Abstract
The induction of strong CD8+ T-cell responses against infectious diseases and cancer has remained a major challenge. Depending on the source of antigen and the infectious agent, priming of CD8+ T cells requires direct and/or cross-presentation of antigenic peptides on major histocompatibility complex (MHC) class I molecules by professional antigen-presenting cells (APCs). However, both pathways show distinct preferences concerning antigen stability. Whereas direct presentation was shown to efficiently present peptides derived from rapidly degraded proteins, cross-presentation is dependent on long-lived antigen species. In this report, we analyzed the role of antigen stability on DNA vaccination and recombinant vaccinia virus (VV) infection using altered versions of the same antigen. The long-lived nucleoprotein (NP) of lymphocytic choriomeningitis virus (LCMV) can be targeted for degradation by N-terminal fusion to ubiquitin or, as we show here, to the ubiquitin-like modifier FAT10. Direct presentation by cells either transfected with NP-encoding plasmids or infected with recombinant VV in vitro was enhanced in the presence of short-lived antigens. In vivo, however, the highest induction of NP-specific CD8+ T-cell responses was achieved in the presence of long-lived NP. Our experiments provide evidence that targeting antigens for proteasomal degradation does not improve the immunogenicity of DNA vaccines and recombinant VVs. Rather, it is the long-lived antigen that is superior for the efficient activation of MHC class I-restricted immune responses in vivo. Hence, our results suggest a dominant role for antigen cross-priming in DNA vaccination and recombinant VV infection.
INTRODUCTION
Vaccination strategies for inducing CD8+ T-cell responses against intracellular pathogens and cancer are based on the major histocompatibility complex class I (MHC-I)-restricted presentations of antigenic peptides by professional antigen-presenting cells (APCs) (37). In these specialized immune cells, two classes of MHC-I-restricted antigen presentation pathways exist in parallel that both can be utilized for immunotherapy and vaccination. Proteins that are expressed within the APCs are usually degraded via the ubiquitin/proteasome system (23), and the generated peptides are translocated into the endoplasmic reticulum (ER) via the transporter associated with antigen processing (TAP). Peptides are then loaded onto MHC-I molecules before they enter the secretory pathway to be presented on the cell surface. This pathway is referred to as the direct-presentation pathway (5). In contrast to this, APCs can also acquire exogenous antigens that are either derived from expression by nonprofessional cells or pathogens or are delivered as particulate vaccines. The mechanism facilitating this way of antigen presentation was discovered by M. Bevan in 1976 and was later termed cross-presentation (5, 9). Since both direct and cross-presentation can lead to MHC-I presentation of antigenic peptides and the priming of naïve cytotoxic CD8+ T cells, they are both interesting targets for vaccinations aiming at specific CD8+ T-cell induction. However, the two pathways favor different antigen properties, especially protein stability. Generation of peptides for direct presentation was shown to strongly depend on the formation of defective ribosomal products (DRiPs) (51). Cotranslational misfolding and rapid proteasomal targeting leads to an increased generation of peptides, which can be loaded onto MHC class I molecules. In contrast to this, there are studies indicating that cross-presentation depends on stable antigens that are not immediately targeted for proteasomal degradation (7, 16). These biases for differences in antigen stability have to be taken into account when aiming at the induction of CD8+ T-cell responses by vaccination. However, for many vaccines the relative contributions of direct and cross-presentation have not yet been elucidated. Therefore, it is hard to predict whether stable or unstable antigens are able to increase the immunogenicity of a particular vaccine.
DNA vaccination is a promising approach to induce MHC class I-restricted immune responses (27). Despite a relatively low intrinsic immunogenicity, it has the advantage of combining low-cost production with easy handling and the independence of a functional cold chain (12). In recent years, DNA vaccines against various targets were investigated and published (8). The optimization of target vectors with improved immunogenicity is currently a focus of interest. To induce CD8+ T-cell responses after intramuscular (i.m.) injection of DNA constructs, encoded antigens can be directly presented by transfected APCs. If this does not occur, antigens expressed by muscle and fibroblast cells have to be cross-presented by professional APCs (47). Depending on the contribution of both pathways, modification of antigen stability therefore could provide beneficial immunogenicity for a vaccination approach using DNA.
