Abstract
We have examined the underlying mechanism of hepatitis C virus (HCV)-mediated IFITM1 regulation. IFITM1 is a potential target of miR-130a. Our results demonstrated that miR-130a expression was significantly higher in HCV-infected hepatocytes and liver biopsy specimens than in controls. Introduction of anti-miR-130a in hepatocytes increased IFITM1 expression. Hepatocytes stably expressing IFITM1 reduced HCV replication. Together, these results suggested that HCV infection of hepatocytes upregulates miR-130a and that use of anti-miR-130a may have potential for restriction of HCV replication.
TEXT
Hepatitis C virus (HCV) is a hepatotropic flavivirus in the Hepacivirus genus and contains a linear, positive-strand RNA molecule of ∼9,500 nucleotides (7). HCV sequence divergence indicates several genotypes and a series of subtypes for the virus (13). An estimated 200 million people worldwide and 4 million people in the United States are infected with HCV (19, 21). Although current treatment is effective in >50% of HCV genotype 1-infected individuals (19), there is an urgent need to identify viral or cellular molecules that can be used as novel targets for therapy.
MicroRNAs (miRNAs) are a class of small noncoding RNAs that are processed by Dicer from precursors with a characteristic hairpin secondary structure (1). Hundreds of evolutionarily conserved miRNAs have been identified in plants, animals, and viruses (2, 3). miRNAs are transcribed genes processed to single-stranded regulatory RNA of ∼22 nucleotides (9). In mammalian cells, miRNAs affect gene silencing via both translational inhibition and mRNA degradation. An individual miRNA is capable of regulating dozens of distinct mRNAs, and together, the >1,000 human miRNAs are believed to modulate more than one-third of the mRNA species encoded in the genome (8, 9). Dysregulation of miRNA occurs frequently in a variety of diseases, including those of the liver (20). Understanding miRNAs modulated by HCV infection in hepatocytes may help in developing effective treatment strategies.
Interferon-induced transmembrane (IFITM) proteins are small (∼130 amino acids), with two transmembrane domains separated by a highly conserved cytoplasmic domain (10). IFITM proteins are induced by type I and II interferons (IFN), and most cells express a basal level of one or more of these proteins. The antiviral function of IFITM proteins has been shown from studies with influenza A virus, West Nile virus, dengue virus, vesicular stomatitis virus, and human immunodeficiency virus (5, 11, 14). We have shown recently that IFITM1 expression was significantly modulated in HCV-infected or IFN-pretreated, HCV-infected hepatocytes compared to that in IFN-treated hepatocytes (16). Transient expression of IFITM1 displayed anti-HCV activity (16), although little is known about the mechanism of its action. In this study, we have shown that HCV infection enhances miR-130a expression, which, in turn, inhibits endogenous IFITM1 expression.
miR-130a expression is enhanced in HCV-infected cells.
In silico (microRNA.org) prediction suggested that several microRNAs have potential targets at the IFITM1 3′ untranslated region (UTR), and miR-130a is one of them (mirSVR score is −0.0716). To verify the status of miR-130a in HCV infection, quantitative PCR was performed for HCV-infected and mock-treated hepatocytes. Total RNA was isolated using TRIzol reagent, and cDNA was synthesized using random hexamers and a SuperScript III reverse transcriptase kit (Invitrogen). Real-time PCR was performed with the cDNA for RNA quantitation using the TaqMan universal PCR master mix and 6-carboxyfluorescein minor groove binder probe for IFITM1, HCV, and for the endogenous control glyceraldehyde-3-phosphate dehydrogenase (GAPDH), as described previously (12, 16). The fold changes of specific mRNA in cells under different conditions compared to levels for mock-treated cells are presented after normalizing data to those of GAPDH mRNA expression. For detection of miR-130a, cDNA was synthesized using the TaqMan microRNA reverse transcription kit (Invitrogen) with a miRNA-specific primer (assay identification number [ID] 000454). miRNA levels were determined in the 7500 real-time system in conjunction with the gene-specific TaqMan assay kit. U6 (assay ID 001973) was used as an endogenous control for normalization. The expression level of miR-130a was significantly higher (∼8-fold) in HCV-infected hepatocytes than in the control hepatocytes (Fig. 1A). RNA from HCV-infected liver biopsy specimens also displayed enhanced miR-130a expression compared to control liver biopsy specimens (not infected with HCV or HBV) (Fig. 1B and C). As expected, IFITM1 expression was not significantly different between the groups of liver biopsy specimens (Fig. 1D).
Fig 1.
