Abstract
Viral genome-linked proteins (VPgs) have been identified in several single-stranded positive-sense RNA virus families. The presence of such protein in the family Astroviridae has not been fully elucidated, although a putative VPg coding region in open reading frame 1a (ORF1a) of astrovirus with high amino acid sequence similarity to the VPg coding region of Caliciviridae has been previously identified. In this work we present several experimental findings that show that human astrovirus (HAstV) RNA encodes a VPg essential for viral infectivity: (i) RNase treatment of RNA purified from astrovirus-infected cells results in a single protein of 13 to 15 kDa, compatible with the predicted astrovirus VPg size; (ii) the antibody used to detect this 13- to 15-kDa protein is specifically directed against a region that includes the putative VPg coding region; (iii) the 13- to 15-kDa protein detected has been partially sequenced and the sequence obtained is contained in the computationally predicted VPg; (iv) the protein resulting from this putative VPg coding region is a highly disordered protein, resembling the VPg of sobemo-, calici- and potyviruses; (v) proteolytic treatment of the genomic RNA leads to loss of infectivity; and (vi) mutagenesis of Tyr-693 included in the putative VPg protein is lethal for HAstV replication, which strongly supports its functional role in the covalent link with the viral RNA.
INTRODUCTION
Viral genome-linked proteins (VPgs) are virus-encoded small proteins that are covalently linked to the 5′ terminus of many RNA viral genomes through a phosphodiester bond. Since the discovery of such a protein in the poliovirus genome, several animal and plant viruses have been reported to bear proteins covalently linked to their genomes (45). Among viruses with single-stranded positive-sense RNA (+ssRNA) genomes, VPgs have been described for animal viruses (Picornaviridae, Caliciviridae, and Dicistroviridae families), plant viruses (Comoviridae, Potyviridae, and Luteoviridae families, and Sobemovirus genus), and fungal viruses (Barnaviridae family) (36, 45). During the viral life cycle of these viruses, VPg performs multiple functions, playing a role in key processes such as genome replication, viral protein synthesis, and potentially genome encapsidation (12, 26).
A common feature of VPgs is that they are rich in basic amino acids [mostly Lys (K), Gly (G), Thr (T), and Arg (R)], which favors the interaction with the negatively charged RNA (30). For covalent binding of VPg to RNA, picornaviruses use the hydroxyl group of a conserved Tyr residue situated near the N terminus of VPg. Poty- and caliciviruses also use a Tyr residue, while comoviruses are reported to exploit a Ser residue (45). In the case of sobemoviruses, the residue for RNA linkage is not conserved within the genera and it appears to be species specific (37). Another common feature of VPgs is their intrinsic disorder. Recently, VPgs of some sobemoviruses and potyviruses were reported to be “natively unfolded proteins” (14, 21, 47), lacking both secondary and tertiary structures and existing as a dynamic ensemble of conformations at physiological conditions. Using computational predictions, disordered domains have been described in VPgs of several caliciviruses, such as rabbit hemorrhagic disease virus, vesicular exanthema of swine virus, Sapporo virus, Manchester virus, and Norwalk virus (21). This property has been postulated to be one of the factors that enable the functional diversity of VPgs (21).
Human astroviruses (HAstV) are recognized as common viral pathogens causing gastroenteritis in children (33). Astroviruses are nonenveloped +ssRNA viruses that belong to the family Astroviridae, which includes both mammalian and avian viruses. The HAstV genome is 6.8 kb in length, is polyadenylated, and contains three overlapping open reading frames (ORFs) and two untranslated regions. The nonstructural proteins (nsPs), which include a viral protease and an RNA-dependent RNA-polymerase (RdRp), are synthesized as polyproteins from ORF1a and ORF1b, respectively, using the genome RNA as the template, while the structural proteins are expressed from a subgenomic RNA produced after replication that contains ORF2 (33). The absence of a methyltransferase-coding region and the similarity of astrovirus RdRp with other polymerases of Koonin's supergroup I (which includes VPg-containing viruses, such as picornaviruses, caliciviruses, and certain plant viruses) suggest the possibility that the HAstV genome, as well as the subgenomic RNA, may be linked to a VPg protein on its 5′ end (1, 27, 29). Consistent with this idea, treatment of HAstV RNA with proteinase K abolishes the recovery of infectious viruses after transfection of wild-type RNA (50). Based on sequence comparisons between human and animal astroviruses with some members of the Caliciviridae, Picornaviridae, and Potyviridae families, a putative VPg coding region has been mapped downstream of the protease motif, coinciding partially with the nuclear localization signal (NLS), and Tyr-693 at the conserved TEEEY-like motif has been postulated to be the residue responsible for the covalent linkage to viral RNA (1, 29). Moreover, two conserved amino acid motifs characteristic of the N-terminal end of calicivirus VPg [KGK(N/T)K and (D/E)EY] can also be identified in astrovirus sequences (1, 8). With respect to the boundaries of this putative VPg, Al-Mutairy et al. (1) suggested a potential N-terminal proteolytic cleavage site [Q(K/A)] located upstream, either immediately next to, or 1 to 2 amino acid residues from the KGK(N/T) K motif and a C-terminal proteolytic cleavage site [Q(P/A/S/L)] between 92 and 143 amino acid residues downstream of the N-terminal cleavage site. Although the presence of VPg in the HAstV genome has been speculated for many years (27) and many observations suggest that HAstV may encode a VPg protein, its presence has not been experimentally proven yet. The main goal of our study was to identify and characterize the genome-linked protein of HAstV.
MATERIALS AND METHODS
Cells and viruses.
The human colon adenocarcinoma cell line CaCo2 was grown in Eagle's minimum essential medium (MEM) supplemented with 10% fetal bovine serum (FBS) and was used to propagate a cell culture-adapted strain of HAstV serotype 4 (HAstV-4) as well as to recover mutant viruses. CaCo2 cells were infected as previously described (38) with some modifications. Briefly, cell monolayers were washed twice with phosphate-buffered saline (PBS) and inoculated with viral stocks pretreated with 10 μg/ml of trypsin (GIX, Sigma) for 30 min at 37°C. After a 1-h adsorption at 37°C, a MEM overlay supplemented with 5 μg/ml of trypsin (for virus recovery) or 2% FBS (for immunofluorescence analysis) was added. Baby hamster kidney cells (BHK-21) and human hepatocellular carcinoma cells (Huh7.5.1) were grown in MEM supplemented with 10% FBS and were used to transfect HAstV RNA.
Detection of genome-linked proteins in HAstV RNA preparations.
Infected or mock-infected CaCo2 cells grown on 25 cm2 flasks were trypsinized at 24 h postinfection and pelleted at 3,000 × g for 10 min. After being washed with PBS, RNA from pelleted cells was extracted using GenElute Mammalian Total RNA miniprep kit (Sigma) and digested with 100 U of RNase I (Ambion) for 1 h at 37°C. The resulting preparations were separated by SDS-PAGE and subjected to blotting. Proteins were electrophoretically transferred to nitrocellulose membranes, and after being blocked with 5% skim milk–TBS buffer [50 mM Tris-HCl (pH 7.6) and 100 mM NaCl], membranes were probed with anti-nuclear addressing region (anti-NAR) serum kindly provided by M. J. Carter from the School of Biomedical and Molecular Sciences, University of Surrey, United Kingdom (52), which recognizes a region of about 30% of the C-terminal end of nsP1a polyprotein (residues 643 to 940 of strain A2/88 Newcastle, corresponding to the region spanning the protease motif and the end of ORF1a). Detection was performed using a horseradish peroxidase-conjugated anti-rabbit antibody and the enzyme reaction was developed with a SuperSignal West Femto kit (Pierce).
