Abstract
Co-evolving positions within protein sequences have been used as spatial constraints to develop a computational approach for modeling membrane protein structures.
Keywords: Co-evolution, membrane proteins, mutual information, protein sequence, structure
In 1973, Christian Anfinsen and colleagues postulated that the information required for a protein to fold into its native structure is encoded in its amino acid sequence. Almost 40 years have passed, but determining the three-dimensional structure of a large protein solely from its amino acid sequence remains a formidable challenge. In the past decade and a half, remarkable progress in the development of cost-effective high-throughput sequencing technologies has led to an explosion of genomic sequence information from evolutionarily diverse organisms. Exploiting this information with statistical tools and constraints derived from evolutionary principles has begun to show promise toward determining protein structure from sequence.
Recently, there has been increasing excitement in predicting protein structure by using co-evolving sites within protein sequences as a structural constraint [1-6]. These studies have demonstrated how improved statistical tools and the growing amount of sequence information hold enormous potential to predict the three-dimensional structure of proteins (Box 1). Along these lines, Hopf et al. [7] have developed a computational approach that exploits protein sequences to predict membrane protein structures. They demonstrate that the three-dimensional structure of α-helical membrane proteins can be determined with very good accuracy using their algorithm EVfold_membrane [7]. Their approach identifies positions in protein sequences that show correlated patterns of mutation and uses this information as a structural constraint to model membrane protein structures [7].
Ab initio structure prediction of membrane proteins
Reliable prediction of structures could have a major impact on our understanding of membrane protein function. This is underscored by the fact that less than 1% of the structures in the Protein Data Bank are of integral membrane proteins despite these comprising over 20% of all genes in mammalian genomes. Membrane proteins are physiologically crucial given their function as a vital communication interface between the intracellular and extracellular environments, and between the cytosol and diverse membrane-bound organelles. Hence, many membrane proteins are pharmacologically important and are potential drug targets. While efforts in structural genomics have led to the elucidation of structures of numerous soluble proteins, determining the structure of membrane proteins remains challenging due to difficulties involved in their expression, purification and crystallization. Thus, any approach that provides insights into structures of membrane proteins will be very useful in explaining their function.
Unlike soluble proteins, membrane proteins predominantly adopt α-helical (as seen in G-protein-coupled receptors) or β-barrel (as seen in porins) structures due to the constraints posed by the lipid environment. Membrane proteins with α-helical structure dominate most cell membranes, while those with β-barrel structure are confined to the outer membrane of bacteria and eukaryotic organelles. Due to the limited structural diversity seen in the membrane protein fold compared to soluble proteins, the former lends itself better to ab initio structure prediction.
In previous approaches, the topology of membrane proteins has been predicted using machine-learning approaches that are trained on lipid exposure and residue contact information [8] and energy-based folding methods that incorporate the knowledge of helix packing constraints [9]. These methods are limited in their accuracy of structure prediction and by the size of membrane proteins that can be investigated [7]. Additionally, there are inherent limitations in machine-learning approaches, which are dictated by the amounts of high-resolution structural data that are already available, and in energy-based methods, which typically require large computational resources. Compared with these methods, the algorithm developed by Hopf et al. [7] uses no a priori structural knowledge and appears to perform better than the previous methods in terms of determining models of larger membrane proteins (up to 14 transmembrane helices). Other benefits of the approach are that it shows increased accuracy, incorporates the increasing abundance of sequence information, and requires relatively low computational power.
