Abstract
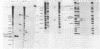
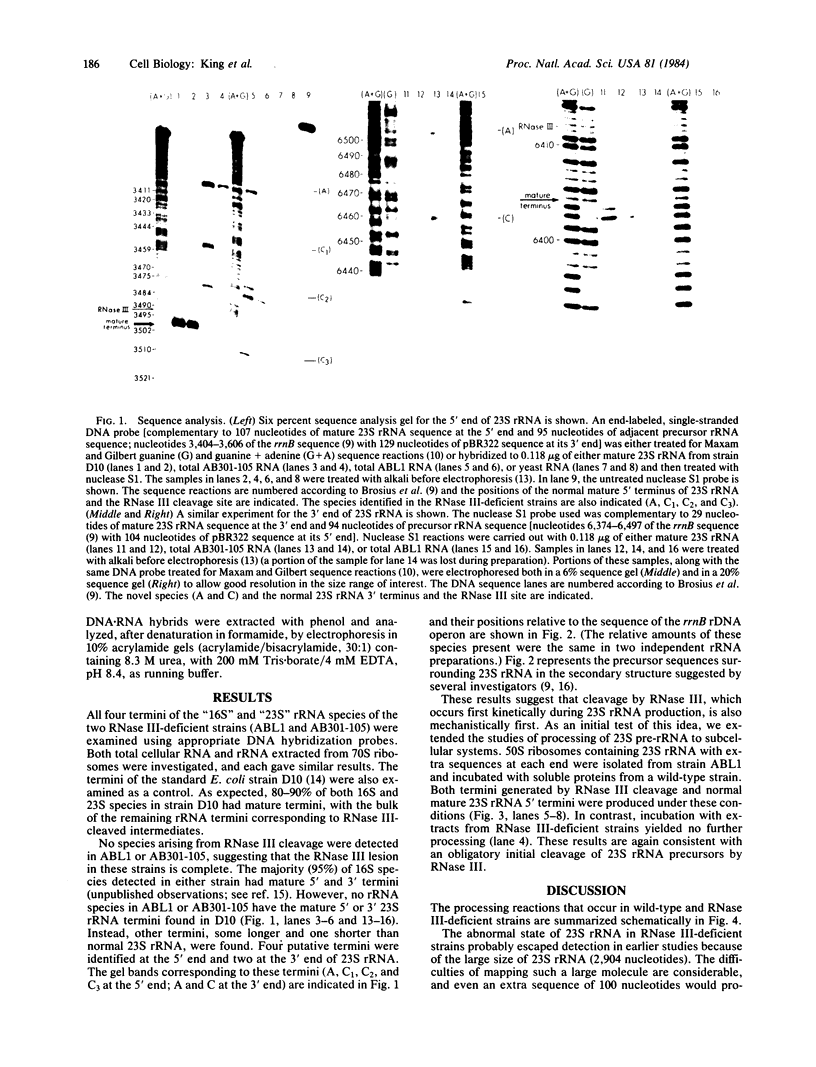
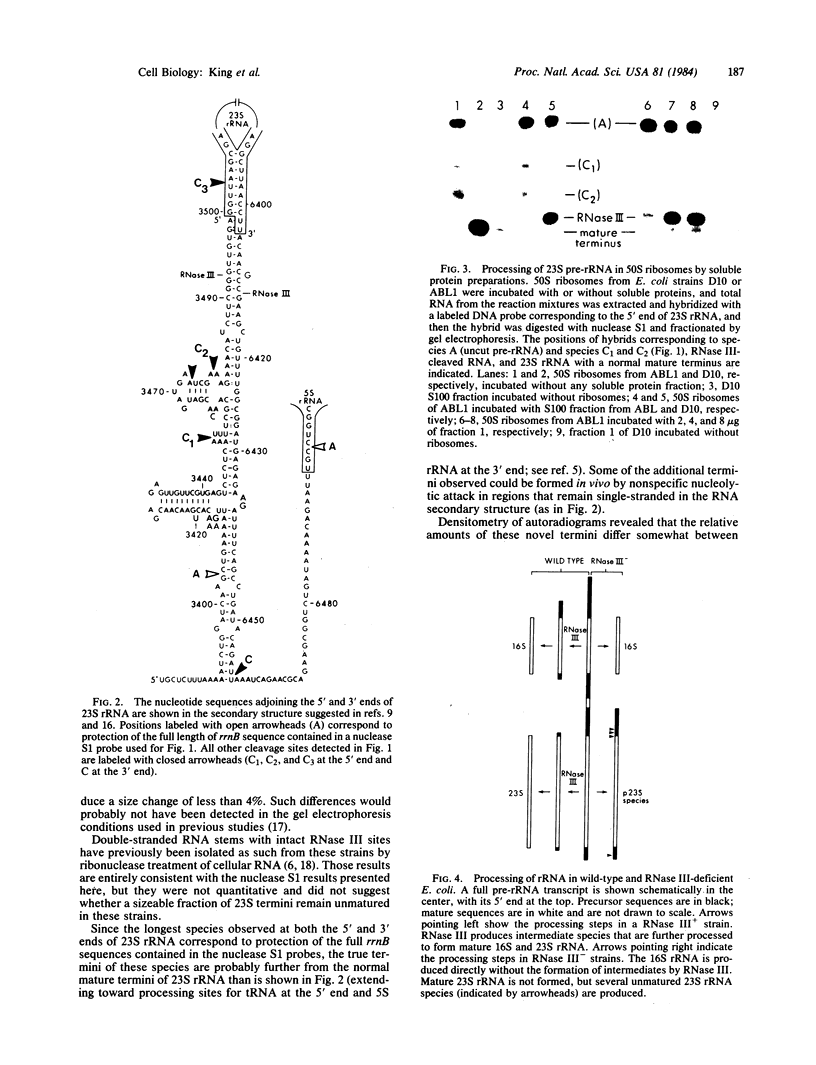
RNase III makes the initial cleavages that excise Escherichia coli precursor 16S and 23S rRNA from a single large primary transcript. In mutants deficient in RNase III, no species cleaved by RNase III are detected and the processing of 23S rRNA precursors to form mature 23S rRNA fails entirely. Instead, 50S ribosomes are formed with rRNAs up to several hundred nucleotides longer than mature 23S rRNA. Unexpectedly, these aberrant subunits function well enough to participate in protein synthesis and permit cell growth. Consistent with the inference that RNase III cleavages are absolutely required for 23S rRNA maturation, when 50S ribosomes from a strain deficient in RNase III were incubated with a ribosome-free extract from a RNase III+ strain, rRNA species processed by RNase III and species with normal mature 23S rRNA termini were produced.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berk A. J., Sharp P. A. Spliced early mRNAs of simian virus 40. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1274–1278. doi: 10.1073/pnas.75.3.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Structure of the adenovirus 2 early mRNAs. Cell. 1978 Jul;14(3):695–711. doi: 10.1016/0092-8674(78)90252-0. [DOI] [PubMed] [Google Scholar]
- Bowman L. H., Rabin B., Schlessinger D. Multiple ribosomal RNA cleavage pathways in mammalian cells. Nucleic Acids Res. 1981 Oct 10;9(19):4951–4966. doi: 10.1093/nar/9.19.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bram R. J., Young R. A., Steitz J. A. The ribonuclease III site flanking 23S sequences in the 30S ribosomal precursor RNA of E. coli. Cell. 1980 Feb;19(2):393–401. doi: 10.1016/0092-8674(80)90513-9. [DOI] [PubMed] [Google Scholar]
- Brosius J., Dull T. J., Sleeter D. D., Noller H. F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981 May 15;148(2):107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- Dahlberg A. E., Dahlberg J. E., Lund E., Tokimatsu H., Rabson A. B., Calvert P. C., Reynolds F., Zahalak M. Processing of the 5' end of Escherichia coli 16S ribosomal RNA. