Abstract
Arenaviruses can cause severe hemorrhagic fever diseases in humans, with limited prophylactic or therapeutic measures. A small RING-domain viral protein Z has been shown to mediate the formation of virus-like particles and to inhibit viral RNA synthesis, although its biological roles in an infectious viral life cycle have not been directly addressed. By taking advantage of the available reverse genetics system for a model arenavirus, Pichinde virus (PICV), we provide the direct evidence for the essential biological roles of the Z protein's conserved residues, including the G2 myristylation site, the conserved C and H residues of RING domain, and the poorly characterized C-terminal L79 and P80 residues. Dicodon substitutions within the late (L) domain (PSAPPYEP) of the PICV Z protein, although producing viable mutant viruses, have significantly reduced virus growth, a finding suggestive of an important role for the intact L domain in viral replication. Further structure-function analyses of both PICV and Lassa fever virus Z proteins suggest that arenavirus Z proteins have similar molecular mechanisms in mediating their multiple functions, with some interesting variations, such as the role of the G2 residue in blocking viral RNA synthesis. In summary, our studies have characterized the biological roles of the Z protein in an infectious arenavirus system and have shed important light on the distinct functions of its domains in virus budding and viral RNA regulation, the knowledge of which may lead to the development of novel antiviral drugs.
INTRODUCTION
Several arenaviruses, including Lassa fever virus (LASV), can cause severe and lethal hemorrhagic fever diseases in humans. LASV causes endemic infections in West Africa with estimated 500,000 cases resulting in ∼5,000 deaths annually (16). Except for a Junin virus vaccine, no licensed vaccine for human usage is currently available for prevention of pathogenic arenavirus infections. Therapeutic options are limited and depend mainly on supportive cares. Ribavirin, a nonspecific antiviral compound, has shown some levels of efficacy only if it is administered at an early stage of viral infection when the symptoms are insidious (15). Immune therapy such as the transfusion of immune plasma has also been used to treat Argentine hemorrhagic fever (8).
Arenaviral bisegmented RNA genomes encode four proteins that include the glycoprotein precursor GPC, the nucleoprotein NP, the RNA-dependent RNA polymerase protein L, and the matrix protein Z (2). The Z protein is a small 15-kDa RING domain protein with multiple functions. It plays a structural role by forming a matrix layer in between the lipid membranes and the NP density layer within viral virions (17, 21). It has been reported to have a self-budding activity and can incorporate viral ribonucleoproteins and glycoproteins into virus-like particles (VLPs) (18, 23). In addition, the Z protein can strongly inhibit viral RNA synthesis (6) by directly locking the L polymerase protein in a catalytically inactive state (12) and therefore is postulated to play an important role in regulating viral genome replication and transcription. Recently, the Z proteins of New World pathogenic arenaviruses Junin, Machupo, Guanarito, and Sabia virus have also been shown to inhibit RIG-1-dependent innate immune response (9).
Many efforts have been made to understand the molecular mechanisms of the Z protein. It contains an N-terminal myristylation site G2, a central RING zinc-finger domain, and C-terminal late (L) domain(s). The G2 myristylation site has been shown to be essential for its membrane association and for self-budding activity (19, 24). The L domain(s) mediates Z budding activity by interacting with certain components of the cellular endosomal sorting complex required for transport (ESCRT) pathway (18, 23, 25). Z-mediated inhibition of lymphocytic choriomeningitis virus (LCMV) promoter-driven reporter RNA synthesis requires the structural integrity of the RING domain but not the N-terminal residues 1 to 16 or the C-terminal residues 79 to 90 (5).
Despite extensive works, no study has yet examined the biological roles of the Z protein in the context of infectious arenaviral life cycle. By taking advantage of our recently developed reverse genetics system for Pichinde virus (PICV) (14), a nonpathogenic arenavirus that can cause Lassa fever-like symptoms in infected guinea pigs (1, 11, 27, 28), we have undertaken an effort to examine the biological roles of each of the conserved residues and domains within the Z protein. Our results have demonstrated that most of the conserved residues are absolutely essential for virus replication, including the G2 residue, the zinc-binding C and H residues of the RING domain, and the invariant L and P residues at the C terminus of the protein. The C-terminal L domain can tolerate di-codon changes to certain extents, albeit resulting in much reduced viral infectivity. Additional functional characterizations of the LASV and PICV Z proteins have provided some important insights into the molecular mechanisms of Z protein in supporting infectious virus replication.
MATERIALS AND METHODS
Cells and viruses.
BSRT7-5 cells, which stably express the T7 RNA polymerase, were obtained from K. Conzelmann (Ludwig-Maximilians-Universität, Munich, Germany) and cultured in minimal essential medium supplemented with 10% fetal bovine serum (FBS), 1 μg of Geneticin per ml, and 50 μg of penicillin-streptomycin/ml. Baby hamster kidney cells BHK21 and Vero cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% FBS and 50 μg of penicillin-streptomycin/ml. Human kidney epithelial 293T cells were grown in DMEM supplemented with 10% FBS and 50 μg of penicillin-streptomycin/ml. Recombinant PICV viruses were amplified in BHK21 cells, and the infectious virus titer was determined by plaque assay in Vero cells as described previously (13).
Plasmids.
