Abstract
UV-A/blue light acts to regulate a number of physiological processes in higher plants. These include light-driven chloroplast movement and phototropism. The NPH1 gene of Arabidopsis encodes an autophosphorylating protein kinase that functions as a photoreceptor for phototropism in response to low-intensity blue light. However, nph1 mutants have been reported to exhibit normal phototropic curvature under high-intensity blue light, indicating the presence of an additional phototropic receptor. A likely candidate is the nph1 homologue, npl1, which has recently been shown to mediate the avoidance response of chloroplasts to high-intensity blue light in Arabidopsis. Here we demonstrate that npl1, like nph1, noncovalently binds the chromophore flavin mononucleotide (FMN) within two specialized PAS domains, termed LOV domains. Furthermore, when expressed in insect cells, npl1, like nph1, undergoes light-dependent autophosphorylation, indicating that npl1 also functions as a light receptor kinase. Consistent with this conclusion, we show that a nph1npl1 double mutant exhibits an impaired phototropic response under both low- and high-intensity blue light. Hence, npl1 functions as a second phototropic receptor under high fluence rate conditions and is, in part, functionally redundant to nph1. We also demonstrate that both chloroplast accumulation in response to low-intensity light and chloroplast avoidance movement in response to high-intensity light are lacking in the nph1npl1 double mutant. Our findings therefore indicate that nph1 and npl1 show partially overlapping functions in two different responses, phototropism and chloroplast relocation, in a fluence rate-dependent manner.
Light is an important environmental factor controlling plant growth and development. In particular, wavelengths in UV-A (320–390 nm) and blue (390–500 nm) regions of the electromagnetic spectrum act to regulate a range of different plant responses. These processes include de-etiolation, photoentrainment of the circadian clock, floral initiation, phototropic curvature, chloroplast relocation, and stomatal opening (1–3). Much of our understanding of blue light perception in higher plants has come from the isolation of blue-light-response mutants of Arabidopsis thaliana. Indeed, molecular genetic studies have shown that the effects of blue light on plant development are mediated by at least four different blue-light receptors in Arabidopsis: cryptochrome 1 (cry1), cryptochrome 2 (cry2), phototropin (nph1, for non-phototropic hypocotyl 1), and the npl1 (nph1-like 1) protein.
The phototropin photoreceptor, nph1, mediates both root and hypocotyl phototropism in response to low-fluence-rate unilateral blue light (<1 μmol⋅m−2⋅s−1) (4, 5). Nph1 is a 120-kDa plasma-membrane-associated protein that contains a serine/threonine kinase domain located within its C terminus. Furthermore, the N-terminal region of nph1 contains a repeated motif of 110 aa, designated LOV1 and LOV2, that belong to the PAS domain (found in PER, ARNT, and SIM proteins) superfamily. PAS domains are found in a variety of proteins and are reported to mediate protein–protein interactions, and to function as internal sensors of oxygen, redox potential, and light (6). The PAS domains of nph1 are more closely related to a subset of proteins within the PAS domain superfamily that are regulated by light, oxygen, or voltage (hence LOV). Recent molecular characterization has shown that recombinant nph1 noncovalently binds the chromophore flavin mononucleotide (FMN) and undergoes autophosphorylation in response to blue light irradiation (7). The light-dependent kinase activity of nph1 is considered to play an important role in mediating phototropic signaling (1, 2). Recent biochemical and photochemical studies have shown that both LOV domains of nph1 bind FMN and undergo a self-contained photocycle (8, 9). Light sensing by LOV1 and LOV2 appears to occur by means of the formation of a stable adduct between the FMN chromophore and the conserved cysteine residue within the LOV domain. The recently obtained crystal structure of the LOV2 domain from Adiantum phy3 is also consistent with the formation of an adduct at the C(4a) position of the isoalloxazine ring of the FMN chromophore (10). It has therefore been proposed that the light-driven reactions of the nph1 LOV domains result in a conformational change of the apoprotein, which in turn leads to activation of the receptor kinase.
Recent physiological studies have shown that whereas nph1 mutants lack phototropism in response to low fluence rates of blue light (<1 μmol⋅m−2⋅s−1), they exhibit normal hypocotyl phototropism under high fluence rates of blue light (1–100 μmol⋅m−2⋅s−1) (11). These observations indicate the presence of an additional phototropic receptor in Arabidopsis. It is unlikely that the cryptochromes play a major role in regulating phototropic curvature in Arabidopsis, as recently suggested (12), because cry1 cry2 double mutants retain phototropic responsiveness to blue light (13). A likely candidate for a second phototropic receptor is the nph1 homologue, npl1.
In higher plants, the cellular localization of chloroplasts is dependent on both light quality and light intensity (14). Light-induced chloroplast movement in Arabidopsis is regulated by blue light and can be separated into two separate responses, depending on light intensity: an accumulation response to low light intensities, which helps maximize light capture for photosynthesis, and an avoidance response to high light intensities, which ameliorates the potentially damaging effects of excess light energy (15, 16). npl1 has recently been shown to regulate the avoidance movement of chloroplasts in response to high-intensity blue light (17, 22). Nevertheless, while npl1 appears to mediate the chloroplast avoidance response, the blue light receptor(s) responsible for the chloroplast accumulation response has remained unknown.
