen starvation (62), it is highly useful as a baseline locus to monitor global changes in histone acetylation.
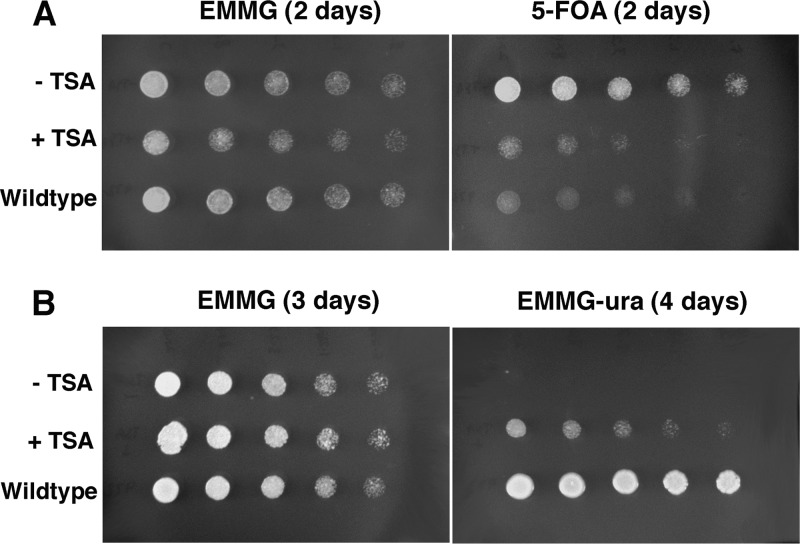
Deletion of hat1 significantly raised the degree of histone acetylation in subtelomeric chromatin at lysine 12 of H4; no change occurred at the fus1 locus (Fig. 6). Increased acetylation at the ura4 gene was also observed at lysines 8 and 16 in the H4 tail domain (Fig. 7). Thus, the loss of ura4 silencing, as measured by the inability to grow in the presence of 5-FOA, is accompanied by alterations of histone posttranslational modifications indicative of a more “active” chromatin configuration. To further test the relationship between histone acetylation and telomeric silencing, S. pombe cells possessing the telomeric marker but wild type for hat1 were grown for several generations in the presence of the deacetylase inhibitor trichostatin A (TSA). Cells were then washed and plated in the absence of TSA. Strikingly, TSA treatment caused a loss of telomeric silencing similar to that seen in hat1Δ cells, which persisted in the absence of the deacetylase inhibitor (Fig. 8). TSA has also been shown to cause derepression at centromeres (27) and the mating type loci (36) in S. pombe, further underscoring the antagonism between silencing and histone hyperacetylation. Our results now represent the first evidence obtained from any organism that Hat1 can independently regulate telomeric silencing, and they suggest an intimate link between the acetylation of new H4 and the proper assembly of heterochromatin.
Fig 6.
hat1 deletion results in an increase in H4 acetylation at the subtelomeric ura4 DNA sequence relative to that in the wild-type strain. Chromatin immunoprecipitation assays were performed on the FY1872 (ura4-telomeric marker) and KTP36 (ura4-telomeric marker, hat1Δ) strains using antibodies against H4 acetylated at K12 (H4-K12ac) (A) and H4 acetylated at K5 and/or K12 (H4-K5/K12ac) (B). Control immunoprecipitations were performed with rabbit IgG (A) or rabbit normal immune serum (RNIS) (B). Reverse transcription-PCRs (RT-PCRs) were performed using primers specific for ura4 and fus1. The average net CT difference between the hat1Δ and wild-type strains was used to calculate the change (enrichment) of immunoprecipitated acetylated histones at the ura4 or fus1 DNA sequence in the hat1Δ strain relative to the wild type.
Fig 7.
hat1 deletion results in an increase in H4-K8/K16 acetylation at the subtelomeric ura4 DNA sequence. Chromatin immunoprecipitation assays were performed on the FY1872 (ura4-telomeric marker) and KTP36 (ura4-telomeric marker, hat1Δ) strains using antibodies against H4 acetylated at K8 and/or K16. Results were analyzed as for Fig. 6.
Fig 8.