Site-directed mutations were introduced into the Z gene on plasmid DNA by QuikChange PCR-mediated mutagenesis according to the manufacturer's instructions (Stratagene). After sequence verification, the DNA fragments containing the desired mutation in the PICV Z gene were subcloned into a plasmid encoding the full-length L segment (14). To generate PICV Z protein expression constructs, wild-type (WT) or mutant PICV gene was amplified from the respective plasmid of the full-length L segment using primers containing hemagglutinin (HA) or Myc tag at the C terminus, and cloned into the pCAGGS expression vector. The C-terminal HA-tagged LASV Z was also similarly cloned into the pCAGGS expression vector. Mutations were introduced into the pCAGGS-LASV-Z by PCR mutagenesis and verified by sequence analysis. Primary sequences of all primers can be provided upon request.
Generation of infectious PICV viruses from plasmid transfection.
Recombinant PICV viruses were generated using the reverse genetics system as described previously (14). The L-segment-encoding plasmid of either WT or mutant Z gene was transfected, along with the S-segment-encoding plasmid into BSRT7-5 cells. At various time points, supernatants were collected for plaque assay in order to quantify the amount of infectious viral particles. After plaque purification, viruses were amplified in BHK-21 cells. Mutations in the recombinant viruses after amplification were verified by sequencing the reverse transcription-PCR (RT-PCR) products. All rescued mutant viruses have been sequence confirmed.
Growth curve analysis.
Cells were seeded in six-well plates at 90 to 100% confluence and infected (in triplicates) with viruses at a multiplicity of infection (MOI) of 0.01 for 1 h at 37°C. After a washing step with phosphate-buffered saline, a fresh aliquot of medium was added to the culture. At different time points postinfection, aliquots of the supernatant were harvested for plaque assaying on Vero cells.
Z budding assay.
293T cells were transfected with HA-tagged LASV or PICV Z plasmid for 48 h. Cell pellets were collected and lysed by radioimmunoprecipitation assay (RIPA) buffer to prepare cell lysates. The Z self-budding particles were concentrated from the supernatants by polyethylene glycol (PEG) precipitation. In brief, supernatants were centrifuged at 14,000 rpm for 10 min. The budded VLPs in the cleared supernatants were precipitated with 8% PEG 8000–0.5 M sodium chloride, collected after centrifugation at 14,000 rpm for 15 min, and lysed by denaturing or nondenaturing RIPA buffer. Cell lysates and the budded VLPs were analyzed on a 15% sodium dodecyl sulfate (SDS)-polyacrylamide gel and detected for Z proteins by Western blotting with the anti-HA antibody.
NP incorporation into Z-induced particles.
293T cells were transfected with the myc-tagged NP expression vector, with or without the WT or mutant HA-tagged Z expression vector. After 48 h, cell lysates and the budded VLPs were prepared as described above in the Z budding assay, separated on a SDS-polyacrylamide gel, and analyzed by Western blotting for NP and Z using anti-myc and anti-HA antibodies, respectively.
RESULTS
Rescue of recombinant PICV Z mutant viruses using a reverse genetics system.
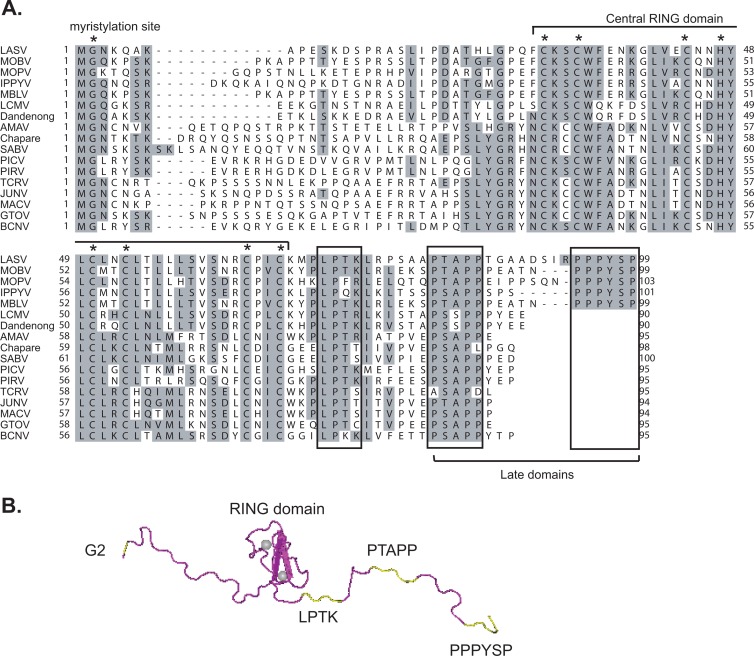
Alignment of all available arenavirus Z sequences in GenBank has revealed multiple conserved residues, including the G2 myristylation site, the invariant CHCC and CCCC zinc-binding residues within the central RING domain, the conserved LP(TK) motif, and the C-terminal proline-rich late (L) domains (Fig. 1). Previous mutational studies have revealed important roles for the G2 residue and the L domains in the Z-mediated virus budding assay (18, 19, 23–25) and for the RING domain in regulating viral RNA synthesis (5). Capul et al. have examined the functional roles of the conserved residues in VLP formation and infectivity (3). However, their biological roles in the context of infectious arenavirus replication have never been examined. We have developed a reverse genetics system for PICV (14), a nonpathogenic arenavirus whose infection of guinea pigs serves as a convenient small-animal model for arenavirus-induced hemorrhagic fever diseases (1, 11, 27, 28). Except for some sequence variations in the C-terminal L domains, both PICV and LASV Z proteins share the same invariant G2, the zinc-binding C/H residues within the RING domain, and the LPTK residues (Fig. 1). LASV Z contains two L domains at the C terminus, PTAPP and PPPYSP, separated by eight residues, whereas PICV Z protein contains the sequence PSAPPYEP, which appears to consist of two overlapping L domains PSAPP and PYEP.