Here we demonstrate that recombinant npl1 functions as a blue light receptor kinase that undergoes light-dependent autophosphorylation. We also show that the LOV domains of npl1, like those of nph1, bind FMN and undergo initial photochemistry that is indicative of the formation of a flavin-cysteinyl adduct. Physiological analysis of a nph1 npl1 double mutant indicate that nph1 and npl1 share functions in two different blue light responses, hypocotyl phototropism and chloroplast relocation. Hence, nph1 and npl1 represent a previously unrecognized family of flavin-based blue light receptors in Arabidopsis.
Materials and Methods
Heterologous Expression in Insect Cells.
The coding sequence of Arabidopsis NPL1 (18) was inserted into the EcoRI site of the baculovirus transfer vector pAcHLT-A (PharMingen) and transfected into Sf9 (Spodoptera frugiperda) insect cells (PharMingen) in accordance with the instructions of the supplier. Recombinant baculovirus was titered by end-point dilution and used to infect Sf9 insect cells. Expression of recombinant nph1 and npl1 was carried out as described (7).
Western Blot Analysis.
Soluble protein samples (10 μg) prepared from insect cells expressing either nph1 or npl1 were boiled in SDS sample buffer, resolved on an SDS/polyacrylamide gel (7.5%), and used for Western blotting. Western blots were analyzed with anti-His antibody (1/5000-fold dilution, Santa Cruz Biotechnology) by using the color development method (Promega) with anti-rabbit IgG conjugated to alkaline phosphatase as the secondary antibody.
In Vitro Phosphorylation Analysis.
Soluble protein extracts isolated from insect cells expressing either nph1 or npl1 were used for in vitro phosphorylation analysis performed as described (7) with minor modifications. Radiolabeled ATP ([γ-32P]ATP (111 TBq/mmol; Amersham Pharmacia Biotech) was diluted 5-fold with unlabeled ATP (10 μM) and 1.25 μl was used for each phosphorylation reaction (total volume 10 μl). Protein samples (10 μg) were prepared in phosphorylation buffer (37.5 mM Tris⋅HCl, pH 7.5/5.3 mM MgSO4/150 mM NaCl/1 mM EGTA/1 mM DTT/5 mM ɛ-aminocaproic acid/1 mM benzamidine/1× complete protease inhibitor mixture, Roche Molecular Biochemicals) containing 0.5% Triton X-100. Samples were irradiated with white light, at a total fluence of 30,000 μmol⋅m−2 in the presence of radiolabeled ATP. Reactions were allowed to proceed at room temperature for 2 min and were stopped by adding 10 μl of 2× SDS sample buffer [110 mM Tris⋅HCl, pH 6.8/4% SDS/20% (vol/vol) glycerol/10% (vol/vol) 2-mercaptoethanol/0.008% bromophenol blue].
Heterologous Expression in Escherichia coli.
Expression of nph1 and npl1 LOV domains in E. coli was carried out as described (8). The npl1 LOV1 domain expression construct extends from amino acid residue 125 to residue 262 and the npl1 LOV2 domain expression construct extends from amino acid residue 371 to residue 512. Each peptide contains a calmodulin-binding protein affinity tag fused to the N terminus.
Spectral Analysis.
Light-minus-dark difference spectra for nph1 and npl1 LOV domains were obtained with a Hewlett Packard 8452A diode array spectrophotometer.
Measurement of Phototropic Curvature.
Hypocotyl curvatures were assayed as described (11). The nph1-101 mutant is in the Landsberg erecta (Ler) background and the npl1-1 mutant is in the Wassilewskija (WS) background. Seeds were planted in square Petri dishes containing 1.5% agar medium as described (11). To induce hypocotyl curvature, 3-day-old etiolated seedlings were irradiated for 12 hr with unilateral blue light supplied by light-emitting diode (LED) blue light lamps (maximum wavelength of 470 nm with a bandwidth of 30 nm). The fluence rate of the light source was adjusted by using blue filters (film no. 72, Tokyo Butai Shoumei, Tokyo). Seedlings were photographed for curvature measurement after the illumination period.
Slit Assay to Measure Chloroplast Relocation.
