Abstract
The goal of this minireview is to provide an overview of varicella-zoster virus (VZV) phylogenetics and phylogeography when placed in the broad context of geologic time. Planet Earth was formed over 4 billion years ago, and the supercontinent Pangaea coalesced around 400 million years ago (mya). Based on detailed tree-building models, the base of the phylogenetic tree of the Herpesviridae family has been estimated at 400 mya. Subsequently, Pangaea split into Laurasia and Gondwanaland; in turn, Africa rifted from Gondwanaland. Based on available data, the hypothesis of this minireview is that the ancestral alphaherpesvirus VZV coevolved in simians, apes, and hominins in Africa. When anatomically modern humans first crossed over the Red Sea 60,000 years ago, VZV was carried along in their dorsal root ganglia. Currently, there are five VZV clades, distinguishable by single nucleotide polymorphisms. These clades likely represent continued VZV coevolution, as humans with latent VZV infection left Arabia and dispersed into Asia (clades 2 and 5) and Europe (clades 1, 3, and 4). The prototype VZV sequence contains nearly 125,000 bp, divided into 70 open reading frames. Generally, isolates within a clade display >99.9% identity to one another, while members of one clade compared to a second clade show 99.8% identity to one another. Recently, four different VZV genotypes that do not segregate into the previously defined five clades have been identified, a result indicating a wider than anticipated diversity among newly collected VZV strains around the world.
INTRODUCTION
Within the broad herpesvirology arena, varicella-zoster virus (VZV) research has rarely been the most prolific. One notable recent exception is whole-genome sequencing and phylogenetics. In this minireview, data that support an out-of-Africa model of VZV evolution and phylogeography are presented. Further, the herpesvirus phylogenetic data are presented in the broadest context, based on a geologic time scale centered around the supercontinent Pangaea in the Paleozoic era. The goal of the minireview is to demonstrate that VZV genomics not only provides fascinating insight into coevolution of viruses with the earliest living forms—for example, invertebrate herpesviruses extant at the time of Pangaea were neurotropic—but also reinforces our knowledge about the migrations of humankind from Arabia throughout Asia, Europe, and the Americas.
(An abbreviated version of this minireview was presented by the author at the Workshop on Phylogenetics of Infectious Diseases—with a Focus on DNA Viruses, sponsored by the Institute for Mathematical Sciences at the National University of Singapore in October 2011, organized by Julian Tang.)
THE BIG BANG AND THE EMERGENCE OF PANGAEA
The Big Bang occurred over 13 billion years ago. Planet Earth was formed about 4.5 billion years ago, as the galaxies cooled. The universal common ancestor of life may have arisen over 3 billion years ago. Subsequent diversification of early living forms accelerated during the Cambrian period over 500 million years ago (mya), when oceans covered the globe (39). Examples of these early marine forms are the contemporary invertebrates called mollusks. Over the next 200 million years, the emerging land masses coalesced around the equator to form the supercontinent of Pangaea, which remained intact for another 100 to 200 million years. Then Pangaea was slowly pulled apart as the underlying tectonic plates moved. Around 175 mya, an initial major separation of Pangaea led to a northern supercontinent called Laurasia and a southern supercontinent called Gondwanaland. Subsequently, around 150 mya, Gondwanaland split further into the current land masses of Africa, South America, India, Australia, and Antarctica (13). The final major break occurred around 60 mya, when Laurasia split into North America and Eurasia. Continental drift continues today, e.g., Australia is moving northward at 5 cm per year. Similarly, the eastern horn of Africa continues moving northeastward, leaving behind the Great Rift Valley, where the bone fossils of Lucy Australopithecus were found in November 1974 (Fig. 1).
Fig 1.

Illustration of Lucy and Tembo Australopithecus based on the hominin fossils recovered in East Africa. These hominins lived over 3 mya in the current country of Ethiopia in East Africa (22, 23). Based on phylogenetic data presented in this minireview, Australopithecans were infected with ancestral VZV. Illustration prepared by and reproduced with permission of Michael Hagelberg, Institute of Human Origins, Arizona State University, Tempe, AZ.
The Great Rift Valley is a great gorge extending 4,500 miles from the Red Sea Rift beneath the Red Sea (Gulf of Arabia) across modern Ethiopia and Kenya, created by a tectonic plate that is slowly pulling the horn of Africa northeastward, thus exposing fossil evidence of life millions of years before contemporary times. The concept that primordial VZV first arose in an ancestral primate in Africa was considered in 1999 (18).
Because herpesviruses are ancient species, a more detailed knowledge of the geologic epochs can both clarify and illuminate the likely timetable of herpesvirus evolution (4). The Paleozoic era extends from 550 to 251 mya, beginning with the Cambrian period and ending with the Permian period. The Permian Basin in Texas is named after a large deposit of rocks from the Permian period. The Permian period was notable for the appearance of the supercontinent Pangaea. On Pangaea, amniotes (vertebrates) evolved into ancestral groups of synapsids (mammals) and sauropsids (reptiles and birds). The Permian period was followed by the Mesozoic era, which included three periods from 250 mya to 65 mya. The Triassic period (250 to 200 mya) has become well known because of the emergence of the dinosaurs. The Triassic period was followed by the Jurassic period (199 to 145 mya). At the beginning of this period, Pangaea had separated into Laurasia and Gondwanaland, while at the end of this period, Gondwanaland would separate into South America, Africa, and Antarctica. The Jurassic period was followed by the Cretaceous period (145 to 65 mya). The latter period ended when the asteroid struck the Caribbean and the temperature of the planet declined. The dinosaurs disappeared and the mammals emerged.