Protein identification by mass spectrometry.
Infected CaCo2 cells (from 10 175-cm2 flasks) were trypsinized at 24 h postinfection and pelleted at 3,000 × g for 10 min. After being washed with PBS, RNA from pelleted cells was extracted using an RNAqueous Midiprep kit (Ambion). Eluted RNA (1 ml) was precipitated with lithium chloride and resuspended in a final volume of 50 μl, prior to RNase I (Ambion) treatment with 1,000 U for 1 h at 37°C. The resulting preparation was separated by SDS-PAGE. The gel was then stained with silver nitrate (AgNO3), and the protein band of interest was excised and in-gel digested with trypsin (sequencing grade modified, Promega) in the automatic Investigator ProGest robot (Genomic Solutions). Tryptic peptides were then extracted from the gel matrix with 10% formic acid and acetonitrile, dried in a speed vac and analyzed by on-line liquid chromatography–nanoelectrospray ionization–tandem mass spectrometry (Cap-LC-nano-ESI-Q-TOF) (CapLC, Micromass-Waters) at the Proteomics Platform of Barcelona Science Park, University of Barcelona (a member of the ProteoRed network). Data were generated in PKL file format and submitted for database searching in the MASCOT server.
Disorder prediction.
Folding predictions were carried out using two software packages: the neural network predictor PONDR VL-XT (44) and FoldIndex (bioportal.weizmann.ac.il/fldbin/findex.) (39), which predict to what extent a given protein sequence is intrinsically unfolded. The analyses were performed using default values.
Analysis of proteolytical digestion of HAstV RNA over viral infectivity.
Total RNA from HAstV-infected CaCo2 cells was used to transfect BHK-21 cells. Prior to transfection, purified RNA was subjected to proteolytical digestion with 400 μg/ml of proteinase K (PK) (Sigma) for 1 h at 55°C in 10 mM Tris-HCl (pH 8), 1 mM EDTA and 0.5% sodium dodecyl sulfate (SDS). As a control, equivalent amounts of RNA were also treated as described above but with the omission of PK. After digestion, PK-treated and untreated RNAs were ethanol precipitated and quantified by spectrophotometry and by a one-step quantitative real-time reverse transcription-PCR (qRT-PCR) targeting a conserved region in HAstV ORF1b as previously described (46). The 91-bp target region of the HAstV genome incorporated into the pGEM plasmid was used to make a standard curve to quantify astrovirus RNA molecules. One day before transfection, cells were seeded on glass coverslips pretreated with poly-l-lysine (Sigma) in 6-well plates so that cells were approximately 60% confluent on the day of transfection. Cells were transfected using FuGENE HD Transfection Reagent (Roche) by following the manufacturer's instructions with some modifications. Briefly, the transfection mixture was prepared by diluting 5 μg of RNA in 100 μl of Opti-MEM I Reduced-Serum Medium (Invitrogen) and adding 10 μl of FuGENE HD Transfection Reagent. After a 25-min incubation at room temperature, the mixture was added to cells previously washed twice with PBS and maintained in 2 ml Opti-MEM I Reduced-Serum Medium. Every transfection experiment included a negative control where cells were mock transfected in the absence of RNA. Evidence for astrovirus replication was monitored by detecting viral proteins by immunofluorescence at 24 h posttransfection and viral RNA by RT-PCR at 12 and 24 h posttransfection (see below).
Construction of VPg mutants.
To introduce mutations in the VPg coding region, the pAVIC infectious clone kindly provided by S. Matsui (Stanford University) was used as starting material (11). This plasmid contains the full-length cDNA of the HAstV-1 Oxford strain, from which infectious RNA can be generated using T7 RNA polymerase and a cap analogue. A BglII-XbaI fragment of pAVIC (positions 1657 to 3156), containing the putative VPg coding region, was inserted into the pCITE-2a vector. Mutations were introduced into the VPg coding sequence in plasmid pCITE-2a by site-directed mutagenesis using the QuikChange mutagenesis protocol (Stratagene) and the oligonucleotides shown in Table 1. Putative mutants were screened by sequence analysis, and the mutated BglII-XbaI fragments were reinserted in the pAVIC plasmid. Full-length mutant and wild-type HAstV cDNAs were obtained from the corresponding plasmids by PCR using the Expand Long Range dNTPack kit (Roche). The primers used in the PCR were 5′ end modified to contain the T7 promoter sequence and 3′ end modified to contain a 43-mer poly(T) tail. The T7-dT-amplified DNAs were then used as templates for in vitro transcription reactions with the mMessage mMachine transcription system (Ambion) to obtain wild-type and modified capped transcripts containing the engineered mutations, i.e., Y693A, Y693S, Y720A, Y723A, Y728A, Y729A, Y747A, and L701I. Following treatment of the transcription mixture with DNase, the RNA was purified by lithium chloride precipitation, quantified by spectrophotometry and analyzed on agarose gels.
Table 1.
Oligonucleotides used for construction of VPg mutants using the QuikChange mutagenesis protocol (Stratagene)
| Mutation | Sense | 5′–3′ sequence |
|---|---|---|
| Y693A | + | CTTCTTACTGAGGAAGAGGCTCGAGAACTCTTAGAGAAAGG |
| Y693A | − | CCTTTCTCTAAGAGTTCTCGAGCCTCTTCCTCAGTAAGAAG |
| Y720A | + | GGTGAGAGGTCTGGCGCCCCTGACTATGATGATG |
| Y720A | − | CATCATCATAGTCAGGGGCGCCAGACCTCTCACC |
| Y723A | + | GGTCTGGCTACCCTGACGCTGATGATGAAGATTACTATG |
| Y723A | − | CATAGTAATCTTCATCATCAGCGTCAGGGTAGCCAGACC |
| Y728A | + | CCCTGACTATGATGATGAAGATGCCTATGATGAAGATGATGATGG |
| Y728A | − | CCATCATCATCTTCATCATAGGCATCTTCATCATCATAGTCAGGG |
| Y729A | + | GATGATGAAGATTACGCTGATGAAGATGATGATGGCTGGGG |
| Y729A | − | CCCCAGCCATCATCATCTTCATCAGCGTAATCTTCATCATC |
| Y747A | + | GGGGAATGGTTGGTGATGATGTAGAATTTGATGCTACTGAAGTG |
| Y747A | − | CACTTCAGTAGCATCAAATTCTACATCATCACCAACCATTCCCC |
| L701I | + | CGAGAACTCTTAGAGAAAGGTATAGACCGTGAGACATTCC |
| L701I | − | GGAATGTCTCACGGTCTATACCTTTCTCTAAGAGTTCTCG |
| Y693S | + | CTTCTTACTGAGGAAGAGTCTCGAGAACTCTTAGAGAAAGG |
| Y693S | − | CCTTTCTCTAAGAGTTCTCGAGACTCTTCCTCAGTAAGAAG |
Analysis of VPg mutations over viral replication and rescue of mutant viruses.