HCV infection enhances miR-130a expression. (A) Immortalized human hepatocytes (IHH) were infected with HCV genotype 1a for 72 h. RNA was isolated, and miR-130a was measured by real-time PCR. U6 RNA was used as an internal control. The results from three independent experiments are presented. (B) Total RNA was extracted from HCV-infected and control liver biopsy specimens (not infected with HCV or HBV). miR-130a expression was analyzed by real-time PCR, and U6 RNA was used as an internal control. A comparison of miR-130a levels in control (n = 8) and HCV-infected (n = 9) liver tissues is shown. (C) Results from individual specimens of panel B are shown. HCV loads from each specimen are mentioned on the top of each bar. (D) IFITM1 expression from control (n = 6) and HCV-infected (n = 6) liver biopsy specimens after normalization with internal control 18S RNA is shown.
Knockdown of miR-130a upregulates IFITM1 in hepatocytes.
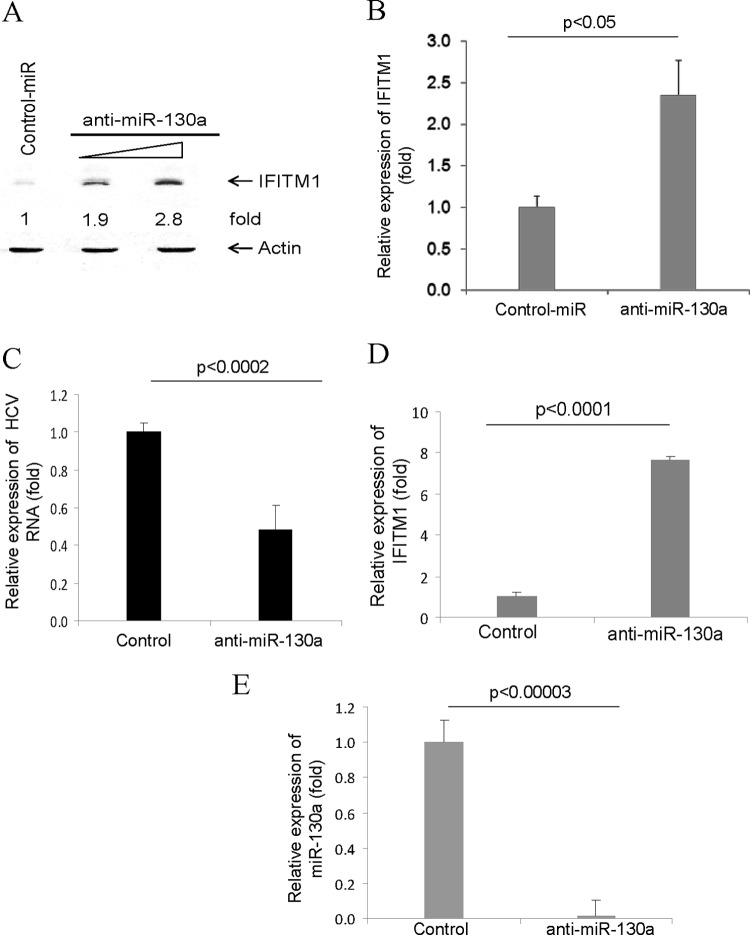
We examined whether miR-130a knockdown enhances IFITM1 expression in hepatocytes. For this, we performed a Western blot analysis in Huh7 cells transfected with 20 nM or 50 nM anti-miR-130a. A significant upregulation of IFITM1 expression compared to that of the control was observed after normalization with the housekeeping protein actin (Fig. 2A). We also examined IFITM1 mRNA levels using similar conditions. Quantitative reverse transcription-PCR (qRT-PCR) data revealed that Huh7 cells transfected with anti-miR-130a display enhanced expression of IFITM1 mRNA compared to that of the control (Fig. 2B). Together, these data suggested that IFITM1 is a target of miR-130a.
Fig 2.
Knockdown of miR-130a enhances IFITM1 expression in hepatocytes. (A) Huh7 cells were treated with a control or anti-miR-130a. Cell lysates were prepared after 48 h of transfection, and IFITM1 expression was analyzed by a Western blot using a specific antibody. The blots were reprobed with an antibody to actin for a comparison of protein loads. The fold change of IFITM1 after normalization with actin is shown and compared with the control-treated sample. (B) IFITM1 mRNA was measured from control and miR-130a knockdown cells as described in Fig. 1. The results are the means of three independent experiments. (C and D) Huh7.5 cells were infected with HCV and were treated with the control or anti-miR-130a. HCV and IFITM1 mRNA levels were measured by real-time PCR and presented after normalization with GAPDH. The results shown are from two independent experiments with three replicates. (E) HCV-infected cells transfected with the control or anti-miR-130a. Cells were examined for miR-130a expression by qRT-PCR. Results after normalization with U6 as an internal control are presented.