A similar situation is observed for viral vectors that are used for immunotherapy and vaccination. Genetically modified strains of vaccinia virus (VV), the effective protective vaccine against smallpox, were developed as tools to induce immune responses against recombinant antigens (31). Even though anti-VV responses are dominated by CD4+ T-cell and B-cell responses, the broad spectrum of infected cell types offers interesting opportunities to also induce class I-restricted responses via direct and cross-presentation. Infection with VV in vivo also targets APCs, which could allow induction of VV-induced CD8+ T-cell responses by direct presentation. Modification of antigenic stability therefore might be used to enhance MHC class I-restricted responses.
In this study, we investigated the role of protein stability on MHC class I presentation after DNA vaccination and infection with recombinant VV (rVV). As a model antigen, we choose the long-lived nucleoprotein (NP) of the murine lymphocytic choriomeningitis virus (LCMV). LCMV is a frequently used model to study antiviral immune responses. It belongs to the arenavirus group and consists of two structural proteins, the NP and the glycoprotein (GP). Infections with LCMV induce strong NP- and GP-specific CD8+ T-cell responses in mice. The LCMV proteins were used as model antigens to study direct and cross-presentation (3, 34). Importantly, for the LCMV NP it was shown that cross- but not direct presentation is dependent on the long-lived form of the antigen and is independent of neosynthesis. Additionally, in this system, DRiPs were published to be the major antigen source for direct presentation (7).
Antigen stability and protein degradation in general are dependent on a complex degradation machinery that maintains protein homeostasis in the cell. Generally, proteins that are supposed to be degraded via the proteasome are conjugated to the 8-kDa protein ubiquitin via a ubiquitin-conjugating enzyme cascade (20). This conjugation leads to proteasomal recognition of the substrate and to its degradation. Besides ubiquitin, there is a family of proteins called ubiquitin-like modifiers that also can be specifically conjugated to target proteins. However, from all ubiquitin-like modifiers, only the HLA-F-adjacent transcript 10 (Fat10; 18 kDa) is, like ubiquitin, able to target proteins for proteasomal degradation (21). In this study, we tried to use ubiquitin-NP as well as Fat10-NP fusion proteins to shorten the half-life of the LCMV NP model antigen. This approach allowed us to investigate the role of antigen stability on immune induction after DNA vaccination and recombinant VV infection. We show for the first time that N-terminal fusion of Fat10 to a viral nucleoprotein leads to a reduction in protein stability, as reported for ubiquitin. Further, we provide evidence that protein stability is a critical parameter that can strongly influence the outcome of a specific immunization approach. Whereas direct presentation after transfection or infection with recombinant VV of cell lines in vitro was increased when introducing short-lived NP-fusion proteins, this was not observed for DNA vaccination and recombinant VV infection in vivo, where stable antigen showed the highest CD8+ T-cell activation. In addition, we found differences in the immunogenicity of constructs fused to either ubiquitin or FAT10, even though their metabolic stability was similar. Our data indicate that targeting antigens for proteasomal degradation must not necessarily improve a vaccination protocol, as has been proposed previously (38).
MATERIALS AND METHODS
Mice, cells, and media.