MARINE HERPESVIRUSES BEFORE PANGAEA
Herpesviruses infect invertebrates such as the oyster and the abalone, marine creatures of the Cambrian period. This family of herpesviruses has been named Malacoherpesviridae (9). Based on the hypothesis of coevolution, therefore, these viruses likely evolved during the Paleozoic era, at least 100 million years before ancestral vertebrate herpesviruses on Pangaea. The herpesvirus of the bivalve oyster has a DNA genome consisting of 207,439 bp, divided into 124 open reading frames (ORFs). The genome is housed in an icosahedral capsid about 116 nm in diameter with a triangulation number of 16, characteristic features of a herpesvirus.
However, in marked contrast to vertebrate herpesviruses, the capsid lacks pentons at the 12 vertices (9). Further analysis of the genome structure suggests that it includes long and short unique regions, four repeat regions, and an additional internal unique region. This structure is represented as follows: TR(L)–U(L)–IR(L)–X–IR(S)–U(S)–TR(S). Among the ORFs are genes resembling vertebrate herpesviral DNA polymerase, ribonucleotide reductase, helicase, primase, and the ATPase subunit of terminase. Similar findings were reported after the sequencing of the genome of the abalone mollusk; in particular, abalone herpesvirus ORFs include those resembling oyster virus terminase and polymerase protein sequences (45).
The abalone herpesvirus is of further interest because it is neurotropic. This neurotropism was identified because infection with this herpesvirus leads to ganglioneuritis and eventual necrosis of the nervous tissue, followed by death of the abalone (45). Microscopic examination disclosed virus-induced lesions in the cerebral and buccal ganglia of the abalone. Thus, neurotropism of herpesviruses has existed for 500 million years.
HERPESVIRUS AT THE TIME OF PANGAEA
Within the order Herpesvirales, there are two other families that arose after Malacoherpesviridae (31). These include Alloherpesviridae, viruses of amphibians and fish, and Herpesviridae, viruses of reptiles, birds, and mammals. Further, Herpesviridae are divided into three subfamilies, called alpha-, beta-, and gammaherpesviruses. In turn, the alphaherpesvirus group includes human herpes simplex virus 1 (HSV-1), HSV-2, and VZV; betaviruses includes human cytomegalovirus (CMV), human herpesvirus 6A (HHV-6A), HHV-6B, and HHV-7; and gammaherpesviruses includes human Epstein-Barr virus (EBV) and HHV-8, also called Kaposi's sarcoma-associated herpesvirus (KSHV). Based on detailed tree-building schemas, initially with simpler neighbor-joining techniques and later with more sophisticated maximum-likelihood and Bayesian models, the base of the phylogenetic tree of the Alphaherpesviridae subfamily has been estimated at 400 mya.
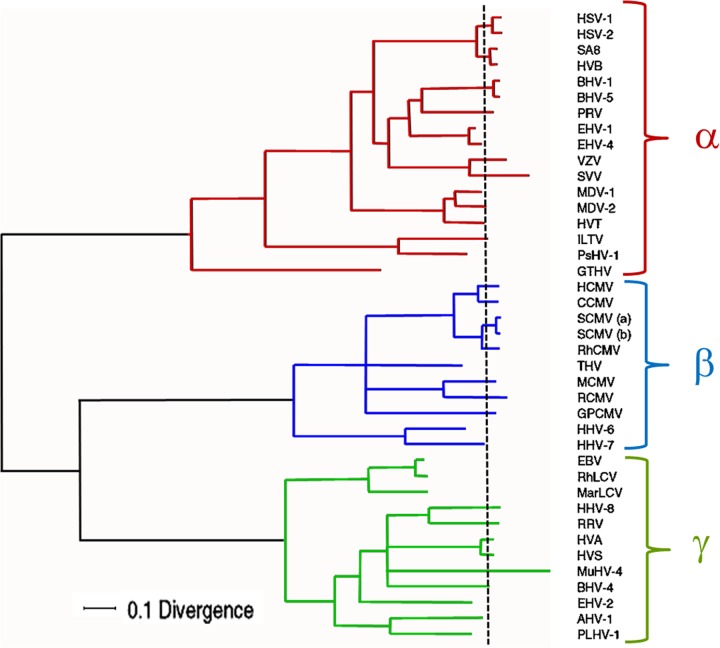
A comprehensive phylogenetic tree has been constructed based on the alignment of amino acid sequences for six genes common to 40 herpesviral species within the alpha-, beta- and gammaherpesvirus subfamilies (Fig. 2). The genes include the orthologs of VZV ORF28 (DNA polymerase subunit), ORF29 (single-stranded DNA binding protein), ORF30 (DNA packaging terminase subunit 2), ORF31 (glycoprotein gB), ORF40 (major capsid protein), and ORF42 (DNA packaging terminase subunit 1). The equivalent HSV-1 genes are the UL30, UL29, UL28, UL27, UL19, and UL15 genes, respectively. Examination of the branching and relative scale within each of the three subfamilies illustrated in Fig. 2 continues to reinforce evidence for coevolution of many individual herpesviral species and their hosts.
Fig 2.