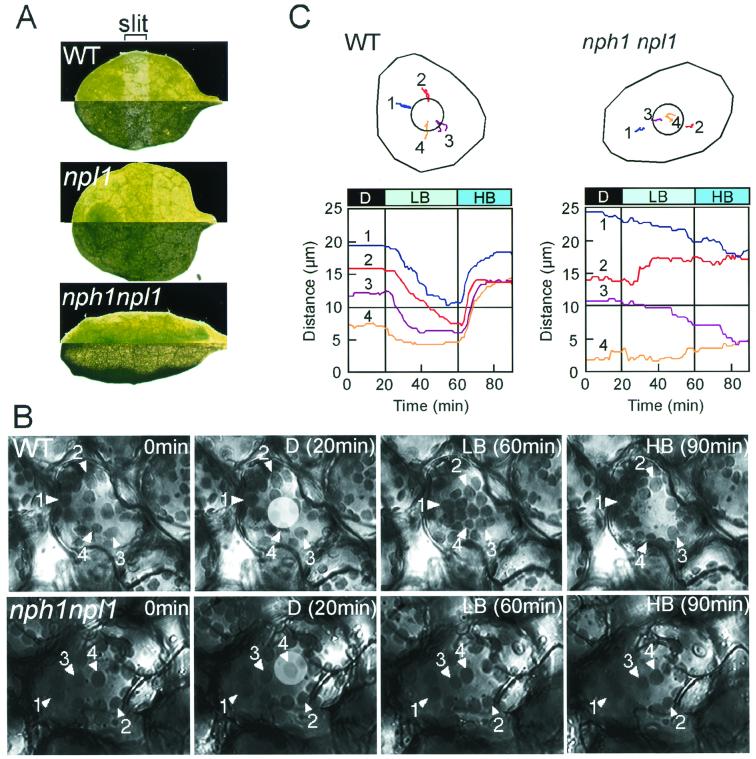
The slit assay method used was carried out as described (17). Leaves were covered with a black plate with open slits (2 mm in width) and irradiated with the blue light source described above (30-nm half-bandwidth centered at 470 nm) at a fluence rate of 100 μmol⋅m−2⋅s−1 for 60 min before photographing. Photographs were taken in dark and bright microscopic fields and are shown in Fig. 3A (upper and lower half of each leaf, respectively).
Figure 3.
Light-activated chloroplast relocation in wild-type plants and the nph1npl1 double mutant. (A) Slit assay for chloroplast relocation. Leaves from wild-type (Columbia), npl1-101, and the nph1npl1 double mutant were partially irradiated with high-intensity blue light for 1 hr. Photographs taken in dark (upper section) and bright (lower section) fields of vision are shown in each case. (B) A series of images monitoring chloroplast relocation in single mesophyll cells from wild-type (Columbia) plants and the nph1npl1 double mutant. Chloroplast accumulation movement was induced by continuous microbeam irradiation with low-intensity blue light (LB, 2 μmol⋅m−2⋅s−1) from the 20th to 60th minute after the onset of the experiment (D, without blue light irradiation). Chloroplast avoidance movement was induced with high-intensity blue light (HB, μmol⋅m−2⋅s−1) from the 60th to the 90th min. The microbeam (20 μm in diameter) used can be seen as a light circle in each image taken after 20 min. (C) Movement tracks of individual chloroplasts. The movement of each chloroplast numbered in B was traced during the experiment and is shown in Upper. Circles in the cells represent the irradiated areas. Distances (μm) between the beam center and each chloroplast were also recorded and are shown in Lower. The 10-μm distance represents the range of the irradiated area (the radius of the blue light microbeam).
Microbeam Assay of Chloroplast Relocation.
Chloroplast relocation in a single cell was measured directly by using a blue light microbeam (17-nm half-bandwidth centered at 449 nm) as described (19). The entire cell was irradiated with red light at 120 μmol⋅m−2⋅s−1 during the recording period to enhance the chloroplast movement as described (19).
Results
Light-Dependent Autophosphorylation of npl1.
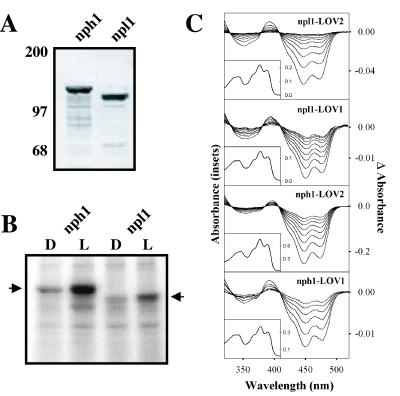
To characterize the molecular properties of npl1 in the absence of other plant proteins, we expressed the npl1 protein in insect cells transfected with recombinant baculovirus containing the NPL1 coding sequence. This approach was previously used to analyze the biochemical and photochemical properties of the nph1 photoreceptor (7). Similar to the expression pattern found for nph1 (7), the majority of recombinant npl1 expressed in insect cells was found to be insoluble (data not shown). However, a small amount of npl1 protein could be detected in the soluble extracts isolated from insect cells. Because recombinant npl1 was expressed with a 6×His affinity tag fused to the N terminus, soluble protein could be visualized by Western analysis (Fig. 1A). A similar level of expression was also observed for nph1 (ref. 7; Fig. 1A). Heterologous nph1 and npl1 proteins are slightly higher in molecular mass than their native counterparts (≈125 kDa and 110 kDa, respectively) because of the inclusion of the N-terminal affinity peptide sequences.
Figure 1.
