Abstract
Hepatitis E virus (HEV) infections are responsible for chronic hepatitis in immunocompromised patients, and this can evolve to cirrhosis. Like all RNA viruses, HEV exists as a mixture of heterogeneous viruses defining quasispecies. The relationship between the genetic heterogeneity described as a quasispecies, cytokine secretion, and the outcome of acute hepatitis in immunocompromised patients remains to be elucidated. We cloned and sequenced the region encoding the M and P capsid domains of HEV from eight solid-organ transplant (SOT) patients with acute HEV infection who subsequently cleared the virus and from eight SOT patients whose infection became chronic. We analyzed the cytokines and chemokines in the sera of these SOT patients by multianalyte profiling. The nucleotide sequence entropy and genetic distances were greater in patients whose infections became chronic. A lower Ka/Ks ratio was associated with the persistence of HEV. The patients who developed chronic infection had lower serum concentrations of interleukin-1 (IL-1) receptor antagonist and soluble IL-2 receptor. Increased concentrations of the chemokines implicated in leukocyte recruitment to the liver were associated with persistent infection. Those patients with chronic HEV infection and progressing liver fibrosis had less quasispecies diversification during the first year than patients without liver fibrosis progression. Great quasispecies heterogeneity, a weak inflammatory response, and high serum concentrations of the chemokines involved in leukocyte recruitment to the liver in the acute phase were associated with persistent HEV infection. Slow quasispecies diversification during the first year was associated with rapidly developing liver fibrosis.
INTRODUCTION
Hepatitis E virus (HEV) infections are a major cause of acute hepatitis in developing countries and are an emerging health problem in industrialized countries due to zoonotic transmission (6). HEV is a nonenveloped hepatotropic virus with an ∼7.2-kb, single-stranded, positive-sense, 5′-capped RNA genome. It consists of short 5′- and 3′-untranslated regions (UTRs) and three partially overlapping open reading frames (ORFs), namely, ORF1, ORF2, and ORF3 (34). The capsid protein, encoded by ORF2, contains 3 linear domains: S, M, and P (10, 40, 41). Variations in the ORF2 domains could influence cellular or humoral immune responses. The M domain contains T cell epitopes (1). It is also a potential receptor binding site, as it contains a sequence that is strictly conserved among all genotypes (1, 10). The P domain forms dimeric spikes on the surface of the capsid (41) and contains neutralization epitopes (28, 30).
HEV infection is responsible for chronic hepatitis in solid-organ transplant (SOT) patients, and these infections can cause cirrhosis (8, 17, 18). The evolution of an HEV infection to chronicity seems to be related, at least in part, to the intensity of the immunosuppressive therapy used. Indeed, reducing the dose of immunosuppressant given to SOT patients can lead to clearance of the virus (14). The mechanisms responsible for persistence of the virus and for differences in the course of fibrosis during HEV infection are largely unknown but are thought to be a complex interplay between virus diversity and the host immune response. The simultaneous presence of several closely related virus variants that are commonly described as quasispecies (22) may enable the virus to circumvent diminished host immune defenses, leading to a chronic infection.
The host immune response to viruses involves the secretion of cytokines and chemokines to regulate innate or adaptive effector functions (9). Cytokines are secreted proteins that regulate the immune response by modulating the activation, proliferation, and differentiation of targeted cells (4). Chemokines are chemotactic cytokines that regulate the recruitment of leukocytes (26). They play a crucial role in inflammatory processes and host defense (26). These molecules have emerged as key players in host defense mechanisms. Immunocompetent patients with acute hepatitis E have high concentrations of interleukin-1β (IL-1β) in serum, suggesting that this cytokine is implicated in the disease (32). But little is known about the immune response and cytokine secretion in immunocompromised patients. However, it was shown recently that recovery from an HEV infection depends on the responses of multispecific T cells to HEV and the secretion of gamma interferon (IFN-γ) (33).
This study was designed to investigate SOT patients and determine the relationship between virus heterogeneity, host cytokine profiles, and the outcome of the acute phase of hepatitis E. We also monitored the relationship between quasispecies diversification in patients with chronic infection and the progression of liver fibrosis for a year.
MATERIALS AND METHODS
Patients and samples.
We studied 16 SOT patients who underwent transplantation between January 2004 and June 2009 at the Toulouse University Hospital, France. We included 8 patients who cleared the virus within 6 months following the acute phase and 8 patients who developed chronic HEV infection. An acute HEV infection was defined by elevated liver transaminase activity and the presence of HEV RNA in the plasma. A chronic infection was defined by the presence of HEV RNA in the plasma for more than 6 months. Other causes of hepatitis were excluded by serological and molecular tests for hepatitis A virus, hepatitis B virus, hepatitis C virus (HCV), Epstein-Barr virus, and cytomegalovirus. Drug-related causes of abnormal liver function test results were also ruled out by patient history. Serum samples were collected during the acute phase for all patients and 1 year later for patients whose infection had become chronic. Samples were stored frozen at −80°C.
Each patient underwent an exhaustive clinical and laboratory examination at the time of the acute phase. This included a clinical examination, examination of the immunosuppressive drugs used, and assessment of alanine and aspartate aminotransferase (ALT and AST, respectively), γ-glutamyl transpeptidase (γGT), and alkaline phosphatase (AP) activities. Serological analyses were performed with EIAgen HEV IgG and EIAgen HEV IgM kits (Adaltis, InGen, France) as recommended by the manufacturer. The serum concentration of HEV RNA was measured by real-time PCR as previously described (23). The genotype was determined by sequencing a 189-nucleotide fragment within the ORF2 gene (24). The sequences were compared to reference HEV strains (GenBank) as previously reported (24).
Those subjects whose acute hepatitis E resolved did not differ from those who developed chronic HEV infection in terms of age at HEV infection, gender, immunosuppressive drugs used, HEV genotype, and serum HEV RNA concentration (Table 1). In contrast, the patients whose acute hepatitis was resolved had higher ALT activities.