Fig 1.
Conserved residues/domains of the Z protein. (A) Sequence alignment of arenavirus Z proteins. The G2 myristylation site and the invariant CCCH/CCCC residues within the central RING domain are indicated by asterisks (*). The conserved LP(TK) residues and the C-terminal late domains are boxed. (B) The three-dimensional model of LASV Z protein is shown in tube form (magenta, PDB accession no. 2KO5), with the G2 residue, LPTK motif, and the two late domains (PTAPP and PPPYSP) shown in yellow. The central RING domain binds two zinc cations that are represented by gray balls.
We introduced alanine substitution at each of the respective conserved residues of the PICV Z protein in the L segment of the reverse genetic system (Table 1). The mutations consist of one to four amino acid substitutions targeting the G2 myristylation site (G2A), the central RING domain (C38A/C41A, C51A/H54A/C57A/C50A, and C71A), the LPTK site [LPTK(78-81)AAAA and individual mutations], and the L domains (P88A/S89A, P90A/P91A, and E94A/P95A). To generate recombinant viruses, plasmids encoding L and S segments were transfected into cells that constitutively express the T7 polymerase. At various time points posttransfection (48, 72, 96, and 120 h), supernatants were collected for plaque assay to determine the amount of infectious virus particles generated (Table 1). In the WT control, infectious recombinant viruses were produced as early as 48 h posttransfection (hpt) and reached a titer of ∼107 PFU/ml at 120 hpt. The successful rate of rescuing WT viruses from this PICV reverse genetics system is 100%, based on our more than a hundred experiences thus far. In contrast, despite multiple attempts, most Z mutants were unable to generate any infectious virus even after 120 hpt, suggesting an essential role for these conserved residues in the arenaviral life cycle. These lethal mutations include G2A (M32), mutations at the C or H of the RING domain (M24, M26, and M28), and those of the invariant L78 or P79 (M30, M914, and M916). In contrast, mutations at the less conserved residues T80 (M918) and K81 (M40) produced viable viruses at lowered titers. Dicodon mutations targeting proline residues of the L domains (PSAPPYEP) led to significantly delayed and reduced production of recombinant viruses. Compared to WT, both P88A/S89A (M34) and P90A/P91A (M36) generated ∼3 log fewer viruses, whereas E94A/P95A (M38) produced ∼ 4 log fewer viruses at 120 hpt (Table 1). Sequence analysis of the RT-PCR products of these mutant viruses were carried out to verify that no WT revertant viruses were recovered. In short, we have shown that most of the invariant residues of the Z protein play indispensable roles in arenavirus biology, such as the G2 myristylation site, the C/H residues within the RING domain, and the LP residues, and that the proline residues within the L domain play important and partially overlapping roles in virus replication.
Table 1.
Infectious titers of the recombinant PICV Z mutants generated by reverse genetics systema
| WT or mutant | Z mutation | Infectious titers (PFU/ml) |
|||
|---|---|---|---|---|---|
| 48 hpt | 72 hpt | 96 hpt | 120 hpt | ||
| WT | WT | 2,600 | 4.8E + 04 | 7.8E + 05 | 1.2E + 07 |
| M32 | G2A | 0 | 0 | 0 | 0 |
| M24 | C38A/C41A | 0 | 0 | 0 | 0 |
| M26 | C51A/H54A/C57A/C60A | 0 | 0 | 0 | 0 |
| M28 | C71A | 0 | 0 | 0 | 0 |
| M34 | P88A/S89A | 0 | 140 | 1,600 | 4,400 |
| M36 | P90A/P91A | 0 | 240 | 1,800 | 4,000 |
| M38 | E94A/P95A | 0 | 0 | 160 | 720 |
| M30 | LPTK(78–81)/AAAA | 0 | 0 | 0 | 0 |
| M914 | L78A | 0 | 0 | 0 | 0 |
| M916 | P79A | 0 | 0 | 0 | 0 |
| M918 | T80A | 12 | 680 | 3,000 | 2.4E + 04 |
| M40 | K81A | 120 | 2,600 | 4.2E + 04 | 3.6E + 05 |
Recombinant PICV strains were generated by the reversegenetics systems (14), in which a plasmid encoding thefull-length L segment of either the WT or the respective mutant Z gene, alongwith a plasmid encoding the S segment, were transfected into BSRT7-5cells. Supernatants collected at 48, 72, 96, and 120 h posttransfection (hpt)were subjected to plaque assay to determine the titer of infectious virus. Theresults shown are representative of three independent experiments.
In vitro growth kinetic analysis of PICV Z mutants.
We plaque purified the five viable recombinant Z mutants—M34 (P88A/S89A), M36 (P90A/P91A), M38 (E94A/P95A), M918 (T80A), and M40 (K81A)—and verified their mutational status by sequence analysis. Compared to WT, K81A produced much smaller plaques, whereas all four of the other mutants produced pinhead-sized plaques (Fig. 2A), suggesting that these mutants have severe growth defects. To quantitatively compare their growth kinetics, we carried out a growth curve analysis of WT and mutant viruses in Vero cells at an MOI of 0.01 (Fig. 2B). K81A and T80A grew less well than WT by 0.5 and 1 log, respectively. All three L domain mutants (P88A/S89A, P90A/P91A, and E94A/P95A) had similar growth kinetics that were at least 2-log less than for the WT. In summary, mutations at the less conserved T80 and K81 residues reduced viral growth, and those in the L domain caused even greater growth defects.
