Abstract
Objective
Acute respiratory infection (ARI) is the major cause of morbidity and mortality in children worldwide. Human respiratory syncytial virus (HRSV) is main viral agent of ARI in infants and young children in terms of effect and prevalence. The aim of this study was to investigate HRSV genotypes during one season in Iran.
Methods
In this cross-sectional study, 107 throat swabs were collected from children less than 5 years of age with acute respiratory infection from October to December 2009. The respiratory samples were obtained from several provinces: Tehran, Isfahan, Hamadan, Zanjan, Kordestan, Lorestan and West Azarbayjan, and were tested for G protein gene of HRSV by RT-PCR.
Findings
Of the 107 respiratory samples, 24 (22.42%) were positive for HRSV, of which 16 (66.6%) belonged to subgroup A and 8 (33.4%) to subgroup B. Phylogenetic analysis revealed that subgroup A strains fell in two genotypes GA1 and GA2, whereas subgroup B strains clustered in genotype BA.
Conclusion
This study revealed that multiple genotypes of HRSV cocirculated during the season 2009 in Iran. Also subgroup A strains were more prevalent than subgroup B strains, and genotype GA1 was predominant during the season.
Keywords: Respiratory Syncytial Virus, Seasonal Infection, Respiratory Tract Infections, Genotype, Iran
Introduction
Human respiratory syncytial virus (HRSV) is the most important infectious agent of serious acute respiratory illnesses in infants and young children worldwide[1, 2]. HRSV causes brochiolitis and pneumonia during the first few months of life in the late fall and winter[1]. It is responsible for 18000–75000 hospitalizations and 90–1900 deaths in the United States annually[3], whereas HRSV is responsible for 27–96% of hospitalized cases caused by viral infections in developing countries[4]. All infants under 2 years of age have experienced at least one HRSV infection. HRSV is also recognized as an important respiratory pathogen in individuals with cardiopulmonary disease, immunocompromised patients and the elderly[5].
HRSV, a member of the genus Pneumovirus, family Paramixoviridae, is an enveloped virus with nonsegmented negative-strand RNA genome[6]. At least 11 proteins are encoded by the HRSV genome. The attachment G glycoprotein and F protein are important HRSV antigens that stimulate the protective response[7].
HRSV strains have been divided into two subgroups A and B on the basis of the reactions with monoclonal antibodies directed against the G protein[8]. Sequencing studies have revealed that the G protein is the most variable HRSV protein both between and within the two subgroups[9]. The G protein gene consists of two variable regions in ectodomain separated by a central, conserved motif[1]. Most frequently, HRSV genotype classification makes use of the second variable region in the G protein gene. Also most studies of molecular epidemiology of HRSV are based on the nucleotide sequencing of this region in the G protein[9].
The aim of the present study was to investigate HRSV genotypes in Iran by RT-PCR for second variable region of the G protein, because understanding of HRSV genotypes distribution patterns will be beneficial for the development of effective vaccines or antiviral therapy.
Subjects and Methods
In this cross-sectional study, 107 throat swabs were collected from children aged less than 5 years suffering from acute respiratory symptoms, such as wheezing, cough, and fever from October to December 2009.
The respiratory samples were obtained from hospital and sentinel sites from several provinces: Tehran, Isfahan, Hamadan, Zanjan, Kordestan, Lorestan, and Wet Azarbayjan by Ministry of Health and Medical Education (MOHME). Detailed clinical histories were collected on admission and in all cases parents or guardians signed an informed consent. All respiratory specimens were transported to the Department of Virology, Tehran University of Medical Sciences, and were stored at −80°C until further examination.
This study was approved by the medical faculty ethics committee of Tehran University of Medical Sciences.
RNA extraction and cDNA synthesis
The viral RNA to be used as a template for cDNA synthesis was extracted directly from throat swabs by using the high pure nucleic extraction kit (Roche Diagnostic, Manheim, Germany). The extracted viral RNA was dissolved in 50 µL of elution buffer. For cDNA synthesis, 17.5 µL of extracted RNA added to the reaction mixture, consisting of 6 µL RT buffer, 2.5 µL dNTP, 2.5 µL random hexamer primers, 1 µL RT enzyme of Moloney murine leukemia virus, and 0.5 µL RNase inhibitor in a total volume of 30 µL, and incubated at 37°C for 45 min.