Treatment with trichostatin A results in the loss of telomeric silencing. Wild-type (972) and telomeric marker (FY1872) S. pombe strains were cultured in medium containing (+TSA) or lacking (−TSA) trichostatin A and allowed to grow for ∼3 generations at 30°C. Cells were washed and then grown at 30°C in the absence of TSA on EMMG (2 to 3 days), 5-FOA (2 days), and EMMG-ura (4 days). Spot cultures represent 2.5-fold serial dilutions.
DISCUSSION
Our results demonstrate that, as in other systems (51, 59, 82), the Hat1 acetyltransferase in fission yeast is associated with an orthologue of RbAp46/48, which in the case of S. pombe is Mis16. Mis16 can also form a separate complex with Mis18 to effect CENP-A loading at centromeres (38). Unlike HAT2 in S. cerevisiae, mis16 is an essential gene (this report and reference 38). Given that hat1Δ cells are viable, the lethality of mis16Δ is most likely due to the role of Mis16 in centromere assembly and chromosome segregation (38). RbAp46 and/or p48 is also involved in centromere assembly in human cells (38), as well as in Drosophila (where p48 participates in the deposition of the centromeric H3 variant CenH3 [30]).
The acetylation of lysines 5 and 12 by Hat1 in vitro has long been established (8, 14, 22, 24, 46; reviewed in references 61, 68, 75, and 82). More recent experiments in vertebrate (chicken) cells have indicated that Hat1 acetylates “cytosolic” histone H4 in vivo, in the conserved K5/K12 pattern (12). There is also evidence that Hat1 from S. cerevisiae acetylates both sites in vivo (64), in contrast to results obtained in vitro with the budding yeast holoenzyme, which acetylates only K12 (59). The Hat1 complex from S. pombe acetylated H4 at lysines 5 and 12 (the sites acetylated in newly synthesized H4). A low level of acetylation at lysine-8 (but not lysine-16) was also detected when recombinant H4 was used as a substrate. It is presently uncertain whether this reflects a true Hat1 activity in vivo. However, it is worth noting that H4 associated with native CAF-1 from human cells shows a low degree of acetylation at lysine-8, in addition to acetylation at K5 and K12 (81). Almost all of the acetyl-K8 was in the diacetylated H4 isoform (81), which may explain why the K5/K12-diacetylated peptide is not appreciably acetylated by Hat1 in vitro.
Deleting the gene coding for Hat1 has minimal effects on cell growth and/or chromatin assembly (12, 14, 45, 59, 70), although a slight increase in G1 cells has been observed (87). However, the loss of Hat1 has been linked to defects in DNA double-strand break repair (12, 14, 66, 67) and the abrogation of telomeric silencing in yeast (references 44 and 63 and this report). In S. cerevisiae, derepression at telomeres is observed only when the deletion of HAT1 is accompanied by mutations of specific acetylatable lysines in the H3 N-terminal domain (44, 63). In S. pombe, loss of Hat1 is itself sufficient to cause a dramatic decrease in telomeric silencing. Thus, the functional redundancy between H3 acetylation and Hat1 observed in S. cerevisiae is absent in S. pombe. The fact that deleting or mutating CAF-1 subunits reduces telomeric silencing in budding yeast (28, 41, 47) provides evidence that replication-coupled histone deposition is integral to the maintenance of silent chromatin. Our results now suggest that the proper diacetylation of new H4 is a critical element in this process. Notably, acetylation of H4 at lysine-12, a hallmark of Hat1 function, has been linked to gene silencing in yeast and Drosophila (17, 44, 74, 80).
In contrast to the effects on telomeric silencing, deletion of hat1 did not cause loss of silencing at the silent mating type locus or at any centromeric region tested. In fact, silencing in hat1Δ mutants became slightly more pronounced at the central core region. It is possible that the loss of Hat1 frees a greater proportion of Mis16 to associate with Mis18 (as we find no evidence for the association of Mis18 with Hat1), thereby facilitating CENP-A deposition at the central core region (23, 38, 78). Silencing at S. pombe centromeres involves several mechanisms, including histone H3 methylation, RNAi, and the deposition of Swi6 (HP1) (11, 18, 20, 35, 49, 56). Multiple elements also contribute to the assembly and maintenance of telomeres and adjacent subtelomeric regions (including telomerase, CAF-1, telomere-specific and heterochromatin proteins, RNAi, and the regulation of histone modifications [4, 16, 28, 29, 41, 47, 55, 77, 84]). Our results provide the first evidence that the steps required for silencing centromeric chromatin act independently of Hat1 and that herein lies a fundamental difference between the telomeric and centromeric silencing pathways in S. pombe.