Fig 2.
Rescue of recombinant PICV viruses with wild-type or mutant Z proteins. (A) Plaque morphology of viable recombinant PICV viruses encoding WT or the respective Z mutant proteins. (B) Determination of viral growth kinetics of recombinant viruses by growth curve analysis. Vero cells were infected with WT or the respective PICV mutants at an MOI of 0.01. Virus titers in the supernatants at various time points postinfection were quantified by plaque assay. The results shown are averages and standard deviations from at least three independent experiments.
Effect of mutations on the Z self-budding activity.
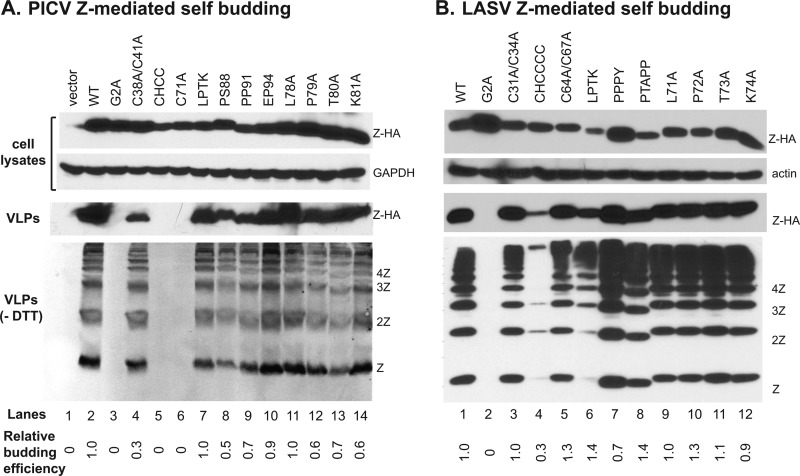
A major known function of the Z protein is to drive the virus budding process at the cellular membrane (18). We therefore examined whether any of the Z mutations affect budding. To do this, we constructed vectors to express HA-tagged WT and mutant PICV Z proteins. After plasmid transfection, cell lysates were prepared and analyzed by Western blotting. WT and all mutant Z proteins were readily expressed, albeit with some variations at the protein levels as detected by anti-HA antibody (Fig. 3A, top panels). The supernatants were collected, cleared of cellular debris by centrifugation, and precipitated with PEG to concentrate the VLPs, which were analyzed by Western blotting with the anti-HA antibody in both reducing (VLPs) and nonreducing (VLPs, no dithiothreitol [−DTT]) conditions in order to detect Z monomeric and multimeric formations (Fig. 3A, bottom panel). The relative budding efficiency for each Z mutant (shown at the bottom of Fig. 3A) was determined by normalizing the levels of the Z proteins in the VLPs to those in the cells, followed by a comparison of mutant to WT (set at 1.0). Consistent with a previous report (19), we found that WT Z proteins could bud out of the cells efficiently (Fig. 3A, VLPs, lane 2), whereas mutation of the G2 myristylation residue (G2A) completely abrogated self-budding activity (lane 3). The three RING domain mutations (CHCC, C71A, and C38A/C41A) either completely lost (lanes 5 and 6) or significantly reduced the self-budding activities (lane 4). The loss-of-function in budding activity of the G2A and RING domain mutations likely explains their inability to support fully infectious viruses (Table 1). In contrast, dicodon mutations within the C-terminal L domain of PICV (P88A/S89A, P90A/P91A, and E94A/P95A) resulted in little to no decrease in budding activity (relative budding efficiency varies from 0.5 to 0.9) (Fig. 3A, lanes 8 to 10), which correlates with the phenotypes of these L domain mutants in viral viability and growth (Table 1 and Fig. 2). Interestingly, individual or combined mutations of the LPTK residues had little effect on Z budding activities (Fig. 3A, lanes 7, 11 to 14), despite the inability to rescue viruses from the L78A or P79A mutation (Table 1), indicating that the two invariant residues have essential functions other than virus budding.
Fig 3.
Effects of mutations at the conserved residues on Z-mediated self-budding activity. (A) Mutational analysis of the PICV Z protein on self-budding. 293T cells were transfected with either WT or the respective PICV Z mutant expression vectors. Expressions of HA-tagged Z proteins (Z-HA), along with β-actin (actin) as a loading control in the cell lysates, were analyzed by Western blotting. Virus supernatants were collected, precipitated by PEG8000, separated by 15% reducing (VLPs) and nonreducing (VLPs, −DTT) gels, and analyzed by Western blotting. Z budding efficiency was quantified by the amount of Z proteins in the released VLPs normalized by the expression level in the cell lysates and is shown as the percentage of WT Z protein budding (set as 1.0). (B) Similar budding assay was conducted on the WT and mutant LASV Z proteins.