Biochemical and photochemical properties of Arabidopsis npl1. (A) Western blot analysis of nph1 and npl1 expressed in insect cells. Soluble protein fractions prepared from insect cells expressing either nph1 or npl1 were probed with anti-His antibody. Positions of molecular mass markers are indicated on the left in kilodaltons. (B) Autoradiograph showing the in vitro light-dependent phosphorylation of soluble protein fractions prepared from insect cells expressing either nph1 or npl1 (indicated by arrows). All manipulations were carried out under dim red light. Samples were given a mock irradiation, D, or irradiated with white light, L, at a total fluence of 30,000 μmol⋅m−2. Arrows on the left and right indicate the approximate molecular masses of recombinant nph1 and npl1 (125 kDa and 110 kDa, respectively). (C) Absorption spectra (Insets) and light-minus-dark difference spectra (main panels) of oat nph1 and Arabidopsis npl1 expressed and purified from E. coli. The difference spectra show dark recovery to the ground state after a light flash and were taken at 1-s intervals after the light flash, except for nph1 LOV2, for which the time interval was 5 s.
When expressed in insect cells, nph1 undergoes light-dependent autophosphorylation and exhibits spectral properties that are consistent with nph1 functioning as a photoreceptor for phototropism (7). We therefore investigated whether recombinant npl1 expressed in insect cells could be phosphorylated in response to irradiation in a similar manner. Insect cells expressing either nph1 or npl1 were grown in complete darkness, and soluble protein fractions were harvested under dim red light and used for in vitro phosphorylation analysis. Autoradiography demonstrated that recombinant npl1, like nph1, becomes phosphorylated after a brief irradiation (Fig. 1B). Interestingly, the overall level of autophosphorylation of npl1 appears to be significantly lower in comparison to nph1, suggesting that npl1 may possess fewer sites for autophosphorylation. Whether nph1 and npl1 contain some common target sites for autophosphorylation awaits further investigation. Nevertheless, these findings demonstrate that npl1, like nph1, undergoes autophosphorylation in response to light, indicating that nph1 and npl1 appear to share similar biochemical activities.
LOV Domains of npl1 Bind FMN.
Because the LOV domains of nph1 function as binding sites for the chromophore FMN (8), we investigated whether the LOV domains of npl1 also bind FMN. The LOV domains of npl1 were expressed in E. coli as calmodulin-binding peptide fusions and subsequently purified by calmodulin-affinity chromatography (8). In each case, the purified LOV domains of npl1 were found to be highly fluorescent and bind a flavin chromophore. As shown in Fig. 1C (Insets), the absorption spectra of npl1 LOV1 and LOV2 are almost indistinguishable from the LOV1 and LOV2 domains of nph1, with absorption maxima in both the UV-A and blue regions of the spectrum. The absorption spectra of npl LOV1 and LOV2 also exhibit a substantial degree of fine structure between 400 and 500 nm, characteristic of the nph1 LOV domains. These findings strongly suggest that the LOV domains of npl1, like those of nph1, bind a flavin species (8). Indeed, the chromophore bound to both npl1 LOV1 and LOV2 when released by ethanol denaturation was subsequently identified as FMN by TLC (ref. 8; data not shown). Hence, npl1, like nph1, is a dual-chromophoric photoreceptor binding an FMN chromophore within each of its two LOV domains.
LOV Domains of npl1 Are Photoactive.
Given the above results, it therefore seems likely that npl1 and nph1 function by means of the same initial photochemistry. To examine this hypothesis, we used a diode array spectrophotometer to monitor possible light-induced absorption changes exhibited by the npl1 LOV domains. In the case of nph1, irradiation of either LOV1 or LOV2 with a brief pulse of light results in a rapid loss in absorbance as shown by the light-minus-dark difference spectra in Fig. 1C (main panels). For each LOV domain, the reaction is fully reversible in the absence of light, and after many seconds it decays back to the initial ground state. The initial ground state and the photoproduct share three isosbestic points, at 330, 375, and 410 nm, respectively (Fig. 1C; ref. 9). These light-induced spectral characteristics correspond to the formation of a covalent adduct between the FMN chromophore and the side chain of a conserved active-site cysteine residue within the LOV domain (9, 20). Similar light-induced absorbance changes were observed for the LOV domains of npl1, indicating that npl1 operates through the same initial photochemistry, involving the formation of an FMN C(4a)-cysteinyl adduct. The active-site cysteine required for this reaction is conserved in both npl1 LOV1 and LOV2 (9). Taken together, the above results demonstrate that nph1 and npl1 are very similar with respect to structure, photochemistry, and activity.
Hypocotyl Phototropism Is Severely Impaired in the nph1 npl1 Double Mutant.