PCR primer sequences
For the external PCR, GPA (nt511-530, 5'- GAAGTGTTCAACTTTGTACC-3') for subgroup A and GPB (nt 494-515, 5'-AAGATGATTACCATTTTG AAGT-3') for subgroup B were used as forward primers[10]. Hemi-nested PCR was carried out with subgroup A-specific forward primer, nRSAG (nt 539-558, 5'- TATGCAGCAACAATCCAACC-3'), and subgroup B-specific forward primer, nRSBG (nt 512-531, 5'-GTGGCAACAATCAACTCTGC-3')[11]. In the external and hemi-nested PCR, primer F1 (nt 3-22, 5'-CAACTCCATTGTTATTTGCC-3') was used as reverse primer for both subgroups A and B[11].
PCR
The external PCR for the amplification of the G protein was performed by adding 10 µL of the synthesized cDNA to 40 µL of the reaction mixture containing 23 µL distilled water, 5 µL 10X PCR buffer, 4 µL dNTP, 2.5 µL MgCl2, 2.5 µL forward primer, 2.5 µL reverse primer, and 0.5 µL of Taq DNA polymerase.
Amplification conditions consisted of 2 min at 95°C, 30 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 2 min, and a final extension at 72°C for 7 min[11]. Five µL of external PCR product was used for hemi-nested PCR with the same conditions. All PCR were run with positive and negative controls. The external and hemi-nested PCR amplicons were 450 and 400 bp, respectively. Both subgroups A and B were analyzed by electrophoresis on a 1.5% agarose gel and visualized under UV light.
DNA sequencing
The hemi-nested primers, nRSAG for subgroup A and nRSBG for subgroup B, were used as forward primers and F1 was used as the reverse primer for sequence determination. The purified PCR products were sequenced in the forward and the reverse directions in an ABI PRISM 310 genetic analyzer (PE Applied BioSystems Inc., Foster City, CA) by using an ABI PRISM BigDye Terminator cycle sequencing ready reaction kit (PE Applied BioSystems Inc).
Phylogenetic analysis
The nucleotide sequences obtained from second variable region of the G protein gene were aligned with HRSV sequences from GenBank database by using the CLUSTAL X program (version 1.83). Phylogenetic analysis was carried out by the neighbor-joining method in TREECON software. Pairwise distances between and within the genotypes were calculated with Kimura 2 parameters. All HRSV sequences were submitted to the GenBank database; the accession numbers are HM063447-HM063470.