In a recent report it was shown that in human cells Hat1 preferentially acetylates H4 in H3.1/H4 dimers (relative to H3.3/H4 dimers) and that Hat1 depletion affects the association of H3.1/H4 with importin 4 (87). However, in another study it was observed that Hat1 depletion in HeLa cells did not cause accumulation of H3/H4 dimers in the cytoplasm, indicating a redundancy in import processes (21). In this regard it is worth noting that in both budding and fission yeasts, the sole H3 subtype is equivalent to the replacement variant H3.3 (53), which, unlike H3.1, can be deposited through the HIRA pathway (33, 42, 86). Our own results establish that Hat1 deletion in S. pombe does not depress silencing at centromeres or the silent mating type locus or alter histone modifications at the fus1 gene, arguing against the global disruption of chromatin organization.
Loss of Hat1 caused a significant increase in the acetylation of subtelomeric chromatin at multiple acetylatable sites in the H4 N-terminal domain. Although perhaps counterintuitive, this is consistent with the loss of transcriptional silencing and with the encroachment of an active chromatin structure into the subtelomere. In line with this, telomeric silencing was also lost by pretreatment with the deacetylase inhibitor trichostatin A (Fig. 8). A similar loss of silencing was previously observed at S. pombe centromeres and the mating type loci after TSA treatment (27, 36). Other studies have shown that the MYST family histone acetyltransferase Mst2 helps to negatively regulate telomeric silencing in S. pombe (32) and that Esa1 (another MYST member and a component of the NuA4 complex) actively acetylates telomeric H4 in budding yeast (88).
It remains formally possible that the loss of Hat1 causes an increase in nucleosome density preferentially at telomeres (and not globally), thereby accounting for the specific rise in acetylation that we describe. However, this would be inconsistent with our observed increase in transcription of the subtelomeric ura4 marker, as gene activation in S. pombe correlates with decreased nucleosome occupancy, especially at promoter regions (10, 48). Moreover, HAT1 deletion has no effect on the nucleosome repeat length (i.e., histone density) of newly replicated chromatin in vertebrate cells (12). It is therefore not unreasonable to propose that the loss of Hat1 provides an opportunity for the anomalous hyperacetylation of subtelomeric chromatin, which interferes with the normal silencing pathway. In future studies it will be of interest to define the other elements involved in the derepression of telomeric chromatin and the increase in telomeric acetylation.
Supplementary Material
ACKNOWLEDGMENTS
We thank Robin Allshire and Hugh Cam for the gifts of S. pombe strains.
This work was supported by Public Health Service grants GM46226 to C.S.H. and GM35837 to A.T.A. from the National Institute of General Medical Sciences.