Table 1.
Features of hepatitis E virus-infected solid-organ transplant recipients in the acute phasea
| Patient parameter | Value |
P value | |
|---|---|---|---|
| Patients without chronic infection (n = 8) | Patients with chronic infection (n = 8) | ||
| Gender (no. of males/no. of females) | 6/2 | 6/2 | NS |
| Age (yr) | 47 (32–58) | 50 (42–55) | NS |
| No. of kidney transplant recipients/no. of liver transplant recipients | 7/1 | 6/2 | NS |
| No. of patients receiving drug | |||
| Cyclosporine | 3 | 0 | NS |
| Tacrolimus | 2 | 6 | NS |
| m-TOR inhibitors | 3 | 2 | NS |
| MPA | 5 | 6 | NS |
| Steroids | 6 | 5 | NS |
| Azathioprine | 0 | 1 | NS |
| Time (mo) between transplantation and HEV infection | 56.5 (36–94) | 27.5 (20.5–60) | NS |
| HEV RNA concn (log copies/ml) | 6.30 (6.22–6.72) | 6.52 (5.73–7.01) | NS |
| No. of patients with HEV genotype 3c/3e/3f | 0/1/7 | 1/0/7 | NS |
| No. of patients positive for anti-HEV IgG/IgM | 1/8 | 1/7 | NS |
| Enzyme activity (IU/liter) | |||
| ALT | 259 (238–340) | 157(129–167) | 0.03 |
| AST | 107 (89–173) | 78 (57–108) | NS |
| γGT | 280 (120–1,165) | 118 (89–141) | NS |
| AP | 410 (298–1,247) | 258 (198–290) | NS |
| Total bilirubin (μmol/liter) | 14 (12–43) | 15 (13–18) | NS |
| Cell count (mm−3) | |||
| White blood cells | 5,400 (4,845–8,255) | 7,130 (4,630–7,260) | NS |
| Lymphocytes | 1,337 (903–1,878) | 1,042 (742–1,428) | NS |
| CD4+ cells | 511 (317–765) | 397 (282–683) | NS |
| CD8+ cells | 434 (247–664) | 445 (305–508) | NS |
| CD19+ cells | 72 (37–135) | 121 (28–208) | NS |
Data are medians (interquartile ranges), unless otherwise indicated. ALT, alanine aminotransferase; AST, aspartate aminotransferase; γGT, γglutamyl transpeptidase; AP, alkaline phosphatase; IgG, immunoglobulin G; IgM, immunoglobulin M; MPA, mycophenolic acid; m-TOR, mammalian target of rapamycin; NS, not significant.
The grade and stage of liver fibrosis were assessed according to the Metavir classification of liver biopsy specimens (3). The 8 patients who evolved to chronic HEV infection included 4 who had progressing liver fibrosis (fibrosers) with a 2-point increase of the Metavir score between the first and second biopsies, and 4 whose liver fibrosis had not progressed (nonfibrosers), defined by similar Metavir scores for the two biopsy specimens. The 2 groups did not differ in terms of age at HEV infection, gender, immunosuppressive drugs used, HEV genotype, and serum HEV RNA concentration.
Cloning and sequencing of ORF2. (i) HEV RNA extraction, HEV cDNA synthesis, and PCR.
RNA was isolated and purified using a QIAamp viral RNA kit (Qiagen, Courtaboeuf, France). A reverse transcriptase PCR (RT-PCR) was used to amplify 1,100 nt of the ORF2 region encoding the M and P domains. The RT-PCR was performed with the SuperScript III OneStep RT-PCR system (Invitrogen, Cergy-Pontoise, France), with the following cycling conditions: 30 min at 50°C, 2 min at 94°C, and 50 cycles of 30 s at 94°C, 30 s at 55°C, and 2 min 30 s at 68°C. The primers used were 5′-GAGCTTGAATTYAGRAATTTGACCCC-3′ for the ORF2 sense direction and 5′-TTTTTTTTTTTTCCGGGGRGCGMGRAACC-3′ for the ORF2 antisense direction. PCR fragments were analyzed by agarose gel electrophoresis and extracted using Qiaquick PCR purification (Qiagen, Courtaboeuf, France) as specified by the manufacturer.
(ii) Plasmid cloning.
The purified PCR products were quantified by spectrophotometry; 10 ng of cDNA was directly ligated into 10 ng of TOPO TA cloning vector (Invitrogen, Cergy-Pontoise, France). Recombinant plasmids were used to transform Escherichia coli competent cells according to the manufacturer's protocol, and transformants were grown on ampicillin plates. Twenty clones were randomly selected for sequencing. Template resampling was avoided because the number of original templates was far greater than the number of templates sequenced (25).
(iii) Nucleotide sequencing.
The cDNAs of the 20 previously selected clones were amplified under the following conditions: 10 min at 94°C followed by 35 cycles of 15 s at 94°C, 30 s at 55°C, and 1 min 30 s at 68°C. The amplification was performed using the Expand High Fidelity PCR system (Roche, Mannheim, Germany). The primers used were for the ORF2 sense and antisense directions. The PCR products were extracted using Qiaquick PCR purification (Qiagen, Courtaboeuf, France) and sequenced on both strands by the dideoxy chain termination method (Prism Ready Reaction AmpliTaq Fs and dye deoxy primers; Applied Biosystems, Paris, France) on an ABI 3130XL high-throughout capillary DNA analyzer (Applied Biosystems, Foster City, CA).
The primers used were 5′-GAGCTTGAATTYAGRAATTTGACCCC-3′ and 5′-GACAGAATTRATTTCGTCGGCTGG-3′ for the ORF2 sense direction and 5′-CCCTTRTCCTGCTGNGCATTCTCGACAGA-3′ for the ORF2 antisense direction. Electrophoregram data were analyzed using Sequencher 4.8 (Gene Codes Corporation).
(iv) Calculation of genetic complexity and diversity.