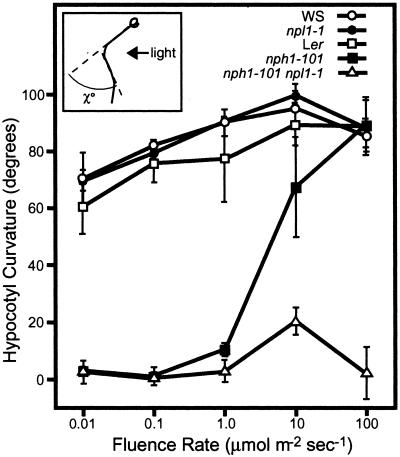
Recently, we reported that whereas null mutants of nph1 lack phototropism in response to low fluence rates of unilateral blue light (<1 μmol⋅m−2⋅s−1), hypocotyl curvature is normal under high fluence rates (1–100 μmol⋅m−2⋅s−1) (11), indicating the presence of an additional phototropic receptor in Arabidopsis. From the above biochemical characterization, it is likely that npl1 acts to function as a second phototropic receptor. However, mutants at the NPL1 locus exhibit normal phototropism when exposed to both low and high fluence rates of blue light (Fig. 2). Given that functional redundancy between phytochrome and cryptochrome photoreceptors is known to exist for a number of different plant responses (21), we examined the curvature response of a nph1 npl1 double mutant. A double mutant was constructed from two null alleles, nph1-101 (nonsense mutation at residue 120) (11) and npl1-1/cav1-5 (mutation by T-DNA insertion into exon 9) (17).
Figure 2.
Hypocotyl phototropism in etiolated wild-type, nph1, npl1, and nph1npl1 mutant seedlings of Arabidopsis. Hypocotyl curvatures of 3.5-day-old seedlings were measured as indicated by χ° in the Inset. Curvatures were measured after a 12-hr exposure to unilateral blue light at the fluence rates indicated. Hypocotyl curvatures of 9 to 16 seedlings were measured in each case and average curvatures were calculated. Values shown represent the average of the three independent experiments. Error bars represent the mean ± the standard deviation.
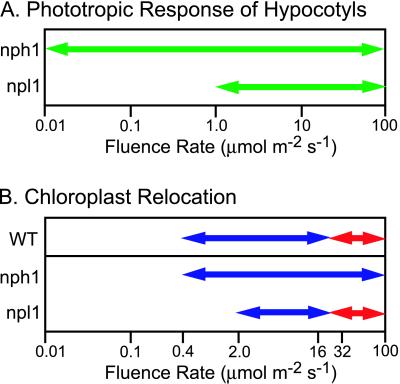
The nph1 npl1 double mutant was found to exhibit impaired hypocotyl phototropism in response to both low and high fluence rates of blue light (Fig. 2). However, a significant degree of curvature was observed in the nph1 npl1 double mutant at a fluence of 10 μmol⋅m−2⋅s−1, suggesting that a third phototropic receptor may exist in Arabidopsis. No curvature response was observed in the double mutant at a fluence rate of 100 μmol⋅m−2⋅s−1 (Fig. 2), a fluence rate at which both of the single nph1 and npl1 mutants show strong phototropic curvature. These findings confirm the hypothesis that npl1 functions as a phototropic receptor in Arabidopsis and indicate that nph1 and npl1 share redundant functions in mediating phototropic curvature in response to unilateral blue light at fluence rates from 1 to 100 μmol⋅m−2⋅s−1 (see Fig. 4A). In the absence of functional nph1, npl1 acts to control hypocotyl phototropism at fluence rates of 1 μmol⋅m−2⋅s−1 or higher (Fig. 4A). In contrast, nph1 is responsible for hypocotyl phototropism over the entire range of fluence rates examined (0.01–100 μmol⋅m−2⋅s−1).
Figure 4.
Proposed photosensitivities required for nph1 and npl1 function. (A) The range of fluence rates effective for nph1 and npl1 action in hypocotyl phototropism. (B) The range of fluence rates expected to be effective for nph1 and npl1 function in chloroplast relocation. Wild-type plants typically show a chloroplast accumulation response to blue light from 0.4 to between 16 and 32 μmol⋅m−2⋅s−1 (blue arrow) and an avoidance response to blue light of 32 μmol⋅m−2⋅s−1 or higher (red arrow). The fluence rate boundary between the accumulation and the avoidance responses is estimated to fall between 16 and 32 μmol⋅m−2⋅s−1 (19). On the basis of our findings with the nph1npl1 double mutant in addition to earlier studies (17, 19), nph1 appears to mediate chloroplast accumulation movement to light at fluence rates from 0.4 to 40 μmol⋅m−2⋅s−1, or higher (blue arrow). npl1 regulates the light-activated chloroplast accumulation at fluence rates from 2 μmol⋅m−2⋅s−1 to between 16 and 32 μmol⋅m−2⋅s−1 (blue arrow). The light-induced chloroplast avoidance movement is mediated by npl1 at a fluence rate of 32 μmol⋅m−2⋅s−1 or higher (red arrow).
The nph1 npl1 Double Mutant Lacks Detectable Light-Induced Chloroplast Relocation.