To validate the above results in a different arenavirus Z protein, similar alanine substitutions were introduced into the HA-tagged LASV Z protein expression vector. These mutations target the G2 site (G2A), the RING domain (C31A/C34A, CHCCCC, and C64A/C67A), the LP(TK) site (LPTK→AAAA and individual mutant), and the L domain (PPPY and PTAPP). WT and all mutant Z proteins were efficiently expressed, albeit with some variations (Fig. 3B, top panels). The PEG-precipitated VLPs were analyzed in both reducing (VLPs) and nonreducing (VLPs, −DTT) conditions (Fig. 3B, bottom panel). Most mutations including those at the LPTK site and the L domains had little effects on Z budding efficiency, except for the G2A mutation that completely abolished budding and a major CHCCCC mutation in the RING domain that significantly reduced budding efficiency. The LPTK→AAAA mutation appeared to decrease the level of Z monomers and dimers in the budded VLP samples (Fig. 3B, bottom panel, lane 4), but nonetheless did not affect the overall relative budding efficiency (∼ 1.4) due to its correspondingly lowered protein expression level in the cytoplasm.
Overall, mutational analyses of the conserved residues of both PICV and LASV Z proteins have produced similar results on the self-budding activity with some variations observed for the RING domain mutants. The RING domain of PICV Z protein seems to play an important role in Z budding, as three RING domain mutants had little or no budding activity (Fig. 3A, lanes 4 to 6). In contrast, the RING domain of LASV Z protein seems to play a less important role in Z budding, since only a 6-residue change (CHCCCC) could lead to a notable decrease in budding (Fig. 3B, lane 4), while two other dicodon mutations (C31A/C34A and C64A/C67A) produced no effects (Fig. 3B, lanes 3 and 5). For both PICV and LASV Z proteins, the G2 myristylation site plays an essential role in Z budding activity, the C-terminal L domains have redundant functions, and the LP(TK) site is not at all required in this process.
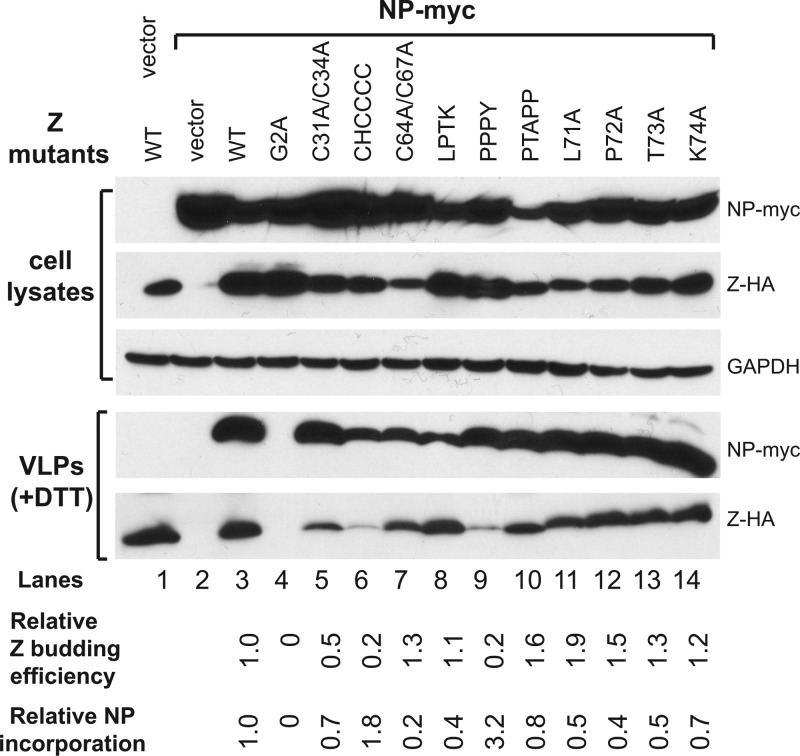
Effect of mutations on NP incorporation into VLPs.
It has been demonstrated that NP can be incorporated into the Z-induced VLPs possibly through specific NP-Z interactions (3, 4). We examined whether Z mutations affect NP incorporation into VLPs by cotransfecting 293T cells with LASV NP-myc expression vector together with either WT or mutant Z-HA vectors. Similar results were observed for PICV Z mutants (data not shown). Cell lysates and VLPs in the supernatants were analyzed for NP and Z by Western blotting with anti-myc and anti-HA antibodies, respectively (Fig. 4). The relative Z budding efficiency was determined as described above, and the relative NP incorporation efficiency was calculated by the level of NP expression normalized to the level of Z protein in the VLPs. NP was not detected in the PEG precipitates from cells expressing NP alone, suggesting that, in contrast to Z, NP does not have a self-budding activity (Fig. 4, lanes 1 to 2). However, NP was readily detected in VLPs when the WT Z protein was coexpressed (Fig. 4, lane 3), demonstrating that NP can be efficiently incorporated into Z-induced VLPs. Compared to the Z budding activity in the absence of NP as shown in Fig. 3B, the presence of NP did not significantly alter the budding behavior of most of the Z mutants, except that two RING domain mutants (C31A/C34A and CHCCCC) and one L domain mutant (PPPY) showed lower budding activity (relative budding efficiencies of 0.5, 0.2, and 0.2, respectively) (Fig. 4, lanes 5, 6, and 9); the reason for this is unclear. Since G2A mutant did not form VLPs, NP was not detected in the PEG precipitate (Fig. 4, lane 4). On the other hand, NP was present at various levels in the VLPs induced by each of the other Z mutants (Fig. 4, lanes 5 to 14), indicating that none of those Z mutants can completely disrupt NP incorporation into VLPs. A closer examination, however, identifies several Z mutants with reduced relative NP incorporation efficiency. For example, the RING domain mutant C64A/C67A significantly decreased the relative NP incorporation level by 80% (Fig. 4, lane 7). The LPTK→AAAA mutation and three individual mutations (L71A, P72A, and T73A) all reduced NP incorporation by 50 to 60% (Fig. 4, lanes 8 and lanes 11 to 13), while the K74A mutation reduced it by 30% (Fig. 4, lane 14). In contrast, the CHCCCC RING domain mutant and the PPPY late domain mutant increased the relative NP incorporation efficiencies by 2- and 3-fold (Fig. 4, lanes 6 and 9), as a result of their substantially lowered levels of Z budding but nonetheless normal amounts of incorporated NPs. In short, we have shown that none of the conserved residues in the Z protein is essential for incorporating NP into VLPs but that the LPTK site, and possibly, the RING domain, may play a more important role in this process.
