Abstract
Intrinsic immunity is a first-line intracellular defense against virus infection, and viruses have evolved mechanisms to counteract it. During herpes simplex virus (HSV) infection, nuclear domain 10 (ND10) components localize adjacent to incoming viral genomes and generate a repressive environment for viral gene expression. Here, we found that the ND10 component, alpha-thalassemia/mental retardation syndrome X-linked (ATRX) protein, is predicted to be a target of HSV-1 miR-H1 and HSV-2 miR-H6. These microRNAs (miRNAs) share a seed sequence and are abundant during lytic infection. Mimics of both miRNAs could deplete endogenous ATRX, and an miR-H1 mimic could repress the expression of a reporter linked to the 3′ untranslated region of ATRX mRNA, identifying a cellular mRNA targeted by an HSV miRNA. Interestingly, ATRX protein and its mRNA were depleted in cells lytically infected with HSV, and ATRX protein was also depleted in cells infected with human cytomegalovirus. However, infection with an HSV-1 mutant lacking miR-H1 still resulted in ATRX depletion. This depletion was sensitive to a proteasome inhibitor and was largely ablated by a deletion of the gene encoding the immediate-early ICP0 protein. Additionally, a deletion of the gene encoding the tegument protein Vhs ablated most of the depletion of ATRX mRNA. Thus, HSV is equipped with multiple mechanisms to limit the expression of ATRX. As ATRX is implicated in repression of lytic viral gene expression, our results suggest roles for these different mechanisms during various phases of HSV infection.
INTRODUCTION
Hosts have evolved a myriad of mechanisms to control infection by viruses, while viruses have evolved counteracting mechanisms, often redundant, to prevail. The adaptive and innate immune systems, which require complex signaling pathways to activate and mobilize their effectors, can effectively control virus infection. Recently, the concept of intrinsic immunity (also called intrinsic antiviral defense) has emerged. Intrinsic immunity provides an immediate antiviral defense mediated by constitutively expressed factors whose activity does not depend on a signaling cascade (reviewed in references 6 and 98).
One set of such host factors includes proteins organized within discrete nuclear substructures known as nuclear domain 10 (ND10; also called promyelocytic leukemia protein [PML] nuclear bodies) that repress the initial transcription of herpesviruses (1, 9, 24, 50, 51, 64, 69, 70, 85–88, 96). ND10s harbor multiple cellular proteins, including PML, speckled protein of 100 kDa (Sp100), death domain-associated protein (hDaxx), and alpha-thalassemia/mental retardation syndrome X-linked (ATRX) protein (34, 58). Of these, ATRX is a member of the switch 2, sucrose nonfermenting 2 (SWI2/SNF2) family of helicases/ATPases. Mutations in the ATRX gene are associated with an X-linked mental retardation and alpha-thalassemia syndrome (63). ATRX, like other members of the SWI2/SNF2 family, is involved in different biological processes, including transcription regulation, cell cycle regulation, and mitotic chromosome segregation (reviewed in references 11 and 63) and chromatin remodeling (27, 94). ATRX forms a chromatin-remodeling complex with hDaxx (81, 97). It is involved in the deposition of the H3.3 histone variant at telomeres, and it is important for telomere integrity and for repression of gene expression at telomeres (17, 28, 31, 48, 95). ATRX also associates with several other chromatin factors involved in transcriptional repression, such as heterochromatin protein 1a (HP1a) (55), a nonhistone component of chromatin, and EZH2 (10), a member of the polycomb group family.
During herpesvirus infections, ND10 constituents are recruited to novel ND10 structures that localize adjacent to incoming parental herpesvirus genomes (21, 25). Subsequently, this association and ND10 integrity are disrupted. In the case of herpes simplex virus 1 (HSV-1), the disruption entails the activity of the immediate early (IE) protein ICP0, which is required for the dispersal of ND10 proteins, including ATRX, and the degradation of PML and Sp100 (7, 12, 50, 53, 54, 59, 61). In cells infected with ICP0-null viruses, replication is impaired and ND10 structures associate with the viral genomes much longer (20, 68, 77). It has been proposed that this association generates a repressive environment for viral transcription due to the activity of ND10 components. Consistent with this concept, replication of ICP0-null viruses can be partially rescued in cells depleted of PML, Sp100, hDaxx, or ATRX (23, 24, 50).
Like many other herpesviruses, HSV-1 and HSV-2 express numerous microRNAs (miRNAs), some of which are conserved between these two viruses (15, 38, 82–84, 91–93). Thus far, only a few targets of HSV miRNAs have been identified, and most of these are encoded from the strand opposite of that of an miRNA and thus are entirely complementary to the miRNA (37, 83, 84, 91). HSV-1 miR-H1, the first HSV miRNA discovered, is expressed abundantly during productive (lytic) infection (15, 38, 42, 91, 92). Interestingly, no positional homolog of HSV-1 miR-H1 has been detected in HSV-2-infected cells. Nonetheless, the seed sequence of HSV-2 miR-H6 (the positional homolog of HSV-1 miR-H6, which is encoded complementary to HSV-1 miR-H1), is identical to the seed sequence of HSV-1 miR-H1, which suggests that these two miRNAs might represent functional homologs and regulate the same genes (38).
To investigate the role of miR-H1, we utilized bioinformatics tools to predict potential targets of this HSV-1 miRNA and identified ATRX as a likely candidate. Our efforts to test this prediction led us to find that HSV-1 is equipped with multiple mechanisms, including miR-H1, to repress expression of ATRX, an effector of host intrinsic immunity.
MATERIALS AND METHODS
Cells and viruses.
African green monkey Vero cells (ATCC; CCL-81), human embryonic kidney HEK-293 cells (ATCC; CRL-1573), human foreskin fibroblast (HFF) cells (ATCC; CRL-1684), and human sarcoma U2OS cells (ATCC; HTB-96) were maintained at 37°C in Dulbecco's modified Eagle's medium (Mediatech, Inc.) supplemented with 5% newborn bovine serum (NBS), 10% fetal bovine serum (FBS), 10% FBS, or 5% NBS and 5% FBS, respectively (Sigma-Aldrich). HSV-1 wild-type (wt) strain KOS; the HSV-1 strain KOS vhs mutant UL41NHB and its rescued derivative, UL41NHB-R, generously provided by D. Leib (Dartmouth Medical School) (78); and the thymidine kinase (TK)-negative mutant of HSV-2 strain 186syn+, 186ΔKpn (35), generously provided by D. Knipe (Harvard Medical School), were propagated and titrated on Vero cells as described previously (14). The HSV-1 strain KOS ICP0-negative mutant 7134 and its rescued derivative 7134R (8), generously provided by D. Knipe, were propagated and titrated on U2OS cells as previously described (8). Human cytomegalovirus (HCMV) strain AD169 was propagated and titrated on HFF cells as previously described (39). To construct the mutant ΔH1/H6, we used a bacterial artificial chromosome (BAC) containing the wt HSV-1 strain KOS genome in which Cre-expressing vector sequences flanked by a pair of loxP sites are deleted from the viral genome by Cre recombinase upon transfection into cells, leaving a 36-bp in-frame sequence (34-bp loxP plus 2 bp) within sequences encoding a loop on the surface of the viral TK structure. A more detailed description of the BAC will be presented in a separate paper (I. Jurak C. Cui, A. Pearson, A. Griffiths, P. A. Schaffer, and D. M. Coen, unpublished data). Mutations were introduced into the BAC using two-step Red-mediated recombination (90). Briefly, PCR constructs were generated that contained the sequences of interest (50 nucleotides surrounding each side of the sequence targeted for deletion), with an inserted positive selection marker (kanamycin or zeocin resistance) adjoining an I-SceI site and a short duplication of the sequences of interest. Duplicated sequences allowed intramolecular recombination after cleavage by I-SceI, leaving no additional sequences other than the mutation. The mutagenesis introduced a novel HindIII site to facilitate identification of the recombinants. The primers used to generate ΔH1/H6 are listed in Table S1, which can be found at https://coen.med.harvard.edu. The integrity of BAC DNA and the presence of the introduced mutation were verified by restriction digestion and sequencing, respectively.
Transfection assays of miRNA mimics.