Wild type and modified capped transcripts containing the engineered mutations, i.e., Y693A, Y693S, Y720A, Y723A, Y728A, Y729A, Y747A, and L701I, were used to transfect both BHK-21 and Huh7.5.1 cells, essentially as described above. In both cases, astrovirus replication was monitored by detecting viral proteins by immunofluorescence at 24 h posttransfection. To recover mutant viruses, Huh7.5.1 cells were incubated for an additional 48 h in the presence of 5 μg/ml of trypsin before being subjected to 3 cycles of freeze/thawing and centrifuged at 3,000 × g to discard cell debris. Lysates from mock- and RNA-transfected Huh7.5.1 cells were then used for sequential passaging on CaCo2 cells previously treated with 10 μg/ml of trypsin for 30 min at 37°C. Astrovirus replication in infected CaCo2 cells was evaluated by immunofluorescence, and the recovered viruses were examined by RT-PCR and sequence analysis using the ABI Prism BigDye Terminator Cycle Sequencing Ready reaction kit (Applied Biosystems).
Immunofluorescence.
Cells were rinsed twice with PBS and fixed with 3% paraformaldehyde in PBS for 20 min at room temperature. Permeabilization was performed for 10 min at room temperature with 0.5% Triton X-100 in 20 mM glycine-PBS. After being washed, cells were blocked for 1 h at room temperature in 20 mM glycine-PBS containing 10% FBS. Cells were incubated overnight at 4°C with a 1:10,000 dilution of the monoclonal antibody (MAb) 8E7 (kindly provided by P. Sanders at R-Biopharm AG), which recognizes astroviral structural proteins (23). An indocarbocyanine 3-conjugated anti-mouse antibody was used as the secondary antibody at a 1:5,000 dilution. After immunofluorescence labeling, cells were stained with 1 μg/ml of DAPI (4′,6′-diamidino-2-phenylindole) for 15 min at room temperature. Mounting was done in Mowiol, and cells were visualized under a fluorescence microscope (Olympus BX61). The percentage of transfected cells expressing viral proteins was estimated by counting the number of positive cells and the total number of cells from 4 different fields per sample.
Detection of HAstV RNA by conventional RT-PCR.
The effect of proteolytical treatment of viral RNA with PK on viral replication in transfected BHK-21 cells was evaluated by RT-PCR. HAstV RNA was detected using the previously described primers A1 and A2 (18, 51), which amplify a fragment of ORF1a, using the A1 forward primer in the RT reaction in order to detect mainly antigenomic RNA. Amplified products were analyzed on a 1.5% agarose gel and detected by ethidium bromide staining.
RESULTS
Identification and sequencing of a viral protein covalently linked to HAstV RNA.
To analyze the presence of a VPg protein linked to the HAstV genome, total RNA (together with proteins covalently associated with it) was isolated from HAstV-4-infected or mock-infected CaCo2 cells and treated with RNase I. Analysis of the treated samples by Western blotting with anti-NAR serum, which is directed against the C-terminal end of the nsP1a polyprotein, containing the putative VPg coding region, revealed a protein copurified with the RNA with an estimated molecular mass in the range of 13 to 15 kDa, which is very close to the expected molecular mass of the product resulting from the VPg coding region suggested by Al-Mutairyet et al. (11 kDa) (1) (Fig. 1A). No bands were detected in samples extracted from mock-infected cells either with or without RNase digestion or from samples extracted from infected cells without RNase treatment, where proteins linked to RNA remained at the top of the resolving gel. When analyzing total cell extracts instead of purified RNA samples, two main proteins of ∼32 and ∼17 kDa reacted with the anti-NAR antibody, as well as a high molecular mass protein (Fig. 1A), consistent with data presented before (52). Whether the ∼17-kDa protein detected in cell extracts corresponds to a precursor protein of the 13- to 15-kDa protein bound to the RNA or to a protein with a different pattern of posttranslational modifications remains to be elucidated.
Fig 1.
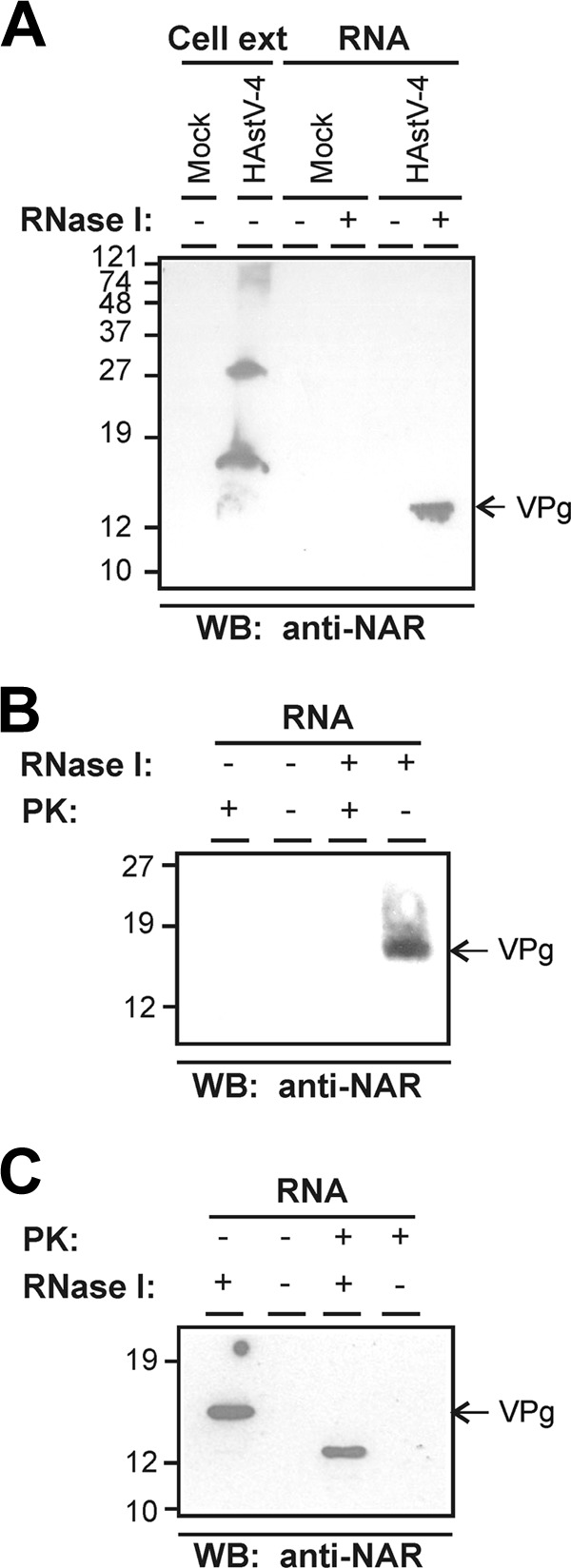
Identification of a viral protein covalently associated with HAstV RNA. (A) Western blotting (WB) analysis of HAstV-4-infected CaCo2 cell extracts and proteins that were copurified with RNA isolated from mock-infected and infected cells at 24 h postinfection and stained with anti-NAR antiserum. RNA-copurified proteins were analyzed before and after treatment with RNase I. (B) Proteolytical digestion of the RNA-associated protein was evaluated by incubation with proteinase K (PK). RNA samples purified from infected cells were mock treated or treated first with RNase I and subsequently incubated with or without PK. Complete digestion of the 13- to 15-kDa protein was confirmed by WB using the anti-NAR antibody. (C) Susceptibility of the VPg protein to PK digestion before the RNase I treatment. Samples treated first with PK and second with RNase I were analyzed by WB with the anti-NAR antibody. Molecular size marker positions in kilodaltons (kDa) are indicated on the left of each panel. Slight mobility pattern differences between the VPg protein after RNase treatment in panels A, B, and C may be explained by the erratic electrophoretic mobility of disordered proteins and/or the efficiency of the RNase treatment.