Findings
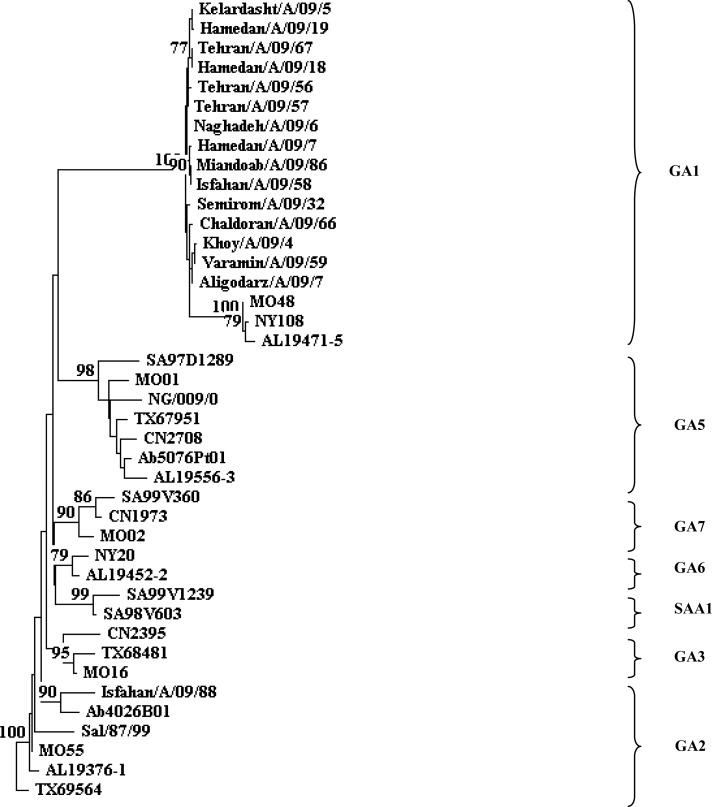
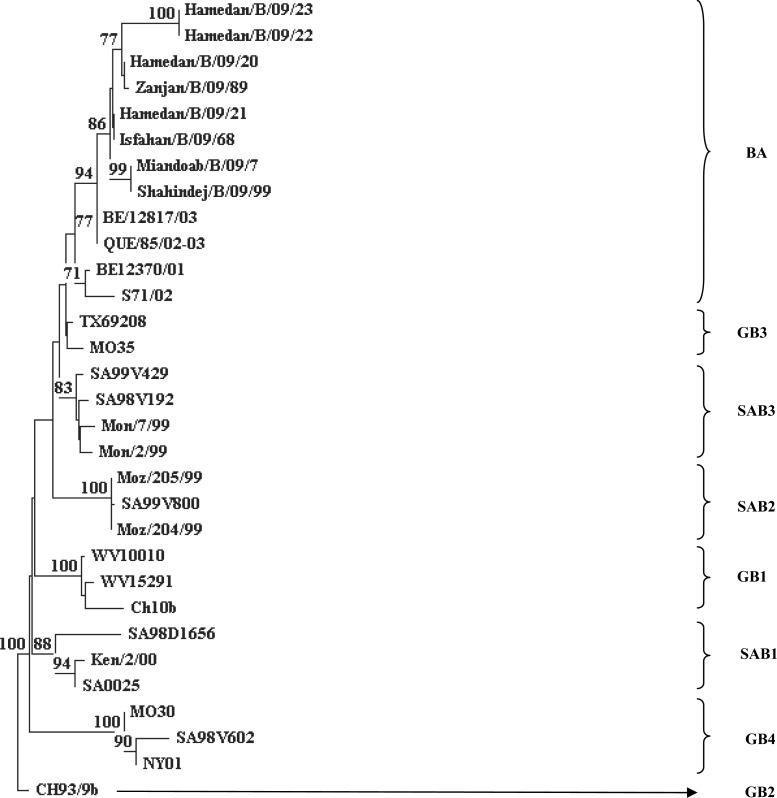
A total number of 107 respiratory specimens from children with acute respiratory infection were examined. Of these 24 (22.24%) were positive for HRSV, of which 16 (66.6%) belonged to subgroup A and 8 (33.4%) to subgroup B. Phylogenetic analysis revealed that subgroup A strains fell in two clusters, 15 strains in genotype GA1, and 1 strain in genotype GA2 (Fig. 1). All subgroup B strains clustered in a genotype BA with a 60-nucleotide insertion in the second variable region of the G protein (Fig. 2).
Fig. 1.
Phylogenetic tree constructed with the HRSV subgroup A nucleotide sequences of based on the second variable region of the G gene. The nucleotide sequences were aligned with the CLUSTAL X (version 1/83), and phylogenetic tree were prepared with TREECON software with a neighbor-joining algorithm. Pairwise distances between and within the genotypes were calculated with Kimura 2 parameters. Iranian isolates indicated by city/subtype/year/sample number. The following reference sequences were used to construct tree: MO48, AL19471-5, and NY108 (GA1); Ab4026B01, TX69564, MO55, and Sal/87/99 (GA2); MO16,CN2395, and TX68481 (GA3); SA97D1289, MO01, CN2708, TX67951, AL19556-3, Sal/173/99, Ab5076Pt01, and NG/009/02 (GA5); AL19452-2 and NY20 (GA6); MO02, SA99V360, and CN1973 (GA7); SA98V603and SA99V1239 (SAA1).
Fig. 2.
Phylogenetic tree constructed with the HRSV subgroup B nucleotide sequences of based on the second variable region of the G gene. The nucleotide sequences were aligned with the CLUSTAL X (version 1/83), and phylogenetic tree were prepared with TREECON software with a neighbor-joining algorithm. Pairwise distances between and within the genotypes were calculated with Kimura 2 parameters. Iranian isolates indicated by city/subtype/year/sample number. The following reference sequences were used to construct tree: WV10010, WV15291, and CH10b (GB1); CH93-9b (GB2); MO35, and TX69208(GB3);MO30, NY01, and SA98V602 (GB4); SA98D1656, SA0025, and Ken/2/00 (SAB1); SA99V800, Moz/204/99, and Moz/205/99 (SAB2); SA99V429, SA98V192, Mon/7/99, and Mon/2/99 (SAB3): S71/02, QUE/85/02-03, BE/12370/01, and BE/12817/03 (BA).