Recent genetic studies have also demonstrated that npl1 functions to regulate the avoidance response of chloroplasts to high-intensity light in Arabidopsis (17, 22). The cav1 mutant (defective in chloroplast avoidance) lacks the chloroplast avoidance movement and the CAV1 gene was found to encode npl1. While the cav1 mutant lacks the chloroplast avoidance movement, it retains normal chloroplast accumulation movement toward cellular regions illuminated with low-intensity light, indicating that a separate photoreceptor controls this response in Arabidopsis. By contrast, the nph1 null mutant nph1-5 has been shown to exhibit both light-induced chloroplast accumulation and avoidance movements (19). To investigate whether nph1 and npl1 exhibit functionally redundant roles in regulating chloroplast movement, we examined chloroplast relocation in the nph1 npl1 double mutant. The chloroplast avoidance movement can be detected rather simply by using the strip assay (17), whereby leaves are partially irradiated with high-intensity blue light (100 μmol⋅m−2⋅s−1) and subsequently viewed under a light microscope. With this assay, the irradiated slit area of a wild-type leaf changed from green to pale green within approximately 1 hr (Fig. 3A). To illustrate the response in more detail, photographs from a dark (upper section) and bright (lower section) field of vision are shown. In contrast to wild type, the irradiated slit area appeared darker than the nonirradiated area in the npl1 single mutant. These findings are consistent with our previous observations, in that mutants lacking npl1 exhibit an impaired chloroplast avoidance response but retain normal chloroplast accumulation movement even under high light conditions (17). The nph1 npl1 double mutant, on the other hand, failed to show any detectable change in green color between the irradiated and nonirradiated slit area, suggesting that the double mutant is impaired in both chloroplast accumulation and avoidance movement. Leaves from the nph1 npl1 double mutant seemed to curl somewhat downward, making them appear narrower than those of wild-type plants.
The above observations were confirmed by monitoring the movement of individual chloroplasts in mesophyll cells over a 90-min period by using a video recording system (Fig. 3 B and C). For these studies, a blue light microbeam (20-μm diameter) was used to elicit chloroplast relocation in a specific part of the cell. Chloroplasts in wild-type cells moved toward the area irradiated with low-intensity blue light (LB, 2 μmol⋅m−2⋅s−1), which is characteristic of the accumulation response. When subsequently irradiated with high-intensity blue light (HB, 40 μmol⋅m−2⋅s−1), chloroplasts moved away from the irradiated area, characteristic of the avoidance response. Consistent with the results of the slit assay, the nph1 npl1 double mutant failed to show any directed movement of chloroplasts in response to irradiation with either LB or HB. For example, over the entire 90-min period, chloroplasts numbered 1 and 3 in the double mutant (indicated with white arrowheads) moved toward/into the irradiated area, whereas chloroplast 4 remained within the irradiated area. Chloroplast 2 moved away from the irradiated area (Fig. 3C). Thus, both the chloroplast accumulation and avoidance movements are lacking in the nph1 npl1 double mutant. Hence, nph1 and npl1 appear to function in a redundant manner to regulate the light-induced chloroplast accumulation movement in Arabidopsis (Fig. 4B). However, the avoidance response is mediated only by npl1. In the absence of functional npl1, nph1 acts to mediate light-induced chloroplast accumulation over a broad range of fluence rates (0.4–100 μmol⋅m−2⋅s−1). When nph1 is lacking, npl1 functions to regulate light-induced chloroplast accumulation over a narrower range of fluence rates. On the basis of previous studies, we estimate npl1 fluence rate-dependence for chloroplast accumulation to range from 2 μmol⋅m−2⋅s−1 to somewhere between 16 and 32 μmol⋅m−2⋅s−1 (Fig. 4B).
Discussion
In plants, hypocotyl phototropism and chloroplast relocation are important to optimize light capture for photosynthesis. For these processes, plants use blue light to sense light direction and light intensity. In each case, the effects of blue light are mediated by nph1 and npl1 photoreceptors. It is evident from the work presented here that these two photoreceptors share similar molecular structure, chromophore complement, photochemistry, and early biochemistry. nph1 and npl1 therefore represent a previously undescribed class of blue light receptors that appear to be ubiquitous throughout the plant kingdom (23). On the basis of the functional roles of nph1 and npl1 in phototropism, we now classify these proteins as members of the phototropin family of photoreceptors.
The work presented here in conjunction with our previous studies (9) demonstrates that phototropin photoreceptors, nph1 and npl1, operate through an initial photochemistry that is unique among higher plant photoreceptors. Whether, npl1, like nph1, is associated with the plasma membrane requires further investigation. Our present hypothesis is that light sensing by the LOV domains results in a conformational change of the photoreceptor apoprotein, which in turn leads to activation of the kinase domain and an initiation of phototropin signal transduction. As yet, it is unclear whether autophosphorylation plays a role in receptor signaling and/or receptor desensitization. Nevertheless, kinase activity appears to be functionally important for phototropin signaling because the npl1 mutant allele cav1-2 contains a single amino acid substitution within the serine/threonine kinase domain (17). To date, it is unknown whether the phototropins, in addition to autophosphorylating, phosphorylate an interacting substrate. Since the LOV domains belong to the PAS domain superfamily, it is possible that they may function as protein–protein interaction sites. Alternatively, nph1 and npl1, like phyA and phyB, may bind a substrate by means of its C-terminal kinase extension (24–26). Potential downstream signaling mechanisms may involve the regulation of calcium, since nph1 has recently been shown to mediate a blue light-dependent increase in cytosolic calcium (27). A regulation of cytosolic calcium has also been shown to play a role in chloroplast relocation (28).