Phylogenetic tree for the family Herpesviridae. A phylogenetic tree was constructed based on an alignment of amino acid sequences for six shared genes from 40 different herpesviral species within all three subfamilies. The genes included the orthologs of VZV ORF28, ORF29, ORF30, ORF31, ORF40, and ORF42. Initial tree evaluation utilized a Bayesian Monte Carlo Markov chain process. The root of the tree was estimated as the midpoint between the mean tip positions of terminal branches in the alphaherpesviruses (α) and those in the betaherpesviruses (β) plus gammaherpesviruses (γ). The mean tip position is marked with a vertical dashed line. A divergence scale is shown at the bottom of the figure. Abbreviations for the 40 viruses are as follows: HSV-1, herpes simplex virus 1; HSV-2, herpes simplex virus 2; SA-8, simian agent 8; HVB, herpesvirus B; BHV-1, bovine herpesvirus 1; BHV-5, bovine herpesvirus 5; PRV, pseudorabies virus; EHV-1, equid herpesvirus 1; EHV-4, equid herpesvirus 4; VZV, varicella-zoster virus; SVV, simian varicella virus; MDV-1, Marek's disease virus type 1; MDV-2, Marek's disease virus type 2; HVT, herpesvirus of turkeys; ILTV, infectious laryngotracheitis virus; PsHV-1, psittacid herpesvirus 1; GTHV, green turtle herpesvirus; HCMV, human cytomegalovirus; CCMV, chimpanzee cytomegalovirus; SCMV (a) and SCMV (b), simian cytomegalovirus; RhCMV, rhesus cytomegalovirus; THV, tupaiid herpesvirus; MCMV, murine cytomegalovirus; RCMV, rat cytomegalovirus; HHV-6, human herpesvirus 6; HHV-7, human herpesvirus 7; EBV, Epstein-Barr virus; RLV, rhesus lymphocryptovirus; MarLCV, marmoset herpesvirus; HHV-8, human herpesvirus 8; RRV, rhesus rhadinovirus; HVA, herpesvirus ateles; HVS, herpesvirus saimiri; MuHV-4, murid herpesvirus 4; BHV-4, bovine herpesvirus 4; EHV-2, equid herpesvirus 2; AHV-1, alcelaphine herpesvirus 1; PLHV-1, porcine herpesvirus 1. Reprinted from Virus Research (31) with permission of the publisher.
As noted in the geologic time scale, 400 mya is a remarkable period coincident with the coming together of Pangaea. Therefore, primordial ancestors of the Herpesviridae likely arose on or around this supercontinent, within their synapsid and sauropsid hosts (Fig. 3). As an example, the green turtle herpesvirus is an ancient alphaherpesvirus that is estimated to have arisen 239 mya, before Pangaea was split asunder (30). This virus continues to flourish within the green sea turtle populations in both the Pacific and Atlantic Oceans (16, 42). On the other hand, HSV-1, HSV-2, and VZV are estimated to have arisen less than 120 mya. Again, the latter time period is of great interest because it follows the rift that separated Africa from the supercontinent of Gondwanaland (25). In other words, if the time scales of the phylogenetic tree are even correct in the first approximation, VZV could only have arisen in Africa. Furthermore, it is possible, perhaps even probable, that the primordial VZV arose in a primate rather than an earlier life form.
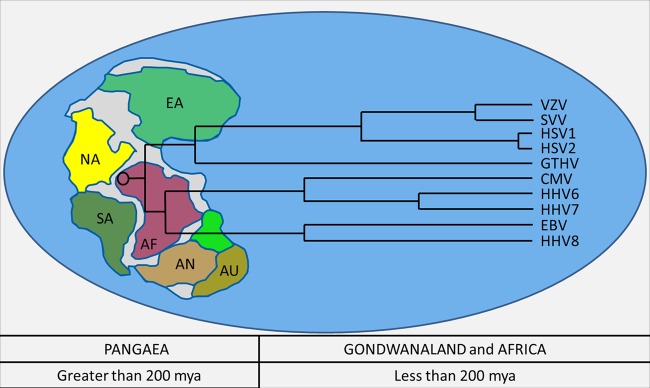
Fig 3.
The age of Pangaea and the emergence of herpesviruses. Based on data provided in several reports cited in this minireview, the herpesviruses arose 400 mya in ancestral synapsids and sauropsids, during or even before formation of the supercontinent of Pangaea. Among the most ancient viral species are the GTHV strains, which infect green sea turtles (Chelonia mydas) (16, 42). This reptilian alphaherpesvirus continues to be found in green turtles in the Atlantic and Pacific Oceans (26). By analogy, it is likely that similarly ancient beta- and gammaherpesviruses existed within sauropsids at the time of Pangaea, as illustrated in the tree. Current human herpesviruses arose after Pangaea had separated into Gondwanaland and subsequently into Africa (<200 mya). Abbreviations within Pangaea: EA, Europe/Asia; NA, North America; SA, South America; AF, Africa; AN, Antarctica; and AU, Australia.
VZV IN AFRICA
If primordial VZV arose in an African primate 70 mya, the early primates certainly were tiny creatures who lived in family groups in the trees. Because of their small size, fossil evidence is scant. However, primatologists suggest that early primates likely resembled lemurs currently found in Madagascar off the southern African coast (28). One example is a Malagasy dwarf lemur. These miniature primates weigh about 60 g and live among family groups in trees. As primates evolved to assume more simian features around 35 mya in Africa, Aegyptopithecus is a candidate common ancestor preceding the divergence of Old World monkeys and the great apes. Aegyptopithecus weighed about 4 kg, lived in trees, and ate fruits. Based on the out-of-Africa model, both of these primates would have harbored varicella viruses.
Around 30 mya, the simians diverged from the great apes. One of the strongest arguments for an African origin of human varicella virus is Old World simian varicella virus (SVV). SVV bears an uncanny genomic resemblance to human VZV, with 70% identity (15). Further, the absence of SVV from New World monkeys suggests that the most recent common ancestor arose in Africa after Gondwanaland had separated into Africa and South America some 120 mya. The SVV genome contains 124,138 bp, divided into the following segments: TR(L)–U(L)–IR(L)–IR(S)–U(S)–TR(S). The genome encodes 69 ORFs.
Around 8 mya, the hominins diverged from the great apes. The well-known Lucy Australopithecus lived in the Great Rift Valley of East Africa over 3 mya (Fig. 1). Subsequently, Homo erectus replaced Australopithecus. Around 150,000 years ago (150 kya), the most recent common ancestor of anatomically modern humans emerged, among whom one of the notable founders within a phylogenetic tree is sometimes called African Eve or Mitchondrial Eve (38). Because of a bottleneck during the exodus across the southern Red Sea, all humankind outside Africa is a descendant of a single mitochondrial haplotype L3 and its descendants M and N (7, 52). This remarkable discovery has greatly facilitated tracing the migrations of human populations out of Arabia (21, 33).