Sensitivity of the 13- to 15-kDa protein to proteolytical digestion was analyzed by treating viral RNA purified from infected cells with PK. RNA samples extracted from infected cells were treated with RNase I as described above and further treated with PK, resulting in a complete degradation of the 13- to 15-kDa protein (Fig. 1B). No protein bands were detected in the controls untreated with RNase I, since undegraded RNA does not enter into the gel. However, when PK digestion was performed prior to RNase I treatment, a partially degraded protein of 12 kDa was detected (Fig. 1C). This partial digestion of the 13- to 15-kDa protein could be explained by a certain degree of protection of the protein against proteolysis by its covalent linkage to RNA.
To confirm the identity of the 13- to 15-kDa protein as the predicted VPg, the protein band was excised from a silver-stained gel (Fig. 2A) and analyzed by combined liquid chromatography-tandem mass spectrometry, with previous trypsin treatment. An additional band of 27 kDa corresponding to the RNase I used to digest the RNA was also detected. Sequencing of the 13- to 15-kDa protein allowed the identification of a peptide of 15 amino acids in length (amino acids 699 to 713 of the nsP1a polyprotein, i.e., KGLDRETFLDLIDRI), which represents 16% of the full-length predicted VPg sequence (Fig. 2B). Although we failed to obtain the complete sequence with the exact boundaries of VPg, the molecular weight of the detected protein is consistent with the cleavage sites predicted by Al-Mutairy et al. (1).
Fig 2.
Silver staining of HAstV VPg and protein identification by mass spectrometry. (A) Total RNA (and proteins associated with it) isolated from cultures of HAstV-4-infected CaCo2 cells at 24 h postinfection was analyzed by SDS-PAGE and silver staining after being treated with RNase I. (B) The 13- to 15-kDa protein band was excised and treated with trypsin for protein identification by Cap-LC-nano-ESI-Q-TOF. The 15-mer peptide identified is indicated in bold over the full length of the predicted HAstV VPg amino acid sequence.
Astrovirus VPg is predicted to be an intrinsically disordered protein (IDP).
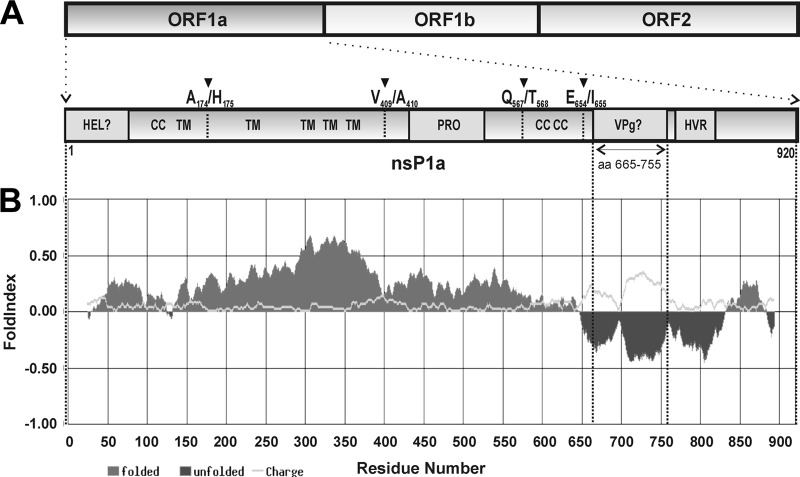
VPgs of several viruses have been described to be intrinsically disordered proteins (14, 21, 47). Considering this common feature, we proceeded to computationally analyze the disorder degree of predicted astrovirus VPg. Full-length polyproteins of HAstV-4 (GenBank accession no. AY720891) were submitted to disorder prediction with FoldIndex software program (39). As shown in Fig. 3, prediction clearly shows that the putative VPg, located between amino acids 665 and 755 of HAstV nsP1a polyprotein, lies in the unfolded region of the polyprotein, in contrast to other astroviral proteins. Similar results were obtained with the predictor PONDR, and results did not change when the VPg sequence was analyzed in isolation (data not shown). Except for the structural polyprotein, where the N-terminal and C-terminal regions were predicted to be unfolded regions (data not shown), the VPg sequence was the only relatively large region within the HAstV genome with a high disorder score. Interestingly, this feature was conserved among all other mammal and avian astroviruses that have been completely sequenced (data not shown).
Fig 3.
Predicted disorder of HAstV nsP1a polyprotein. (A) Schematic representation of the HAstV genome organization and nsP1a polyprotein (enlarged fragment). The predicted transmembrane helices (TM), protease motif (PRO), predicted helicase domain (HEL), coiled-coil structures (CC), predicted genome-linked protein (VPg) and hypervariable region (HVR) are indicated. Arrowheads refer to putative identified proteolytic cleavage sites (33). (B) FoldIndex prediction for HAstV nsP1a polyprotein containing the predicted VPg domain between amino acids 665 and 755.
VPg protein is essential for viral RNA infectivity.
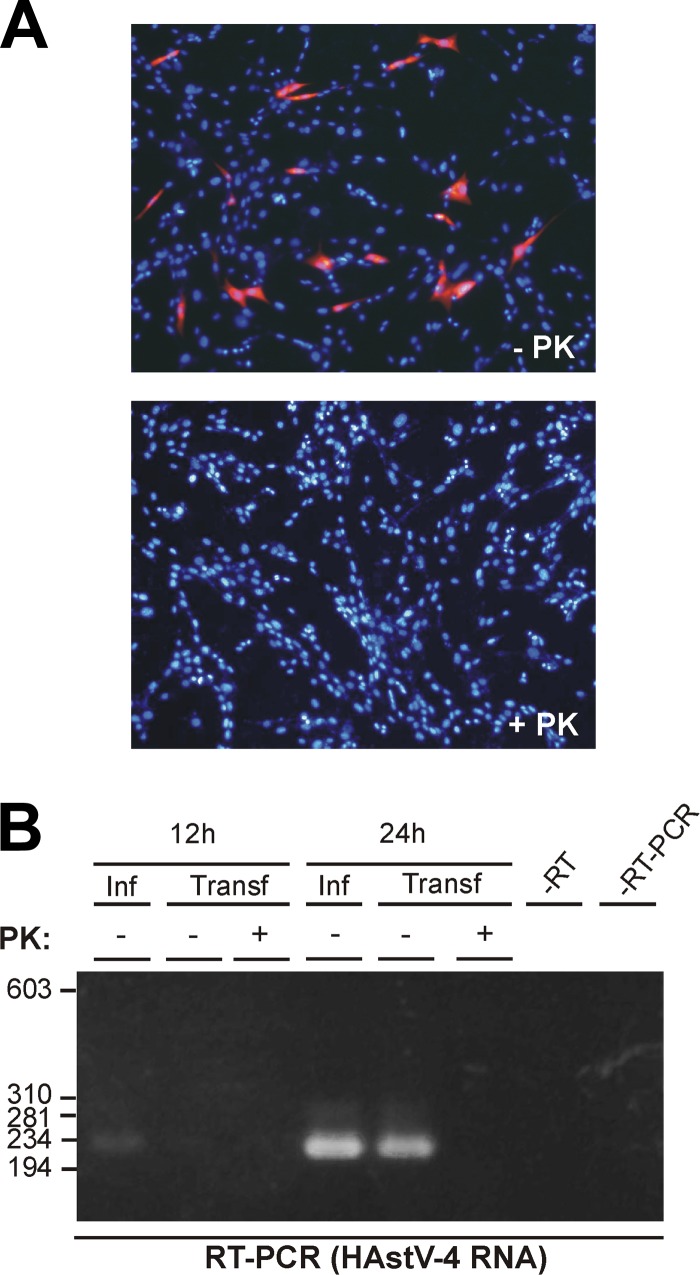
Total RNA isolated from HAstV-4-infected CaCo2 cells was used to transfect BHK-21 cells after treatment with or without PK. HAstV replication in transfected cells was analyzed by immunofluorescence using the 8E7 MAb, which is directed against astroviral structural proteins, and by detection of the viral replicating RNA by conventional RT-PCR. While, in the absence of PK treatment, approximately 5% of transfected cells expressed viral structural proteins, PK treatment completely abolished viral expression (Fig. 4A), suggesting that the covalent linkage of VPg to the viral genome plays a pivotal role in infectivity. Proteolytical treatment of viral RNA also resulted in the abolishment of viral RNA transcription, since astrovirus RNA (mostly replicating negative-sense RNA) was detected by RT-PCR using the forward primer in the RT reaction only in cells transfected with PK-untreated RNA (Fig. 4B). Nevertheless, a direct role of VPg on viral transcription cannot be assumed from these experiments, since loss of RNA replication could be due to abolishment of expression of viral proteins, including nonstructural proteins involved in viral RNA synthesis.