The results presented here indicate that the phototropins, like the cryptochromes and the phytochromes, exhibit functional redundancy in regulating light-mediated processes in Arabidopsis. nph1 and npl1 photoreceptors appear to mediate both phototropism and chloroplast accumulation movement to low intensities of blue light. nph1 is the primary photoreceptor mediating phototropism in Arabidopsis, because npl1 single mutants exhibit normal phototropic curvature in response to unilateral blue light (Fig. 2). In the absence of nph1, npl1 mediates hypocotyl phototropism in response to only high fluence rates of unilateral blue light (1–100 μmol⋅m−2⋅s−1). Therefore, the overlap in function between nph1 and npl1 photoreceptors occurs only at these higher fluence rates (Fig. 4A). Our findings also indicate that nph1 and npl1 function in a fluence rate-dependent manner to regulate phototropism. npl1 appears to function at high fluence rates of blue light (>1 μmol⋅m−2⋅s−1), whereas nph1 appears to exhibit a broad fluence rate-dependence, mediating the phototropic response at both low (0.01–1 μmol⋅m−2⋅s−1) and high fluence rates (>1 μmol⋅m−2⋅s−1).
Similarly, nph1 and npl1 exhibit, in part, functionally redundant roles in regulating the movement response of chloroplasts toward low-intensity blue light. The chloroplast avoidance response to high-intensity blue light, however, is mediated only by npl1. nph1 appears to be the primary photoreceptor mediating the chloroplast accumulation movement response. The npl1 mutant shows an accumulation response at 2 μmol⋅m−2⋅s−1 (17) or lower (data not shown), and also at 40 μmol⋅m−2⋅s−1 or higher (17). Thus, in the absence of npl1, nph1 mediates chloroplast accumulation movement at fluence rates from 0.4 to 40 μmol⋅m−2⋅s−1 or higher (Fig. 4B), spanning two orders of magnitude. In the nph1 null mutant nph1-5, the chloroplast accumulation response is somewhat retarded, and is induced only by light of 2 μmol⋅m−2⋅s−1 or higher (19). Hence, npl1 regulates the accumulation response at fluence rates between 2 and 32 μmol⋅m−2⋅s−1. As a result, the overlap in function between nph1 and npl1 in regulating chloroplast accumulation occurs only over this range of fluence rates. Therefore, as in phototropism, npl1 appears to operate predominantly under high fluence rates of blue light, whereas the nph1 photoreceptor functions in response to a broad range of fluence rates to mediate light-induced chloroplast accumulation in Arabidopsis. It will now be important to establish why two such very similar photoreceptors appear to exhibit different photosensitivities. Further detailed biochemical and photochemical analysis of the nph1 and npl1 proteins and their isolated LOV domains will help to resolve this issue.
Taken together, the phototropins, nph1 and npl1, represent a previously unrecognized family of blue light receptors in Arabidopsis and function to regulate both phototropism and light-mediated chloroplast relocation. Further analysis of phototropin-deficient mutants will determine whether nph1 and npl1 play a role in regulating other blue-light-activated processes in Arabidopsis. A goal for future research will be to understand how nph1 and npl1, on one hand, activate a process requiring cell–cell communication (phototropism) and, on the other, a cell-autonomous process (chloroplast relocation).
Acknowledgments
We are grateful to Prof. Roberto Bogomolni for helpful discussions and providing the diode array spectrophotometer facilities. This work was supported by funds from the Japan Society for the Promotion of Science to T.S., and from PRESTO (Precursory Research for Embryonic Science and Technology) and the Japan Science and Technology Corporation to T.K. This work was also funded by Grants-in-Aid for Scientific Research on Priority Areas (Grant 10182101) of the Japanese Ministry of Education, Science, Sports and Culture in Japan (MIXT), the Human Frontier Science Program, the Mitsubishi Foundation to K.O., by a U.S. National Science Foundation Grant (9601164) to W.R.B., and by grants from PROBRAIN (Program for Promotion of Basic Research Activities for Innovative Biosciences), the Mitsubishi Foundation, Grant-in-Aid for International Scientific Research (Joint Research 10044214), and Scientific Research (B, 09440270) from MIXT to M.W.