Footnotes
Published ahead of print 6 July 2012
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1. Ai X, Parthun MR. 2004. The nuclear Hat1p/Hat2p complex: a molecular link between type B histone acetyltransferases and chromatin assembly. Mol. Cell 14:195–205 [DOI] [PubMed] [Google Scholar]
- 2. Allis CD, Chicoine LG, Richman R, Schulman IG. 1985. Deposition-related histone acetylation in micronuclei of conjugating Tetrahymena. Proc. Natl. Acad. Sci. U. S. A. 82:8048–8052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Allshire RC, Nimmo ER, Ekwall K, Javerzat JP, Cranston G. 1995. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 9:218–233 [DOI] [PubMed] [Google Scholar]
- 4. Almeida R, Buscaino A, Allshire RC. 2006. Molecular biology: silencing unlimited. Curr. Biol. 16:R635–R638 [DOI] [PubMed] [Google Scholar]
- 5. Alvarez F, et al. 2011. Sequential establishment of marks on soluble histones H3 and H4. J. Biol. Chem. 286:17714–17721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Annunziato AT, Seale RL. 1983. Histone deacetylation is required for the maturation of newly replicated chromatin. J. Biol. Chem. 258:12675–12684 [PubMed] [Google Scholar]
- 7. Annunziato AT, Hansen JC. 2000. Role of histone acetylation in the assembly and modulation of chromatin structures. Gene Expr. 9:37–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Annunziato AT. 2012. Assembling chromatin: the long and winding road. Biochim. Biophys. Acta 1819:196–210 [DOI] [PubMed] [Google Scholar]
- 9. Bahler J, et al. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943–951 [DOI] [PubMed] [Google Scholar]
- 10. Bai L, Morozov AV. 2010. Gene regulation by nucleosome positioning. Trends Genet. 26:476–483 [DOI] [PubMed] [Google Scholar]
- 11. Bannister AJ, Kouzarides T. 2011. Regulation of chromatin by histone modifications. Cell Res. 21:381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Barman HK, et al. 2006. Histone acetyltransferase 1 is dispensable for replication-coupled chromatin assembly but contributes to recover DNA damages created following replication blockage in vertebrate cells. Biochem. Biophys. Res. Commun. 345:1547–1557 [DOI] [PubMed] [Google Scholar]
- 13. Benson LJ, et al. 2006. Modifications of H3 and H4 during chromatin replication, nucleosome assembly, and histone exchange. J. Biol. Chem. 281:9287–9296 [DOI] [PubMed] [Google Scholar]
- 14. Benson LJ, et al. 2007. Properties of the type B histone acetyltransferase Hat1-H4 tail interaction, site preference, and involvement in DNA repair. J. Biol. Chem. 282:836–842 [DOI] [PubMed] [Google Scholar]
- 15. Blackwell JS, Wilkinson ST, Mosammaparast N, Pemberton LF. 2007. Mutational analysis of H3 and H4N termini reveals distinct roles in nuclear import. J. Biol. Chem. 282:20142–20150 [DOI] [PubMed] [Google Scholar]
- 16. Blasco MA. 2007. The epigenetic regulation of mammalian telomeres. Nat. Rev. Genet. 8:299–309 [DOI] [PubMed] [Google Scholar]
- 17. Braunstein M, Sobel RE, Allis CD, Turner BM, Broach JR. 1996. Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol. Cell. Biol. 16:4349–4356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Burrack LS, Berman J. 2012. Flexibility of centromere and kinetochore structures. Trends Genet. 28:204–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cam HP, et al. 2005. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat. Genet. 37:809–819 [DOI] [PubMed] [Google Scholar]
- 20. Cam HP, Chen ES, Grewal SIS. 2009. Transcriptional scaffolds for heterochromatin assembly. Cell 136:610–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Campos EI, et al. 2010. The program for processing newly synthesized histones H3.1 and H4. Nat. Struct. Mol. Biol. 17:1343–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chang L, et al. 1997. Histones in transit: cytosolic histone complexes and diacetylation of H4 during nucleosome assembly in human cells. Biochemistry 36:469–480 [DOI] [PubMed] [Google Scholar]
- 23. Chen ES, Saitoh S, Yanagida M, Takahashi K. 2003. A cell cycle-regulated GATA factor promotes centromeric localization of CENP-A in fission yeast. Mol. Cell 11:175–187 [DOI] [PubMed] [Google Scholar]