Fig 4.
Effects of proteolytical digestion on HAstV RNA infectivity. RNA isolated from HAstV-infected CaCo2 cells, treated or untreated with proteinase K (PK), was used to transfect BHK-21 cells. Viral replication was analyzed by detecting viral structural proteins by immunofluorescence analysis at 24 h posttransfection using the 8E7 MAb (magnification, ×100), (A) and replicating viral RNA by RT-PCR at 12 h and 24 h posttransfection (B). RNAs isolated from infected CaCo2 cells at 12 h and 24 h postinfection were used as positive controls of RT-PCR. Molecular size marker positions in base pairs (bp) are indicated on the left of the corresponding panel.
As a control prior to transfection, purified HAstV RNA was quantified by qRT-PCR in order to ensure transfection with equal amounts of genome copy numbers. No difference in titers of HAstV-4 RNA genome copies in the absence and presence of PK [(5.318 ± 0.22) × 109 and (4.953 ± 0.18) × 109 HAstV-4 RNA copies/μg of RNA, respectively] was observed.
Mutagenesis of Tyr-693 in the VPg protein is lethal for HAstV replication.
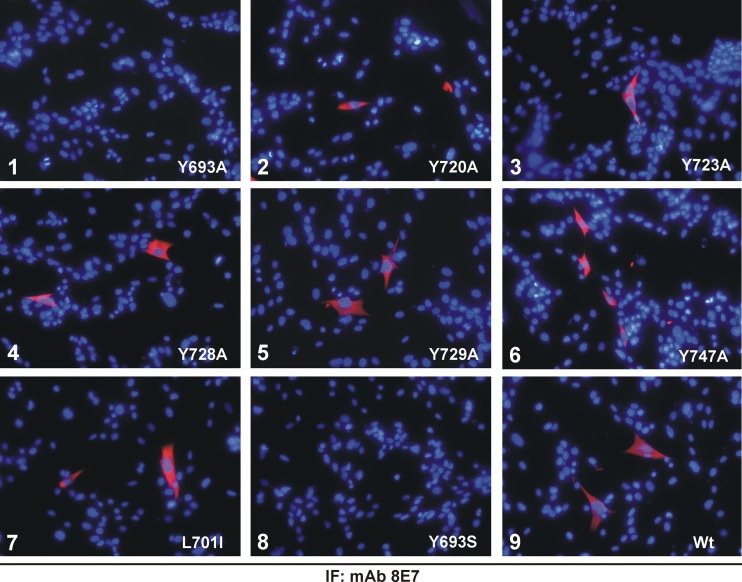
A mutational analysis of the VPg region of HAstV was performed to determine whether Tyr-693, predicted as the potential residue involved in covalent linkage to viral RNA (1, 29), is essential for protein function. Putative HAstV VPg contains, in addition to Tyr-693, five Tyr residues, at positions 720, 723, 728, 729, and 747, that could potentially link VPg to viral RNA (positions are numbered according to GenBank accession no. L23513). When comparing HAstV sequences to other astrovirus and calicivirus sequences, it can be seen that the only Tyr residue which is 100% conserved among all isolates, including both mammalian and avian astroviruses as well as caliciviruses, is Tyr-693, which is contained within the (D/E)EY motif (Fig. 5). Tyr-723 and Tyr-747 are also highly conserved, but some astrovirus and calicivirus strains show semiconservative substitutions to Trp or Phe residues. Finally, while Tyr-720 shows strong conservation only among mammalian astroviruses, Tyr-728 and Tyr-729 show a minor degree of conservation. All these Tyr residues were mutated to Ala using the infectious HAstV-1 clone. Mutation of Leu-701 to Ile was used as a control mutation. Capped transcripts obtained from wild-type and mutated plasmids were first used to transfect BHK-21 cells, and astrovirus replication was monitored by immunofluorescence using the 8E7 MAb. Cells expressing astrovirus capsid proteins were detected in cells transfected with the Y720A, Y723A, Y728A, Y729A, and Y747A constructs, as well as with the wild-type and control constructs (L701I) (Fig. 6). Transfection with the Y693A mutant did not show positive cells by immunofluorescence, showing that Tyr-693 is essential for virus replication. In no case were mock-transfected cells found to express astroviral structural proteins (data not shown).
Fig 5.
Multiple alignment of astrovirus (AstV) and calicivirus (CV) sequences in the region predicted for astrovirus VPg. Sequence alignment was performed using the ClustalW2 tool (31). The following representative strains were included in the analysis (GenBank accession numbers are indicated in parentheses): human astrovirus type 1 strain Oxford (HAstV-1) (L23513), human astrovirus type 4 strain Dresden (HAstV-4) (AY720891), human astrovirus MLB-1 (MLB-1) (FJ222451), bovine astrovirus B76/HK (BAstV) (HQ916316), porcine astrovirus 5 strain 33/USA (PAstV) (JF713711), human astrovirus VA-1 (VA-1) (NC_013060), ovine astrovirus (OAstV) (NC_002469), mink astrovirus (MAstV) (AY179509), turkey astrovirus 2 (TAstV-2) (NC_005790), duck astrovirus 1 strain DA93 (DAstV) (FJ919228), and chicken astrovirus (CAstV) (NC_003790) for AstVs and Norwalk virus (HuCV/NV) (NP_786948), Lordsdale virus (HuCV/Lordsdale) (X86557), feline calicivirus (FCV) (NP_783307), and rabbit hemorrhagic disease virus (RHDV) (NP_740330) for CVs. Complete and high-rate amino acid identity (including conservative and semiconservative substitutions) are visualized by a gray shading. Gaps are denoted by dashes. Conserved KGK(N/T)K and (D/E)EY motifs are marked by boxes. The six Tyr residues at positions 693, 720, 723, 728, 729, and 747 (numbered according to the HAstV-1 Oxford reference strain [L23513]) which were changed to Ala are indicated with arrowheads. The Tyr at position 693 was also changed to Ser, and mutation of Leu-701 to Ile was used as a control mutation.
Fig 6.