Nucleotide sequences were aligned using Clustal X 1.83. We quantified the complexity of the HEV strain in the region of interest by calculating the Shannon entropy as follow: S = −Σpiln(pi), where pi is the frequency of each sequence in the virus quasispecies (39). The normalized entropy, Sn, was calculated at the nucleotide level as follows: Sn = S/ln(N), where N is the total number of sequences analyzed. We quantified diversity as the mean genetic distance calculated for all pairs of nucleotide sequences, using MEGA 4.0.2 (21). The calculation was based on a Kimura two-parameter distance matrix. The mean and standard error of the mean (SEM) within-sample genetic distances were calculated for the acute-phase quasispecies in each of the 16 patients.
The rates of intrasample synonymous substitutions per synonymous site (Ks) and nonsynonymous substitutions per nonsynonymous site (Ka) were calculated by the method of Nei and Gojobori with the Jukes-Cantor correction for multiple substitutions (13), using MEGA 4.0.2 (21). The Ka/Ks ratio is an indicator of the strength of the positive (>1) or negative (<1) selection pressure on a quasispecies (42).
Multiplex cytokine analysis.
A human cytokine 25-plex panel (Invitrogen, Cergy-Pontoise, France) was used to analyze 25 cytokines, chemokines, and markers of T cell activation in serum samples according to the manufacturer's protocol. The samples were measured using the antibody bead mix in duplicate with a biotinylated detection antibody followed by streptavidin-phycoerythrin. This kit can quantify IL-1β, IL-2, IL-4, IL-5, IL-6, IL-7, IL-10, IL-12 (p40 and p70), IL-13, IL-15, IL-17, tumor necrosis factor alpha (TNF-α), granulocyte-macrophage colony-stimulating factor (GM-CSF), IFN-α, IFN-γ, IL-1 receptor antagonist (IL-1Ra), soluble IL-2 receptor (sIL-2R), and the following chemokines: CXCL8, interferon-inducible protein 10 (IP-10), monocyte chemotactic protein 1 (MCP-1), monokine induced by gamma interferon (MIG), macrophage inflammatory protein 1α (MIP-1α), MIP-1β, regulated upon activation, normal T cell expressed and secreted (RANTES), and eotaxin.
The plate was read with a Luminex 200 instrumentation system (Luminex Corporation, Austin, TX). Data were collected for 100 beads per cytokine from each well. The raw data (mean fluorescence intensities) were processed with Xponent 3.1 software (Xmap technology; Luminex) to obtain concentrations.
Statistical analysis.
All statistical calculations were performed using Stata software (version 9.2; Stata Corp.). The characteristics of individuals were compared using Fisher's exact test for categorical variables (type of immunosuppressive treatments, presence of anti-HEV IgM and IgG). The Mann-Whitney test was used to compare quantitative variables, including genetic parameters. P values of <0.05 were considered statistically significant. When sample sizes were smaller than 5, no statistical analysis was performed. Correlations between quantities of cytokines and AST/ALT activities or Ka/Ks ratios were estimated by calculating Spearman's rank correlation coefficient.
Nucleotide sequence accession numbers.
The sequences determined in this study have been deposited in the GenBank database under accession numbers JQ763600 to JQ764559.
RESULTS
Analysis of HEV quasispecies and relationship to the outcome of acute infection.
The distribution of HEV quasispecies during the acute phase of HEV infection was analyzed by cloning of an ORF2 fragment encoding the M and P domains of the capsid protein.
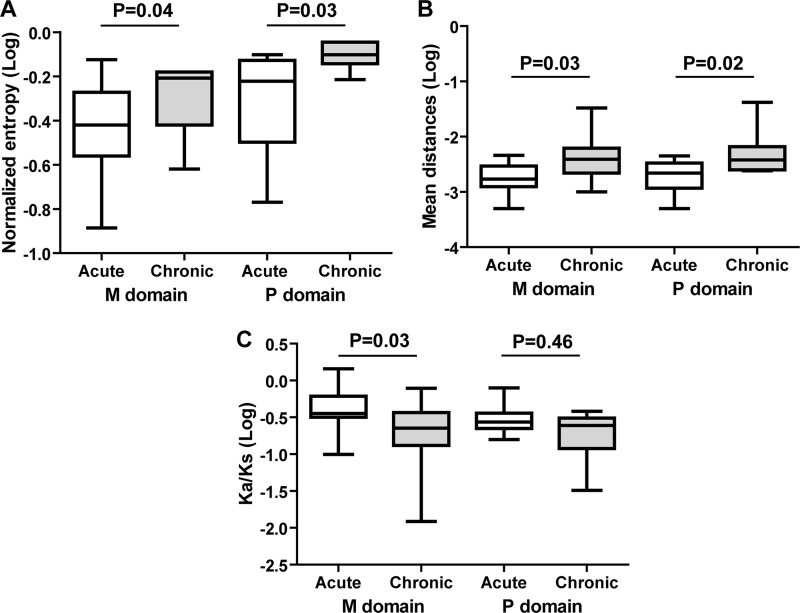
The nucleotide sequence entropy (complexity) of the region encoding the M domain was higher in patients whose infection became chronic (median [interquartile range], 0.62 [0.46 to 0.66]) than in patients who cleared the virus (0.34 [0.28 to 0.45]; P = 0.04). The complexity of the region encoding the P domain was also higher in patients whose infection became chronic (0.77 [0.71 to 0.86]) than in patients who cleared the virus (0.58 [0.38 to 0.73]) (P = 0.03) (Fig. 1A).
Fig 1.
Box plot representation of quasispecies parameters for patients who did or did not develop chronic HEV infection. White boxes, acute infection (patients who cleared the virus); gray boxes, chronic infection (patients with chronic infection). (A) Mean values for nucleotide entropy (complexity); (B) mean values for nucleotide genetic distance (diversity); (C) mean values for Ka/Ks ratio. The Mann-Whitney test was used to compare differences between the 2 groups.