ZOSTER AS A SURVIVAL STRATEGY
Since early hominin societies in Africa are considered to have been composed of small hunter gatherer populations, the dual diseases of varicella and zoster provide a survival strategy. Varicella is the only human herpesvirus to be transmitted primarily by aerosolization. In a small family group or even a large clan of 100 or more individuals, it is likely that all nonimmune individuals would contract varicella during the same short period if exposed to a single infected hominin. Therefore, without latency and reactivation, the virus could not survive, since all susceptible hominins would have been infected. However, by entering a latent state in the neurons and reactivating decades later, the virus can re-emerge as zoster. As noted above, a striking observation is that the property of neurotropism (although not latency) has been documented in the most ancient of herpesviruses residing within the abalone.
Since zoster is also contagious, the virus is spread horizontally from the elderly human with zoster to all individuals born in the family group or clan since the last episode of varicella decades earlier. Presumably through this mechanism of latency and reactivation, VZV persisted over the millennia within relatively isolated populations as they left Africa and entered the Middle East, before continuing on to Asia and subsequently to Europe. A perfect example of this zoster-to-varicella mode of transmission was described among the population on the remote island of Tristan da Cunha, a British protectorate in the South Atlantic Ocean midway between South Africa and Argentina. On this island with a population of around 200 people, varicella only occurs among the inhabitants after an elderly citizen develops zoster (49). As discussed in more detail below, no genetic differences have been discovered between sequences from varicella and zoster viruses; in other words, the virus that undergoes latency appears genetically identical to the virus that emerges from latency decades later (41, 47).
For as-yet-unknown evolutionary reasons, VZV is the outlier with regard to mode of transmission to children within earlier cultures. Most other human herpesviruses are spread via saliva from mother to young child, or grandmother to child, when the mother (or grandmother) reactivates a latent infection. Often, this vertical transmission occurs in the first years of life, as is the case for HSV-1, CMV, HHV-6, and EBV (6, 17, 27). Premastication of solid food by mothers before feeding the food to an infant is a common mode of transmission of these viruses, although several other common child-rearing practices involving maternal saliva have been described (6). In addition to saliva, CMV is spread vertically through breast milk, since in a vast majority of CMV-seropositive women, the virus is reactivated during lactation (32). Transmission of HHV-7 and KSHV also occurs within family groups, presumably through exchange of saliva, although the children are often older when they acquire their primary infection (17, 57). An apparent exception to the model of saliva transmission from mother-to-child is HSV-2, which is commonly spread by sexual contact in adolescence and adulthood from an infected partner to an uninfected partner. Even with HSV-2, however, some infants can acquire a perinatal infection vertically from an HSV-2 infected mother, who has a reactivation around time of parturition (55). Thus, the newborn infant is infected on the mucosal surfaces of the mouth, nares, or eyes. In the era before antiviral therapy, a frequent outcome for the infected newborn was death. Nevertheless, some infected infants have only a mild or asymptomatic HSV-2 infection and easily survive into adulthood with latent HSV-2 infection. In short, VZV is not usually spread by vertical transmission routes common to the other human herpesviruses.
DISPERSAL OF VZV INTO EUROPE AND ASIA
After humankind with their latent VZV had emigrated out of Africa and into Arabia, there were subsequent migrations which have been measured by analysis of several genetic population markers, including those found on the Y chromosome and mitochondrial DNA (mtDNA) (2). Although 60 kya is commonly stated as the migration date for the out-of-Africa event, recent evidence suggests earlier migrations may have occurred between 80 to 100 kya. Part of this speculation is based on evidence that humans first colonized southeast Asia 60 kya. Humans also migrated northward into west and central Asia and then dispersed westward into Europe 30 to 40 kya. Based on further Y-chromosome analyses, the migrations within Europe involved complicated patterns. Some speculate that there were two major migrations, around 40 and 10 kya. Others find evidence for expansion, retreat, and re-expansion of the European populations. In both scenarios, a major expansion occurred 6 to 10 kya.
Similar genomic analyses within the Japanese population have shown that the contemporary population arose from two migrations (20). Those in the first migration over 20 to 30 kya arrived via land bridges from Asia, while those in the second migration arrived by sea from Korea over 2 kya. Of note, the origins of the first migration have been traced to ancestral humans in Southeast Asia, while the second migration began in central Asia.
The time period of 6 to 10 kya is of special interest because this time period overlaps with the origin of the Proto-Indo-European (PIE) language (1). The PIE culture likely arose on the Caspian steppe north of the Black Sea, in current eastern Ukraine and southern Russia. The PIE languages then spread westward and southward. During the same time period, an agricultural culture replaced a hunter-gatherer culture. Thus, sufficiently large communities would have arisen for transmission of VZV infection primarily by periodic outbreaks of varicella among children. Because smallpox (variola) was also present, the actual name varicella was selected, likely during Roman times, as an irregular diminutive form of variola, i.e., varicella was once considered a less virulent variant of variola.