Effect of VPg point mutations on viral replication. Immunofluorescence analysis of astrovirus replication on BHK-21 cells transfected with capped transcripts derived from the wild-type (Wt) plasmid (pAVIC) or the corresponding plasmids containing the engineered mutations in the VPg coding region (panel 1: Y693A; panel 2: Y720A; panel 3: Y723A; panel 4: Y728A; panel 5: Y729A; panel 6: Y747A; panel 7: L701I; panel 8: Y693S; panel 9: Wt). Detection of astroviral structural proteins on transfected BHK-21 cells was performed at 24 h posttransfection using the 8E7 MAb. Magnification, ×200.
Since the efficiency of the reverse genetics system described using BHK-21 cells is low (11), the method described by Velázquez-Moctezuma et al. using Huh7.5.1 cells (50) was used for the rescue of infectious viruses. Immunofluorescence analysis was also used to monitor viral replication in Huh7.5.1 cells after transfection, and results were similar to those obtained with BHK-21 cells (data not shown). Virions recovered from transfected Huh7.5.1 cells were amplified by subsequent passages in CaCo2 cells. Again, only the HAstV Y720A, Y723A, Y728A, Y729A, and Y747A mutants, as well as the wild-type and control constructs (L701I), could be rescued and further passaged in CaCo2 cells, but mutation Y693A was lethal. The rescued viruses were analyzed by RT-PCR, and sequence analysis confirmed the presence of the engineered mutation in the RNA genome. Interestingly, it is of note that the Y728A, Y729A, and L701 mutants were rescued to high titers during early passages, showing no differences from wild-type virus, while the Y720A, Y723A, and Y747A mutants, which contain mutations at Tyr residues that are more conserved (Fig. 5), were harder to rescue to high titers (data not shown).
Additionally, it was also examined whether replacement of Tyr-693 by Ser, with its available hydroxyl group, could render infectious viruses, but this mutation was lethal as well and no structural protein expression was detected by immunofluorescence in the original transfection (Fig. 6) or in subsequent passages on CaCo2 cells.
DISCUSSION
Although the presence of a viral genome-linked protein in HAstV and its genomic localization has been speculated and suggested by several authors in the past (1, 27, 29, 50), its existence has not yet been experimentally demonstrated. In the current study, we discuss several experimental findings that support the presence of such a protein in the HAstV genome. Analysis of the proteins copurified with astrovirus RNA by Western blotting with an antibody against the C-terminal region of astrovirus nsP1a, containing the putative VPg region, revealed a protein of 13 to 15 kDa. Direct sequencing of this protein confirmed that it contains at least 16% of the central part of the computationally predicted VPg (1). Although the complete amino acid sequence of the protein was not obtained due to technical limitations, i.e., getting a large amount of such a small protein, the molecular mass of the detected band was very close to that expected for the predicted VPg (1), which is 11 kDa. In addition, anti-1a778–792 antiserum, which is directed against a synthetic peptide (from residue 778 to 792) contained within the hypervariable region (HVR) of nsP1a (16), did not react with the 13- to 15-kDa protein (data not shown), which is consistent with the predicted C-terminal cleavage site published by Al-Mutairy et al. (1) at residue Q755.
The genomic localization of astrovirus VPg (protease-VPg-polymerase) is similar to that of the VPgs of the sobemovirus-like supergroup (sobemo-, luteo- and barnaviruses) and differs from that of the VPgs of the picornavirus-like supergroup (picorna-, calici-, poty-, como-, and dicistroviruses), in which the VPg coding region is located upstream of the viral protease motif (45). HAstV VPg is located downstream of the protease motif close to the 3′ end of ORF1a and is included in the C-terminal nsP1a protein (16). Data using the anti-1a778–792 antibody against the HVR region showed that the C-terminal nsP1a protein accumulates in the perinuclear region, in association with the endoplasmic reticulum and the viral RNA (16). Whether this colocalization with viral RNA takes place before or after the proteolytic cleavage that releases the VPg protein is still unknown and needs to be further studied. However, due to the high hydrophilic amino acid content of VPg, it is likely that the C-terminal nsP1a protein or even a larger protein could function as a hydrophobic precursor, directing VPg to cellular membranes prior to the proteolytic cleavage necessary to release mature VPg, similar to what occurs with poliovirus, in which precursor 3AB is a hydrophobic membrane-bound protein that acts as a VPg donor in membranous replication complexes for RNA synthesis (41).
VPg protein detected on SDS-PAGE gels shows a molecular mass slightly higher than expected (13 to 15 kDa versus 11 kDa), and this could be due to several causes. Since purified proteins are digested with RNase prior to electrophoresis, the slight increase in molecular mass could be due to a small number of residual nucleotides linked to the protein. Alternatively, it could also be due to posttranslational modifications of the protein, such as phosphorylation. In fact, the C-terminal nsP1a protein expressed in insect cells is modified posttranslationally by phosphorylation (10), and although phosphorylated residues were not specifically identified, some of the potential phosphorylation sites are located within the putative VPg region, specifically the Tyr at positions 693, 728, 729 and 747 (17). Indeed, phosphorylation has been described as a posttranslational modification of sobemo- and potyviral VPgs (37, 40). Moreover, the C-terminal nsP1a protein interacts with itself to form oligomers and with the viral RdRp (10). Interestingly, the oligomerization and polymerase interaction domains map between residues 744 and 777 and residues 655 and 743 of nsP1a polyprotein (10), respectively, which lie partially in the first case and almost completely in the second case in the putative VPg region (amino acids 665 to 755). These potential multiple binding capacities of astrovirus VPg have also been described for VPg proteins or their precursors in other viruses, such as poliovirus and potyvirus (9, 12, 26, 41, 53), and may be related to its disordered structure. Using computational predictions, we found that HAstV VPg, like sobemo-, poty- and calicivirus' VPgs (14, 21, 47), is an IDP. A larger plasticity of disordered proteins would combine high specificity with low affinity to ensure faster association and dissociation rates, enable transient binding of numerous structurally distinct targets, provide the ability to overcome steric restrictions, and facilitate the participation in multiple biological processes, such as cell cycle control, transcriptional and translational regulation, membrane fusion and transport, and signal transduction (48). Since many viruses show a restricted genome size, they rely on the multifunctionality of their proteins and have been shown to contain the highest proportion of proteins with conserved predicted disordered regions, compared to archaea, bacteria and eukaryota (5). Interestingly, intrinsic disorder has been shown to be important for protein phosphorylation, and it is hypothesized that phosphorylation occurs predominantly within IDP regions (24). As reversible modifications, phosphorylations are well known to regulate many processes of +ssRNA virus life cycles, including replication (25). Phosphorylation of IDPs seems to be important for their structural and functional regulation, since both disorder to order and order to disorder transitions have been observed to follow the phosphorylation event (28), with conformational changes often affecting protein function. Based on these considerations, a regulation of VPg function dependent on its phosphorylation status could be proposed.