- 24. Chicoine LG, Schulman IG, Richman R, Cook RG, Allis CD. 1986. Nonrandom utilization of acetylation sites in histones isolated from Tetrahymena: evidence for functionally distinct H4 acetylation sites. J. Biol. Chem. 261:1071–1076 [PubMed] [Google Scholar]
- 25. Dohke K, et al. 2008. Fission yeast chromatin assembly factor 1 assists in the replication-coupled maintenance of heterochromatin. Genes Cells 13:1027–1043 [DOI] [PubMed] [Google Scholar]
- 26. Ejlassi-Lassallette A, Mocquard E, Arnaud MC, Thiriet C. 2011. H4 replication-dependent diacetylation and Hat1 promote S-phase chromatin assembly in vivo. Mol. Biol. Cell 22:245–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ekwall K, Olsson T, Turner BM, Cranston G, Allshire RC. 1997. Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell 91:1021–1032 [DOI] [PubMed] [Google Scholar]
- 28. Enomoto S, Mccunezierath PD, Geraminejad M, Sanders MA, Berman J. 1997. RLF2, a subunit of yeast chromatin assembly factor-I, is required for telomeric chromatin function in vivo. Genes Dev. 11:358–370 [DOI] [PubMed] [Google Scholar]
- 29. Enomoto S, Berman J. 1998. Chromatin assembly factor I contributes to the maintenance, but not the re-establishment, of silencing at the yeast silent mating loci. Genes Devel. 12:219–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Furuyama T, Dalal Y, Henikoff S. 2006. Chaperone-mediated assembly of centromeric chromatin in vitro. Proc. Nat. Acad. Sci. U. S. A. 103:6172–6177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Glowczewski L, Waterborg JH, Berman JG. 2004. Yeast chromatin assembly complex 1 protein excludes nonacetylatable forms of histone H4 from chromatin and the nucleus. Mol. Cell. Biol. 24:10180–10192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gomez EB, Espinosa JM, Forsburg SL. 2005. Schizosaccharomyces pombe mst2(+) encodes a MYST family histone acetyltransferase that negatively regulates telomere silencing. Mol. Cell. Biol. 25:8887–8903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Greenall A, et al. 2006. Hip3 interacts with the HIRA proteins Hip1 and Slm9 and is required for transcriptional silencing and accurate chromosome segregation. J. Biol. Chem. 281:8732–8739 [DOI] [PubMed] [Google Scholar]
- 34. Grewal SI, Klar AJ. 1997. A recombinationally repressed region between mat2 and mat3 loci shares homology to centromeric repeats and regulates directionality of mating-type switching in fission yeast. Genetics 146:1221–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Grewal SI. 2010. RNAi-dependent formation of heterochromatin and its diverse functions. Curr. Opin. Genet. Dev. 20:134–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Grewal SIS, Bonaduce MJ, Klar AJS. 1998. Histone deacetylase homologs regulate epigenetic inheritance of transcriptional silencing and chromosome segregation in fission yeast. Genetics 150:563–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gutz H, Heslot H, Leupold U, Loprieno N. 1974. Schizosaccharomyces pombe, p 395–446. In King RC. (ed), Handbook of genetics. Plenum Press, New York, NY [Google Scholar]
- 38. Hayashi T, et al. 2004. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell 118:715–729 [DOI] [PubMed] [Google Scholar]
- 39. Heckman DS, et al. 2001. Molecular evidence for the early colonization of land by fungi and plants. Science 293:1129–1133 [DOI] [PubMed] [Google Scholar]
- 40. Jackson V, Shires A, Tanphaichitr N, Chalkley R. 1976. Modifications to histones immediately after synthesis. J. Mol. Biol. 104:471–483 [DOI] [PubMed] [Google Scholar]
- 41. Kaufman PD, Kobayashi R, Stillman B. 1997. Ultraviolet radiation sensitivity and reduction of telomeric silencing Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev. 11:345–357 [DOI] [PubMed] [Google Scholar]
- 42. Kaufman PD, Cohen JL, Osley MA. 1998. Hir proteins are required for position-dependent gene silencing in Saccharomyces cerevisiae in the absence of chromatin assembly factor I. Mol. Cell. Biol. 18:4793–4806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Keeney JB, Boeke JD. 1994. Efficient targeted integration at leu1-32 and ura4-294 in Schizosaccharomyces pombe. Genetics 136:849–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kelly TJ, Qin S, Gottschling DE, Parthun MR. 2000. Type B histone acetyltransferase Hat1p participates in telomeric silencing. Mol. Cell. Biol. 20:7051–7058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kleff S, Andrulis ED, Anderson CW, Sternglanz R. 1995. Identification of a gene encoding a yeast histone H4 acetyltransferase. J. Biol. Chem. 270:24674–24677 [DOI] [PubMed] [Google Scholar]