To analyze the effect of miRNAs on endogenous protein expression, HEK-293 cells were transfected sequentially three times, every 4 h, with 5 pmol of synthetic mimics of HSV-1 miR-H1 (5′GAUGGAAGGACGGGAAGUGGA3′), miR-H1-mut (5′GACUCGAGGACGGGAAGUGGA3′), HSV-2 miR-H6 (5′AAUGGAAGGCGAGGGGAUGCAG3′), and human miR-25 (5′CAUUGCACUUGUCUCCGGUCUGA3′) (Qiagen) using Lipofectamine 2000 (Invitrogen) according to the manufacturer's recommendation. Samples for Western blot analysis were harvested 72 h later. To analyze the effect of miRNAs on luciferase reporter gene expression, the complete 3′ untranslated region (UTR) of the ATRX gene was amplified using two PCRs (based on NCBI reference sequence NM_000489.3; primers used for the PCR amplification are provided in Table S2, which can be found at https://coen.med.harvard.edu) and cloned into pMIR-REPORT, a firefly luciferase expression plasmid (Ambion), resulting in pMIR-3′UTR ATRX. The predicted miR-H1 target sites within the 3′UTR were mutated using a QuikChange kit (Stratagene) according to the manufacturer's instructions, to generate pMIR-3′UTR ATRX-mut (primers used for mutagenesis are given in Table S2, which can be found at https://coen.med.harvard.edu). Sequences of the cloned 3′UTR and the introduced mutations were verified by sequencing of the entire 3′UTR in both plasmids. HEK-293 cells, seeded at the density of 3 × 104 cells/well in a 96-well plate, were cotransfected with the 0.1 μg luciferase expression constructs, along with 0.1 μg of pMIR-REPORT beta-galactosidase reporter control vector and 0.1, 0.5, or 2 pmol of miR-H1, miR-H1-mut, or miR-H6 mimics, respectively. At 24 or 48 h after transfection, luciferase activity was measured using the ONE-Glo Luciferase assay system according to the manufacturer's instructions (Promega) and normalized to beta-galactosidase activity measured using the Beta-Glo assay system, according to the manufacturer's instructions (Promega). Signals were detected using the Wallac 1420 VICTOR2 plate reader (Perkin Elmer) at the Institute of Chemistry and Cell Biology Longwood screening facility (Harvard Medical School).
Western blotting, antibodies, and inhibitors.
Transfected, mock-infected, or infected cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (20 mM Tris-HCl [pH 7.5], 300 mM NaCl, 1% sodium deoxycholate, 1% NP-40, 0.1% SDS, 1× Complete protease inhibitor cocktail [Roche]). Proteins were separated using 4 to 20% miniprotean TGX precast gels (Bio-Rad), and Western blotting was performed as previously described (36) using mouse monoclonal antibodies to detect ICP0 (Abcam; clone 5H7), ICP4 (Abcam; 10F1), gC (Fitzgerald; M701139), ICP27 (Virusys; H1113), and actin (Abcam; 8226) and BMPR2 (Abcam; MM0060-9A10) and rabbit polyclonal antibodies to detect TK (generously provided by Bill Summers, Yale University), PML (Abcam), and ATRX (Santa Cruz Biotechnology; H-300). In some experiments, cells were treated with the protein synthesis inhibitor cycloheximide (100 μg/ml; Sigma), the RNA transcription inhibitor actinomycin D (1 μg/ml; Sigma), the proteasome inhibitor MG132 (10 μM; Calbiochem), the lysosome inhibitor NH4Cl (10 mM; Sigma), or the viral DNA synthesis inhibitor acyclovir (20 μM; Sigma) 30 min prior to and during the course of infection.
Northern blotting.
For the detection of mRNAs, HEK-293 cells were mock or HSV infected, and total RNA was extracted at the times after infection indicated in the figures using TRIzol reagent (Invitrogen), according to the manufacturer's protocol. RNA samples, together with RiboRuler high-range RNA ladder (Fermentas), were separated in 1% agarose gels using The NorthernMax-Gly kit (Ambion), stained with ethidium bromide, transferred to a BrightStar-Plus positively charged membrane (Ambion) by downward capillary transfer using a TurboBlotter system (Whatman), cross-linked to the membrane using UV light, and prehybridized in ULTRAhyb buffer solution (Ambion) for ≥30 min at 68°C. ATRX, ICP0, and actin riboprobes labeled with [32P]UTP (PerkinElmer) were generated by in vitro RNA transcription using a MAXIScript T7 kit from the linearized plasmid templates p-actin, p-ATRX, and p-ICP0, according to the manufacturer's protocol (Ambion). p-Actin contains 140 nucleotides of exon 4 of the human beta-actin gene (nucleotides 682 to 821 based on NM_001101) inserted in pIDTBblue (Integrated DNA Technologies), p-ATRX contains 248 nucleotides of exon 9 of the ATRX gene (nucleotides 2838 to 2885; NM_138270.2), and p-ICP0 contains a 333-bp BamHI-XhoI fragment derived from ICP0 cDNA (60), inserted in pSL-301 (Invitrogen). The radiolabeled probes were added to the prehybridization solution and incubated overnight at 68°C. After hybridization, blots were washed two times for 10 min in low-stringency wash solution 1 (Ambion) at room temperature and two times for 20 min in high-stringency wash solution 2 (Ambion) at 68°C and exposed to a phosphorimager screen. Signals were detected using the Bio-Rad personal molecular imager system. For detection of miRNAs, cells were infected and harvested at various hours postinfection (hpi), and small enriched RNA was prepared and analyzed as previously described (38). Single-stranded DNA oligonucleotides (Integrated DNA Technologies) used for detection of the miRNAs are listed in Table S3, which can be found at https://coen.med.harvard.edu.
Stem-loop qRT-PCR.
Quantitative reverse transcription-PCR (qRT-PCR) was performed as previously described (42) with some modifications. Briefly, HEK-293 cells seeded in 12-well plates (1 × 106 cells/well) were mock infected or infected with 7134 or 7134R (multiplicity of infection [MOI] of 1). Total RNA was extracted at 18 hpi using TRIzol reagent (Invitrogen), according to the manufacturer's protocol, and assayed for HSV-1 miR-H1 and human let-7a, which was used as a normalization control using the conditions previously described (42). Data were analyzed in terms of the fold increase of miR-H1 in infected cells relative to the background signal from mock-infected cells, normalized to the values for let-7a.
RESULTS
Seed sequence homologs HSV-1 miR-H1 and HSV-2 miR-H6 target cellular ATRX.
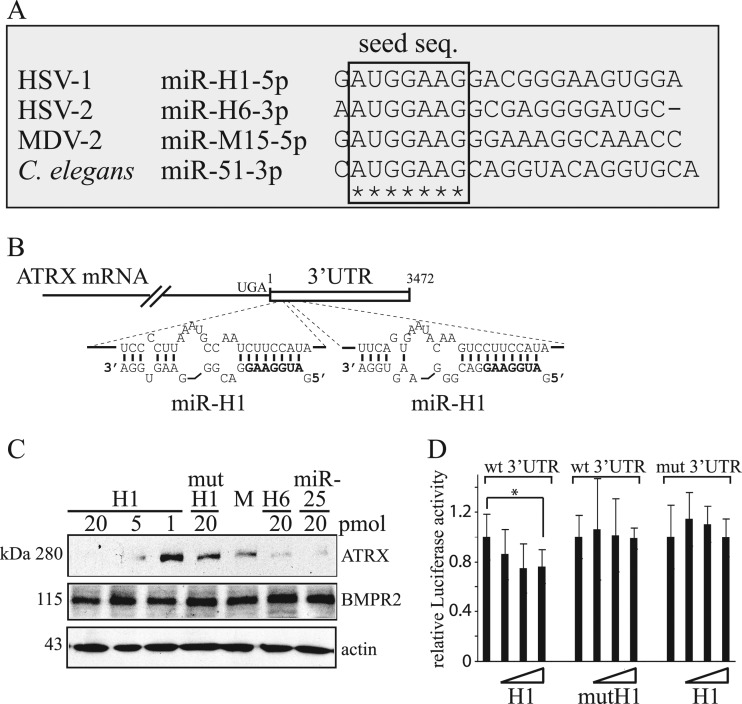
HSV-1 miR-H1 and its HSV-2 counterpart, miR-H6, have identical seed sequences and are abundantly expressed during lytic infection, suggesting that these miRNA have common targets (38). As a first step in identifying such targets, we asked if the seed sequence of miR-H1 can be found in other miRNAs. Interestingly, in the miRNA database, miRBase (www.mirbase.org), we found that miR-M15* (99), expressed by another herpesvirus, Marek's Disease virus type 2 (MDV-2), and miR-51* (45), expressed by the nematode C. elegans, have seed sequences identical to HSV-1 miR-H1 and HSV-2 miR-H6 (Fig. 1A). This finding, together with a lack of predicted conserved binding sites for the seed sequence within the HSV-1 and HSV-2 genomes (I. Jurak and D. M. Coen, unpublished data), led us to hypothesize that HSV-1 miR-H1 and HSV-2 miR-H6 target cellular transcripts. To test this hypothesis, we used TargetScan5.1 software, which predicts targets by searching for the presence of conserved sites that match the seed region of miRNAs, considering matches to annotated human 3′ UTRs and their orthologs (26, 29, 47). We found 227 human mRNAs with conserved sequences complementary to the seed sequence of miR-H1 (see Table S4, which can be found at https://coen.med.harvard.edu). Among these, mRNAs for ATRX protein and bone morphogenetic protein receptor 2 (BMPR2) were the top two candidates. These two mRNAs each have two predicted HSV-1 miR-H1 binding sites (the sites in ATRX mRNA are shown in Fig. 1B). As discussed above, ATRX is a constituent of ND10s and has a role in intrinsic immunity against herpesviruses (22, 50, 51). BMPR2 belongs to the bone morphogenetic protein (BMP) receptor family of transmembrane serine/threonine kinases, which are members of the transforming growth factor beta (TGF-β) superfamily (16) and have a critical role in heart, neural, bone, and cartilage development (13, 32, 52, 56, 89). Little is known about roles of BMPR2 in herpesvirus infection.