VPg proteins of other +ssRNA viruses have been related to both translation initiation of the viral genome and protein-primed RNA replication processes (12, 26). In the case of caliciviruses, removal of VPg by proteolytical treatment of RNA dramatically reduces both the infectivity (15, 22) and translation of RNA in vitro (22), suggesting a role in viral protein expression. Similar observations have been described for other viruses, such as nepo- and sobemoviruses (20, 49). Additional evidence for the involvement of calicivirus VPg in translation initiation has been provided by interactions between VPg and eIF3 and eIF4E and eIF4GI (4, 6, 7, 13). Besides playing a role in translation, calicivirus VPg may act as a primer during viral RNA replication, since it has been shown that it can be nucleotidylylated by the viral RdRp or its precursor (2, 19, 32) and that the RdRp can synthesize VPg-primed RNA in vitro (43). In contrast, in the case of picornaviruses, it is clear that VPg plays a role in the initiation of RNA replication, while is dispensable for translation. Participation of VPg on viral transcription has been extensively described for poliovirus, in which an uridylylated form of VPg produced by the 3Dpol is thought to play a role in priming both negative- and positive-strand synthesis (41). Since the picornavirus genome translation is dependent on an internal ribosome entry site (IRES), proteolytic treatment of RNA neither leads to a loss of infectivity nor has an effect on its translatability in vitro (41). Data here presented show that PK treatment of HAstV RNA completely abolishes infectivity, as measured by both viral protein synthesis by immunofluorescence and production of viral RNA molecules in transfected cells, and this highlights a strong similarity to the calicivirus strategy, where VPg is essential for genome translation. Consistently, HAstV VPg's size and amino acid sequence are more similar to calicivirus VPgs than picornavirus VPgs, which are relatively smaller proteins (approximately 2 to 6 kDa). If HAstV VPg was not required for translation of the genomic RNA molecules released in the cytoplasm after cell entry, transfection of RNAs that have been treated with PK would have resulted in both expression of viral proteins and viral replication. Although other observations, such as the requirement of a cap analogue at the 5′ end of RNAs transcribed in vitro for their infectivity after transfection and the lack of identification of an IRES sequence within the HAstV genome, also support the hypothesis that HAstV VPg may interact with the cellular machinery to initiate translation, additional studies on genomic RNA translation should be performed in vitro in order to confirm this idea.
Since it has been previously shown that the astrovirus RdRp interacts with a domain of nsP1a protein which matches almost completely VPg (10), we could speculate that astrovirus VPg gets nucleotidylylated and also plays a role in viral transcription, similarly to what occurs in poliovirus, caliciviruses, and other VPg-containing viruses. Although further studies should be performed to determine the specific role of astrovirus VPg on virus replication, our results clearly show that its function is dependent on Tyr-693, which is conserved among all human and animal astroviruses studied to date. As demonstrated for poliovirus (42), potyvirus (35), calicivirus (34), and comovirus (3), mutation of the Tyr or Ser residue responsible for the linkage to RNA or a change in its position within VPg results in a lethal phenotype. In the present study, we demonstrate that mutagenesis of Tyr-693 to Ala, and even its replacement by Ser, which also provides a hydroxyl group, is lethal for astrovirus replication, strongly supporting the functional role of this residue in forming the covalent linkage to viral RNA. Since transfection of cells with capped Y693A and Y693S mutant transcripts does not produce infectious viruses, our hypothesis is that VPg may be required to initiate the synthesis of all types of viral RNAs produced within cells during infection. Under these experimental conditions, initial capped transcripts would have been translated upon cell entry, but VPg proteins synthesized with mutated Tyr-693 residue would have been unable to assist in the synthesis of nascent RNAs. Finally, our results also indicate that Tyr residues at positions 720, 723, and 747, although not critical for virus infectivity, may also be important for replication efficiency. Characterization of HAstV VPg protein will not only provide insights into the replication cycle of this human pathogen but also help to identify novel unique antiviral targets.
ACKNOWLEDGMENTS
C. Fuentes was recipient of an FI fellowship from the Generalitat de Catalunya. This work was supported in part by grants 2005SGR00966 and 2009SGR00024 and the Biotechnology Reference Network of the Generalitat de Catalunya.
We acknowledge the technical expertise of the Barcelona Science Park of the University of Barcelona. We are grateful to M. J. Carter, S. M. Matsui, and P. Sanders for providing reagents.
Footnotes
Published ahead of print 11 July 2012
REFERENCES
- 1. Al-Mutairy B, Walter JE, Pothen A, Mitchell DK. 2005. Genome prediction of putative genome-linked viral protein (VPg) of astroviruses. Virus Genes 31:21–30 [DOI] [PubMed] [Google Scholar]
- 2. Belliot G, Sosnovtsev SV, Chang KO, McPhie P, Green KY. 2008. Nucleotidylylation of the VPg protein of a human norovirus by its proteinase-polymerase precursor protein. Virology 374:33–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Carette JE, et al. 2001. Mutational analysis of the genome-linked protein of cowpea mosaic virus. Virology 290:21–29 [DOI] [PubMed] [Google Scholar]
- 4. Chaudhry Y, et al. 2006. Caliciviruses differ in their functional requirements for eIF4F components. J. Biol. Chem. 281:25315–25325 [DOI] [PubMed] [Google Scholar]
- 5. Chen JW, Romero P, Uversky VN, Dunker AK. 2006. Conservation of intrinsic disorder in protein domains and families: I. A database of conserved predicted disordered regions. J. Proteome Res. 5:879–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Daughenbaugh KF, Fraser CS, Hershey JW, Hardy ME. 2003. The genome-linked protein VPg of the Norwalk virus binds eIF3, suggesting its role in translation initiation complex recruitment. EMBO J. 22:2852–2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Daughenbaugh KF, Wobus CE, Hardy ME. 2006. VPg of murine norovirus binds translation initiation factors in infected cells. Virol. J. 3:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dunham DM, Jiang X, Berke T, Smith AW, Matson DO. 1998. Genomic mapping of a calicivirus VPg. Arch. Virol. 143:2421–2430 [DOI] [PubMed] [Google Scholar]
- 9. Fellers J, Wan J, Hong Y, Collins GB, Hunt AG. 1998. In vitro interactions between a potyvirus-encoded, genome-linked protein and RNA-dependent RNA polymerase. J. Gen. Virol. 79:2043–2049 [DOI] [PubMed] [Google Scholar]
- 10. Fuentes C, Guix S, Bosch A, Pintó RM. 2011. The C-terminal nsP1a protein of human astrovirus is a phosphoprotein that interacts with the viral polymerase. J. Virol. 85:4470–4479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Geigenmüller U, Ginzton NH, Matsui SM. 1997. Construction of a genome-length cDNA clone for human astrovirus serotype 1 and synthesis of infectious RNA transcripts. J. Virol. 71:1713–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goodfellow I. 2011. The genome-linked protein VPg of vertebrate viruses—a multifaceted protein. Curr. Opin. Virol. 1:355–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goodfellow I, et al. 2005. Calicivirus translation initiation requires an interaction between VPg and eIF4E. EMBO Rep. 6:968–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Grzela R, et al. 2008. Virulence factor of potato virus Y, genome-attached terminal protein VPg, is a highly disordered protein. J. Biol. Chem. 283:213–221 [DOI] [PubMed] [Google Scholar]