Knockdown of miR-130a inhibits HCV RNA replication.
We next examined the effect of miR-130a on HCV replication and IFITM1 expression using Huh7.5 cells infected with HCV genotype 2a (multiplicity of infection [MOI], 0.1). Cells were transfected with the control or anti-miR-130a 2 days postinfection, and RNA was isolated 48 h posttransfection. HCV replication was reduced in miR-130a knockdown cells compared to that in control-transfected cells (Fig. 2C). The IFITM1 mRNA level was higher in miR-130a knockdown cells than in the control (Fig. 2D). A significant inhibition of miR-130a was observed following anti-miR-130a treatment (Fig. 2E). Similar results were obtained when cells harboring an HCV subgenomic replicon were treated with anti-miR-130a (data not shown).
IFITM1 is a direct target of miR-130a.
To identify the underlying mechanisms of how miR-130a downregulates IFITM1, we performed a luciferase reporter assay. For this, the 3′ UTR of IFITM1 (with a potential binding site of miR-130a or its mutant) was cloned into pMIR-Report luciferase vector (Ambion). Cells were cotransfected with the wild-type or mutant 3′ UTR of IFITM1 and mimic miR-130a. Cell lysates were prepared 48 h posttransfection for the luciferase assay as described previously (17, 18). The results demonstrated that miR-130a significantly repressed the wild-type IFITM1 3′ UTR directly, as evident from decreased luciferase activity (Fig. 3A and B). On the other hand, the mutant 3′ UTR of IFITM1 luciferase activity was unaffected by mimic miR-130a.
Fig 3.
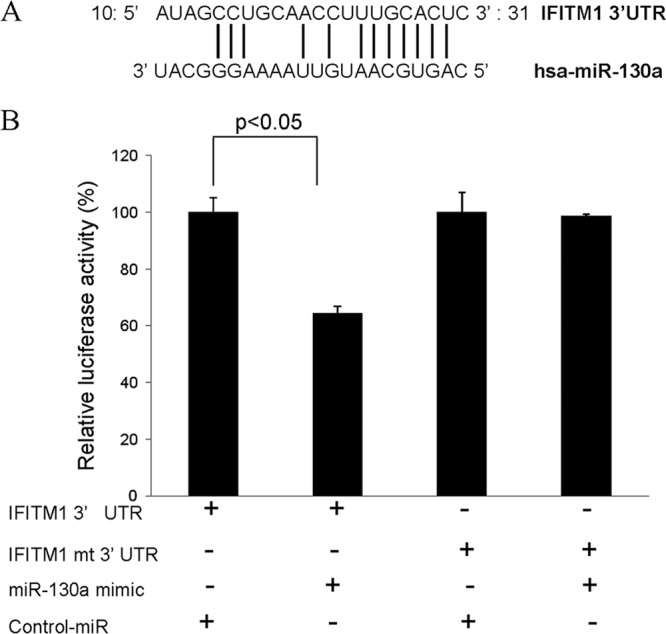
miR-130a targets IFITM1 in hepatocytes. (A) Computational analysis of the IFITM1 3′ UTR revealed a single putative miR-130a binding site. The miR-130a target region of IFITM1 (GenBank accession number NM_003641.3) is indicated. (B) Luciferase assay showing decreased reporter activity after cotransfection of the IFITM1 3′ UTR and mimic miR-130a. On the other hand, the mutant 3′ UTR of IFITM1 had no detectable effect on reporter activity. The results are the means ± the standard errors from three independent experiments.
Exogenous expression of IFITM1 without the 3′ UTR rescues the miR-130a effect.
We have shown previously that transient expression of IFITM1 has anti-HCV activity. We therefore hypothesized that HCV infection upregulates miR-130a expression, which, in turn, blocks the increase of IFITM1 expression, resulting, in part, in establishment of persistent infection. To further verify our hypothesis, we generated a stable Huh7 cell line expressing IFITM1 cDNA (without the 3′ UTR). Control or Huh7-IFITM1 cells were infected with HCV genotype 2a for 72 h. Cellular RNA was isolated, and HCV RNA was quantitated as previously described (12, 16). HCV replication was inhibited in Huh7-IFITM1-infected cells compared to that in control cells (Fig. 4A). Similar results were obtained when IFITM1 (without the 3′ UTR) was introduced in HCV-infected hepatocytes (16). As expected, miR-130a expression levels remained unchanged in both control and experimental cells (Fig. 4B). Together, these results suggested that the effect of miR-130a can be rescued by IFITM1 without the 3′ UTR.