The mean within-sample genetic distance (diversity) of the region encoding the M domain was greater for the 8 chronically infected patients (0.0039 [0.0025 to 0.0050]) than for the patients who cleared the virus (0.0016 [0.0013 to 0.0025]) (P = 0.03). The diversity of the region of the virus genome encoding the P domain was also greater in the 8 chronically infected patients (0.0039 [0.0030 to 0.0050]) than in the patients who cleared the virus (0.0020 [0.0015 to 0.0026]) (P = 0.02) (Fig. 1B).
The Ka/Ks ratio of the M domain was lower for the patients whose hepatitis became chronic (0.18 [0.12 to 0.29]) than for the patients who cleared the virus (0.44 [0.34 to 0.58]) (P = 0.03), but the Ka/Ks ratios of the P domain of the viruses in the 2 groups were not different for patients whose infection became chronic and those who cleared the virus (0.20 [0.15 to 0.29] versus 0.28 [0.23 to 0.32]) (P = 0.46) (Fig. 1C).
Levels of cytokines, markers of T cell activation, and chemokines and relationship to the acute-phase outcome.
We compared the concentrations of cytokines and chemokines in the patients who cleared the virus and those whose HEV infection became chronic (Table 2). Two cytokines (IFN-γ and IL-2) whose concentrations in 80% or more of the samples were below the limit of detection were removed from this analysis.
Table 2.
Serum cytokine concentrations during the acute phase in patients who cleared the virus and patients who developed chronic infectiona
| Cytokine or chemokine | Concn (pg/ml) |
P value | |
|---|---|---|---|
| Patients without chronic HEV infection (n = 8) | Patients with chronic HEV infection (n = 8) | ||
| Inflammatory cytokines | |||
| IL-1Ra | 1,463.9 (1,313.2–1,637.6) | 1,067.1 (885.3–1,237.0) | 0.05 |
| TNF-α | 37.8 (28.6–52.7) | 23.7 (22.5–29.8) | 0.07 |
| IL-1β | 37.9 (15.1–70.1) | 31.9 (14.7–44.3) | NS |
| IL-6 | 21.5 (19.4–41.1) | 24.9 (18.3–27.2) | NS |
| IL-17 | 82.8 (73.5–101.4) | 95.8 (63.8–131.4) | NS |
| IFN-α | 212.9 (196.5–249.1) | 252.9 (220.5-288-9) | NS |
| Th1/Th2 cytokines | |||
| IL-12 | 538.1 (473.3–668.1) | 522.9 (504.7–578.1) | NS |
| IL-4 | 14.9 (9.4–33.1) | 13.2 (10.9–27.2) | NS |
| IL-5 | 4.4 (4.3–6.6) | 8 (4.2–11.1) | NS |
| IL-10 | 21.1 (12.8–30.1) | 21.1 (20.0–42.9) | NS |
| IL-13 | 51.7 (28.8-185-9) | 72.9 (52.3–95.4) | NS |
| Growth and cellular cytokines | |||
| IL-7 | 55.1 (47.2–79.3) | 93 (60.3–128.2) | NS |
| IL-15 | 91.5 (69.8–113.0) | 117 (106.5–148.8) | NS |
| GM-CSF | 80.5 (20.0–193.2) | 26.5 (20.0–58.0) | NS |
| Marker of T cell activation | |||
| sIL-2R | 957.7 (717.5–1116.8) | 391.5 (272.0–788.0) | 0.05 |
| Chemokines | |||
| RANTES | 6,500.4 (5,019–8,334.5) | 11,800 (10,056.2–13,055.0) | 0.02 |
| MIP-1α | 129.1 (117.0–160.3) | 181.2 (135.3–330.2) | 0.07 |
| MIP-1β | 146.7 (96.3–216.7) | 248.8 (210.1-306-5) | 0.04 |
| MCP-1 | 547.9 (484.5–702.1) | 1,210.8 (966.3–1,879.0) | 0.01 |
| CXCL8 | 30.9 (21.0–143.7 | 226.9 (146.5–404.2) | 0.02 |
| MIG | 398.4 (304.6–505.6) | 209 (160.2–309.0) | NS |
| IP-10 | 283.6 (261.3–341.5) | 353 (142.1–568.6) | NS |
| Eotaxin | 397.4 (373.5–472.0) | 528.2 (446.8–675.7) | NS |
Data are medians (interquartile ranges). GM-CSF, granulocyte-macrophage colony-stimulating factor; IFN-α, alpha interferon; IFN-γ, gamma interferon; IL-1Ra, IL-1 receptor antagonist; sIL-2R, soluble IL-2 receptor; IP-10, interferon-inducible protein 10; MCP-1, monocyte chemotactic protein 1; MIG, monokine induced by interferon gamma; MIP-1α, macrophage inflammatory protein 1α; MIP-1β, macrophage inflammatory protein 1β; TNF-α, tumor necrosis factor alpha; RANTES, regulated upon activation, normal T cell expressed and secreted.