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3598–3602. doi: 10.1073/pnas.75.8.3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlberg A. E., Peacock A. C. Studies of 16 and 23 S ribosomal RNA of Escherichia coli using composite gel electrophoresis. J Mol Biol. 1971 Jan 14;55(1):61–74. doi: 10.1016/0022-2836(71)90281-6. [DOI] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. T7 early RNAs and Escherichia coli ribosomal RNAs are cut from large precursor RNAs in vivo by ribonuclease 3. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3296–3300. doi: 10.1073/pnas.70.12.3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegenheimer P., Apirion D. Precursors to 16S and 23S ribosomal RNA from a ribonuclear III-strain of Escherichia coli contain intact RNase III processing sites. Nucleic Acids Res. 1980 Apr 25;8(8):1873–1891. doi: 10.1093/nar/8.8.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegenheimer P., Apirion D. Processing of procaryotic ribonucleic acid. Microbiol Rev. 1981 Dec;45(4):502–541. doi: 10.1128/mr.45.4.502-541.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesteland R. F. Isolation and characterization of ribonuclease I mutants of Escherichia coli. J Mol Biol. 1966 Mar;16(1):67–84. doi: 10.1016/s0022-2836(66)80263-2. [DOI] [PubMed] [Google Scholar]
- Ginsburg D., Steitz J. A. The 30 S ribosomal precursor RNA from Escherichia coli. A primary transcript containing 23 S, 16 S, and 5 S sequences. J Biol Chem. 1975 Jul 25;250(14):5647–5654. [PubMed] [Google Scholar]
- Gitelman D. R., Apirion D. The synthesis of some proteins is affected in RNA processing mutants of Escherichia coli. Biochem Biophys Res Commun. 1980 Oct 16;96(3):1063–1070. doi: 10.1016/0006-291x(80)90060-1. [DOI] [PubMed] [Google Scholar]
- Jacobs K. A., Schlessinger D. Escherichia coli DNA-directed beta-galactosidase synthesis in presence and absence of Ca2+. Biochemistry. 1977 Mar 8;16(5):914–920. doi: 10.1021/bi00624a016. [DOI] [PubMed] [Google Scholar]
- Kindler P., Keil T. U., Hofschneider P. H. Isolation and characterization of a ribonuclease 3 deficient mutant of Escherichia coli. Mol Gen Genet. 1973 Oct 16;126(1):53–59. doi: 10.1007/BF00333481. [DOI] [PubMed] [Google Scholar]
- King T. C., Schlessinger D. S1 nuclease mapping analysis of ribosomal RNA processing in wild type and processing deficient Escherichia coli. J Biol Chem. 1983 Oct 10;258(19):12034–12042. [PubMed] [Google Scholar]
- Lindahl L. Intermediates and time kinetics of the in vivo assembly of Escherichia coli ribosomes. J Mol Biol. 1975 Feb 15;92(1):15–37. doi: 10.1016/0022-2836(75)90089-3. [DOI] [PubMed] [Google Scholar]