Genotype GA1 was detected in 15 (62.5%) HRSV-positive samples, so GA1 was the predominant genotype during the season 2009. In the present study, 21 (87%) positive samples were obtained from children under one year of age.
Discussion
In developing countries one-third of all deaths in young children are due to acute respiratory infections, with HRSV responsible for 27–96% of hospitalized cases caused by a viral infection[4].
Therefore, genetic diversity of HRSV has been studied in many parts of the world including South Africa[12], Belgium[13], India[14], Argentina[15], and Jordan[16].
The studies on genetic variability of HRSV revealed that subgroup A strains should be classified into 8 genotypes (GA1-7,SAA1) and subgroup B strains into 8 genotypes (GB1-4,SAB1-3,BA)[17]. Infectivity of the virus, the development of immunological resistance in the community, and viral genetic drift due to spontaneous mutation may be important in the patterns of seasonal circulation and genetic evolution of HRSV strains[7].
Although several studies on prevalence of HRSV were carried out in Iran[18, 19], the present study has determined HRSV genotypes by sequencing of the second variable region of the G protein during the season 2009. In this study 107 respiratory samples were examined, of which 24 (22.24%) were positive for HRSV. This finding is similar to that found in studies in both developing and industrialized countries: 25.46% in Jordan[20], 18.4% in Malaysia[21], 16/2% in Germany[22], 28% in Brazil[23], 21% in Austria[24] and 27.08% in India[25].
16 (66.6%) HRSV-positive samples belonged to subgroup A and 8 (33.4%) to subgroup B. Therefore, our results revealed that subgroup A strains were the most common finding and subgroup B strains infections appear to have been less frequent during the season 2009. This may be caused by several factors. It has been speculated that strains of subgroup B may cause less severe disease, so may be recognized less often. Another possibility is that infections with B strains may produce longer-lasting subgroup-specific immunity. Also infants infected with viruses of subgroup B strains induce a greater antibody to the glycoprotein G than do their counterparts infected with A strains[26].
Phylogenetic analysis revealed that subgroup A strains fell into two clusters, namely 15 strains in genotype GA1, and 1 strain in genotype GA2. GA1 was predominant genotype during the season 2009, whereas this genotype was obtained In several cities (Isfahan, Hamedan, Tehran, Zanjan, Khoy, Aligodarz, Sanandaj, and Miandoab). We speculate that GA1 was the predominant genotype of HRSV during 2009 in Iran.
All HRSV-positive samples of subgroup B clustered in genotype BA. For the first time, new genotype BA was reported from Buenos Aires (Buenos Aires [BA] virus) in 1999[27]. This genotype appears to be spreading globally and has been reported from India[14], Kenya[28], Japan[29], Spain[30] Canada and United Kingdom[31]. This virus contained the 60-nucleotide insertion in G protein. Antigenic changes in G protein and avoidance of host immune responses maybe the cause of rapid global spread of BA viruses[31]. This is the first report of genotype BA detected in Iran.
In this study, 21 (87.5%) positive samples were obtained from children under one year of age. Hence our results, as those of others[1, 2], revealed HRSV as a major viral agent in infants and young children.
Finally, this study supported that RT-PCR for second variable region of G protein is an effective method for further studies of HRSV genotype designation in Iran. Also our results may be useful for designing HRSV vaccines in future as well as developing novel prevention or treatment strategies.
Conclusion
We conclude that multiple genotypes of HRSV (GA1,GA2,BA) cocirculated during the season 2009 in Iran. This study revealed that subgroup A strains were more predominant than subgroup B strains. Also we suggest further studies to determine HRSV genotypes over longer periods of time for better understanding of the distribution patterns of HRSV genotypes in Iran.
Acknowledgment
This study was funded by grant No, 9011 from Tehran University of Medical Sciences. We thank personnel of local and state health departments of Ministry of Health and Medical Education of Iran for assistance in collecting data and respiratory samples.
Conflict of Interest
There were not any conflicts of interest.