Fig 4.
Effects of mutations at the conserved residues of Z on the efficiency of NP incorporated into Z-mediated VLP formations. 293T cells were transfected with WT or mutant Z protein expression vector and with or without the NP expression vector. The amounts of VLPs released into the supernatants were analyzed by Western blot against Z and NP proteins. The relative Z budding efficiency was quantified as described in the legend of Fig. 3. The relative NP incorporation efficiency for each Z mutant was determined by the relative NP levels incorporated into the released VLPs compared to the WT Z control (set as 1.0).
Effect of mutations on inhibiting viral RNA synthesis in minigenome assay.
Arenavirus Z protein has been shown to regulate viral RNA replication and transcription by inhibiting viral RNA synthesis (6). We ask whether any of our Z mutants could affect viral RNA synthesis by using our recently established minigenome system (20). This system consists of the LASV S segment that is devoid of viral gene coding sequences but instead encodes the Renilla luciferase reporter (RLuc) gene in a negative-sense orientation and contains the conserved viral terminal sequences as well as the intergenic region. This 5RLuc3 minigenome is recognized by LASV or PICV polymerase complex that consists of the respective NP and L proteins and can produce up to 5- to 6-log-higher levels of RLuc expression than a control reaction that lacks the L-protein-encoding plasmid (Fig. 5, control versus no Z). As expected, we found that the WT Z protein strongly inhibited viral RNA synthesis in both LASV and PICV minigenomic systems since it effectively reduced the RLuc expression by 2 logs. For both LASV and PICV Z proteins, all RING domain mutations and the four-residue change of the LPTK site completely lost the ability to inhibit viral RNA synthesis, whereas all L domain mutants functioned just like the WT (Fig. 5). Interestingly, whereas the G2A mutant of LASV Z protein still strongly inhibited viral RNA synthesis at the same level as the WT, the same mutation in the PICV Z context completely lost the inhibitory activity (Fig. 5, shown by arrows). In summary, we found that the RING domain and the LPTK residues are essential in regulating viral RNA synthesis and that the G2 residue in different Z proteins may have a differential role.
Fig 5.
Effects of mutations at the conserved residues on Z-mediated inhibition of viral RNA synthesis. LASV Z (left panel) or PICV Z (right panel) proteins of either wild-type or mutant forms were examined for their ability to inhibit viral RNA transcription in the minigenome assay. Arrows show differential effects of G2A mutation between LASV and PICV Z proteins. The results shown are averages of at least three independent experiments with error bars representing standard deviations.
DISCUSSION
Our study provides unequivocal evidence for the essential roles of the invariant residues of arenaviral Z protein in infectious viral replication. These residues, including G2, the zinc-binding C and H residues within the RING domain, and L78 and P79 (numbered according to PICV Z protein), are absolutely conserved among all known Z proteins. Previous studies have demonstrated some of these residues' critical roles in VLP formation and viral RNA regulation (3, 5, 19, 24). Our study, however, establishes their essential roles in the context of infectious virus life cycle, since alanine substitutions at these residues have resulted in nonproductive PICV virus production (Table 1). The proline-rich L domain can tolerate mutations to some levels; however, these mutations have dramatically altered viral plaque formation and significantly reduced viral growth ability, suggesting an important role of the intact L domain in viral replication. Our comprehensive analyses of key residues of arenaviral Z proteins have also provided some novel insights into their mechanistic roles in virus budding, nucleocapsid incorporation, and viral RNA regulation, as discussed below.
An essential biological role of the Z protein in arenavirus infection is the Z-mediated virus budding. From both previous publications (19, 24) and the present study (Fig. 3), the G2 myristylation site is indispensable for the Z protein-mediated budding activity, which most likely explains the vital role of G2 in arenavirus infection (Table 1). Also important in mediating Z budding is its C-terminal L domain(s) (18, 23, 25). The L domain consists of a stretch of conserved amino acid residues that interact with components of the ESCRT pathway to mediate membrane budding. Thus far, three classes of viral L domains have been defined, PT/SAP, LxxLF or YPXL, and PPxY, which interact with Tsg101, ALIX, and NEdd4-like HECT ubiquitin ligases, respectively (10). These L domains are functionally interchangeable (26). Some arenaviral Z proteins, such as LASV Z, contain two separate L domains PTAPP and PPPYSP, whereas others contain a proline-rich region that seems to consist of two overlapping L domains (e.g., PSAPPYEP for PICV) or just one L domain (e.g., PTAPPP for Junin) (Fig. 1). Although dicodon mutation of the Proline residues in PSAPPYEP of PICV Z protein has minor effect on its self-budding activity (Fig. 3A, lanes 8 to 10), possibly due to the overexpression system, they have significantly altered viral plaque morphology and reduced viral growth (Fig. 2), suggesting that the intact L domain(s) of arenaviral Z proteins are required for efficient virus budding and replication.