From sequences to membrane protein structure prediction
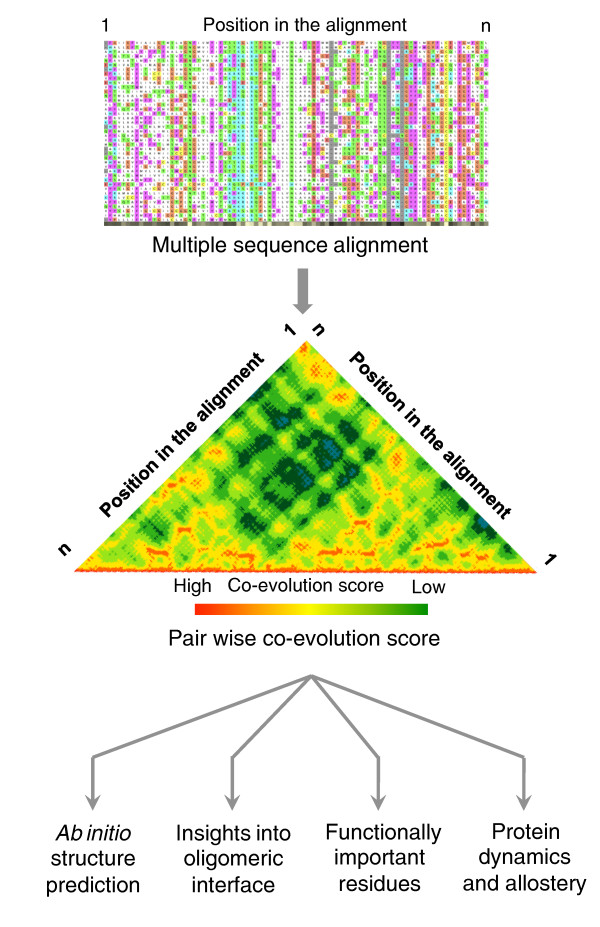
According to Anfinsen's hypothesis, the native protein structure is intrinsically encoded in its sequence. But how is it possible to infer this information solely from sequence data? The EVfold_membrane algorithm developed by Hopf et al. [7] uses a global statistical approach (entropy maximization) and identifies evolutionary coupling (co-evolution) between positions on a sequence from a multiple sequence alignment (Figure 1). The basic principle of the method is that spatially interacting residues tend to co-evolve; thus, co-evolving sites on a sequence can be used as a spatial constraint to predict the three-dimensional structure of a protein. Using the evolutionary coupling between positions as constraints, distance geometry and simulated annealing methods are then used to generate three-dimensional models of proteins, akin to the approach employed in deciphering solution structures of proteins using NMR spectroscopy. The predicted models are assessed and ranked based on various criteria, such as the quality of secondary structure, lipid accessibility of the residues and satisfaction of evolutionary constraints.
Figure 1.
Inferring protein structure, function and dynamics from residue co-evolution. Co-evolving positions within a sequence can be inferred from investigating multiple sequence alignment of homologous protein sequences (Box 1). Such information can be exploited to provide information on spatial proximity of residues and hence to infer three-dimensional structures, residues at the oligomeric interface, amino acids that are functionally important and the presence of potential allosteric communication paths in the structure.
The performance of this algorithm was benchmarked against the experimentally determined structures of 25 membrane proteins from diverse protein families. The predictions showed general agreement with the overall fold and expected deviations in loop regions, unstructured segments in the transmembrane region, and in the orientation of side chains of amino acids. The power of this algorithm was further highlighted by applying the method to predict the structures of 11 membrane proteins with unknown structure. Some of the predicted structures show similarity to folds of known families, thereby making it possible to detect evolutionary relationships between protein families that have significantly diverged.
Beyond membrane protein structure prediction: function and dynamics
Unlike RNA structures, in which pairwise interactions between bases are highly specific, a single residue in a protein typically contacts multiple residues in its proximity to maintain the three-dimensional fold of a protein. This results in complex co-evolution profiles between different positions that may become hard to interpret. In other words, this aspect of proteins leads to the appearance of background noise while computing pairwise co-evolution. This phenomenon makes reliable identification of significant co-evolutionary correlations difficult (Box 1). While there has been significant progress in improving the signal-to-noise ratio over the last years, the methods still have a high false-positive rate, making it difficult to differentiate between direct and indirect interactions between positions in the protein [8]. However, if one can disentangle this information, which is still an area of intense research, it can reveal key molecular aspects of the structure, dynamics and function of the protein.