References
- 1.Holberg CJ, Wright AL, Martinez FD, et al. Risk factors for respiratory syncytial illnesses in the first year of life. Am J Epidemiol. 1991;133(11):1135–51. doi: 10.1093/oxfordjournals.aje.a115826. [DOI] [PubMed] [Google Scholar]
- 2.Collins PL, Chanock RM, Murphy BR. Respiratory syncytial virus. In: Knipe DM, Howley PM, editors. Fields Virology. 4th edn. Philadelphia: Lippincott-Williams and Wilkins; 2001. pp. 1443–86. [Google Scholar]
- 3.Anderson L, Parker R, Strikas R. Association between respiratory syncytial virus outbreaks and lower respiratory tract deaths of infants and young children. J Infect Dis. 1990;161(4):640–46. doi: 10.1093/infdis/161.4.640. [DOI] [PubMed] [Google Scholar]
- 4.Weber M, Mulholland E, Greenwood B. Respiratory syncytial virus infection in tropical and developing countries. Trop Med Int Health. 1998;3(4):268–80. doi: 10.1046/j.1365-3156.1998.00213.x. [DOI] [PubMed] [Google Scholar]
- 5.Falsey AR, Wash EE. Respiratory syncytial virus infections in adults. Clin Microb Rev. 2000;13(3):371–84. doi: 10.1128/cmr.13.3.371-384.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cane PA. Molecular epidemiology of respiratory syncytial virus. Rev Med Virol. 2001;11(2):103–16. doi: 10.1002/rmv.305. [DOI] [PubMed] [Google Scholar]
- 7.Sullender WM. Respiratory syncytial virus genetic and antigenic diversity. Clin Microbiol Rev. 2000;13(1):11–15. doi: 10.1128/cmr.13.1.1-15.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cristina J, Lopez JA, Albo C, et al. Analysis of genetic variability in human respiratory syncytial virus by the RNase. A mismatch cleavage method: subtype divergence and heterogeneity. Virology. 1990;174(1):126–34. doi: 10.1016/0042-6822(90)90061-u. [DOI] [PubMed] [Google Scholar]
- 9.Garcia O, Martin M, Dopazo J, et al. Evolutionary pattern of human respiratory syncytial virus (subgroup A): cocirculating lineages and correlation of genetic and antigenic changes in the G glycoprotein. J Virol. 1994;68(9):5448–59. doi: 10.1128/jvi.68.9.5448-5459.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peret TC, Hall CB, Schnabel KC, et al. Circulation patterns of genetically distinct group A and B strains of human respiratory syncytial virus in a community. J Gen Virol. 1998;79(pt 9):2221–9. doi: 10.1099/0022-1317-79-9-2221. [DOI] [PubMed] [Google Scholar]
- 11.Sato M, Saito R, Sakai T, et al. Molecular epidemiology of respiratory syncytial virus infections among children with acute respiratory symptoms in a community over three seasons. J Clin Microbiol. 2005;43(1):36–40. doi: 10.1128/JCM.43.1.36-40.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Venter M, Madhi SA, Tiemessen CT, et al. Genetic diversity and molecular epidemiology of respiratory syncytial virus over four consecutive seasons in South Africa: Identification of new subgroup A and B genotypes. J Gen Virol. 2001;82(Pt 9):2117–24. doi: 10.1099/0022-1317-82-9-2117. [DOI] [PubMed] [Google Scholar]
- 13.Zlateva KT, Lemey P, Moes E, et al. Genetic variability and molecular evolution of the human respiratory syncytial virus subgroup B attachment G protein. J Virol. 2005;79(14):9157–67. doi: 10.1128/JVI.79.14.9157-9167.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parveen S, Sullender WM, Fowler K, et al. Genetic variability in the G protein gene of group A and B respiratory syncytial viruses from India. J Clin Microbiol. 2006;44(9):3055–64. doi: 10.1128/JCM.00187-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galiano MC, Palomo C, Videla CM, et al. Genetic and antigenic variability of human respiratory syncytial virus (groups a and b) isolated over seven consecutive seasons in Argentina (1995 to 2001) J Clin Microbiol. 2005;43(5):2266–73. doi: 10.1128/JCM.43.5.2266-2273.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaplan NM, Dove W, Abd-Eldayem SA, et al. Molecular epidemiology and disease severity of respiratory syncytial virus in relation to other potential pathogens in children hospitalized with acute respiratory infection in Jordan. J Med Virol. 2008;80(1):168–74. doi: 10.1002/jmv.21067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ostlund MR, Lindell AT, Stenler S, et al. Molecular epidemiology and genetic variability of respiratory syncytial virus (RSV) in Stockholm,2002 –2003. J Med Virol. 2008;80(1):159–67. doi: 10.1002/jmv.21066. [DOI] [PubMed] [Google Scholar]