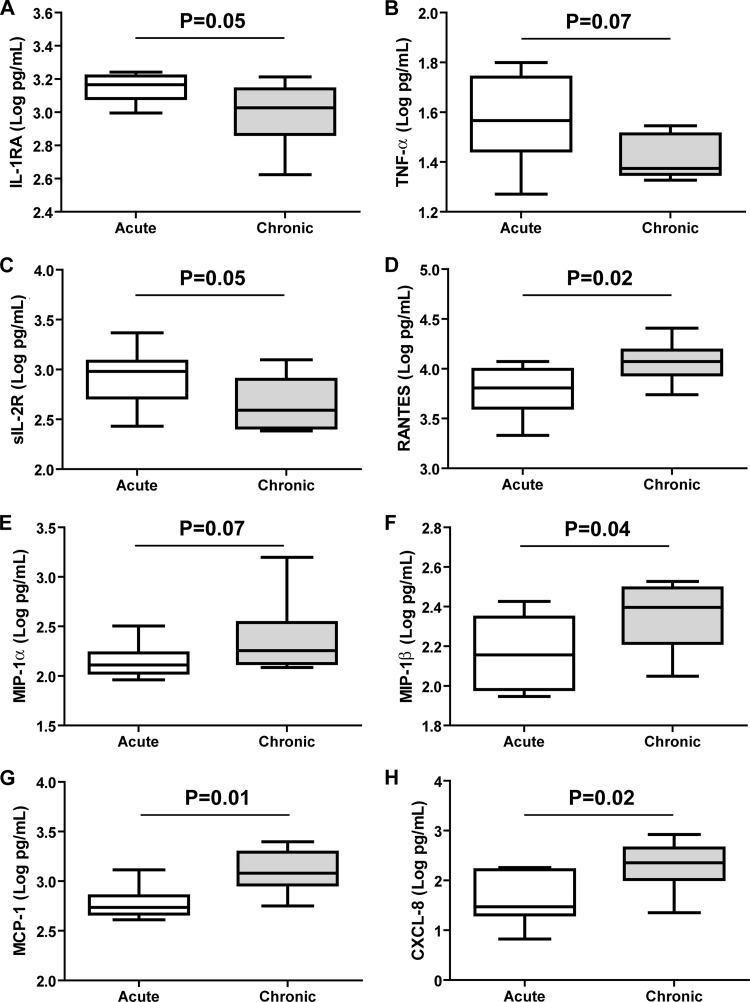
We first examined the cytokines involved in inflammation. The IL-1Ra concentration was higher in patients who cleared the virus than in those whose infection became chronic (P = 0.05) (Fig. 2A). In addition, the TNF-α concentration tended to be higher in patients who cleared the virus (P = 0.07) (Fig. 2B). The concentrations of the other inflammatory cytokines (IL-1β, IL-6, and IL-17) and of IFN-α in patients who cleared the virus and those whose infection became chronic were similar.
Fig 2.
Box plot representation of cytokine/chemokine concentrations in patients who did and did not develop chronic HEV infection. White boxes, acute infection (patients who cleared the virus); gray boxes, chronic infection (patients with chronic infection). (A) IL-1Ra; (B) TNF-α; (C) sIL-2R; (D) RANTES; (E) MIP-1α; (F) MIP-1β; (G) MCP-1; (H) CXCL8. The Mann-Whitney test was used to compare differences between the 2 groups.
The concentrations of Th1 (IL-12) and Th2 (IL-4, IL-5, IL-10, and IL-13) cytokines in patients who cleared the virus and those whose infection became chronic were similar. The concentrations of cytokines implicated in cell differentiation and survival (IL-7, IL-15, and GM-CSF) in the 2 groups were also similar (Table 2).
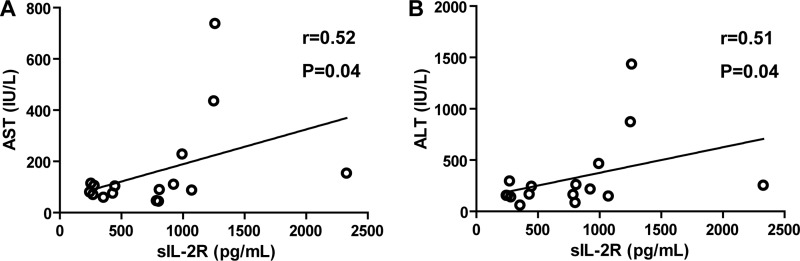
The concentration of sIL-2R, a marker of T cell activation, was higher in patients who cleared the virus than in those whose infection became chronic (P = 0.05) (Fig. 2C). The concentration of sIL-2R was positively correlated with the activities of AST (r = 0.52; P = 0.04) and ALT (r = 0.51; P = 0.04) (Fig. 3A and B), but there was no correlation between the sIL-2R concentration and the Ka/Ks ratios of the M and P capsid domains.
Fig 3.
Correlation between sIL-2R concentrations and AST/ALT activities. Correlations were estimated by calculating Spearman's rank correlation coefficient.
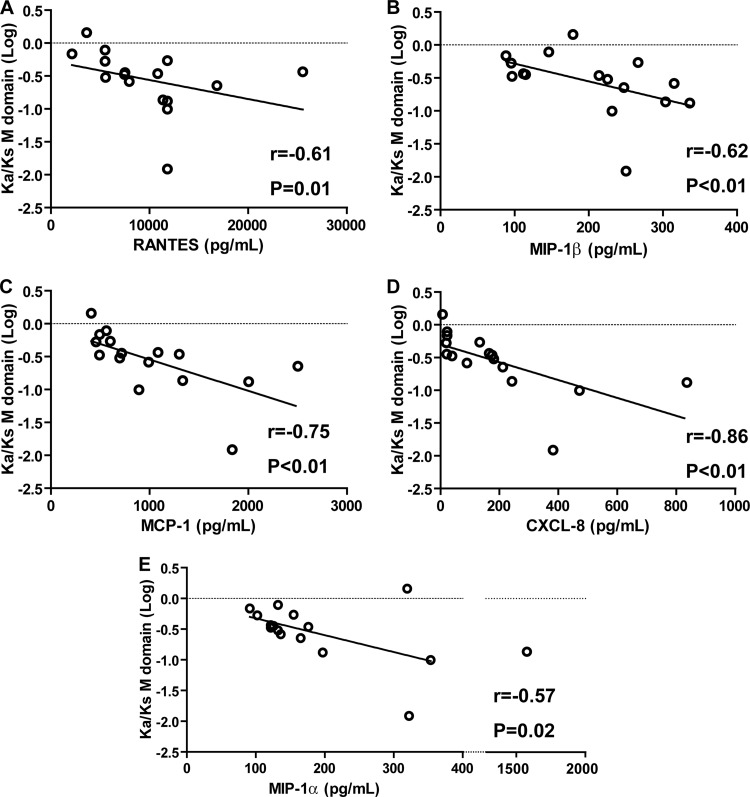
The concentrations of the chemokines RANTES, MIP-1β, MCP-1, and CXCL8 were higher in patients whose HEV infection became chronic (P = 0.02, 0.04, 0.01, and 0.02, respectively) (Fig. 2D, F, G, and H). The concentration of MIP-1α tended to be higher in patients whose infection became chronic (P = 0.07) (Fig. 2E). We found no correlation between the concentrations of RANTES, MIP-1α, MIP-1β, MCP-1, and CXCL-8 and the AST/ALT activities or the Kia/Ks ratio of the P domain. In contrast, the concentrations of these 5 chemokines were negatively correlated with the Ka/Ks ratio of the M domain (Fig. 4). The concentrations of the other chemokines (MIG, IP-10, and eotaxin) were similar in the 2 groups.