Abbreviations
- LOV domain
light, oxygen, or voltage-sensing domain
- nph1
non-phototropic hypocotyl 1
- npl1
nph1-like 1
References
- 1.Short T W, Briggs W R. Annu Rev Plant Physiol Plant Mol Biol. 1994;45:143–171. [Google Scholar]
- 2.Briggs W R, Huala E. Annu Rev Cell Dev Biol. 1999;15:33–62. doi: 10.1146/annurev.cellbio.15.1.33. [DOI] [PubMed] [Google Scholar]
- 3.Lin C. Trends Plant Sci. 2000;5:337–342. doi: 10.1016/s1360-1385(00)01687-3. [DOI] [PubMed] [Google Scholar]
- 4.Liscum M, Briggs W R. Plant Cell. 1995;7:473–485. doi: 10.1105/tpc.7.4.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huala E, Oeller P W, Liscum E, Han I-S, Larsen E, Briggs W R. Science. 1997;278:2120–2123. doi: 10.1126/science.278.5346.2120. [DOI] [PubMed] [Google Scholar]
- 6.Taylor B L, Zhulin I B. Microbiol Mol Biol Rev. 1999;22:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christie J M, Reymond P, Powell G K, Bernasconi P, Raibekas A A, Liscum E, Briggs W R. Science. 1998;282:1698–1701. doi: 10.1126/science.282.5394.1698. [DOI] [PubMed] [Google Scholar]
- 8.Christie J M, Salomon M, Nozue K, Wada M, Briggs W R. Proc Natl Acad Sci USA. 1999;96:8779–8783. doi: 10.1073/pnas.96.15.8779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salomon M, Christie J M, Kneib E, Lempert U, Briggs W R. Biochemistry. 2000;39:9401–9410. doi: 10.1021/bi000585+. [DOI] [PubMed] [Google Scholar]
- 10.Crosson S, Moffat K. Proc Natl Acad Sci USA. 2001;98:2995–3000. doi: 10.1073/pnas.051520298. . (First Published February 27, 2001, 10.1073/pnas.051520298) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sakai T, Wada T, Ishiguro S, Okada K. Plant Cell. 2000;12:225–236. doi: 10.1105/tpc.12.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahmad M, Jarillo J A, Smirnova O, Cashmore A R. Nature (London) 1998;392:720–723. doi: 10.1038/33701. [DOI] [PubMed] [Google Scholar]
- 13.Lascève G, Leymarie J, Olney M A, Liscum E, Christie J M, Vavasseur A, Briggs W R. Plant Physiol. 1999;120:605–614. doi: 10.1104/pp.120.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wada M, Grolig F, Haupt W. J Photochem Photobiol B. 1993;17:3–25. [Google Scholar]
- 15.Yatsuhashi H. J Plant Res. 1996;109:139–146. [Google Scholar]
- 16.Haupt W, Scheuerlein R. Plant Cell Environ. 1990;13:595–614. [Google Scholar]
- 17.Kagawa T, Sakai T, Suetsugu N, Oikawa K, Ishiguro S, Kato T, Tabata S, Okada K, Wada M. Science. 2001;291:2138–2141. doi: 10.1126/science.291.5511.2138. [DOI] [PubMed] [Google Scholar]
- 18.Jarillo J A, Ahmad M, Cashmore A R. Plant Physiol. 1998;117:719. [Google Scholar]
- 19.Kagawa T, Wada M. Plant Cell Physiol. 2000;41:84–93. doi: 10.1093/pcp/41.1.84. [DOI] [PubMed] [Google Scholar]
- 20.Miller S M, Massey V, Ballou D, Williams C H, Jr, Distefano M D, Moore M J, Walsh C T. Biochemistry. 1990;29:2831–2841. doi: 10.1021/bi00463a028. [DOI] [PubMed] [Google Scholar]
- 21.Casal J J. Photochem Photobiol. 2000;71:1–11. doi: 10.1562/0031-8655(2000)071<0001:pcppii>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 22.Jarillo J A, Gabrys H, Capel J, Alonso J M, Ecker J R, Cashmore A R. Nature (London) 2001;410:952–954. doi: 10.1038/35073622. [DOI] [PubMed] [Google Scholar]
- 23.Christie J M, Briggs W R. J Biol Chem. 2001;276:11457–11460. doi: 10.1074/jbc.R100004200. [DOI] [PubMed] [Google Scholar]
- 24.Ni M, Tepperman J M, Quail P H. Cell. 1998;95:657–667. doi: 10.1016/s0092-8674(00)81636-0. [DOI] [PubMed] [Google Scholar]
- 25.Fankhauser C, Yeh K-C, Lagarias J C, Zhang H, Elich T D, Chory J. Science. 1999;284:1539–1541. doi: 10.1126/science.284.5419.1539. [DOI] [PubMed] [Google Scholar]
- 26.Chol G, Yi H, Lee J, Kwon Y-K, Soh M S, Shin B, Luka Z, Hahn T-R, Song P-I. Nature (London) 1999;401:610–613. doi: 10.1038/44176. [DOI] [PubMed] [Google Scholar]
- 27.Baum G, Long J C, Jenkins G I, Trewavas A J. Proc Natl Acad Sci USA. 1999;96:13554–13559. doi: 10.1073/pnas.96.23.13554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tlalka M, Fricker M. Plant J. 1999;20:461–473. doi: 10.1046/j.1365-313x.1999.00621.x. [DOI] [PubMed] [Google Scholar]