VZV CLADES, SNPs, AND PHYLOGEOGRAPHY
In 1986, the VZV Dumas strain from Holland was completely sequenced (8). The VZV genome is the smallest among the human herpesviruses. In an evolutionary sense, therefore, VZV is a minimalist herpesvirus, retaining the smallest complement of ORFs required for survival. Its basic structure is the same as that for SVV. In 2002, the Oka vaccine and parental strains from Japan were sequenced (14). Between 2004 and 2007, the complete sequences of 12 additional North American strains were published (19, 37, 40, 51). In 2008, investigators interested in VZV phylogenetic analysis met in London to survey all available published data on complete VZV genomic sequences (5). By that time, there were over 20 complete VZV sequences in GenBank. Since the publication of the first single nucleotide polymorphism (SNP) analysis of a subgroup of VZV ORFs, it was becoming apparent that VZV genomes could be distinguished on the basis of phylogeography (3, 11, 53). Therefore, at the 2008 VZV nomenclature meeting, the participants agreed to designate each geographic cluster of VZV genotypes a clade. The clades would be numbered on the basis of the time in which the genomic sequences were published (5). Each clade would include one or two prototypic sequences. Thus, clade 1 is annotated by the first VZV strain to be sequenced, namely, Dumas, while clade 2 is typified by the Oka parental and vaccine sequences. Clade 3 includes strains O3-500 and HJO, while clade 4 includes strains DR and 8. Clade 5 is distinguished by strain Ca123.
With regard to phylogeography, VZV clades 1, 3, and 4 are European and North American. Clade 2 is Asian (Japanese), and clade 5 is from the Indian subcontinent (24) (Fig. 4). One point that merits emphasis is that the three European clades can be easily distinguished from members of the Asian clade 2. All three sequencing groups which originally outlined the genotypic differences between VZV strains easily distinguished these clades, even though each group used a different set of SNPs in its analyses. Generally, isolates within a clade display >99.9% identity to one another, while members of one clade compared to members of a second clade show >99.8% identity (40). Otherwise stated, the VZV intraclade diversity was minimal, at approximately 1 in 1,400 bases. The validity of the clade nomenclature has been confirmed by the complete sequencing of an additional 21 VZV genomes by another group of investigators, who were not involved in the 2008 VZV nomenclature meeting (58). They estimated the evolutionary rate to equal 3.9 × 10−9 substitutions per site per year.
Fig 4.
Phylogeography of VZV clades around the world. Based on the data in this minireview, the most likely explanation for VZV dispersal around the world is by VZV coevolution with modern humans as humans with latent varicella infection migrated out of Africa around 60 to 100 kya into Arabia and then onward to Asia and Europe. In 2008, five clades were recognized after a VZV consensus nomenclature meeting (5). Further information about the clades is found in the text. The most recent ancestor to the five VZV clades remains to be defined.
Among the 21 VZV strains members of this group collected in Germany, they also discovered two distinct novel genomic sequences that did not segregate into any of the five clades based on the previously defined criteria; these two strains were tentatively designated genotypes VIII and IX. Their bootstrap analyses with Simplot did identify considerable similarity between clade 4 and the novel genotype IX. Previously, at the nomenclature meeting in 2008, two additional novel VZV genotypes were designated VI and VII (5). All members of the original committee that adopted a consensus nomenclature in 2008 intended that designation of a new clade would include at least two completely sequenced genomes, since a clade by definition cannot consist of one member. Even more importantly, the concept of a viral clade was intended to imply a cluster of similar VZV strains arising from a common ancestor, perhaps the best example being clade 2 strains from Japan. A second meeting of a VZV consensus nomenclature committee may be required to consider a redefinition of the current clades as well as inclusion of any new candidate clades.
VZV GENOTYPES WITH NEW PHENOTYPES
Even though VZV strains can be segregated into distinct clades, viruses within every clade generally have a similar phenotype when isolated and passaged in cultured cells. The first community VZV strain with a phenotype distinguishable from those of the other strains mentioned above was described in 1998 (44). In December 1995, a presumed VZV isolate was obtained in the Minneapolis/St. Paul metropolitan area. The patient was admitted to the hospital with a primary diagnosis of severe varicella infection and a secondary diagnosis of acute lymphatic leukemia. After treatment with intravenous acyclovir, the patient recovered. However, the virology laboratory was unable to confirm the diagnosis of varicella because the viral isolate did not react with the reagent in the anti-VZV rapid diagnostic kit. Because the commercial laboratory could not identify the isolate, the isolate was extensively characterized and named VZV MSP. The investigations demonstrated that the gE protein was present but that the gE protein had lost a B-cell epitope, secondary to a single nucleotide change within codon 150, GAC to AAC, which in turn led to a change in amino acid from aspartic acid to asparagine (43). Additional sequencing demonstrated that VZV MSP belonged to clade 1, similar to VZV Dumas (19).
The mutation within a B-cell epitope on a surface viral glycoprotein closely resembles antigenic drift in influenza virus. In fact, identical switches from aspartic acid to asparagine have previously been identified in the hemagglutinin protein of influenza A virus. Since the discovery of this first gE antigenic variant virus in Minneapolis/St. Paul, MN, a second gE mutant virus was discovered in British Columbia, Canada (50). The Canadian case involved an elderly man with zoster, a fact which indicates that the mutant virus would have been present about 50 years earlier (around the year 1950), when the man had chickenpox as a child. Subsequently, stored VZV isolates from the Karolinska University Hospital in Stockholm, Sweden, were examined, and two gE mutant viruses were discovered (56). Finally, a gE mutant virus was isolated from a child with fatal varicella in Rome, Italy (36). When the histories of these five cases are reviewed, there is a clear indication that the gE mutant viruses were isolated from varicella or zoster cases of sufficient severity that the patients sought medical care or even were admitted for administration of intravenous acyclovir therapy.
The purported increased severity of human cases of infection with gE mutant viruses correlates with the data from the severe combined immunodeficient (SCID) mouse model of VZV infection (34). In human skin explants inserted in the SCID mouse, VZV infection was markedly more severe after infection with the gE mutant virus than that with prototypic wild-type varicella viruses (43). In short, the evidence from the above five cases indicates that immune pressure can select more virulent antigenic variants of VZV that have lost a B-cell epitope. Yet surveillance by my laboratory and others suggests that these gE mutant viruses have not spread from the index cases to others in their environment. In other words, even if more virulent within an infected host, the mutant viruses appear less fit in their transmissibility. Thus, there is no current evidence that these antigenic variant viruses are a major contributor to the SNPs seen within clades.