Fig 1.
HSV-1 miR-H1 and HSV-2 miR-H6 seed sequence conservation and targeting of ATRX mRNA. (A) Sequence alignment of miR-H1, miR-H6, miR-M15, and miR-51 expressed by HSV-1, HSV-2, Marek's disease virus type 2 (MDV-2), and Caenorhabditis elegans, respectively. Identical nucleotides are indicated with stars, and the seed sequence (nucleotides 2 to 8) is indicated within a frame. (B) Schematic representation of ATRX mRNA and its potential targeting by miR-H1. The mRNA coding sequence and 3′UTR are shown as a solid line and an unfilled box, respectively. UGA indicates the stop codon for the coding region, and the numbers indicate the first and the last nucleotide of the 3′UTR sequence. Below are shown expanded views of miR-H1 (lower sequence) binding to sequences within the 3′UTR predicted by RNAhybrid (67). The seed sequence of miR-H1 is shown in bold. (C) Western blot analysis of HEK-293 cells sequentially transfected three times with the indicated amounts of miRNA mimics and analyzed 72 h after transfection. H1, HSV-1 miR-H1; mutH1, mutated miR-H1; M, mock transfected; H6, HSV-2 miR-H6; miR-25, human miR-25. The proteins detected in the analysis and their sizes are indicated to the right and to the left of the panels, respectively. (D) HEK-293 cells were cotransfected with luciferase-expressing and β-galactosidase-expressing constructs and mimics of miRNAs, and luciferase activity was measured 24 h after transfection. The luciferase reporter gene fused to either the 3′UTR of ATRX mRNA or a mutant 3′UTR is indicated as wt 3′UTR or mut 3′UTR, respectively. Wild-type or mutant mimics are indicated as H1 and mutH1, respectively, and the increasing amounts of transfected mimics (0.1, 0.5, and 2 pmol) are shown as unfilled triangles. The experiment was performed in triplicate, and luciferase activity was normalized to the activity of β-galactosidase. The error bars represent standard deviations, and a statistically significant difference is indicated with an asterisk (P = 0.026; t test).
To investigate the computational prediction that HSV-1 miR-H1 and HSV-2 miR-H6 target ATRX and/or BMPR2 mRNAs, we mock-transfected or transfected HEK-293 cells with different amounts of mimics of these two miRNAs, human miR-H25 (which is predicted to target ATRX mRNA by TargetScan5.1, but at different sites), and, as a negative control, miR-H1 with an altered seed sequence. We then analyzed the expression of ATRX, BMPR2, and, as a loading control, actin, by Western blotting (Fig. 1C). Mimics of miR-H1 decreased levels of endogenous ATRX protein in a dose-dependent manner. Similarly, mimics of HSV-2 miR-H6 and human miR-25 also decreased ATRX expression. In contrast, the mutant form of miR-H1 did not meaningfully decrease ATRX levels, relative to those in mock-transfected cells. We did not observe any obvious changes in the levels of BMPR2 (or in our loading control protein, actin) in cells transfected with miRNA mimics (Fig. 1C). Thus, in subsequent experiments, we focused on ATRX.
We next asked if miR-H1 can specifically target the 3′UTR of ATRX mRNA. To this end, we fused a luciferase reporter gene to either the wt 3′UTR of ATRX mRNA or a mutant 3′UTR in which several nucleotides within the predicted seed sequence binding sites were altered (see Fig. S1 at https://coen.med.harvard.edu). The reporter plasmids were cotransfected with either wt or mutant miR-H1 mimics, along with a β-galactosidase reporter plasmid to normalize for transfection efficiency. Luciferase and β-galactosidase activities were measured 24 h after transfection. Consistent with the downregulation of endogenous ATRX, the miR-H1 mimic significantly reduced luciferase activity from the reporter gene fused to the wt 3′UTR of ATRX mRNA (although the reduction was modest, as is typical in such assays), whereas the mutant miR-H1 mimic had little if any effect (Fig. 1D). Similarly, miR-H1 mimics had little if any effect on luciferase activity if the predicted target sites were mutated (Fig. 1D). These results suggest that miR-H1 specifically binds to the predicted target sites within the 3′UTR of ATRX mRNA to downregulate ATRX expression.
ATRX protein levels are downregulated during lytic HSV and HCMV infection.
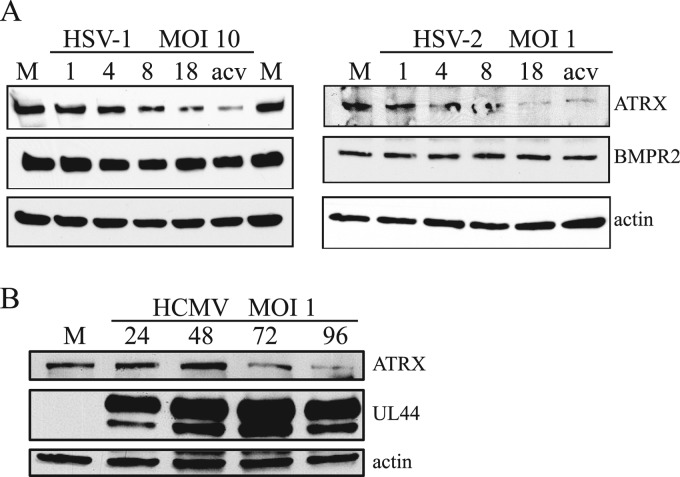
We next asked if levels of ATRX protein might be altered during lytic HSV infection. To address this, we infected Vero cells with wt HSV-1 strain KOS (MOI of 10) or HSV-2 186ΔKpn (MOI of 1). We used a lower MOI for HSV-2 because cells infected at higher MOIs exhibited cytopathic effect very early in infection and completely detached from the surface as early as 10 hpi.
Infected cells were harvested at different times postinfection, and the samples were subjected to Western blot analysis. In cells infected with either virus, ATRX protein levels began to decline between 4 and 8 hpi and were even more greatly reduced by 18 hpi (Fig. 2A). In contrast, we did not observe any obvious changes in BMPR2 (or, as a loading control, actin) levels. The degree of depletion of ATRX by KOS was dependent on MOI (data not shown). A substantial depletion of ATRX protein was observed in other cell types tested, including primary human foreskin fibroblasts (HFFs) and the human embryonic kidney cell line (HEK-293) (see Fig. S2 at https://coen.med.harvard.edu). The depletion was most obvious in HEK-293 cells, in which the ATRX protein level decreased more than 5-fold compared to that of mock-infected cells at 18 hpi based on a dilution series (see Fig. S3 at https://coen.med.harvard.edu). So for subsequent experiments, we used these cells. Regardless, we found variability in the degree of depletion of ATRX from experiment to experiment, which likely is due to the experimental challenge of Western blots of a very large protein (∼280 kDa).
Fig 2.
ATRX protein levels are downregulated during lytic HSV and HCMV infection. (A) Vero cells were mock infected (M) or infected with HSV-1 or HSV-2 at an MOI of 10 or 1, respectively. Samples for the analysis were collected at different hours after infection, indicated above the top panels, and the proteins were analyzed by Western blotting using antibodies against the proteins indicated to the right. M, mock-infected cells; acv, infection in the presence of acyclovir. (B) Similar to panel A, except that HFF cells were mock-infected (M) or infected with HCMV at an MOI of 1.
In addition, we were interested if other members of the herpesvirus family induce depletion of ATRX protein. To address this, we infected HFF cells with HCMV strain AD169 and analyzed ATRX expression at different times postinfection by Western blotting. Interestingly, similar to HSV, in cells infected with HCMV, ATRX protein levels were substantially reduced late in infection (Fig. 2B). These results suggest selection for a function that depletes the ATRX protein among diverse herpesviruses.
Although the kinetics of the reduction in ATRX protein levels coincided with the accumulation of miR-H1 (15, 42), levels of ATRX protein were still substantially reduced in cells infected in the presence of acyclovir, an inhibitor of viral DNA synthesis (Fig. 2A), which inhibits miR-H1 expression (15, 42). This result suggested that miR-H1 cannot be the only factor responsible for the reduction in ATRX.
miR-H1 is not required for reduced ATRX expression in HSV-1-infected cells.