- 46. Kolle D, Sarg B, Lindner H, Loidl P. 1998. Substrate and sequential site specificity of cytoplasmic histone acetyltransferases of maize and rat liver. FEBS Lett. 421:109–114 [DOI] [PubMed] [Google Scholar]
- 47. Krawitz DC, Kama T, Kaufman PD. 2002. Chromatin assembly factor I mutants defective for PCNA binding require Asf1/Hir proteins for silencing. Mol. Cell. Biol. 22:614–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lantermann AB, et al. 2010. Schizosaccharomyces pombe genome-wide nucleosome mapping reveals positioning mechanisms distinct from those of Saccharomyces cerevisiae. Nat. Struct. Mol. Biol. 17:251–257 [DOI] [PubMed] [Google Scholar]
- 49. Lejeune E, Bayne EH, Allshire RC. 2010. On the connection between RNAi and heterochromatin at centromeres. Cold Spring Harbor Symp. Quant. Biol. 75:275–283 [DOI] [PubMed] [Google Scholar]
- 50. Leupold U. 1950. Die Vererbung von Homothallie und Heterothallie bei Schizosaccharomyces pombe. C. R. Trav. Lab. Carlsberg Ser. Physiol. 24:381–480 [Google Scholar]
- 51. Lusser A, et al. 1999. Analysis of the histone acetyltransferase B complex of maize embryos. Nucleic Acids Res. 27:4427–4435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Makowski AM, Dutnall RN, Annunziato AT. 2001. Effects of acetylation of histone H4 at lysines 8 and 16 on activity of the Hat1 histone acetyltransferase. J. Biol. Chem. 276:43499–43502 [DOI] [PubMed] [Google Scholar]
- 53. Malik HS, Henikoff S. 2003. Phylogenomics of the nucleosome. Nat. Struct. Biol. 10:882–891 [DOI] [PubMed] [Google Scholar]
- 54. Manolis KG, et al. 2001. Novel functional requirements for non-homologous DNA end joining in Schizosaccharomyces pombe. EMBO J. 20:210–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Moazed D. 2009. Small RNAs in transcriptional gene silencing and genome defence. Nature 457:413–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Moazed D. 2011. Mechanisms for the inheritance of chromatin states. Cell 146:510–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Moreno S, Klar A, Nurse P. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795–823 [DOI] [PubMed] [Google Scholar]
- 58. Mukhopadhyay A, Deplancke B, Walhout AJ, Tissenbaum HA. 2008. Chromatin immunoprecipitation (ChIP) coupled to detection by quantitative real-time PCR to study transcription factor binding to DNA in Caenorhabditis elegans. Nat. Protoc. 3:698–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Parthun MR, Widom J, Gottschling DE. 1996. The major cytoplasmic histone acetyltransferase in yeast: links to chromatin replication and histone metabolism. Cell 87:85–94 [DOI] [PubMed] [Google Scholar]
- 60. Parthun MR. 2007. Hat1: the emerging cellular roles of a type B histone acetyltransferase. Oncogene 26:5319–5328 [DOI] [PubMed] [Google Scholar]
- 61. Parthun MR. 2012. Histone acetyltransferase 1: more than just an enzyme? Biochim. Biophys. Acta 1819:256–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Petersen J, Weilguny D, Egel R, Nielsen O. 1995. Characterization of fus1 of Schizosaccharomyces pombe: a developmentally controlled function needed for conjugation. Mol. Cell. Biol. 15:3697–36707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Poveda A, et al. 2004. Hif1 is a component of yeast histone acetyltransferase B, a complex mainly localized in the nucleus. J. Biol. Chem. 279:16033–16043 [DOI] [PubMed] [Google Scholar]
- 64. Poveda A, Sendra R. 2008. Site specificity of yeast histone acetyltransferase B complex in vivo. FEBS J. 275:2122–2136 [DOI] [PubMed] [Google Scholar]
- 65. Puig O, et al. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24:218–229 [DOI] [PubMed] [Google Scholar]