VZV RECOMBINATION AND PHYLOGENETICS
The stability of the VZV genome likely is related in part to the small number of replication cycles during primary VZV infection; for example, over a 2-week period, VZV may undergo only 14 replication cycles before the immune response blocks further replication (18). Genomic stability has been firmly documented by full genomic sequencing of the laboratory strain VZV 32 at sequential passages 5, 22, and 72 (51). Virtually no base substitutions were detected after 22 passages in cultured cells, while 30 base substitutions were identified at passage level 72. VZV stability is enhanced by the low number of short sequence repeat (reiteration) regions and other hot spots within the genome, in contrast to HSV and pseudorabies virus (PRV) (48). Yet VZV recombination definitely occurs under conditions of dually infected cultured cells (10). Under the condition of natural infection in humans, recombination presumably would occur when a child contracted primary varicella after exposures to two different people, each of whom was infected with VZV belonging to a different clade. Subsequent VZV replication in the child during the viremia and skin vesicle formation would offer an opportunity for recombination of two genetically different viruses to occur when the same cell was infected.
What is even more interesting is that all investigators agree that recombination occurs, albeit rarely, but the investigators disagree about which recombination events may have already occurred to form the current clades (29, 37, 40, 53, 58). Nevertheless, by carrying out genomic analyses of randomly collected VZV isolates obtained in two non-European countries (Thailand and Brazil) that had extensive immigration of Europeans beginning in the 16th century, it is apparent that recombination can occur within 500 years after circulation of both European and non-European VZV genotypes within the local population (35, 53).
VZV EVOLUTION IN THE 21st CENTURY
When the VZV phylogenetic literature is reviewed in the broad context of geologic events since the creation of supercontinent Pangaea, the simplest explanation for the origin of primordial VZV is within a primate living in Africa. The striking genetic and immunologic similarity between Old World SVV and human VZV is particularly compelling; for example, there is a straightforward experiment demonstrating that immunization of monkeys with human VZV can prevent subsequent infection with SVV (12). Certainly, therefore, a common ancestor to SVV and VZV existed about 30 mya in Africa. Primordial VZV coevolved with the ancestors to anatomically modern humans over the past 7 million years, dependent for its survival in small populations on the dual neuronal adaptations of latency and reactivation as zoster (Fig. 1). Between 100 and 60 kya, VZV continued to coevolve in early humankind in East Africa. The out-of-Africa model presumes a bottleneck at the time of exodus of VZV-harboring anatomically modern humans across the narrow Bab el Mandeb strait of the Red Sea into Arabia. VZV and the other human herpesviruses were in a precarious situation, since the human population had dwindled to less than 10,000 (54). But the population recovered in Arabia, prior to further migrations outward to Asia and Europe. Therefore, VZV clades (present in early humans whose ancestors never left Africa) remain to be discovered within isolated African populations. In one sense, therefore, the phylogeography of the varicella chromosome harbored with human cells can be compared to mtDNA within human cells.
However, if the founder effect of the mitochondrial haplotype surviving the hominins' exodus from Africa is mimicked by latent VZV that left Africa in the same hominins, knowledge about other clades in Africa may not be overly informative with regard to the five clades. In other words, the human genomic bottleneck data suggest the possibility that most contemporary VZV clades could be descendants of a very small number of genotypes present in a very small human population that found its way across the Red Sea into Arabia around 60 to 100 kya. If true, the most recent common ancestor to all worldwide clades found outside Africa may still be latent within an inhabitant of a remote village in Yemen. Based on data about subsequent population dispersals into Europe and Japan, the common ancestor to European VZV clades may have existed 30 to 40 kya; likewise, the common ancestor to Japanese clades may have existed 20 to 30 kya. To extend the mtDNA analogy, VZV clades 1, 3, and 4 may resemble dispersal patterns of one mtDNA haplotype, while VZV clades 2 and 5 may resemble a second mtDNA haplotype. At the same time, VZV complete genome sequencing likely occurred at a propitious time in the late 20th century, when distinct geographic VZV clades within stable human populations could still be defined by a few alleles. With the dramatic increases in emigration of large population groups over the recent decades, however, “nonindigenous” VZV strains will be introduced into most urban populations, as has already been documented in London and Bangkok (46, 53).
ACKNOWLEDGMENTS
I thank Donald Johanson at Arizona State University for his comments about the discovery of Lucy Australopithecus and for the figure representing Lucy. I thank Graham Tipples, Geoffrey Peters, and Shaun Tyler for our collaborative VZV sequencing projects at the National Microbiology Laboratory of Canada. I thank the many herpesvirologists whose manuscripts are cited and who kindly answered questions about their conclusions.
VZV research by the author is supported by NIH grant AI89716.