- 15. Guix S, et al. 2007. Norwalk virus RNA is infectious in mammalian cells. J. Virol. 81:12238–12248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guix S, Caballero S, Bosch A, Pintó RM. 2004. C-terminal nsP1a protein of human astrovirus colocalizes with the endoplasmic reticulum and viral RNA. J. Virol. 78:13627–13636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guix S, Caballero S, Bosch A, Pintó RM. 2005. Human astrovirus C-terminal nsP1a protein is involved in RNA replication. Virology 333:124–131 [DOI] [PubMed] [Google Scholar]
- 18. Guix S, et al. 2002. Molecular epidemiology of astrovirus infection in Barcelona, Spain. J. Clin. Microbiol. 40:133–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Han KR, et al. 2010. Murine norovirus-1 3Dpol exhibits RNA-dependent RNA polymerase activity and nucleotidylylates on Tyr of the VPg. J. Gen. Virol. 91:1713–1722 [DOI] [PubMed] [Google Scholar]
- 20. Harrison BD, Barker H. 1978. Protease-sensitive structure needed for infectivity of nepovirus RNA. J. Gen. Virol. 40:711–715 [Google Scholar]
- 21. Hébrard E, et al. 2009. Intrinsic disorder in viral proteins genome-linked: experimental and predictive analyses. Virol. J. 6:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Herbert TP, Brierley I, Brown TD. 1997. Identification of a protein linked to the genomic and subgenomic mRNAs of feline calicivirus and its role in translation. J. Gen. Virol. 78:1033–1040 [DOI] [PubMed] [Google Scholar]
- 23. Herrmann JE, Hudson RW, Perron-Henry DM, Kurtz JB, Blacklow NR. 1988. Antigenic characterization of cell-cultivated astrovirus serotypes and development of astrovirus-specific monoclonal antibodies. J. Infect. Dis. 158:182–185 [DOI] [PubMed] [Google Scholar]
- 24. Iakoucheva LM, et al. 2004. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 32:1037–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jakubiec A, Jupin I. 2007. Regulation of positive-strand RNA virus replication: the emerging role of phosphorylation. Virus Res. 129:73–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jiang J, Laliberté JF. 2011. The genome-linked protein VPg of plant viruses-a protein with many partners. Curr. Opin. Virol. 1:347–354 [DOI] [PubMed] [Google Scholar]
- 27. Jiang B, Monroe SS, Koonin EV, Stine SE, Glass RI. 1993. RNA sequence of astrovirus: distinctive genomic organization and a putative retrovirus-like ribosomal frameshifting signal that directs the viral replicase synthesis. Proc. Natl. Acad. Sci. U. S. A. 90:10539–10543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Johnson LN, Lewis RJ. 2001. Structural basis for control by phosphorylation. Chem. Rev. 101:2209–2242 [DOI] [PubMed] [Google Scholar]
- 29. Jonassen CM, Jonassen TT, Sveen TM, Grinde B. 2003. Complete genomic sequences of astroviruses from sheep and turkey: comparison with related viruses. Virus Res. 91:195–201 [DOI] [PubMed] [Google Scholar]
- 30. Kitamura N, et al. 1980. The genome-linked protein of picornaviruses. VII. Genetic mapping of poliovirus VPg by protein and RNA sequence studies. Cell 21:295–302 [DOI] [PubMed] [Google Scholar]
- 31. Larkin MA, et al. 2007. ClustalW and ClustalX version 2. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 32. Machín A, Martín Alonso JM, Dalton KP, Parra F. 2009. Functional differences between precursor and mature forms of the RNA-dependent RNA polymerase from rabbit hemorrhagic disease virus. J. Gen. Virol. 90:2114–2118 [DOI] [PubMed] [Google Scholar]
- 33. Méndez E, Arias CF. 2007. Astroviruses, p 981–1000 In Knipe DM, Howlen PM. (ed), Fields virology, vol 1 Lippincott, Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 34. Mitra T, Sosnovtsev SV, Green KY. 2004. Mutagenesis of tyrosine 24 in the VPg protein is lethal for feline calicivirus. J. Virol. 78:4931–4935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Murphy JF, Klein PG, Hunt AG, Shaw JG. 1996. Replacement of the tyrosine residue that links a potyviral VPg to the viral RNA is lethal. Virology 220:535–538 [DOI] [PubMed] [Google Scholar]
- 36. Nakashima N, Shibuya N. 2006. Multiple coding sequences for the genome-linked virus protein (VPg) in dicistroviruses. J. Invertebr. Pathol. 92:100–104 [DOI] [PubMed] [Google Scholar]
- 37. Olspert A, Peil L, Hébrard E, Fargette D, Truve E. 2011. Protein-RNA linkage and post-translational modifications of two sobemovirus VPgs. J. Gen. Virol. 92:445–452 [DOI] [PubMed] [Google Scholar]
- 38. Pintó RM, Díez JM, Bosch A. 1994. Use of the colonic carcinoma cell line CaCo2 for in vivo amplification and detection of enteric viruses. J. Med. Virol. 44:310–315 [DOI] [PubMed] [Google Scholar]
- 39. Prilusky J, et al. 2005. FoldIndex: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics 21:3435–3438 [DOI] [PubMed] [Google Scholar]
- 40. Puustinen P, Rajamäki ML, Ivanov KI, Valkonen JP, Mäkinen K. 2002. Detection of the potyviral genome-linked protein VPg in virions and its phosphorylation by host kinases. J. Virol. 76:12703–12711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Racaniello VR. 2007. Picornaviridae: the viruses and their replication, p 795–838 In Knipe DM, Howlen PM. (ed), Fields virology, vol 1 Lippincott, Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 42. Reuer Q, Kuhn RJ, Wimmer E. 1990. Characterization of poliovirus clones containing lethal and nonlethal mutations in the genome-linked protein VPg. J. Virol. 64:2967–2975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rohayem J, Robel I, KJäger Scheffler U, Rudolph W. 2006. Protein-primed and de novo initiation of RNA synthesis by norovirus 3Dpol. J. Virol. 80:7060–7069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Romero P, et al. 2001. Sequence complexity of disordered protein. Proteins 42:38–48 [DOI] [PubMed] [Google Scholar]
- 45. Sadowy E, Milner M, Haenni AL. 2001. Proteins attached to viral genomes are multifunctional. Adv. Virus Res. 57:185–262 [DOI] [PubMed] [Google Scholar]
- 46. Sano D, Pintó RM, Omura T, Bosch A. 2010. Detection of oxidative damages on viral capsid protein for evaluating structural integrity and infectivity of human norovirus. Environ. Sci. Technol. 44:808–812 [DOI] [PubMed] [Google Scholar]
- 47. Satheshkumar PS, Gayathri P, Prasad K, Savithri HS. 2005. “Natively unfolded” VPg is essential for Sesbania mosaic virus serine protease activity. J. Biol. Chem. 280:30291–30300 [DOI] [PubMed] [Google Scholar]
- 48. Tompa P. 2005. The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett. 579:3346–3354 [DOI] [PubMed] [Google Scholar]
- 49. Veerisetty V, Sehgal OP. 1980. Proteinase K-sensitive factor essential for the infectivity of southern bean mosaic virus ribonucleic acid. Phytopathology 70:282–284 [Google Scholar]
- 50. Velázquez-Moctezuma R, Baños-Lara MR, Acevedo Y, Méndez E. 2012. Alternative cell lines to improve the rescue of infectious human astrovirus from a cDNA clone. J. Virol. Methods 179:295–302 [DOI] [PubMed] [Google Scholar]
- 51. Willcocks MM, Ashton N, Kurtz JB, Cubitt WD, Carter MJ. 1994. Cell culture adaptation of astrovirus involves a deletion. J. Virol. 68:6057–6058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Willcocks MM, Boxall AS, Carter MJ. 1999. Processing and intracellular location of human astrovirus nonstructural proteins. J. Gen. Virol. 80:2607–2611 [DOI] [PubMed] [Google Scholar]
- 53. Yambao ML, Masuta C, Nakahara K, Uyeda I. 2003. The central and C-terminal domains of VPg of Clover yellow vein virus are important for VPg-HCPro and VPg-VPg interactions. J. Gen. Virol. 84:2861–2869 [DOI] [PubMed] [Google Scholar]