The mechanism governing the incorporation of nucleocapsids into arenavirus virions is largely unknown. It has been proposed that the interaction between Z and NP is required to recruit and incorporate the nucleocapsids into budding virions at the cell membrane (4, 7). A recent study suggests that ALIX, a component of ESCRT network that interacts with L domain LxxLF or YPXL, is required for NP incorporation into Mopeia virus Z-induced VLPs by interacting with both Z and NP (22). Our study did not identify a single residue/domain of Z that is indispensable for NP incorporation (Fig. 4), but suggests that the LPTK site, and possibly, the RING domain, may play a more important role.
The central RING domain is a zinc-binding motif with invariant C and H residues that bind two zinc cations. Although we have shown that the zinc-binding residues are absolutely essential for arenavirus replication (Table 1), the functional role(s) of the RING domain in arenavirus biology is less clear. It seems unlikely that the lethal phenotype observed for all RING domain mutations is due to the abolishment of either Z budding or NP incorporation, because these Z mutations show differential effects on Z budding activity (Fig. 3) and NP incorporation efficiency (Fig. 4). On the other hand, all of the RING domain mutations completely abolish the Z protein's ability to inhibit viral RNA synthesis (Fig. 5), although it remains unclear how Z-mediated inhibition of viral RNA synthesis is vital for arenavirus infection. A possible hypothesis is that this inhibition is necessary in order to initiate the viral assembly step. Further studies are required to fully understand the biological significance of Z-mediated viral RNA inhibition in arenavirus life cycle.
We have demonstrated an essential role for L78 or P79 (numbered according to PICV Z protein) in infectious viral production. Using the PICV reverse genetics system, we have shown that both L→A and P→A mutations in the Z protein lead to a complete loss of viral infectivity (Table 1). This is consistent with a recent study by Capul et al. (3) showing that both L→A and P→A mutations of LASV Z protein strongly reduce VLP infectivity. Their functional mechanisms in infectious viral replication, however, are less clear. We and other researchers have shown that mutagenesis of the invariant L78 or P79 residue does not affect Z budding activity (3, 4; the present study). Furthermore, both lethal (L78A and P80A) and nonlethal mutants (T81A and K82A) in the LPTK motif led to an ∼50% decrease in NP incorporation. Therefore, we do not believe that the indispensable role of L and P residues in the viral life cycle is to mediate the NP incorporation into viral particles. On the other hand, we (Fig. 5) and the Capul study (3) have shown that both L→A and P→A mutations abolish Z-mediated inhibition of viral RNA synthesis, which may explain their inability to support infectious viral replication as discussed above for the RING domain mutants. Nevertheless, the exact functional mechanisms of the invariant L and P residues in arenaviral infectious life cycle remain to be fully elucidated.
Finally, our study has characterized the molecular mechanisms of both LASV and PICV Z protein in budding (Fig. 3) and RNA regulation (Fig. 5). Most of the conserved residues/domains have similar functional mechanisms in the different Z proteins, such as the role of G2 in budding, and that of RING domain and LPTK in RNA inhibition. There are, however, some interesting variations. A notable one is the role of G2 in blocking viral RNA synthesis (Fig. 5). The G2 residue is indispensable for PICV Z-mediated viral RNA inhibition but is unnecessary for LASV Z. A previous study has shown that the Z protein of LCMV, another Old World arenavirus, does not require the N-terminal residues 1 to 16 (including G2) to inhibit viral RNA synthesis (5). It remains to be determined whether the G2 residue of other arenavirus proteins plays an important role in viral RNA regulation. In addition, a recent study using Machupo virus polymerase complex in an in vitro polymerase assay has suggested that Z directly interacts with the L polymerase protein to block the early steps of viral RNA synthesis in a species-specific manner (12). Therefore, the functional mechanism of Z in viral RNA regulation requires further characterization in specific arenavirus species.
In summary, our studies have provided evidence for the essential roles of the conserved residues (domains) of the Z protein in the infectious arenavirus life cycle. In addition, our studies have also provided some novel insights into the functional mechanisms of the conserved residues (domains), including the poorly characterized LP(TK) residues, of the Z protein in viral infectivity. These studies may lead to the development of novel antivirals targeting the essential Z protein in order to treat arenavirus-induced diseases.
ACKNOWLEDGMENTS
We thank K. Conzelmann (Ludwig-Maximilians-Universität) for the BSRT7-5 cells.
This study was supported in part by NIH grants R01AI083409 to Y.L. and R56AI091805 and R01AI093580 to H.L.