Footnotes
Published ahead of print 3 July 2012
REFERENCES
- 1. Anthony DA. 2007. The horse, the wheel, and language: how Bronze Age riders from the Eurasian steppes shaped the modern world. Princeton University Press, Princeton, NJ [Google Scholar]
- 2. Atkinson QD, Gray RD, Drummond AJ. 2008. mtDNA variation predicts population size in humans and reveals a major Southern Asian chapter in human prehistory. Mol. Biol. Evol. 25:468–474 [DOI] [PubMed] [Google Scholar]
- 3. Barrett-Muir W, et al. 2003. Genetic variation of varicella-zoster virus: evidence for geographical separation of strains. J. Med. Virol. 70(Suppl. 1):S42–S47 [DOI] [PubMed] [Google Scholar]
- 4. Benton MJ, Donoghue PC. 2007. Paleontological evidence to date the tree of life. Mol. Biol. Evol. 24:26–53 [DOI] [PubMed] [Google Scholar]
- 5. Breuer J, Grose C, Norberg P, Tipples G, Schmid DS. 2010. A proposal for a common nomenclature for viral clades that form the species varicella-zoster virus: summary of VZV Nomenclature Meeting 2008, Barts and the London School of Medicine and Dentistry, 24–25 July 2008. J. Gen. Virol. 91:821–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Butler LM, Neilands TB, Mosam A, Mzolo S, Martin JN. 2010. A population-based study of how children are exposed to saliva in KwaZulu-Natal Province, South Africa: implications for the spread of saliva-borne pathogens to children. Trop. Med. Intern. Health 15:442–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cavalli-Sforza LL, Menozzi P, Piazza A. 1993. Demic expansions and human evolution. Science 259:639–646 [DOI] [PubMed] [Google Scholar]
- 8. Davison AJ, Scott JE. 1986. The complete DNA sequence of varicella-zoster virus. J. Gen. Virol. 67:1759–1816 [DOI] [PubMed] [Google Scholar]
- 9. Davison AJ, et al. 2005. A novel class of herpesvirus with bivalve hosts. J. Gen. Virol. 86:41–53 [DOI] [PubMed] [Google Scholar]
- 10. Dohner DE, Adams SG, Gelb LD. 1988. Recombination in tissue culture between varicella-zoster virus strains. J. Med. Virol. 24:329–341 [DOI] [PubMed] [Google Scholar]
- 11. Faga B, Maury W, Bruckner DA, Grose C. 2001. Identification and mapping of single nucleotide polymorphisms in the varicella-zoster virus genome. Virology 280:1–6 [DOI] [PubMed] [Google Scholar]
- 12. Felsenfeld AD, Schmidt NJ. 1979. Varicella-zoster virus immunizes patas monkeys against simian varicella-like disease. J. Gen. Virol. 42:171–178 [DOI] [PubMed] [Google Scholar]
- 13. Fooden J. 1972. Breakup of Pangaea and isolation of relict mammals in Australia, South America, and Madagascar. Science 175:894–898 [DOI] [PubMed] [Google Scholar]
- 14. Gomi Y, et al. 2002. Comparison of the complete DNA sequences of the Oka varicella vaccine and its parental virus. J. Virol. 76:11447–11459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gray WL, Pumphrey CY, Ruyechan WT, Fletcher TM. 1992. The simian varicella virus and varicella zoster virus genomes are similar in size and structure. Virology 186:562–572 [DOI] [PubMed] [Google Scholar]
- 16. Greenblatt RJ, et al. 2005. Genomic variation of the fibropapilloma-associated marine turtle herpesvirus across seven geographic areas and three host species. J. Virol. 79:1125–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Grose C. 2009. Human herpesviruses 6, 7 and 8, p 2071–2076 In Feigin R, Cherry J, Demmler-Harrison G, Kaplan S. (ed), Textbook of pediatric infectious diseases. Elsevier, Philadelphia, PA [Google Scholar]
- 18. Grose C. 1999. Varicella-zoster virus: less immutable than once thought. Pediatrics 103:1027–1028 [DOI] [PubMed] [Google Scholar]
- 19. Grose C, et al. 2004. Complete DNA sequence analyses of the first two varicella-zoster virus glycoprotein E (D150N) mutant viruses found in North America: evolution of genotypes with an accelerated cell spread phenotype. J. Virol. 78:6799–6807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hammer MF, et al. 2006. Dual origins of the Japanese: common ground for hunter-gatherer and farmer Y chromosomes. J. Hum. Genet. 51:47–58 [DOI] [PubMed] [Google Scholar]
- 21. Ingman M, Gyllensten U. 2001. Analysis of the complete human mtDNA genome: methodology and inferences for human evolution. J. Hered. 92:454–461 [DOI] [PubMed] [Google Scholar]
- 22. Johanson DC, Wong K. 2009. Lucy's legacy: the quest for human origins. Random House, New York, NY [Google Scholar]
- 23. Johanson DC, White TD. 1979. A systematic assessment of early African hominids. Science 203:321–330 [DOI] [PubMed] [Google Scholar]
- 24. Kolesnik M, et al. 2011. Varicella outbreak in Indian students in Magdeburg with detection of the African-Indian VZV clade 5. J. Dtsch. Dermatol. Ges. 9:444–447 [DOI] [PubMed] [Google Scholar]
- 25. Kumar P, et al. 2007. The rapid drift of the Indian tectonic plate. Nature 449:894–897 [DOI] [PubMed] [Google Scholar]
- 26. Lackovich JK, et al. 1999. Association of herpesvirus with fibropapillomatosis of the green turtle Chelonia mydas and the loggerhead turtle Caretta caretta in Florida. Dis. Aquat. Organ. 37:89–97 [DOI] [PubMed] [Google Scholar]
- 27. Loutfy SA, Alam El-Din HM, Ibrahim MF, Hafez MM. 2006. Seroprevalence of herpes simplex virus types 1 and 2, Epstein-Barr virus, and cytomegalovirus in children with acute lymphoblastic leukemia in Egypt. Saudi Med. J. 27:1139–1145 [PubMed] [Google Scholar]
- 28. Martin RD. 1990. Primate origins and evolution. Princeton University Press, Princeton, NJ [Google Scholar]
- 29. McGeoch DJ. 2009. Lineages of varicella-zoster virus. J. Gen. Virol. 90:963–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McGeoch DJ, Gatherer D. 2005. Integrating reptilian herpesviruses into the family Herpesviridae. J. Virol. 79:725–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McGeoch DJ, Rixon FJ, Davison AJ. 2006. Topics in herpesvirus genomics and evolution. Virus Res. 117:90–104 [DOI] [PubMed] [Google Scholar]