- Lowry C. V., Dahlberg J. E. Structural differences between the 16S ribosomal RNA of E. coli and its precursor. Nat New Biol. 1971 Jul 14;232(28):52–54. doi: 10.1038/newbio232052a0. [DOI] [PubMed] [Google Scholar]
- Mangiarotti G., Turco E., Perlo C., Altruda F. Role of precursor 16S RNA in assembly of E. coli 30S ribosomes. Nature. 1975 Feb 13;253(5492):569–571. doi: 10.1038/253569a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Nikolaev N., Birenbaum M., Schlessinger D. 30 S pre-ribosomal RNA of Escherichia coli:primary and secondary processing. Biochim Biophys Acta. 1975 Jul 23;395(4):478–489. doi: 10.1016/0005-2787(75)90071-4. [DOI] [PubMed] [Google Scholar]
- Nikolaev N., Schlessinger D. Binding of ribosomal proteins to 30S preribosomal ribonucleic acid of Escherichia coli. Biochemistry. 1974 Oct 8;13(21):4272–4278. doi: 10.1021/bi00718a005. [DOI] [PubMed] [Google Scholar]
- Nikolaev N., Silengo L., Schlessinger D. Synthesis of a large precursor to ribosomal RNA in a mutant of Escherichia coli. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3361–3365. doi: 10.1073/pnas.70.12.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatsky I. N., Evstafieva A. G., Bystrova T. F., Bogdanov A. A., Vasiliev V. D. Topography of RNA in the ribosome: localization of the 3'-end of the 23 S rna on the surface of the 50 S ribosomal subunit by immune electron microscopy. FEBS Lett. 1980 Dec 29;122(2):251–255. doi: 10.1016/0014-5793(80)80450-9. [DOI] [PubMed] [Google Scholar]
- Silengo L., Nikolaev N., Schlessinger D., Imamoto F. Stabilization of mRNA with polar effects in an Escherichia coli mutant. Mol Gen Genet. 1974;134(1):7–19. doi: 10.1007/BF00332808. [DOI] [PubMed] [Google Scholar]
- Sprague K. U., Steitz J. A. The 3' terminal oligonucleotide of E. coli 16S ribosomal RNA: the sequence in both wild-type and RNase iii- cells is complementary to the polypurine tracts common to mRNA initiator regions. Nucleic Acids Res. 1975 Jun;2(6):787–798. doi: 10.1093/nar/2.6.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöffler-Meilicke M., Stöffler G., Odom O. W., Zinn A., Kramer G., Hardesty B. Localization of 3' ends of 5S and 23S rRNAs in reconstituted subunits of Escherichia coli ribosomes. Proc Natl Acad Sci U S A. 1981 Sep;78(9):5538–5542. doi: 10.1073/pnas.78.9.5538. [DOI] [PMC free article] [PubMed] [Google Scholar]