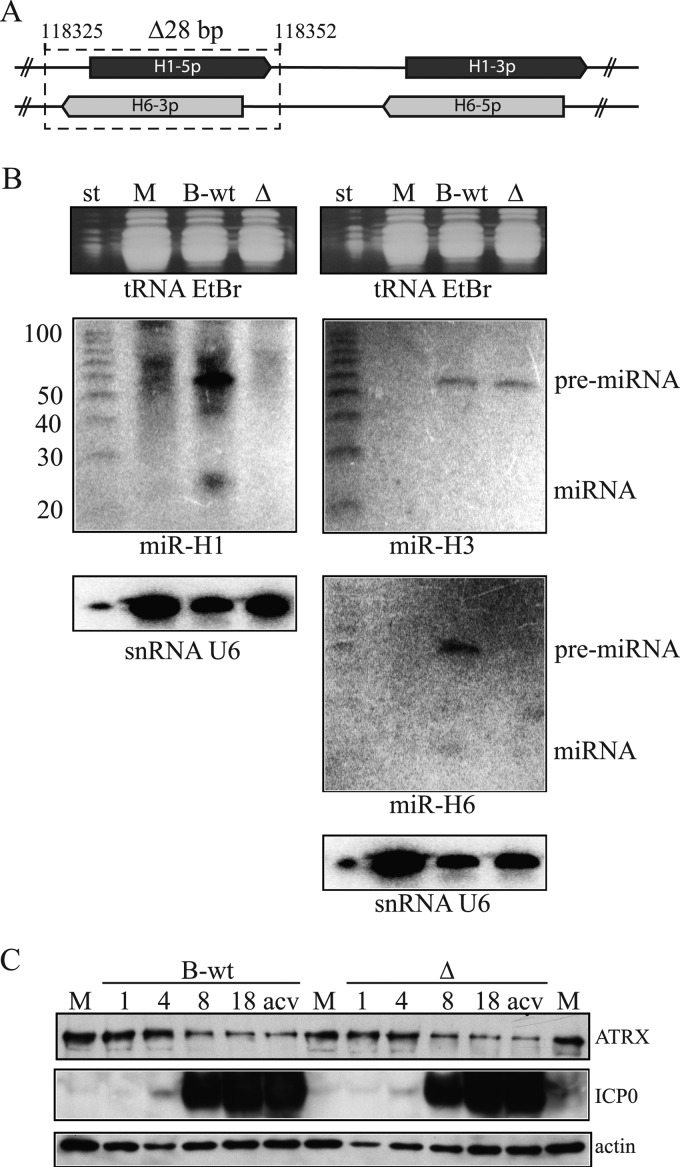
To address whether miR-H1 is necessary for the reduction of ATRX, we generated a mutant virus, ΔH1/H6, that carries a 28-bp deletion spanning the entire miR-H1 coding sequence (Fig. 3A). This deletion also deletes HSV-1 miR-H6, which is encoded from the opposite strand and is complementary to miR-H1. The deletion of the miR-H1/6 sequence in both terminal repeats (TRL and IRL) was confirmed by the expected pattern of DNA fragments after digestion with restriction enzymes and by sequencing of the miR-H1 locus (data not shown). As a control, we generated virus from our parental BAC containing the wt HSV-1 strain KOS genome (B-wt). The replication kinetics of ΔH1/H6 were indistinguishable from those of the control virus in lytically infected Vero or HEK-293 cells (Jurak and Coen, unpublished). We did not detect expression of mature HSV-1 miR-H1 or miR-H6 or their pre-miRNAs in cells lytically infected with the mutant virus (Fig. 3B), whereas these miRNAs were readily detected in B-wt-infected cells. snRNA U6 and tRNAs served as loading controls. In addition, the expression of pre-miR-H3 was similar in B-wt- and mutant-infected cells (Fig. 3B).
Fig 3.
miR-H1 is not required for the depletion of ATRX protein. (A) Schematic of the genomic locus spanning miR-H1 and miR-H6. The double-stranded DNA HSV-1 genome is depicted with solid lines, and the location of the 5p and 3p strands of miR-H1 (dark gray) and miR-H6 (light gray) are shown as arrows. The 28-bp deletion is shown as a dotted line surrounding the miR-H1-5p and miR-H6-3p loci. The numbers indicate the first and the last nucleotides of the deletion. (B) Small enriched RNAs were extracted from mock-infected (M) or B-wt- and ΔH1/H6 (Δ)-infected HEK-293 cells (MOI of 10) and analyzed on two separate polyacrylamide gels (left and right), stained with ethidium bromide (top), and blotted to a membrane for hybridization with probes for the RNAs indicated below each of the panels. After each hybridization and exposure to a phosphorimager, membranes were stripped and sequentially hybridized with each of the remaining probes. The nucleotide sizes of the RNA markers (st) are indicated to the left of the middle panels. The positions of detected pre-miRNAs and miRNAs are indicated to the right of the middle panels. (C) HEK-293 cells were mock infected (M) or infected with wt or ΔH1/H6 (Δ) at an MOI of 10. Samples for the analysis were collected at different times after infection, indicated above the top panels, and the proteins were analyzed by Western blotting using antibodies against the proteins indicated to the right. acv, infection in the presence of acyclovir.
However, in cells infected with ΔH1/H6 virus, ATRX protein levels were depleted in a manner comparable to that observed in wt-infected cells (Fig. 3C). These results, together with the inability of acyclovir to prevent ATRX depletion (Fig. 3A), led us to explore additional mechanisms by which HSV-1 regulates ATRX.
ATRX protein depletion is sensitive to a proteasome inhibitor.
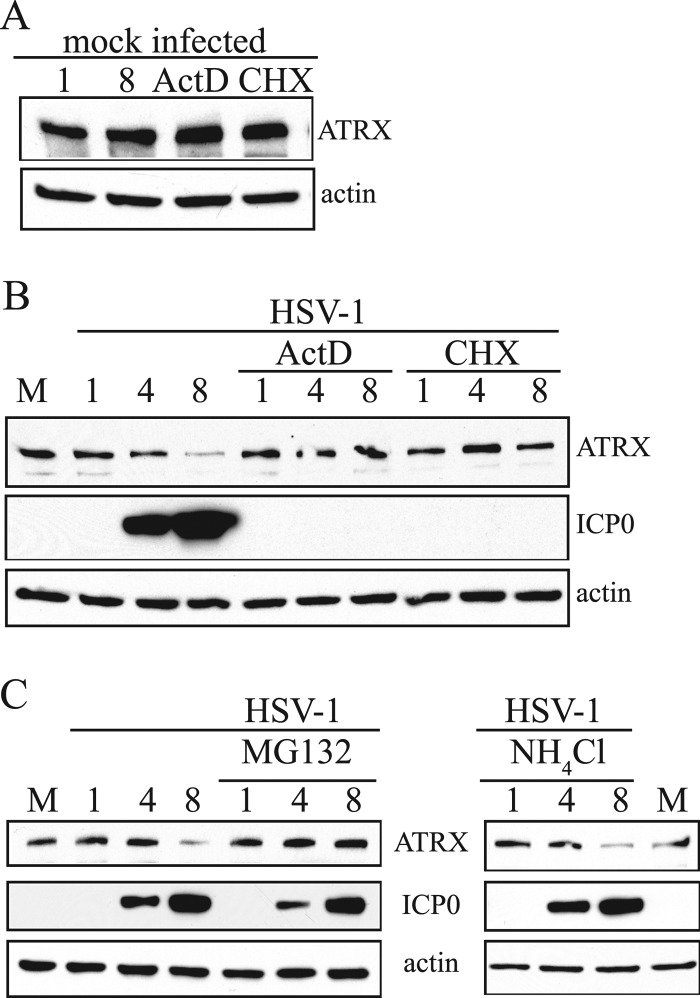
To further investigate the mechanisms for the loss of ATRX protein, we treated uninfected or infected HEK-293 cells with an inhibitor of protein synthesis (cycloheximide [CHX]) or an RNA transcription inhibitor (actinomycin D [ActD]) and analyzed ATRX protein by Western blot analysis. Addition of these inhibitors for 8 h did not meaningfully affect ATRX protein levels in uninfected cells (Fig. 4A). In contrast, addition of ActD or CHX 30 min prior to and for 8 h during HSV-1 infection (MOI of 10) prevented depletion of ATRX (as well as expression of the HSV-1 IE protein ICP0, as expected) (Fig. 4B). These results suggested that ATRX is actively depleted during infection and that this depletion requires synthesis of viral proteins. We next sought to investigate whether proteasome and/or lysosome pathways, two major cellular mechanisms for protein degradation, were involved in ATRX depletion. To this end, we infected HEK-293 cells with wt HSV-1 (MOI of 10) in the presence of 10 μM MG132, a well-established proteasome inhibitor, or 20 mM NH4Cl, which is widely used to inhibit lysosome function, and analyzed ATRX proteins at different times after infection. Addition of MG132 prevented depletion of ATRX, whereas addition of NH4Cl had little effect (Fig. 4C). Thus, our data suggest that depletion of ATRX requires proteasome function.
Fig 4.
ATRX protein depletion is sensitive to a proteasome inhibitor. (A) Proteins extracted from uninfected HEK-293 cells that were either mock treated or treated with actinomycin D (ActD) or cycloheximide (CHX) for 8 h were analyzed by Western blot with antibodies against the proteins indicated to the right of the panels. (B) HEK-293 cells were mock infected, HSV-1 infected (MOI of 10), or HSV-1 infected and treated with ActD or CHX, and samples for the analysis were collected at different hours postinfection, indicated above the top panels, and the proteins were analyzed by Western blotting using antibodies against the proteins indicated to the right. (C) The experiment was performed as described in the legend to panel B, except that cells were treated with MG132 or NH4Cl, as indicated above the panels, and the proteins analyzed are indicated between the panels.
ICP0 is required for depletion of ATRX protein.