- 32. Meier J, et al. 2005. Human cytomegalovirus reactivation during lactation and mother-to-child transmission in preterm infants. J. Clin. Microbiol. 43:1318–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Melé M, et al. 2012. Recombination gives a new insight in the effective population size and the history of the old world human populations. Mol. Biol. Evol. 29:25–30 [DOI] [PubMed] [Google Scholar]
- 34. Moffat JF, et al. 1998. Attenuation of the vaccine Oka strain of varicella-zoster virus and role of glycoprotein C in alphaherpesvirus virulence demonstrated in the SCID-hu mouse. J. Virol. 72:965–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Muir WB, Nichols R, Breuer J. 2002. Phylogenetic analysis of varicella-zoster virus: evidence of intercontinental spread of genotypes and recombination. J. Virol. 76:1971–1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Natoli S, et al. 2006. A novel mutation of varicella-zoster virus associated to fatal hepatitis. J. Clin. Virol. 37:72–74 [DOI] [PubMed] [Google Scholar]
- 37. Norberg P, et al. 2006. Complete-genome phylogenetic approach to varicella-zoster virus evolution: genetic divergence and evidence for recombination. J. Virol. 80:9569–9576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pakendorf B, Stoneking M. 2005. Mitochondrial DNA and human evolution. Annu. Rev. Genom. Hum. Genet. 6:165–183 [DOI] [PubMed] [Google Scholar]
- 39. Peebles PJE. 1980. The large scale structure of the universe. Princeton University Press, Princeton, NJ [Google Scholar]
- 40. Peters GA, et al. 2006. A full-genome phylogenetic analysis of varicella-zoster virus reveals a novel origin of replication-based genotyping scheme and evidence of recombination between major circulating clades. J. Virol. 80:9850–9860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pichini B, Ecker JR, Grose C, Hyman RW. 1983. DNA mapping of paired varicella-zoster virus isolates from patients with shingles. Lancet ii:1223–1225 [DOI] [PubMed] [Google Scholar]
- 42. Quackenbush SL, et al. 2001. Quantitative analysis of herpesvirus sequences from normal tissue and fibropapillomas of marine turtles with real-time PCR. Virology 287:105–111 [DOI] [PubMed] [Google Scholar]
- 43. Santos RA, et al. 2000. Varicella-zoster virus gE escape mutant VZV-MSP exhibits an accelerated cell-to-cell spread phenotype in both infected cell cultures and SCID-hu mice. Virology 275:306–317 [DOI] [PubMed] [Google Scholar]
- 44. Santos RA, Padilla JA, Hatfield C, Grose C. 1998. Antigenic variation of varicella zoster virus Fc receptor gE: loss of a major B cell epitope in the ectodomain. Virology 249:21–31 [DOI] [PubMed] [Google Scholar]
- 45. Savin KW, et al. 2010. A neurotropic herpesvirus infecting the gastropod, abalone, shares ancestry with oyster herpesvirus and a herpesvirus associated with the amphioxus genome. Virol. J. 7:308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sengupta N, et al. 2007. Varicella-zoster-virus genotypes in East London: a prospective study in patients with herpes zoster. J. Infect. Dis. 196:1014–1020 [DOI] [PubMed] [Google Scholar]
- 47. Straus SE, et al. 1984. Endonuclease analysis of viral DNA from varicella and subsequent zoster infections in the same patient. N. Engl. J. Med. 311:1362–1364 [DOI] [PubMed] [Google Scholar]
- 48. Szpara ML, et al. 2011. A wide extent of inter-strain diversity in virulent and vaccine strains of alphaherpesviruses. PLoS Pathog. 7:e1002282 doi:10.1371/journal.ppat.1002282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Taylor-Robinson D, Tyrrell DA. 1963. Virus diseases on Tristan da Cunha. Trans. R. Soc. Trop. Med. Hyg. 57:19–22 [DOI] [PubMed] [Google Scholar]
- 50. Tipples GA, et al. 2002. New variant of varicella-zoster virus. Emerg. Infect. Dis. 8:1504–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tyler SD, et al. 2007. Genomic cartography of varicella-zoster virus: a complete genome-based analysis of strain variability with implications for attenuation and phenotypic differences. Virology 359:447–458 [DOI] [PubMed] [Google Scholar]
- 52. Vigilant L, Stoneking M, Harpending H, Hawkes K, Wilson AC. 1991. African populations and the evolution of human mitochondrial DNA. Science 253:1503–1507 [DOI] [PubMed] [Google Scholar]
- 53. Wagenaar TR, Chow VT, Buranathai C, Thawatsupha P, Grose C. 2003. The out of Africa model of varicella-zoster virus evolution: single nucleotide polymorphisms and private alleles distinguish Asian clades from European/North American clades. Vaccine 21:1072–1081 [DOI] [PubMed] [Google Scholar]
- 54. Wells S. 2002. The journey of man: a genetic odyssey. Random House, New York, NY [Google Scholar]
- 55. Whitley RJ, Nahmias AJ, Visintine AM, Fleming CL, Alford CA. 1980. The natural history of herpes simplex virus infection of mother and newborn. Pediatrics 66:489–494 [PubMed] [Google Scholar]
- 56. Wirgart BZ, Estrada V, Jackson W, Linde A, Grose C. 2006. A novel varicella-zoster virus gE mutation discovered in two Swedish isolates. J. Clin. Virol. 37:134–136 [DOI] [PubMed] [Google Scholar]
- 57. Wojcicki JM. 2003. Traditional behavioural practices, the exchange of saliva and HHV-8 transmission in sub-Saharan African populations. Br. J. Cancer 89:2016–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zell R, et al. 2012. Sequencing of 21 varicella-zoster virus genomes reveals two novel genotypes and evidence of recombination. J. Virol. 86:1608–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]