Fig 4.
Correlation between chemokine concentrations and Ka/Ks ratios of the M domain. (A) RANTES; (B) MIP-1β; (C) MCP-1; (D) CXCL8; (E) MIP-1α. Correlations were estimated by calculating Spearman's rank correlation coefficient.
Analysis of HEV quasispecies and relationship with progressing liver fibrosis.
Four of the 8 patients who developed chronic HEV infection had progressing liver fibrosis (fibrosers), and 4 did not (nonfibrosers).
During the acute phase, the Ka/Ks ratio of the M domain of virus in the nonfibrosers (0.25 [0.14 to 0.47]) was similar to that of virus in the fibrosers (0.16 [0.078 to 0.23]), and the same was true for the P domain (0.27 [0.21 to 0.29] in nonfibrosers and 0.16 [0.12 to 0.21] in fibrosers).
One year after the acute phase, the Ka/Ks ratios of the M domain of virus in the nonfibrosers (0.12 [0.05 to 0.20]) and fibrosers (0.20 [0.13 to 0.27]) were similar. The Ka/Ks ratios of the P domain of virus in the nonfibrosers (0.31 [0.23 to 0.41]) and fibrosers (0.16 [0.14 to 0.20]) were also similar.
We studied the intersample virus diversification (rKa/Ks) during 1 year after the acute phase. The diversification of the M domain in nonfibrosers (0.16 [0.14 to 0.20]) and fibrosers (0.13 [0.08 to 0.19]) were similar. In contrast, the diversification of the P domain in nonfibrosers (0.29 [0.23 to 0.40]) seemed greater than that in fibrosers (0.11 [0.08 to 0.16]), although the small number of patients precluded statistical analysis (Fig. 5).
Fig 5.
Scatterplot representation of quasispecies diversification in patients who did and did not develop liver fibrosis. ●, patients without progressing liver fibrosis (NF); ▲, patients with progressing liver fibrosis (F).
DISCUSSION
It was shown recently that hepatitis E virus can lead to chronic infection in SOT patients, but the virological and immunological factors associated with viral persistence have not yet been elucidated. This study examines the relationship between the heterogeneity of HEV quasispecies, the secretion of various cytokines and chemokines, and the outcome of acute hepatitis.
In the acute phase, the complexity and diversity of the region encoding the M and P capsid domains of the virus were greater in immunocompromised patients who developed chronic infection than in patients who cleared the virus. The great genetic heterogeneity of virus in patients with persistent hepatitis could reflect changes in the host environment, such as the appearance of neutralizing antibodies or intrahepatic cytotoxic T lymphocytes. Viruses with an enhanced ability to persist could emerge under these selective constraints. Similar results have been described for hepatitis C virus quasispecies, where increased diversity in hypervariable region 1 during acute infection was associated with progression to a chronic infection (7).
The Ka/Ks ratio of the virus M domain was lower in patients who developed chronic infection than in those who cleared the virus. Both Ka/Ks ratios were negative, so in patients who developed a chronic HEV infection, the negative selection was stronger. Negative selection promotes conservation of the amino acid sequence; thus, development of a chronic infection is associated with conservation of the amino acid sequence of the M domain. The M domain contains T cell epitopes (1). Thus, the immune pressure mediated by T cells could explain the difference in selection, since the T cell response determines HEV eradication (33). An effective immune response should lead to less conservation of amino acid sequences, which is in line with our observations. The Ka/Ks ratios of the P domain of virus in the 2 groups of patients were similar, perhaps due to the limited number of patients included in our study. Alternatively, the P capsid domain, which is a B cell epitope (28, 30), could not be the key epitope for virus clearance.
We investigated the role of cytokines in the clearance of HEV by comparing the acute-phase serum cytokine profiles of the patients who cleared the virus and those who developed chronic infection. The concentrations of two cytokines produced during the inflammatory process were lower in patients whose HEV infection became chronic than in those who cleared the virus. Lower circulating concentrations of cytokines may reflect a weaker inflammatory response in these patients. IL-1Ra is produced during inflammation. It binds competitively to the IL-1 receptor without activating the IL-1 signaling pathway, thus attenuating the effect of IL-1 (2). IL-1Ra also stimulates IFN-λ1 (IL-29) production by plasmacytoid dendritic cells (27). IFN-λ1 contributes to the antiviral defenses by stimulating the synthesis of 2′-to-5′ oligoadenylate synthase (OAS) and MxA (20). Thus, a lower IL-1Ra concentration could weaken the anti-HEV defenses by diminishing IFN-λ1 secretion. TNF-α is also a key cytokine in inflammation, as it can recruit and activate macrophages, NK cells, and T cells (36). A tendency to lower the TNF-α concentration could weaken the host's anti-HEV defenses by impairing effector recruitment.
The concentration of sIL-2R, which is a marker of the activation of T lymphocytes (31), was also lower in the patients who developed a chronic infection. Thus, HEV is likely to persist in patients who have fewer activated T cells. This may be due to immunosuppressive therapy, which interferes with T cell activation or proliferation (11). The amount of circulating sIL-2R was also positively correlated with the activities of ALT and AST. Higher ALT/AST activities have been reported for patients who cleared the virus (23). Since the virus is not cytolytic, the elimination of infected hepatocytes may depend on the degree of T cell activation. Patients who cleared their HEV infection after transplantation have been found to have a strong, HEV-specific T cell response (33).