C57BL/6 (H-2b) and BALB/c (H-2d) mice were originally obtained from Charles River Laboratories and further bred in the animal facilities of the University of Konstanz. All animals were kept under specific-pathogen-free conditions in accordance with the rules of the veterinarian authority of Regierungspräsidium Freiburg and were used for the experiments at 6 to 12 weeks of age. Primary murine CD8+ T-cell lines were cultured in RPMI 1640, 10% fetal calf serum (FCS), 20 U/ml interleukin-2 (IL-2), 50 μM β-mercaptoethanol, 50 μg/ml gentamicin. The human embryonic kidney cell line HEK293 (19) was maintained in Dulbecco's modified Eagle medium (DMEM), 10% FCS, 100 U/ml penicillin-streptomycin (P-S). The murine fibroblast cell line B8-wt (H-2d) (22) was cultured in Iscove's modified Dulbecco's medium (IMDM), 10% FCS, 100 U/ml P-S. B8-Db cells are B8 cells stably transfected with H2-Db and were cultured in IMDM, 10% FCS, 100 U/ml P-S, 5 μg/ml puromycin. MC57 (H-2b) cells are C57BL/6-derived methylcholanthrene-induced fibrosarcoma cells and were cultured in IMDM, 5% FCS, 100 U/ml P-S (4). Primary murine embryonic fibroblasts were cultured in DMEM, 10% FCS, 100 U/ml P-S. The dendritic cell (DC) line DC2.4 (H-2b) was kept in RPMI 1640, 10% FCS, 100 U/ml P-S (7). The human cell line 143B (TK−) was maintained in MEM, 10% FCS, 100 U/ml P-S, 25 μg/ml bromodeoxyuridine (BrdU) (ATCC line CRL-8303). BSC-40 cells are kidney-derived epithelial cells from Cercopithecus aethiops and were cultured in MEM, 10% FCS, 100 U/ml P-S (ATCC line CRL-2761). Primary peritoneal macrophages were cultured in DMEM, 10% FCS, 100 U/ml P-S. All cell culture media and supplements were obtained from Gibco, Invitrogen.
Generation of NP constructs.
The plasmids pCMV_NP and pCMV_Ub-NP were kindly provided by L. Whitton (Scripps Research Institute) (38). The plasmid pCMV_FAT10-NP, encoding an N-terminal Fat10 fusion protein of the NP, was generated as follows. Mouse Fat10 was amplified by PCR from pBKCMV_HA-FAT10-GFP (kindly provided by G. Schmidke, University of Konstanz), generating an N-terminal XhoI and a C-terminal EcoRI restriction site using the primer pair 5′-TGG TAC CTC GAG ATG GCT TCT GTC CGC ACC-3′ (forward) and 5′-ATA CTA GAA TTC TGC CAC AGT GCA GTG TGT-3′ (reverse), introducing a GG-to-VA mutation at the C-terminal end of the amino acid sequence of Fat10. This mutation protects Fat10 from being cleaved off the substrate by putative de-Fat10-ylating enzymes. NP was amplified by PCR from pCMV_NP using the primer pair 5′-TAT GAT GAA TTC ATG TCC TTG TCT AAG GAA GT-3′ (forward) and 5′-ATC CCC GCG GCC GCT TAG AGT GTC ACA ACA TT-3′ (reverse), introducing an EcoRI and a NotI restriction site. Both fragments were digested with EcoRI and ligated before Fat10-VA-NP was amplified by PCR using the primers Fat10_for and the reversed primer NP_rev. The amplified Fat10-VA-NP construct was then introduced into the XhoI/NotI site of pCMV.
Pulse-chase experiment.