Footnotes
Published ahead of print 3 July 2012
REFERENCES
- 1. Aronson JF, Herzog NK, Jerrells TR. 1994. Pathological and virological features of arenavirus disease in guinea pigs: comparison of two Pichinde virus strains. Am. J. Pathol. 145:228–235 [PMC free article] [PubMed] [Google Scholar]
- 2. Buchmeier MJ, De La Torre JC, Peters CJ. 2007. Arenaviridae: the viruses and their replication, p 1791–1827 In Knipe DM, Howley PM. (ed), Fields virology, 5th ed, vol 2 Lippincott/The Williams & Wilkins Co, Philadelphia, PA [Google Scholar]
- 3. Capul AA, de la Torre JC, Buchmeier MJ. 2011. Conserved residues in Lassa fever virus Z protein modulate viral infectivity at the level of the ribonucleoprotein. J. Virol. 85:3172–3178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Casabona JC, Levingston Macleod JM, Loureiro ME, Gomez GA, Lopez N. 2009. The RING domain and the L79 residue of Z protein are involved in both the rescue of nucleocapsids and the incorporation of glycoproteins into infectious chimeric arenavirus-like particles. J. Virol. 83:7029–7039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cornu TI, de la Torre JC. 2002. Characterization of the arenavirus RING finger Z protein regions required for Z-mediated inhibition of viral RNA synthesis. J. Virol. 76:6678–6688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cornu TI, de la Torre JC. 2001. RING finger Z protein of lymphocytic choriomeningitis virus (LCMV) inhibits transcription and RNA replication of an LCMV S-segment minigenome. J. Virol. 75:9415–9426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Eichler R, et al. 2004. Characterization of the Lassa virus matrix protein Z: electron microscopic study of virus-like particles and interaction with the nucleoprotein (NP). Virus Res. 100:249–255 [DOI] [PubMed] [Google Scholar]
- 8. Enria DA, Briggiler AM, Sanchez Z. 2008. Treatment of Argentine hemorrhagic fever. Antivir. Res. 78:132–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fan L, Briese T, Lipkin WI. 2010. Z proteins of New World arenaviruses bind RIG-I and interfere with type I interferon induction. J. Virol. 84:1785–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Freed EO. 2002. Viral late domains. J. Virol. 76:4679–4687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jahrling PB, Hesse RA, Rhoderick JB, Elwell MA, Moe JB. 1981. Pathogenesis of a Pichinde virus strain adapted to produce lethal infections in guinea pigs. Infect. Immun. 32:872–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kranzusch PJ, Whelan SP. 2011. Arenavirus Z protein controls viral RNA synthesis by locking a polymerase-promoter complex. Proc. Natl. Acad. Sci. U. S. A. 108:19743–19748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lan S, McLay L, Aronson J, Ly H, Liang Y. 2008. Genome comparison of virulent and avirulent strains of the Pichinde arenavirus. Arch. Virol. 153:1241–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lan S, et al. 2009. Development of infectious clones for virulent and avirulent Pichinde viruses: a model virus to study arenavirus-induced hemorrhagic fevers. J. Virol. 83:6357–6362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McCormick JB, et al. 1986. Lassa fever: effective therapy with ribavirin. N. Engl. J. Med. 314:20–26 [DOI] [PubMed] [Google Scholar]
- 16. McCormick JB, Webb PA, Krebs JW, Johnson KM, Smith ES. 1987. A prospective study of the epidemiology and ecology of Lassa fever. J. Infect. Dis. 155:437–444 [DOI] [PubMed] [Google Scholar]
- 17. Neuman BW, et al. 2005. Complementarity in the supramolecular design of arenaviruses and retroviruses revealed by electron cryomicroscopy and image analysis. J. Virol. 79:3822–3830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Perez M, Craven RC, de la Torre JC. 2003. The small RING finger protein Z drives arenavirus budding: implications for antiviral strategies. Proc. Natl. Acad. Sci. U. S. A. 100:12978–12983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Perez M, Greenwald DL, de la Torre JC. 2004. Myristoylation of the RING finger Z protein is essential for arenavirus budding. J. Virol. 78:11443–11448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Qi X, et al. 2010. Cap binding and immune evasion revealed by Lassa nucleoprotein structure. Nature 468:779–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Salvato MS, Schweighofer KJ, Burns J, Shimomaye EM. 1992. Biochemical and immunological evidence that the 11-kDa zinc-binding protein of lymphocytic choriomeningitis virus is a structural component of the virus. Virus Res. 22:185–198 [DOI] [PubMed] [Google Scholar]
- 22. Shtanko O, Watanabe S, Jasenosky LD, Watanabe T, Kawaoka Y. 2011. ALIX/AIP1 is required for NP incorporation into Mopeia virus Z-induced virus-like particles. J. Virol. 85:3631–3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Strecker T, et al. 2003. Lassa virus Z protein is a matrix protein and sufficient for the release of virus-like particles. J. Virol. 77:10700–10705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Strecker T, et al. 2006. The role of myristoylation in the membrane association of the Lassa virus matrix protein Z. Virol. J. 3:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Urata S, Noda T, Kawaoka Y, Yokosawa H, Yasuda J. 2006. Cellular factors required for Lassa virus budding. J. Virol. 80:4191–4195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhadina M, Bieniasz PD. 2010. Functional interchangeability of late domains, late domain cofactors and ubiquitin in viral budding. PLoS Pathog. 6:e1001153 doi:10.1371/journal.ppat.1001153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang L, Marriott K, Aronson JF. 1999. Sequence analysis of the small RNA segment of guinea pig-passaged Pichinde virus variants. Am. J. Trop. Med. Hyg. 61:220–225 [DOI] [PubMed] [Google Scholar]
- 28. Zhang L, Marriott KA, Harnish DG, Aronson JF. 2001. Reassortant analysis of guinea pig virulence of Pichinde virus variants. Virology 290:30–38 [DOI] [PubMed] [Google Scholar]