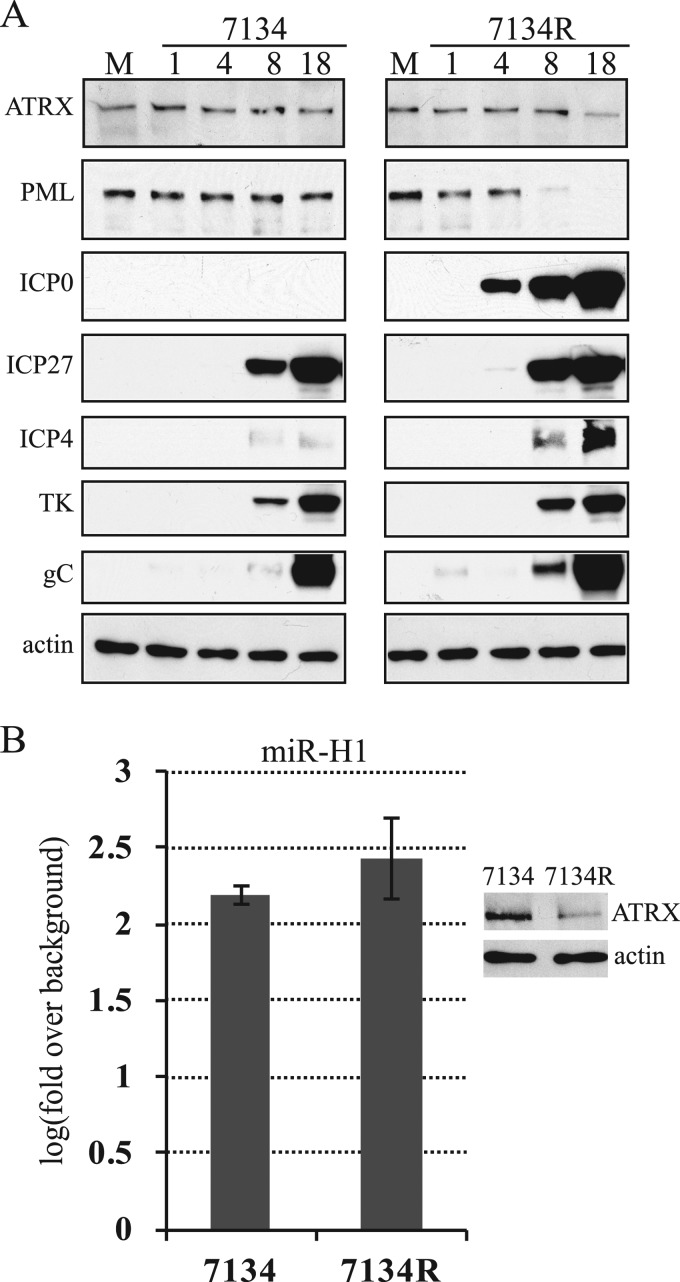
Because certain other cellular proteins, including PML and Sp100, are targeted for proteasome-dependent degradation by HSV in a manner requiring ICP0 (12, 19, 44, 49, 62), we infected HEK-293 cells with 7134, an ICP0-null mutant in which the ICP0 gene is replaced by an lacZ expression cassette (8), and a marker-rescued derivative of 7134, 7134R. Infected cells were harvested at different times after infection and analyzed by Western blotting. To avoid difficulties in interpretation due to major decreases or delays in viral gene expression due to the ICP0 gene mutation, for these experiments we infected cells at an MOI of 1, at which ICP0-deficient viruses exhibit a nearly wild-type phenotype (68). To ensure that infection with mutant virus was reasonably comparable to that with the rescued virus, we analyzed representative IE, E, and L viral proteins throughout the experiment. Although there were reduced levels of ICP4 compared to those in infection with 7134R, we observed IE protein ICP27, E protein TK, and late protein gC amounts roughly similar to those in infection with 7134R (Fig. 5A). As expected, 7134 did not express ICP0, and no depletion of PML was observed, while 7134R did express ICP0 and induced depletion of PML. We asked whether the expression of miR-H1 cells was comparable in 7134R- and in 7134-infected cells by analyzing the expression of this miRNA and the cellular miRNA let-7a, which has been previously shown to be expressed at constant levels during HSV-1 productive infection (42), by qRT-PCR. The mutant virus, 7134, expressed miR-H1 at only slightly reduced levels, ∼90% of 7134R, and this reduction was not statistically significant (Fig. 5B). Notably, we did not observe any meaningful depletion of ATRX protein in cells infected with 7134, whereas in cells infected with 7134R, the depletion was evident at 18 hpi (Fig. 5A). Depletion of ATRX by 7134R occurred later in this experiment than in the experiments shown in other figures, because the MOI was lower in this experiment. Even at higher MOIs, where ICP0+ viruses induced depletion of ATRX at earlier time points, ICP0-deficient viruses were defective for depletion (data not shown). These results suggested that ICP0 is required for the depletion of ATRX, extending the previous observation that ICP0 is required for dispersal of ATRX within the nucleus early in infection (20, 50).
Fig 5.
ICP0 is required for depletion of ATRX protein. (A) HEK-293 cells were mock infected (M) or infected with ICP0-null virus (7134) and its marker-rescued derivative (7134R) at an MOI of 1. Samples for the analysis were collected at different hours after infection, indicated above the top panels, and the proteins were analyzed by Western blotting using antibodies against the proteins indicated to the left. (B) Similar to panel A, except that total RNA and proteins were extracted from mock-, 7134-, and 7134R-infected cells at 18 hpi. The expression of HSV-1 miR-H1 and cellular let-7a were measured by qRT-PCR. Each bar represents log10 mean changes in expression relative to that of a mock-infected control sample and normalized to values of let-7a. The experiment was performed in triplicate, and error bars indicate standard deviations. Differences were not statistically significant (P = 0.275; t test). The panel to the right represents detection of ATRX and actin proteins by Western blotting from the same experiment.
ATRX mRNA is rapidly degraded in lytically infected cells.
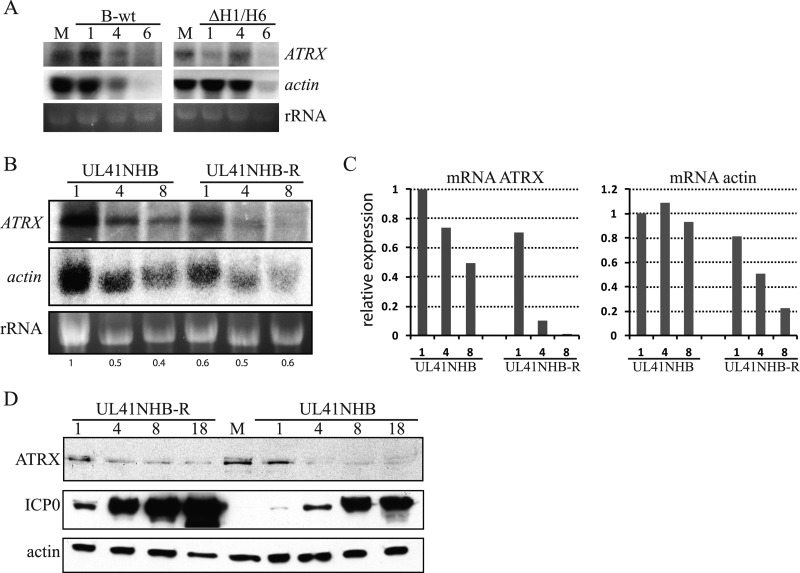
To further characterize the depletion of ATRX in infected cells, we asked if its mRNA is also depleted, and if so whether this depletion requires miR-H1 or Vhs, a well-known HSV-1-encoded endoribonuclease (74). To test this, we infected cells with B-wt, ΔH1/H6, the vhs mutant virus UL41NHB, and its rescued derivative, UL41NHB-R, at an MOI of 5 and harvested cells for Northern blot analysis at different times after infection. Interestingly, we observed that ATRX mRNA levels rapidly decreased, starting at 1 hpi, and were undetectable at 6 to 8 hpi in B-wt-, ΔH1/H6-, or UL41NHB-R-infected cells (Fig. 6A and B), whereas in cells infected with UL41NHB (Fig. 6B), ATRX mRNA was readily detected at 8 hpi. In four independent experiments, we observed higher levels of ATRX mRNA in cells infected with the vhs mutant than with its rescued derivative (not shown). Similarly, levels of actin mRNA were rapidly decreased in cells infected with B-wt, ΔH1/H6, or UL41NHB-R compared to those in cells infected with Vhs-deficient virus (Fig. 6A and B), which is in agreement with previous studies (5, 78). Levels of rRNA were fairly similar in the different samples (Fig. 6A and B), and when the Northern blot signals and rRNA signals in Fig. 6B were compared, it was apparent that both ATRX and actin mRNA decrease more rapidly in UL41NHB-R-infected cells than in UL41NHB-infected cells (Fig. 6C). Interestingly, ATRX mRNA seemed to decay faster than actin mRNA in the absence of vhs. This might indicate a second mechanism for ATRX mRNA destabilization, perhaps related to the large size of this mRNA.