Fig 4.
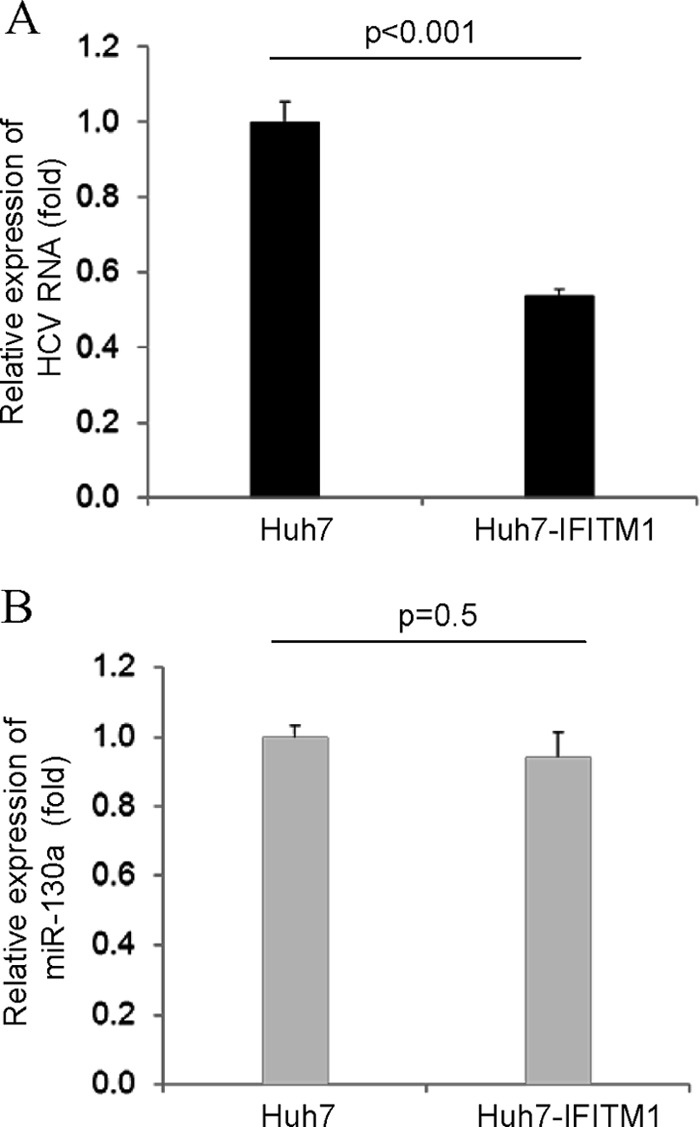
Exogenous expression of IFITM1 rescues the miR-130a effect. Huh7 cells were stably transfected with IFITM1 or vector control, followed by HCV genotype 2a infection. Cells were incubated for 72 h. (A) Total RNA from cells was isolated and analyzed for HCV replication after normalization with GAPDH. (B) miR-130a expression status was examined from total RNA by real-time PCR as described in Fig. 1. The results from two independent experiments with three replicates are shown.
miRNA changes in the liver have been reported in disease processes such as hepatocarcinogenesis and liver fibrosis (4, 6, 20). Recent studies on the roles of miRNAs in several human diseases suggest that the relationship between HCV, miRNAs, and mRNA may provide new insights into host cell response to HCV infection (15). Our results demonstrated that HCV infection upregulates miR-130a and that IFITM1 is a direct target for this miRNA. Knockdown of miR-130a enhanced IFITM1 expression and reduced HCV replication. To our knowledge, this is the first report suggesting a role of a miRNA directly involved in HCV-mediated innate immune response (Fig. 5). Together, these results demonstrated that HCV infection upregulates miR-130a, which, in turn, blocks the increase of IFITM1 expression, resulting, in part, in establishment of persistent infection.
Fig 5.