Analysis of protein stability of different NP constructs was performed by radioactive pulse-chase experiments, which were adopted from reference 40. Therefore, the indicated cell types were either transfected with NP-encoding pCMV constructs using Fugene 6 (Roche) or infected with recombinant VV. Eighteen h after transfection or 3 h after infection, cells were washed two times with phosphate-buffered saline (PBS) and then incubated in medium lacking methionine (modified RPMI 1640 medium; R5713; Sigma) for 1 h. After starvation, radioactively labeled [35S]methionine was added to the cells in a concentration of 0.25 mCi/ml for 1 h. [35S]methionine containing radioactive labeling medium was removed, and cells were washed with PBS and further incubated in medium at 37°C. At the indicated time points, cells were washed with ice-cold PBS and cell pellets were frozen at −20°C. For lysis, pellets were resuspended in lysis buffer (50 mM Tris-HCl, pH 8, 150 mM NaCl, 1 mM EDTA, 2% NP-40, protease inhibitors, in H2O) and incubated for 30 min on ice. Afterwards, cell debris was removed by centrifugation and the radioactivity of the supernatant was quantified using a scintillation counter (Top Count NXT; Packard). Radioactivity values were used to adjust supernatants before NP-specific immunoprecipitation (IP). In a step of preclearance, protein G beads (Sigma) were added to the cell supernatant for 1 h. Afterwards, beads were removed by centrifugation and fresh protein G beads were added in the presence of the NP-specific antibody KL53 (40). Samples were incubated on a rotator at 4°C. After 18 h, beads were washed three times with ice-cold NET-TON buffer (650 mM NaCl, 5 mM EDTA, 50 mM Tris-HCl, pH 8, 0.5% Triton X-100, 1 mg/ml ovalbumin [Sigma]) and NET-T buffer (150 mM NaCl, 5 mM EDTA, 50 mM Tris-HCl, pH 8), respectively. The beads then were resuspended in SDS sample buffer, heated for 5 min at 95°C, and loaded onto 10% SDS-PAGE gels. After electrophoresis, gels were dried and exposed to a radiosensitive photo plate. After 24 to 48 h, radioactive bands were visualized using a phosphorimager (Molecular Imager FX; Bio-Rad).
Generation of specific CD8+ T-cell lines.
Naive C57BL/6 or BALB/c mice at 6 to 8 weeks of age were intravenously (i.v.) infected with 200 PFU LCMV. Starting from 3 weeks after infection, memory mice were used for the generation of NP396-404- and NP118-126-specific CD8+ T-cell lines. Spleens from memory mice were homogenized, and isolated splenocytes were further purified by Ficoll density gradient centrifugation (GE Healthcare). Splenocytes were further cultured in 6-well plates (1.5 × 107 cells/well) and directly pulsed with 10−6 M NP396 or NP118 peptide. Medium was renewed every second day. On day 5, Ficoll gradient centrifugation was repeated to remove dead cells. Remaining splenocytes were washed with PBS and further cultured in the presence of IL-2. NP396 (H2-Db)- and NP118 (H2-Ld)-specific CD8+ T-cell lines were used between days 7 and 9 of culture.
Preparation of primary peritoneal macrophages.
Peritoneal macrophages (pMΦs) were prepared by intraperitoneal (i.p.) injection of 2 ml 3% thioglycolate solution into either C57BL/6 (H-2b) or BALB/c (H-2d) mice. After 3 days, peritoneal cells were washed out of the abdominal cavity using 10 ml of PBS. Cells were cultured for 2 days, and adherent cells were used for transfection and antigen presentation assays.
In vitro antigen presentation assay.
To determine the extent of direct presentation after transfection of NP-encoding plasmids or infection with recombinant VV, activation of CD8+ T-cell lines was measured in an in vitro assay. For transfection, cells were electroporated using Amaxa technology (Lonza). B8-Db cells were harvested and used at a density of 2 × 106 cells per 100 μl. One μg plasmid DNA was used for individual transfections. Electroporation was performed according to the user's manual with the MEF2 kit and program A01 of the Amaxa device. One μg of plasmid encoding green fluorescent protein (GFP) was cotransfected as an internal control to determine transfection efficiency, and cells were cultured in complete medium. After 18 h, cells were harvested and the numbers of GFP+ cells were analyzed by flow cytometry. For analysis of direct presentation after VV infection, indicated cells were incubated with recombinant VV at a multiplicity of infection (MOI) of 10 for 4 h. To determine antigen presentation of NP-derived epitopes, transfected or infected cells were titrated in 96-well plates starting with an initial density of 2 × 105 cells in 100 μl RPMI medium per well. All samples were prepared in duplicate. NP-specific CD8+ T cells (2 × 105) in 100 μl RPMI medium containing brefeldin A in a concentration of 20 μg/ml (2×) were added to each of the wells. After 4 h of incubation at 37°C and 5% CO2, activation of CD8+ T cells was determined by intracellular cytokine staining (ICS) and flow cytometry.