Fig 6.
Vhs is required for depletion of ATRX mRNA. (A) Total RNAs were extracted from mock (M)-, B-wt-, or ΔH1/H6-infected HEK-293 cells (MOI of 5) and separated by agarose gel electrophoresis, stained for RNA with ethidium bromide (bottom panels), and blotted to a membrane for hybridization with probes for the RNAs indicated to the right of the panels. (B) Similar to panel A, except that cells were infected with UL41NHB-R or UL41NHB (MOI of 5). (C) Signal densities from panel B were quantified using ImageJ (NIH) software. Each bar represents the relative expression of the mRNA indicated above the diagrams normalized to rRNA and setting the value from UL41NHB-infected cells at 1 hpi as 1. (D) Proteins were extracted from mock-infected cells (M) or cells infected with UL41NHB-R or UL41NHB at an MOI of 5. Samples for the analysis were collected at the hours after infection indicated above the top panels, and the proteins indicated to the left were analyzed by Western blotting.
Taken together, these results suggest that Vhs, but not miR-H1, has a major role in depletion of ATRX mRNA. Nonetheless, Western blot analysis showed that in cells infected with UL41NHB, ATRX protein was depleted, and that this depletion was comparable to that observed in cells infected with B-wt, ΔH1/H6, or UL41NHB-R (Fig. 6D). Thus, although Vhs has a major role in degradation of ATRX mRNA in lytically infected cells, its contribution to depletion of ATRX protein is rather minor compared to that of ICP0.
DISCUSSION
Viruses have evolved mechanisms to disarm host defense mechanisms and thus ensure efficient replication. In this study, we discovered that HSV-1 miR-H1 and HSV-2 miR-H6, which share a seed sequence, can repress the expression of ATRX, a host factor involved in intrinsic immunity against herpesviruses. To our knowledge, this is the first report identifying a cellular protein whose expression is targeted by HSV miRNAs. However, we found that these miRNAs are not required for most of the observed depletion of ATRX protein and ATRX mRNA in lytically infected cells. Instead, we found that the IE protein ICP0 is required for the reduction of ATRX protein levels, and the tegument protein, Vhs, is required for depletion of ATRX mRNA. Thus, our results show that HSV is equipped with multiple mechanisms to limit the expression of ATRX and alleviate its repression of viral gene expression.
miRNAs encoded by different viruses usually share little or no homology (73, 80); nonetheless, many viruses have independently evolved unrelated miRNAs to target common cellular factors. An example of such convergent evolution is targeting of major histocompatibility complex class I-related chain B by HCMV, Epstein-Barr virus, and Kaposi's sarcoma-associated herpesvirus, by miR-UL112-1, miR-K7, and miR-BART2, respectively (57, 76). These miRNAs do not share seed sequences. Remarkably, we found that the seed sequence of HSV-1 miR-H1 is conserved, not only with HSV-2 miR-H6, but also with miR-M15-5p expressed by the much more distantly related herpesvirus of chickens, MDV-2, suggesting that targeting ATRX by miRNAs might represent another example of convergent evolution.
Using TargetScan5.1, we identified 227 potential human targets of miR-H1. We experimentally evaluated only the top two candidates, ATRX and BMPR2. Although we validated ATRX as a target in our transfection assays, we found no evidence that BMPR2 is a target for these miRNAs. It is not uncommon for computational predictions to fail to be experimentally validated (4, 65, 72). On the other hand, miRNAs often induce only modest changes to the protein output of their targets (3, 30), and we cannot exclude the possibility that BMPR2 levels did change but that these changes were so small that we could not detect them. For that matter, we cannot exclude the possibility that HSV-1 miR-H1 and HSV-2 miR-H6 have other targets. Indeed, we think that is likely and deserves further investigation. Based on recent reports indicating that miRNAs predominantly act to decrease target mRNA levels (30, 71), we assayed for the effects of deletion of miR-H1 on the levels of ATRX mRNA during lytic infection. We found little if any effect, which was almost certainly due to the importance of Vhs for ATRX mRNA degradation. This role of Vhs is unlikely to be specific for ATRX, given its role in degradation of most host mRNAs (43, 75, 79). Regardless, the depletion of ATRX mRNA appears to make little contribution to the depletion of ATRX protein during infection, which is mediated mainly by ICP0.
Our results indicating that ATRX is depleted during HSV-1 infection and that this depletion depends on ICP0 and proteasome function extend the earlier observation that ICP0 is required for the dispersal of ATRX and its interaction partner, hDaxx, from ND10 early in infection (50). ICP0 has been previously reported to be required for the degradation of other components of ND10, PML, and Sp100, which depends on residues required for ubiquitin ligase activity (12, 19). It is not clear if the dispersal of ATRX/hDaxx is an outcome of degradation of structural elements of ND10 scaffold or a direct effect on these proteins. It is interesting that we observed depletion of ATRX later in infection than depletion of PML (Fig. 5). Also, Lukashchuk and Everett found no evidence for direct association of ICP0 and either ATRX or hDaxx using an immunoprecipitation assay (50). This raises the possibility that ATRX degradation is secondary to the degradation of PML. However, there might be only a transient interaction between ICP0 and ATRX, which could be difficult to detect using immunoprecipitation.
Interestingly, we found that ATRX protein levels were depleted not only in HSV-infected cells but also in HCMV-infected cells, suggesting a selection for depletion of this protein among diverse herpesviruses. Similar to other herpesviruses that have evolved a variety of mechanisms to alleviate the repressive function of ND10s (88), HCMV tegument protein pp71 induces displacement of ATRX from ND10 (51) and triggers degradation of hDaxx (33) to allow efficient expression of IE1 protein, which further antagonizes the repressive function of ND10s (41) by abrogating the SUMOylation of PML (2, 46) and downregulating sp100 (85). It is possible that HCMV also employs these mechanisms to deplete ATRX protein. However, we also cannot exclude the possibility that HCMV has independently evolved miRNAs to target ATRX.
Why should HSV be equipped with at least three different mechanisms for repressing the expression of ATRX? A possible answer to this question starts with an intriguing model that has been discussed by Lukashckuk and Everett (50). In this model, the ATRX/hDaxx complex associates with nucleosomes that are assembled on the viral genome and recruits other proteins such as variant histones (17, 28, 48, 66) to generate a repressive environment. During lytic infection, this repression can be alleviated by ICP0. There may nevertheless be a role for HSV-1 miR-H1 late in infection to prevent an inhibitory accumulation of ATRX protein. Detecting such a role may require analysis of ICP0–vhs–miR-H1 triple mutants.
However, HSV also undergoes latent infection during which lytic gene expression, including that of the ICP0 gene and vhs, is mostly repressed. During latency, the ATRX-repressive complex could associate with HSV genomes and contribute to the repression of lytic gene expression. Upon reactivation from latency, unlike lytic infection, there would be no Vhs arriving within an incoming virion and there would be little or no ICP0 present to counteract ATRX. There is evidence, however, for expression of HSV-1 miR-H1 and, especially, HSV-2 miR-H6 during latent infection (38, 42, 82, 92). Additionally, there is evidence that the initial burst of viral gene expression following a reactivation stimulus is not regulated in the same way as during lytic infection (18, 40) (J. M. Pesola M. F. Kramer, J. W. Carbone, C. J. Jahns, and D. M. Coen, unpublished data). Thus, miRNAs that are ordinarily late-gene products might be expressed at relatively high levels during reactivation, even prior to viral DNA synthesis. Thus, during both latency and reactivation, miRNA repression of ATRX expression might aid in derepression of viral gene expression, facilitating rapid reactivation. This would be followed by accumulation of ICP0 to mediate degradation and dispersal of ND10 constituents (PML, Sp100, ATRX), which could lead to the remodeling of chromatin and robust activation of viral promoters and full-blown reactivation. This scenario makes several testable predictions which merit investigation.
ACKNOWLEDGMENTS
We thank David Knipe and David Leib for generous gifts of reagents and helpful discussions, Bill Summers for generously providing antibodies, and David Knipe for a critical reading of the manuscript. We are grateful to the Institute of Chemistry and Cell Biology Longwood for use of their plate reader and Dana-Farber/Harvard Cancer Center DNA Resource Core for DNA sequencing. We also thank members of the Coen lab for advice and support, particularly Jean Pesola for work on assays of ATRX mRNA and help with statistical analysis and Seamus McCarron for technical assistance.
This work was supported by NIH grant RO1 AI26126.