We found that the serum concentrations of five chemokines were higher in those patients whose HEV infection became chronic. This indicates that the persistence of HEV is associated with high concentrations of chemokines. RANTES, MIP-1α, and MIP-1β can recruit T cells bearing CCR5 chemokine receptors to an inflamed liver (5). The involvement of CCR5+ cells in the pathogenesis of hepatitis E was suggested recently (37). MCP-1 promotes the recruitment of monocytes and macrophages to the liver via the CCR2 chemokine receptor (38). CXCL8, a CXCR1 ligand, may be important for recruiting neutrophils (35). CXCL8 also inhibits the antiviral action of IFN-α (19), thus impairing the host's antiviral defenses. It may appear counterintuitive that high serum concentrations of chemokines are negative prognostic markers of viral clearance. Chemokines are supposed to recruit activated lymphocytes to the liver, but high chemokine concentrations during the acute phase of infection may lead to the downregulation of the chemokine receptors on T cells by molecule internalization. This could impair specific T cell migration during a primary infection, thus impairing HEV clearance. Finally, the circulating chemokine concentrations were negatively correlated with the Ka/Ks ratio of the M domain. High chemokine concentrations are associated with low Ka/Ks ratios, which is in line with our observations on the patients who developed chronic infection. However, one limitation of our study is that we compared many parameters, so the false discovery rate cannot be excluded as a factor.
High concentrations of chemokines during the acute phase may be the first step toward the persistence of an HEV infection, but this relationship needs further investigation. A study of the T cells invading the liver during the acute phase of infection could confirm the hypothesis of impaired recruitment, since activated CD8+ T cells were found recently in liver biopsy specimens from HEV-infected patients (29).
Four of the patients whose HEV infection became chronic developed liver fibrosis (fibrosers), while four did not (nonfibrosers). The diversification of the P capsid domain (rKa/Ks ratio) seemed to be lower in the fibrosers than in the nonfibrosers. Thus, the slow diversification of the P capsid domain seems to be associated with progression to liver fibrosis. A study including more patients is needed to confirm these data. In patients chronically infected with HCV, the progression of liver fibrosis was significantly associated with slower HVR-1 diversification. This could indicate the selection of more aggressive variants in fibrosers (12).
Our study was focused on HEV quasispecies and cytokines/chemokines in immunocompromised patients. A compartmentalization of HEV variants in blood and cerebrospinal fluid has been reported for a kidney transplant patient with chronic hepatitis and neurological symptoms (16). Similar investigations could be relevant for immunocompetent patients experiencing resolutive infection or for patients with severe hepatitis E during pregnancy in developing countries or in the context of preexisting chronic liver disease (15).
We concluded that quasispecies heterogeneity in the region encoding the M and P capsid domains during the acute phase of infection is associated with the development of chronic HEV infection. Moreover, the production of low concentrations of inflammatory cytokines and a soluble marker of T cell activation and the synthesis of high concentrations of chemokines during the acute phase seem to be critical for the persistence of HEV infection. Finally, we found that low quasispecies diversification during the first year seems to be associated with the rapid progression of fibrosis.
Footnotes
Published ahead of print 3 July 2012
REFERENCES
- 1. Aggarwal R, et al. 2007. T-cell epitope mapping of ORF2 and ORF3 proteins of human hepatitis E virus. J. Viral Hepat. 14:283–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arend WP, Malyak M, Guthridge CJ, Gabay C. 1998. Interleukin-1 receptor antagonist: role in biology. Annu. Rev. Immunol. 16:27–55 [DOI] [PubMed] [Google Scholar]
- 3. Bedossa P, Poynard T. 1996. An algorithm for the grading of activity in chronic hepatitis C. Hepatology 24:289–293 [DOI] [PubMed] [Google Scholar]
- 4. Borish LC, Steinke JW. 2003. Cytokines and chemokines. J. Allergy Clin. Immunol. 111:S460–S475 [DOI] [PubMed] [Google Scholar]
- 5. Cook DN, et al. 1995. Requirement of MIP-1 alpha for an inflammatory response to viral infection. Science 269:1583–1585 [DOI] [PubMed] [Google Scholar]
- 6. Dalton HR, Bendall R, Ijaz S, Banks M. 2008. Hepatitis E: an emerging infection in developed countries. Lancet Infect. Dis. 8:698–709 [DOI] [PubMed] [Google Scholar]
- 7. Farci P, et al. 2000. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science 288:339–344 [DOI] [PubMed] [Google Scholar]
- 8. Gerolami R, Moal V, Colson P. 2008. Chronic hepatitis E with cirrhosis in a kidney-transplant recipient. N. Engl. J. Med. 358:859–860 [DOI] [PubMed] [Google Scholar]
- 9. Guidotti LG, Chisari FV. 2001. Noncytolytic control of viral infections by the innate and adaptive immune response. Annu. Rev. Immunol. 19:65–91 [DOI] [PubMed] [Google Scholar]
- 10. Guu TS, et al. 2009. Structure of the hepatitis E virus-like particle suggests mechanisms for virus assembly and receptor binding. Proc. Natl. Acad. Sci. U. S. A. 106:12992–12997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hong JC, Kahan BD. 2000. Immunosuppressive agents in organ transplantation: past, present, and future. Semin. Nephrol. 20:108–125 [PubMed] [Google Scholar]