In addition to inferring the structural proximity of specific residues, co-evolution between positions in a sequence can provide information about the conformation dynamics (including allosteric communication and alternative conformations) and molecular function (such as which residues are involved in substrate recognition, regulation and interaction) of proteins (Figure 1). By resolving contradictory distance constraints and investigating residues that co-evolve with a large number of positions, Hopf et al. [7] harnessed this information to identify different conformational states (as shown for membrane proteins glpT and OCTN, which transport glycerol 3-phosphate and organic cations, respectively). This has also provided hints about contacts in homo-oligomerization interfaces (as shown for the ABC transporter MsbA), and has facilitated prediction of functionally important sites (as shown for adiponectin receptor protein 1, AdipoR1). Thus, information on co-evolving positions can be exploited to gain important insights into the dynamics and function of proteins.
Future directions and novel applications
The computational study undertaken by Hopf et al. [7] demonstrates the potential of what one can learn about the structure of membrane proteins from residue co-evolution. While this is an impressive and important landmark in membrane protein structure prediction, there is still scope for improvement. For instance, constraints derived from biochemical experiments on protein function (such as restraining the distances between side chains of substrate binding residues in transporters) could be incorporated. Moreover, the method could be extended further by creating hybrid computational approaches that exploit Rosetta or other classical folding methods to leverage the strengths of constraints both in terms of co-evolution and energy.
In terms of applications, there are a number of problems where one can exploit this method. For instance, a variety of meta-genomics studies from diverse niches are providing us with a deluge of sequence information of several proteins that are completely uncharacterized. In this context, sequence co-evolution-based approaches can provide insights into structures of novel families of predicted proteins, which could be important for biotechnological and protein engineering applications. The algorithm can also be useful in medicine and drug development as a number of membrane proteins are implicated in several human disease conditions and many of them are difficult targets for structural analysis (for example, the cystic fibrosis transmembrane conductance regulator, whose structure remains to be determined). Thus, ab initio structure determination of such medically important membrane proteins could provide structural models to better interpret disease mutations and serve as potential starting points for structure-based drug design. Such models can help identify structurally proximal residues that could be engineered to increase the stability of membrane proteins for expression, making them suitable for crystallization and for structure determination. With major advancements in the field of electron cryomicroscopy, it is now routinely possible to obtain low-resolution maps (8 to 12 Å) of proteins either by single particle reconstruction or electron crystallography. Inferring topology, subunit orientation and the oligomeric arrangement of membrane proteins using this method might provide models to interpret and complement the electron microscopy maps of macromolecular complexes involving membrane proteins.
Conclusions
Presently, structural biology struggles to hold pace with the rapid growth in genome sequencing. Only a fraction of all known sequence families is represented in the Protein Data Bank with at least one structure of its members. Despite the great progress in structure prediction using protein fragments, improved force fields, molecular dynamics simulations and homology modeling, structures of membrane proteins remain widely inaccessible. Computational methods such as EVfold_membrane thus hold the key to accelerating the structure determination of membrane proteins, thereby bridging the widening sequence-structure gap.
Abbreviations
NMR: nuclear magnetic resonance
Competing interests
The authors declare that they have no competing interests.
Box 1. Identification and interpretation of co-evolving positions
Metrics to quantify pairwise co-evolution of positions from multiple sequence alignments
State of the art approaches to obtain pairwise co-evolutionary correlations between positions in a multiple sequence alignment mainly rely on multivariate statistics. Such approaches take into account the frequencies at which certain amino acid residues appear at aligned sites. Among the most popular are Pearson correlation, maximum likelihood approaches, Bayesian statistics, chi-square-like methods and statistical coupling analysis [10]. In particular, approaches from information theory that use mutual information and Shannon entropy have recently been applied to three-dimensional structure prediction with promising results [1-8].
Filtering relevant and significant correlations from high background noise
One main obstacle in quantifying co-evolution is to filter the relevant information from noise that typically appears in the same order of magnitude as the signal seen for significant correlations [10]. This noise can be due to two main sources: (1) statistical noise due to incomplete or unrepresentative sequence sets and random co-variance, and (2) phylogenetic noise arising from the tree-like phylogenetic relationship of homologous sequences. Sophisticated normalization methods such as interdependency mutual information or comparison to test sets with random distributions can significantly enhance the signal to noise ratio [10].