- 66. Qin S, Parthun MR. 2002. Histone H3 and the histone acetyltransferase Hat1p contribute to DNA double-strand break repair. Mol. Cell. Biol. 22:8353–8365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Qin S, Parthun MR. 2006. Recruitment of the type B histone acetyltransferase Hat1p to chromatin is linked to DNA double-strand breaks. Mol. Cell. Biol. 26:3649–3658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Richman R, Chicoine LG, Collini MP, Cook RG, Allis CD. 1988. Micronuclei and the cytoplasm of growing Tetrahymena contain a histone acetylase activity which is highly specific for free histone H4. J. Cell Biol. 106:1017–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rigaut G, et al. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030–1032 [DOI] [PubMed] [Google Scholar]
- 70. Rosaleny LE, Antunez O, Ruiz-Garcia AB, Perez-Ortin JE, Tordera V. 2005. Yeast HAT1 and HAT2 deletions have different life-span and transcriptome phenotypes. FEBS Lett. 579:4063–4068 [DOI] [PubMed] [Google Scholar]
- 71. Ruiz-Carrillo A, Wangh LJ, Allfrey VG. 1975. Processing of newly synthesized histone molecules. Science 190:117–128 [DOI] [PubMed] [Google Scholar]
- 72. Ruiz-Garcia AB, et al. 1998. HAT1 and HAT2 proteins are components of a yeast nuclear histone acetyltransferase enzyme specific for free histone H4. J. Biol. Chem. 273:12599–12605 [DOI] [PubMed] [Google Scholar]
- 73. Sirbu BM, et al. 2011. Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev. 25:1320–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Smith CM, et al. 2002. Heritable chromatin structure: mapping ‘‘memory'' in histones H3 and H4. Proc. Natl. Acad. Sci. U. S. A. 99:16454–16461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sobel RE, Cook RG, Allis CD. 1994. Non-random acetylation of histone H4 by a cytoplasmic histone acetyltransferase as determined by novel methodology. J. Biol. Chem. 269:18576–18582 [PubMed] [Google Scholar]
- 76. Sobel RE, Cook RG, Perry CA, Annunziato AT, Allis CD. 1995. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc. Natl. Acad. Sci. U. S. A. 92:1237–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Sugiyama T, et al. 2007. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell 128:491–504 [DOI] [PubMed] [Google Scholar]
- 78. Takahashi K, Chen ES, Yanagida M. 2000. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science 288:2215–2219 [DOI] [PubMed] [Google Scholar]
- 79. Takahashi K, Takayama Y, Masuda F, Kobayashi Y, Saitoh S. 2005. Two distinct pathways responsible for the loading of CENP-A to centromeres in the fission yeast cell cycle. Philos. Trans. R. Soc. B Biol. Sci. 360:595–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Turner BM, Birley AJ, Lavender J. 1992. Histone H4 isoforms acetylated at specific lysine residues define individual chromosomes and chromatin domains in Drosophila polytene nuclei. Cell 69:375–384 [DOI] [PubMed] [Google Scholar]
- 81. Verreault A, Kaufman PD, Kobayashi R, Stillman B. 1996. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell 87:95–104 [DOI] [PubMed] [Google Scholar]
- 82. Verreault A, Kaufman PD, Kobayashi R, Stillman B. 1998. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr. Biol. 8:96–108 [DOI] [PubMed] [Google Scholar]
- 83. Waterborg JH, Matthews JH. 1984. Patterns of histone acetylation in Physarum polycephalum: H2A and H2B acetylation is functionally distinct from H3 and H4 acetylation. Eur. J. Biochem. 142:329–335 [DOI] [PubMed] [Google Scholar]
- 84. White SA, Allshire RC. 2008. RNAi-mediated chromatin silencing in fission yeast. Curr. Top. Microbiol. Immunol. 320:157–183 [DOI] [PubMed] [Google Scholar]
- 85. Wood V, et al. 2002. The genome sequence of Schizosaccharomyces pombe. Nature 415:871–880 [DOI] [PubMed] [Google Scholar]
- 86. Yamane K, et al. 2011. Asf1/HIRA facilitate global histone deacetylation and associate with HP1 to promote nucleosome occupancy at heterochromatic loci. Mol. Cell 41:56–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Zhang H, Han J, Kang B, Burgess R, Zhang Z. 2012. Human histone acetyltransferase HAT1 preferentially acetylates H4 molecules in H3.1-H4 dimers over H3.3-H4 dimers. J. Biol. Chem. 287:6573–6581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Zhou BO, et al. 2011. Histone H4 lysine 12 acetylation regulates telomeric heterochromatin plasticity in Saccharomyces cerevisiae. PLoS Genet. 7:e1001272 doi:10.1371/journal.pgen.1001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.