Footnotes
Published ahead of print 11 July 2012
REFERENCES
- 1. Adler M, Tavalai N, Muller R, Stamminger T. 2011. Human cytomegalovirus immediate-early gene expression is restricted by the nuclear domain 10 component Sp100. J. Gen. Virol. 92:1532–1538 [DOI] [PubMed] [Google Scholar]
- 2. Ahn JH, Brignole EJ, III, Hayward GS. 1998. Disruption of PML subnuclear domains by the acidic IE1 protein of human cytomegalovirus is mediated through interaction with PML and may modulate a RING finger-dependent cryptic transactivator function of PML. Mol. Cell. Biol. 18:4899–4913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baek D, et al. 2008. The impact of microRNAs on protein output. Nature 455:64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barbato C, et al. 2009. Computational challenges in miRNA target predictions: to be or not to be a true target? J. Biomed. Biotechnol. 2009:803069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Becker Y, Tavor E, Asher Y, Berkowitz C, Moyal M. 1993. Effect of herpes simplex virus type-1 UL41 gene on the stability of mRNA from the cellular genes: beta-actin, fibronectin, glucose transporter-1, and docking protein, and on virus intraperitoneal pathogenicity to newborn mice. Virus Genes 7:133–143 [DOI] [PubMed] [Google Scholar]
- 6. Bieniasz PD. 2004. Intrinsic immunity: a front-line defense against viral attack. Nat. Immunol. 5:1109–1115 [DOI] [PubMed] [Google Scholar]
- 7. Boutell C, Sadis S, Everett RD. 2002. Herpes simplex virus type 1 immediate-early protein ICP0 and its isolated RING finger domain act as ubiquitin E3 ligases in vitro. J. Virol. 76:841–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cai WZ, Schaffer PA. 1989. Herpes simplex virus type 1 ICP0 plays a critical role in the de novo synthesis of infectious virus following transfection of viral DNA. J. Virol. 63:4579–4589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cantrell SR, Bresnahan WA. 2006. Human cytomegalovirus (HCMV) UL82 gene product (pp71) relieves hDaxx-mediated repression of HCMV replication. J. Virol. 80:6188–6191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cardoso C, et al. 1998. Specific interaction between the XNP/ATR-X gene product and the SET domain of the human EZH2 protein. Hum. Mol. Genet. 7:679–684 [DOI] [PubMed] [Google Scholar]
- 11. Carlson M, Laurent BC. 1994. The SNF/SWI family of global transcriptional activators. Curr. Opin. Cell Biol. 6:396–402 [DOI] [PubMed] [Google Scholar]
- 12. Chelbi-Alix MK, de The H. 1999. Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene 18:935–941 [DOI] [PubMed] [Google Scholar]
- 13. Chen D, Zhao M, Mundy GR. 2004. Bone morphogenetic proteins. Growth Factors 22:233–241 [DOI] [PubMed] [Google Scholar]
- 14. Coen DM, Fleming HE, Jr, Leslie LK, Retondo MJ. 1985. Sensitivity of arabinosyladenine-resistant mutants of herpes simplex virus to other antiviral drugs and mapping of drug hypersensitivity mutations to the DNA polymerase locus. J. Virol. 53:477–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cui C, et al. 2006. Prediction and identification of herpes simplex virus 1-encoded microRNAs. J. Virol. 80:5499–5508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Derynck R, Miyazono K. (ed). 2008. TGF-β and the TGF-β family. Cold Spring Harbor Laboratory Press, New York, NY [Google Scholar]
- 17. Drane P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. 2010. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 24:1253–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Du T, Zhou G, Roizman B. 2011. HSV-1 gene expression from reactivated ganglia is disordered and concurrent with suppression of latency-associated transcript and miRNAs. Proc. Natl. Acad. Sci. U. S. A. 108:18820–18824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Everett RD, et al. 1998. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J. Virol. 72:6581–6591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Everett RD, Maul GG. 1994. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 13:5062–5069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Everett RD, Murray J. 2005. ND10 components relocate to sites associated with herpes simplex virus type 1 nucleoprotein complexes during virus infection. J. Virol. 79:5078–5089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Everett RD, Murray J, Orr A, Preston CM. 2007. Herpes simplex virus type 1 genomes are associated with ND10 nuclear substructures in quiescently infected human fibroblasts. J. Virol. 81:10991–11004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Everett RD, Parada C, Gripon P, Sirma H, Orr A. 2008. Replication of ICP0-null mutant herpes simplex virus type 1 is restricted by both PML and Sp100. J. Virol. 82:2661–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Everett RD, et al. 2006. PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. J. Virol. 80:7995–8005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Everett RD, Sourvinos G, Orr A. 2003. Recruitment of herpes simplex virus type 1 transcriptional regulatory protein ICP4 into foci juxtaposed to ND10 in live, infected cells. J. Virol. 77:3680–3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Friedman RC, Farh KK, Burge CB, Bartel DP. 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19:92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gibbons RJ, et al. 1997. Mutations in transcriptional regulator ATRX establish the functional significance of a PHD-like domain. Nat. Genet. 17:146–148 [DOI] [PubMed] [Google Scholar]
- 28. Goldberg AD, et al. 2010. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140:678–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Grimson A, et al. 2007. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell 27:91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Guo H, Ingolia NT, Weissman JS, Bartel DP. 2010. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466:835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Heaphy CM, et al. 2011. Altered telomeres in tumors with ATRX and DAXX mutations. Science 333:425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hogan BL. 1996. Bone morphogenetic proteins: multifunctional regulators of vertebrate development. Genes Dev. 10:1580–1594 [DOI] [PubMed] [Google Scholar]
- 33. Hwang J, Kalejta RF. 2007. Proteasome-dependent, ubiquitin-independent degradation of Daxx by the viral pp71 protein in human cytomegalovirus-infected cells. Virology 367:334–338 [DOI] [PubMed] [Google Scholar]
- 34. Ishov AM, et al. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 147:221–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jones CA, Taylor TJ, Knipe DM. 2000. Biological properties of herpes simplex virus 2 replication-defective mutant strains in a murine nasal infection model. Virology 278:137–150 [DOI] [PubMed] [Google Scholar]
- 36. Jurak I, Brune W. 2006. Induction of apoptosis limits cytomegalovirus cross-species infection. EMBO J. 25:2634–2642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jurak I, Griffiths A, Coen DM. 2011. Mammalian alphaherpesvirus miRNAs. Biochim. Biophys. Acta 1809:641–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jurak I, et al. 2010. Numerous conserved and divergent microRNAs expressed by herpes simplex viruses 1 and 2. J. Virol. 84:4659–4672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kamil JP, Coen DM. 2007. Human cytomegalovirus protein kinase UL97 forms a complex with the tegument phosphoprotein pp65. J. Virol. 81:10659–10668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kim JY, Mandarino A, Chao MV, Mohr I, Wilson AC. 2012. Transient reversal of episome silencing precedes VP16-dependent transcription during reactivation of latent HSV-1 in neurons. PLoS Pathog. 8:e1002540 doi:10.1371/journal.ppat.1002540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Korioth F, Maul GG, Plachter B, Stamminger T, Frey J. 1996. The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp. Cell Res. 229:155–158 [DOI] [PubMed] [Google Scholar]
- 42. Kramer MF, et al. 2011. Herpes simplex virus 1 microRNAs expressed abundantly during latent infection are not essential for latency in mouse trigeminal ganglia. Virology 417:239–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Krikorian CR, Read GS. 1991. In vitro mRNA degradation system to study the virion host shutoff function of herpes simplex virus. J. Virol. 65:112–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kummer M, et al. 2007. Herpes simplex virus type 1 induces CD83 degradation in mature dendritic cells with immediate-early kinetics via the cellular proteasome. J. Virol. 81:6326–6338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lau NC, Lim LP, Weinstein EG, Bartel DP. 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294:858–862 [DOI] [PubMed] [Google Scholar]
- 46. Lee HR, et al. 2004. Ability of the human cytomegalovirus IE1 protein to modulate sumoylation of PML correlates with its functional activities in transcriptional regulation and infectivity in cultured fibroblast cells. J. Virol. 78:6527–6542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lewis BP, Burge CB, Bartel DP. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120:15–20 [DOI] [PubMed] [Google Scholar]
- 48. Lewis PW, Elsaesser SJ, Noh KM, Stadler SC, Allis CD. 2010. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc. Natl. Acad. Sci. U. S. A. 107:14075–14080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lomonte P, Sullivan KF, Everett RD. 2001. Degradation of nucleosome-associated centromeric histone H3-like protein CENP-A induced by herpes simplex virus type 1 protein ICP0. J. Biol. Chem. 276:5829–5835 [DOI] [PubMed] [Google Scholar]
- 50. Lukashchuk V, Everett RD. 2010. Regulation of ICP0-null mutant herpes simplex virus type 1 infection by ND10 components ATRX and hDaxx. J. Virol. 84:4026–4040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lukashchuk V, McFarlane S, Everett RD, Preston CM. 2008. Human cytomegalovirus protein pp71 displaces the chromatin-associated factor ATRX from nuclear domain 10 at early stages of infection. J. Virol. 82:12543–12554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Machado RD, et al. 2006. Mutations of the TGF-beta type II receptor BMPR2 in pulmonary arterial hypertension. Hum. Mutat. 27:121–132 [DOI] [PubMed] [Google Scholar]