Disentangling direct and indirect correlations
Maximum-entropy-based algorithms have recently been successfully applied to co-evolutionary sequence correlation in order to find residue contacts in proteins and have been implemented using a numerical method from mean field theory [7]. Another method is based on calculating the inverse covariance matrix to obtain partial correlations. In this approach, the accuracy of the strongest direct interactions can significantly be improved by sparse inverse covariance estimation, in which the inverse covariance matrix is numerically estimated while iteratively removing weaker interactions. An efficient algorithm known as graphical LASSO has been used to transfer this principle to co-evolutionary correlation problems in order to predict residue contacts [1,4].
Contributor Information
Tilman Flock, Email: tilman.flock@caltech.edu.
AJ Venkatakrishnan, Email: ajv@mrc-lmb.cam.ac.uk.
KR Vinothkumar, Email: vkumar@mrc-lmb.cam.ac.uk.
M Madan Babu, Email: madanm@mrc-lmb.cam.ac.uk.
Acknowledgements
We thank Andrew Deonarine for comments on the draft. The authors thank the MRC for funding their research. MB acknowledges support from HFSP, EraSysBio+ and the EMBO YI programme. We apologise to our colleagues whose work was not cited due to space limitations.
References
- Taylor WR, Jones DT, Sadowski MI. Protein topology from predicted residue contacts. Protein Sci. 2012;21:299–305. doi: 10.1002/pro.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcos F, Pagnani A, Lunt B, Bertolino A, Marks DS, Sander C, Zecchina R, Onuchic JN, Hwa T, Weigt M. Direct-coupling analysis of residue coevolution captures native contacts across many protein families. Proc Natl Acad Sci USA. 2011;108:E1293–1301. doi: 10.1073/pnas.1111471108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks DS, Colwell LJ, Sheridan R, Hopf TA, Pagnani A, Zecchina R, Sander C. Protein 3D structure computed from evolutionary sequence variation. PLoS One. 2011;6:e28766. doi: 10.1371/journal.pone.0028766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Buchan DW, Cozzetto D, Pontil M. PSICOV: precise structural contact prediction using sparse inverse covariance estimation on large multiple sequence alignments. Bioinformatics. 2012;28:184–190. doi: 10.1093/bioinformatics/btr638. [DOI] [PubMed] [Google Scholar]
- Fuchs A, Martin-Galiano AJ, Kalman M, Fleishman S, Ben-Tal N, Frishman D. Co-evolving residues in membrane proteins. Bioinformatics. 2007;23:3312–3319. doi: 10.1093/bioinformatics/btm515. [DOI] [PubMed] [Google Scholar]
- Taylor WR, Sadowski MI. Structural constraints on the covariance matrix derived from multiple aligned protein sequences. PLoS One. 2011;6:e28265. doi: 10.1371/journal.pone.0028265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopf TA, Colwell LJ, Sheridan R, Rost B, Sander C, Marks DS. Three-dimensional structures of membrane proteins from genomic sequencing. Cell. 2012. doi: 10.1016/j.cell.2012.04.012. [DOI] [PMC free article] [PubMed]
- Nugent T, Jones DT. Predicting transmembrane helix packing arrangements using residue contacts and a force-directed algorithm. PLoS Comput Biol. 2010;6:e1000714. doi: 10.1371/journal.pcbi.1000714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth P, Wallner B, Baker D. Prediction of membrane protein structures with complex topologies using limited constraints. Proc Natl Acad Sci USA. 2009;106:1409–1414. doi: 10.1073/pnas.0808323106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner DS, Pirovano W, Pesole G. Correlated substitution analysis and the prediction of amino acid structural contacts. Brief Bioinform. 2008;9:46–56. doi: 10.1093/bib/bbm052. [DOI] [PubMed] [Google Scholar]