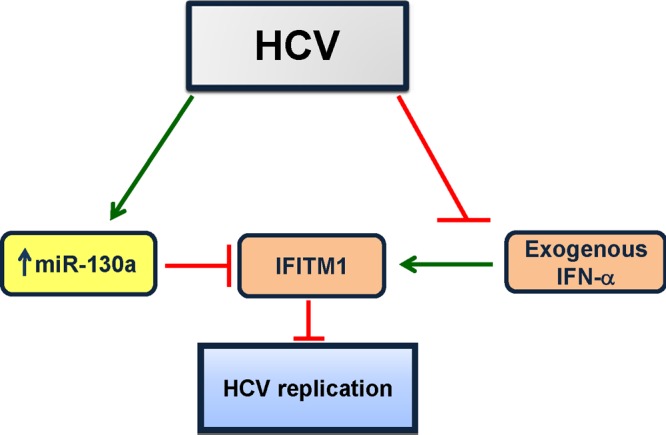
Schematic diagram showing the cross talk among HCV, miR-130a, and IFITM1. Enhanced IFITM1 expression inhibits HCV replication. HCV infection upregulates miR-130a in hepatocytes, which, in turn, inhibits IFITM1 expression. On the other hand, HCV infection inhibits alpha interferon (IFN-α)-induced IFITM1 expression. Arrows denote upregulation, and blunt arrows denote inhibition.
ACKNOWLEDGMENTS
We thank Leonard Grosso for measuring HCV genome copy numbers.
This work was supported by grant 5U54AI057160 (R.R.) to the Midwest Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research and grants DK081817 and AI065535 (R.B.R.) from the National Institutes of Health.
Footnotes
Published ahead of print 11 July 2012
REFERENCES
- 1. Ambros V. 2004. The functions of animal microRNAs. Nature 431:350–355 [DOI] [PubMed] [Google Scholar]
- 2. Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D. 2003. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr. Biol. 13:807–818 [DOI] [PubMed] [Google Scholar]
- 3. Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136:215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bihrer V, et al. 2011. Serum miR-122 as a biomarker of necroinflammation in patients with chronic hepatitis C virus infection. Am. J. Gastroenterol. 106:1663–1669 [DOI] [PubMed] [Google Scholar]
- 5. Brass AL, et al. 2009. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139:1243–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cermelli S, Ruggieri A, Marrero JA, Ioannou GN, Beretta L. 2011. Circulating microRNAs in patients with chronic hepatitis C and non-alcoholic fatty liver disease. PLoS One 6:e23937 doi:10.1371/journal.pone.0023937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Choo QL, et al. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359–362 [DOI] [PubMed] [Google Scholar]
- 8. Farazi TA, Spitzer JI, Morozov P, Tuschl T. 2011. miRNAs in human cancer. J. Pathol. 223:102–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Filipowicz W, Bhattacharyya SN, Sonenberg N. 2008. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 9:102–114 [DOI] [PubMed] [Google Scholar]
- 10. Friedman RL, Manly SP, McMahon M, Kerr IM, Stark GR. 1984. Transcriptional and posttranscriptional regulation of interferon-induced gene expression in human cells. Cell 38:745–755 [DOI] [PubMed] [Google Scholar]
- 11. Jiang D, et al. 2010. Identification of five interferon-induced cellular proteins that inhibit West Nile virus and dengue virus infections. J. Virol. 84:8332–8341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kanda T, et al. 2006. Generation of infectious hepatitis C virus in immortalized human hepatocytes. J. Virol. 80:4633–4639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kuiken C, Simmonds P. 2009. Nomenclature and numbering of the hepatitis C virus. Methods Mol. Biol. 510:33–53 [DOI] [PubMed] [Google Scholar]
- 14. Lu J, et al. 2011. The IFITM proteins inhibit HIV-1 infection. J. Virol. 85:2126–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Peng X, et al. 2009. Computational identification of hepatitis C virus associated microRNA-mRNA regulatory modules in human livers. BMC Genomics 10:373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Raychoudhuri A, et al. 2011. ISG56 and IFITM1 proteins inhibit hepatitis C virus replication. J. Virol. 85:12881–12889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ru P, Steele R, Hsueh EC, Ray RB. 2011. Anti-miR-203 upregulates SOCS3 expression in breast cancer cells and enhances cisplatin chemosensitivity. Genes Cancer 2:720–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Steele R, Mott JL, Ray RB. 2010. MBP-1 upregulates miR-29b that represses Mcl-1, collagens, and matrix-metalloproteinase-2 in prostate cancer cells. Genes Cancer 1:381–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tavis JE, Donlin MJ, Aurora R, Fan X, Di Bisceglie AM. 2011. Prospects for personalizing antiviral therapy for hepatitis C virus with pharmacogenetics. Genome Med. 3:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Varnholt H, et al. 2008. MicroRNA gene expression profile of hepatitis C virus-associated hepatocellular carcinoma. Hepatology 47:1223–1232 [DOI] [PubMed] [Google Scholar]
- 21. Yang JD, Roberts LR. 2010. Hepatocellular carcinoma: a global view. Nat. Rev. Gastroenterol. Hepatol. 7:448–458 [DOI] [PMC free article] [PubMed] [Google Scholar]