- 53. Maul GG, Everett RD. 1994. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J. Gen. Virol. 75:1223–1233 [DOI] [PubMed] [Google Scholar]
- 54. Maul GG, Guldner HH, Spivack JG. 1993. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate early gene 1 product (ICP0). J. Gen. Virol. 74:2679–2690 [DOI] [PubMed] [Google Scholar]
- 55. McDowell TL, et al. 1999. Localization of a putative transcriptional regulator (ATRX) at pericentromeric heterochromatin and the short arms of acrocentric chromosomes. Proc. Natl. Acad. Sci. U. S. A. 96:13983–13988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Morrell NW. 2010. Role of bone morphogenetic protein receptors in the development of pulmonary arterial hypertension. Adv. Exp. Med. Biol. 661:251–264 [DOI] [PubMed] [Google Scholar]
- 57. Nachmani D, Stern-Ginossar N, Sarid R, Mandelboim O. 2009. Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe 5:376–385 [DOI] [PubMed] [Google Scholar]
- 58. Negorev D, Maul GG. 2001. Cellular proteins localized at and interacting within ND10/PML nuclear bodies/PODs suggest functions of a nuclear depot. Oncogene 20:7234–7242 [DOI] [PubMed] [Google Scholar]
- 59. Negorev DG, Vladimirova OV, Ivanov A, Rauscher F, III, Maul GG. 2006. Differential role of Sp100 isoforms in interferon-mediated repression of herpes simplex virus type 1 immediate-early protein expression. J. Virol. 80:8019–8029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pan D, Coen DM. 2012. Quantification and analysis of thymidine kinase expression from acyclovir-resistant g-string insertion and deletion mutants in herpes simplex virus-infected cells. J. Virol. 86:4518–4526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Parkinson J, Everett RD. 2000. Alphaherpesvirus proteins related to herpes simplex virus type 1 ICP0 affect cellular structures and proteins. J. Virol. 74:10006–10017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Parkinson J, Lees-Miller SP, Everett RD. 1999. Herpes simplex virus type 1 immediate-early protein vmw110 induces the proteasome-dependent degradation of the catalytic subunit of DNA-dependent protein kinase. J. Virol. 73:650–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Picketts DJ, et al. 1996. ATRX encodes a novel member of the SNF2 family of proteins: mutations point to a common mechanism underlying the ATR-X syndrome. Hum. Mol. Genet. 5:1899–1907 [DOI] [PubMed] [Google Scholar]
- 64. Preston CM, Nicholl MJ. 2006. Role of the cellular protein hDaxx in human cytomegalovirus immediate-early gene expression. J. Gen. Virol. 87:1113–1121 [DOI] [PubMed] [Google Scholar]
- 65. Rajewsky N. 2006. microRNA target predictions in animals. Nat. Genet. 38(Suppl):S8–S13 [DOI] [PubMed] [Google Scholar]
- 66. Ratnakumar K, et al. 2012. ATRX-mediated chromatin association of histone variant macroH2A1 regulates alpha-globin expression. Genes Dev. 26:433–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. 2004. Fast and effective prediction of microRNA/target duplexes. RNA 10:1507–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sacks WR, Schaffer PA. 1987. Deletion mutants in the gene encoding the herpes simplex virus type 1 immediate-early protein ICP0 exhibit impaired growth in cell culture. J. Virol. 61:829–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Saffert RT, Kalejta RF. 2007. Human cytomegalovirus gene expression is silenced by Daxx-mediated intrinsic immune defense in model latent infections established in vitro. J. Virol. 81:9109–9120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Saffert RT, Kalejta RF. 2006. Inactivating a cellular intrinsic immune defense mediated by Daxx is the mechanism through which the human cytomegalovirus pp71 protein stimulates viral immediate-early gene expression. J. Virol. 80:3863–3871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Selbach M, et al. 2008. Widespread changes in protein synthesis induced by microRNAs. Nature 455:58–63 [DOI] [PubMed] [Google Scholar]
- 72. Sethupathy P, Megraw M, Hatzigeorgiou AG. 2006. A guide through present computational approaches for the identification of mammalian microRNA targets. Nat. Methods 3:881–886 [DOI] [PubMed] [Google Scholar]
- 73. Skalsky RL, Cullen BR. 2010. Viruses, microRNAs, and host interactions. Annu. Rev. Microbiol. 64:123–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Smiley JR. 2004. Herpes simplex virus virion host shutoff protein: immune evasion mediated by a viral RNase? J. Virol. 78:1063–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sorenson CM, Hart PA, Ross J. 1991. Analysis of herpes simplex virus-induced mRNA destabilizing activity using an in vitro mRNA decay system. Nucleic Acids Res. 19:4459–4465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Stern-Ginossar N, et al. 2007. Host immune system gene targeting by a viral miRNA. Science 317:376–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Stow ND, Stow EC. 1986. Isolation and characterization of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate early polypeptide Vmw110. J. Gen. Virol. 67:2571–2585 [DOI] [PubMed] [Google Scholar]
- 78. Strelow LI, Leib DA. 1995. Role of the virion host shutoff (vhs) of herpes simplex virus type 1 in latency and pathogenesis. J. Virol. 69:6779–6786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Taddeo B, Roizman B. 2006. The virion host shutoff protein (UL41) of herpes simplex virus 1 is an endoribonuclease with a substrate specificity similar to that of RNase A. J. Virol. 80:9341–9345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Takane K, Kanai A. 2011. Vertebrate virus-encoded microRNAs and their sequence conservation. Jpn. J. Infect. Dis. 64:357–366 [PubMed] [Google Scholar]
- 81. Tang J, et al. 2004. A novel transcription regulatory complex containing death domain-associated protein and the ATR-X syndrome protein. J. Biol. Chem. 279:20369–20377 [DOI] [PubMed] [Google Scholar]
- 82. Tang S, Bertke AS, Patel A, Margolis TP, Krause PR. 2011. Herpes simplex virus 2 microRNA miR-H6 is a novel latency-associated transcript-associated microRNA, but reduction of its expression does not influence the establishment of viral latency or the recurrence phenotype. J. Virol. 85:4501–4509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Tang S, et al. 2008. An acutely and latently expressed herpes simplex virus 2 viral microRNA inhibits expression of ICP34.5, a viral neurovirulence factor. Proc. Natl. Acad. Sci. U. S. A. 105:10931–10936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Tang S, Patel A, Krause PR. 2009. Novel less-abundant viral microRNAs encoded by herpes simplex virus 2 latency-associated transcript and their roles in regulating ICP34.5 and ICP0 mRNAs. J. Virol. 83:1433–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Tavalai N, Adler M, Scherer M, Riedl Y, Stamminger T. 2011. Evidence for a dual antiviral role of the major nuclear domain 10 component Sp100 during the immediate-early and late phases of the human cytomegalovirus replication cycle. J. Virol. 85:9447–9458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Tavalai N, Papior P, Rechter S, Leis M, Stamminger T. 2006. Evidence for a role of the cellular ND10 protein PML in mediating intrinsic immunity against human cytomegalovirus infections. J. Virol. 80:8006–8018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Tavalai N, Papior P, Rechter S, Stamminger T. 2008. Nuclear domain 10 components promyelocytic leukemia protein and hDaxx independently contribute to an intrinsic antiviral defense against human cytomegalovirus infection. J. Virol. 82:126–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Tavalai N, Stamminger T. 2008. New insights into the role of the subnuclear structure ND10 for viral infection. Biochim. Biophys. Acta 1783:2207–2221 [DOI] [PubMed] [Google Scholar]
- 89. Thomson JR, et al. 2000. Sporadic primary pulmonary hypertension is associated with germline mutations of the gene encoding BMPR-II, a receptor member of the TGF-beta family. J. Med. Genet. 37:741–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Tischer BK, von Einem J, Kaufer B, Osterrieder N. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40:191–197 [DOI] [PubMed] [Google Scholar]
- 91. Umbach JL, et al. 2008. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 454:780–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Umbach JL, Nagel MA, Cohrs RJ, Gilden DH, Cullen BR. 2009. Analysis of human alphaherpesvirus microRNA expression in latently infected human trigeminal ganglia. J. Virol. 83:10677–10683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Umbach JL, et al. 2010. Identification of viral microRNAs expressed in human sacral ganglia latently infected with herpes simplex virus 2. J. Virol. 84:1189–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Villard L, et al. 1997. Determination of the genomic structure of the XNP/ATRX gene encoding a potential zinc finger helicase. Genomics 43:149–155 [DOI] [PubMed] [Google Scholar]
- 95. Wong LH, et al. 2010. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 20:351–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Woodhall DL, Groves IJ, Reeves MB, Wilkinson G, Sinclair JH. 2006. Human Daxx-mediated repression of human cytomegalovirus gene expression correlates with a repressive chromatin structure around the major immediate early promoter. J. Biol. Chem. 281:37652–37660 [DOI] [PubMed] [Google Scholar]
- 97. Xue Y, et al. 2003. The ATRX syndrome protein forms a chromatin-remodeling complex with Daxx and localizes in promyelocytic leukemia nuclear bodies. Proc. Natl. Acad. Sci. U. S. A. 100:10635–10640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Yan N, Chen ZJ. 2012. Intrinsic antiviral immunity. Nat. Immunol. 13:214–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Yao Y, et al. 2007. Marek's disease virus type 2 (MDV-2)-encoded microRNAs show no sequence conservation with those encoded by MDV-1. J. Virol. 81:7164–7170 [DOI] [PMC free article] [PubMed] [Google Scholar]