- 18.Naghipour M, Cuevas LE, Bakhshinejad T, et al. Human Bocavirus in Iranian children with acute respiratory infections. J Med Virol. 2007;79(5):539–43. doi: 10.1002/jmv.20815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Milani M. Respiratory syncytial virus infection among young children with acute respiratory infection. Acta Medica Iranica. 2003;41(4):269–72. [Google Scholar]
- 20.Bdour S. Respiratory syncytial virus subgroup A in hospitalized children in Zarqa, Jordan. Ann Trop Paediatr. 2001;21(3):253–61. doi: 10.1080/02724930120077844. [DOI] [PubMed] [Google Scholar]
- 21.Chan PW, Chew FT, Tan TN, et al. Seasonal variation in respiratory syncytial virus chest infection in the tropics. Pediatr Pulmonol. 2002;34(1):47–51. doi: 10.1002/ppul.10095. [DOI] [PubMed] [Google Scholar]
- 22.Weigl JA, Puppe W, Schmitt HJ. Seasonality of respiratory syncytial virus-positive hospitalizations in children in Kiel, Germany, over a 7-year period. Infection. 2002;30(4):186–92. doi: 10.1007/s15010-002-2159-1. [DOI] [PubMed] [Google Scholar]
- 23.Checon RE, Siqueira MM, Lugon AK, et al. Seasonal pattern of respiratory syncytial virus in a region with a tropical climate in southeastern Brazil. Am J Trop Med Hyg. 2002;67(5):490–1. doi: 10.4269/ajtmh.2002.67.490. [DOI] [PubMed] [Google Scholar]
- 24.Resch B, Gusenleitner W, Muller W. The impact of respiratory syncytial virus infection: A prospective study in hospitalised infants younger than 2 years. Infection. 2002;30(4):193–7. doi: 10.1007/s15010-002-2122-1. [DOI] [PubMed] [Google Scholar]
- 25.Chakravarti A, Kashyap B. Respiratory syncytial virus in lower respiratory tract infections. Iran J Ped. 2007;17(2):123–8. [Google Scholar]
- 26.Hall CB, Walsh EE, Schnabel KC, et al. Occurrence of groups A and B of respiratory syncytial virus over 15 years: associated epidemiologic and clinical characteristics in hospitalized and ambulatory children. J Infect Dis. 1990;162(6):1283–90. doi: 10.1093/infdis/162.6.1283. [DOI] [PubMed] [Google Scholar]
- 27.Trento A, Galiano M, Videla C, et al. Major changes in the G protein of human respiratory syncytial virus isolates introduced by a duplication of 60 nucleotides. J Gen Virol. 2003;84(Pt 11):3115–20. doi: 10.1099/vir.0.19357-0. [DOI] [PubMed] [Google Scholar]
- 28.Scott PD, Ochola R, Ngama M, et al. Molecular epidemiology of respiratory syncytial virus in Kilifi district, Kenya. J Med Virol. 2004;74(2):344–54. doi: 10.1002/jmv.20183. [DOI] [PubMed] [Google Scholar]
- 29.Kuroiwa Y, Nagai K, Okita L, et al. A phylogenetic study of human respiratory syncytial viruses group A and B strains isolated in two cities in Japan from1980 –2002. J Med Virol. 2005;76(2):241–7. doi: 10.1002/jmv.20348. [DOI] [PubMed] [Google Scholar]
- 30.Trento A, Casas I, Calderon A, et al. Ten years of global evolution of the human respiratory syncytial virus BA genotype with a 60-nucleotide duplication in the G protein gene. J Virol. 2010;84(15):7500–12. doi: 10.1128/JVI.00345-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trento A, Viegas M, Galiano M, et al. Natural history of human respiratory syncytial virus inferred from phylogenetic analysis of the attachment (G) glycoprotein with a 60-nucleotide duplication. J Virol. 2006;80(2):975–84. doi: 10.1128/JVI.80.2.975-984.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]