ICS.
ICS was performed to detect intracellular accumulation of gamma interferon (IFN-γ) as a measure of CD8+ T-cell activation. Samples were centrifuged, and 50 μl TriColor anti-mouse CD8a (1:200 in PBS; Invitrogen) was added to each well on ice and incubated for 15 min in the dark. After samples were washed with ice-cold PBS, 50 μl of 3% paraformaldehyde (Acros Organics) solution was added for 5 min at room temperature. Afterwards, samples were again washed with PBS. Staining for IFN-γ was performed by adding 50-μl fluorescein isothiocyanate (FITC) anti-mouse IFN-γ (1:1,000; kind gift from M. Basler, University of Constance) in PBS, 1% saponin. The staining was incubated overnight at 4°C. Before acquisition by flow cytometry (FACScan; BD), samples were washed, resuspended in PBS, and kept on ice in the dark.
DNA immunization.
NP-expressing pCMV plasmids for DNA vaccination were generated in Escherichia coli TOP10 F′ and purified using a plasmid purification kit (Midi plasmid kit; Sigma). Plasmid concentration was determined by a spectrophotometer (NanoVue; GE Healthcare) and adjusted to 2 μg/μl. One hundred μg plasmid DNA in 50 μl was injected and electroporated into each of the two hind legs of C57BL/6 mice intramuscularly (i.m.) using an in vivo electroporation device (kindly provided by Ichor Medical Systems, San Diego, CA). DNA immunization was repeated two times after 14 and 28 days. On day 7 after the last boost, mice were sacrificed and splenocytes prepared. The NP396-specific immune response was quantified by ICS.
Generation of rVV.
Recombinant vaccinia virus (rVV) expressing the LCMV NP was kindly provided by R. Zinkernagel (Zürich University, Switzerland) (39). rVV expressing ubiquitin-NP (Ub-NP) and Fat10-NP fusion proteins were generated as follows. NP sequences were introduced into SalI/NotI restriction sites of the vaccinia transfer plasmid pSC11-S-B-A-K-N (kindly provided by Bernhard Moss, NIAID NIH), generating the transfer plasmids pSC11-S-B-A-K-N_Ub-NP and pSC11-S-B-A-K-N_Fat10-VA-NP. Both constructs were amplified by PCR from pCMV-Ub-NP/pCMV-Fat10-VA-NP using the primer pair 5′-TAT GAT GTC GAC ACT CTA GAG GAT CCG GTA C-3′ (forward) and 5′-ATC CCC CCA TGG TTA GAG TGT CAC AAC ATT-3′ (reverse), introducing restriction sites for 5′SalI and 3′NotI. Generation of rVV was adapted from reference 44. Briefly, subconfluent 143B (TK−) cells were infected with wild-type VV strain WR at an MOI of 0.1. After 2 h, unbound virus was removed by addition of fresh medium. After vaccinia virus transfer, plasmids were transfected into TK− cells using Fugene 6 (Sigma). After 48 h, medium was removed and cells were embedded in 1% agarose in MEM containing 10% FCS, 25 μg/ml BrdU, 300 μg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (Sigma). Plaques that appeared blue within 6 h were picked and resuspended in 500 μl MEM, 2% FCS. Isolated viral samples were frozen 3 times at −70°C and then used to reinfect subconfluent TK− cells. Purification of plaques was repeated until in the three following reinfections only blue plaques could be detected. After the last isolation, rVV were used to infect BCS-40 cells to generate large amounts of virus. The viral titer of each rVV was determined, and aliquots were frozen at −80°C.