- 12. Izopet J, et al. 2000. Longitudinal analysis of hepatitis C virus replication and liver fibrosis progression in renal transplant recipients. J. Infect. Dis. 181:852–858 [DOI] [PubMed] [Google Scholar]
- 13. Jukes TH, Cantor TR. 1969. Evolution of protein molecules, p 21–132 In Munro IHN. (ed), Mammalian protein metabolism. Academic Press, Inc, New York, NY [Google Scholar]
- 14. Kamar N, et al. 2010. Influence of immunosuppressive therapy on the natural history of genotype 3 hepatitis-E virus infection after organ transplantation. Transplantation 89:353–360 [DOI] [PubMed] [Google Scholar]
- 15. Kamar N, et al. 2012. Hepatitis E. Lancet 379:2477–2488 [DOI] [PubMed] [Google Scholar]
- 16. Kamar N, et al. 2010. Hepatitis E virus-induced neurological symptoms in a kidney-transplant patient with chronic hepatitis. Am. J. Transplant. 10:1321–1324 [DOI] [PubMed] [Google Scholar]
- 17. Kamar N, et al. 2008. Hepatitis E virus-related cirrhosis in kidney- and kidney-pancreas-transplant recipients. Am. J. Transplant. 8:1744–1748 [DOI] [PubMed] [Google Scholar]
- 18. Kamar N, et al. 2008. Hepatitis E virus and chronic hepatitis in organ-transplant recipients. N. Engl. J. Med. 358:811–817 [DOI] [PubMed] [Google Scholar]
- 19. Khabar KS, et al. 1997. The alpha chemokine, interleukin 8, inhibits the antiviral action of interferon alpha. J. Exp. Med. 186:1077–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kotenko SV, et al. 2003. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat. Immunol. 4:69–77 [DOI] [PubMed] [Google Scholar]
- 21. Kumar S, Tamura K, Nei M. 1994. MEGA: molecular evolutionary genetics analysis software for microcomputers. Comput. Appl. Biosci. 10:189–191 [DOI] [PubMed] [Google Scholar]
- 22. Lauring AS, Andino R. 2010. Quasispecies theory and the behavior of RNA viruses. PLoS Pathog. 6:e1001005 doi:10.1371/journal.ppat.1001005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Legrand-Abravanel F, et al. 2010. Characteristics of autochthonous hepatitis E virus infection in solid-organ transplant recipients in France. J. Infect. Dis. 202:835–844 [DOI] [PubMed] [Google Scholar]
- 24. Legrand-Abravanel F, et al. 2009. Hepatitis E virus genotype 3 diversity, France. Emerg. Infect. Dis. 15:110–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu SL, et al. 1996. HIV quasispecies and resampling. Science 273:415–416 [DOI] [PubMed] [Google Scholar]
- 26. Luster AD. 1998. Chemokines—chemotactic cytokines that mediate inflammation. N. Engl. J. Med. 338:436–445 [DOI] [PubMed] [Google Scholar]
- 27. Megjugorac NJ, Gallagher GE, Gallagher G. 2010. IL-4 enhances IFN-lambda1 (IL-29) production by plasmacytoid DCs via monocyte secretion of IL-1Ra. Blood 115:4185–4190 [DOI] [PubMed] [Google Scholar]
- 28. Meng J, et al. 2001. Identification and characterization of the neutralization epitope(s) of the hepatitis E virus. Virology 288:203–211 [DOI] [PubMed] [Google Scholar]
- 29. Prabhu SB, et al. 2011. Study of cellular immune response against hepatitis E virus (HEV). J. Viral Hepat. 18:587–594 [DOI] [PubMed] [Google Scholar]
- 30. Riddell MA, Li F, Anderson DA. 2000. Identification of immunodominant and conformational epitopes in the capsid protein of hepatitis E virus by using monoclonal antibodies. J. Virol. 74:8011–8017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rubin LA, et al. 1985. Soluble interleukin 2 receptors are released from activated human lymphoid cells in vitro. J. Immunol. 135:3172–3177 [PubMed] [Google Scholar]
- 32. Srivastava R, et al. 2007. Cellular immune responses in acute hepatitis E virus infection to the viral open reading frame 2 protein. Viral Immunol. 20:56–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Suneetha PV, et al. 2012. Hepatitis E virus (HEV)-specific T-cell responses are associated with control of HEV infection. Hepatology 55:695–708 [DOI] [PubMed] [Google Scholar]
- 34. Tam AW, et al. 1991. Hepatitis E virus (HEV): molecular cloning and sequencing of the full-length viral genome. Virology 185:120–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Taub DD, Anver M, Oppenheim JJ, Longo DL, Murphy WJ. 1996. T lymphocyte recruitment by interleukin-8 (IL-8). IL-8-induced degranulation of neutrophils releases potent chemoattractants for human T lymphocytes both in vitro and in vivo. J. Clin. Invest. 97:1931–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tracey KJ, Cerami A. 1994. Tumor necrosis factor: a pleiotropic cytokine and therapeutic target. Annu. Rev. Med. 45:491–503 [DOI] [PubMed] [Google Scholar]
- 37. TrehanPati N, et al. 2011. Gene expression profiles of T cells from hepatitis E virus infected patients in acute and resolving phase. J. Clin. Immunol. 31:498–508 [DOI] [PubMed] [Google Scholar]
- 38. Wald O, Weiss ID, Galun E, Peled A. 2007. Chemokines in hepatitis C virus infection: pathogenesis, prognosis and therapeutics. Cytokine 39:50–62 [DOI] [PubMed] [Google Scholar]
- 39. Wolinsky SM, et al. 1996. Adaptive evolution of human immunodeficiency virus-type 1 during the natural course of infection. Science 272:537–542 [DOI] [PubMed] [Google Scholar]
- 40. Xing L, et al. 2010. Structure of hepatitis E virion-sized particle reveals an RNA-dependent viral assembly pathway. J. Biol. Chem. 285:33175–33183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yamashita T, et al. 2009. Biological and immunological characteristics of hepatitis E virus-like particles based on the crystal structure. Proc. Natl. Acad. Sci. U. S. A. 106:12986–12991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang Z, Bielawski JP. 2000. Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 15:496–503 [DOI] [PMC free article] [PubMed] [Google Scholar]