For the generation of enhanced GFP (eGFP) expressing recombinant vaccinia virus, the insert eGFP (GenBank accession number U55762) was isolated from pEGFP-N2 (Clontech) by digestion with SmaI and NotI, followed by blunting. The blunted insert was cloned into the SmaI-digested vector pSC11.3 (32). Recombinant vaccinia virus (rVV) was generated by homologous recombination over the thymidine kinase gene, followed by four rounds of plaque purification using blue/white selection as described previously (30, 32). rVV-eGFP was propagated on BSC40 cells, and its titers were determined.
Virus titer determination.
The titer of rVV-infected BSC-40 cell lysates was analyzed after 6 freeze-thaw cycles of cell suspensions. One day prior to the experiment, BSC-40 cells were plated in 24-well plates to reach confluence the next day. Lysates of rVV-infected BCS-40 cells were titrated into 24 wells in 1:10 dilutions, starting with an initial dilution of 1:1,000. In the case of the ex vivo determination of viral titers in ovaries of infected mice, ovaries were taken on day 4 after infection and physically disrupted by dounce homogenization in MEM, 10% FCS. Suspensions were frozen and thawed 6 times. Titrated amounts of lysate were added to 24 wells of BSC-40 cells starting with pure lysate. After infection, BCS-40 cells were cultured in MEM, 10% FCS, 100 U/ml P-S and incubated for 24 to 48 at 37°C, 5% CO2. Afterwards medium was removed and plaques visualized by addition of 0.5% crystal violet solution for 1 h. Plates then were washed in a water bath, and plaques were counted to calculate the number of PFU in the stock lysate.
Determination of viral infection in vivo by flow cytometry.
C57BL/6 mice were injected i.p. with 3 × 106 PFU rVV-eGFP in 200 μl PBS. Mice were euthanized after 15 or 23 h, and spleens were isolated and single-cell suspensions prepared. Approximately 106 spleen cells were stained with anti-I-Ab (MHC-II) PacificBlue and anti-CD11c APC (BioLegend) in 100 μl fluorescence-activated cell sorting (FACS) buffer (PBS, 2% FCS, 20 mM EDTA, 0.05% NaN3) for 15 min at 4°C. Cells were washed twice with 2 ml FACS buffer, and the eGFP signal was measured after gating on CD11c+ MHC-II+ dendritic cells. Data were acquired with a CyAn ADP9 flow cytometer (Beckman Coulter) and analyzed with FlowJo software. To determine the percentage of infected cells 4 h after incubation with all other recombinant VV, the indicated samples were harvested, washed with PBS, and fixed using 50 μl of 3% paraformaldehyde (Acros Organics) solution for 5 min at room temperature. Afterwards, samples were again washed with PBS, and staining for intracellular vaccinia virus was performed by adding 50 μl FITC anti-vaccinia polyclonal antibody (ab19979; 1:100; Abcam) in PBS, 1% saponin and incubating the samples for 1 h at 4°C. Before data acquisition by flow cytometry (FACScan; BD Biosciences), samples were washed, resuspended in PBS, and kept on ice in the dark.
Assessment of immune responses in vaccinia virus-infected mice.
For analysis of immune responses in mice during VV infection, C57BL/6 or BALB/c mice were injected intraperitoneally with 2 × 106 PFU wild-type or recombinant VV. Noninjected mice served as controls. On day 8 (direct response) or 21 (memory response) after infection, mice were sacrificed and splenocytes prepared and restimulated with 10−6 M either B8R20-27, NP396-404, or NP118-126 peptide in the presence of 10 μg/ml brefeldin A for 5 h. Afterwards the activation of CD8+ T cells was quantified by ICS and flow cytometry.
RESULTS
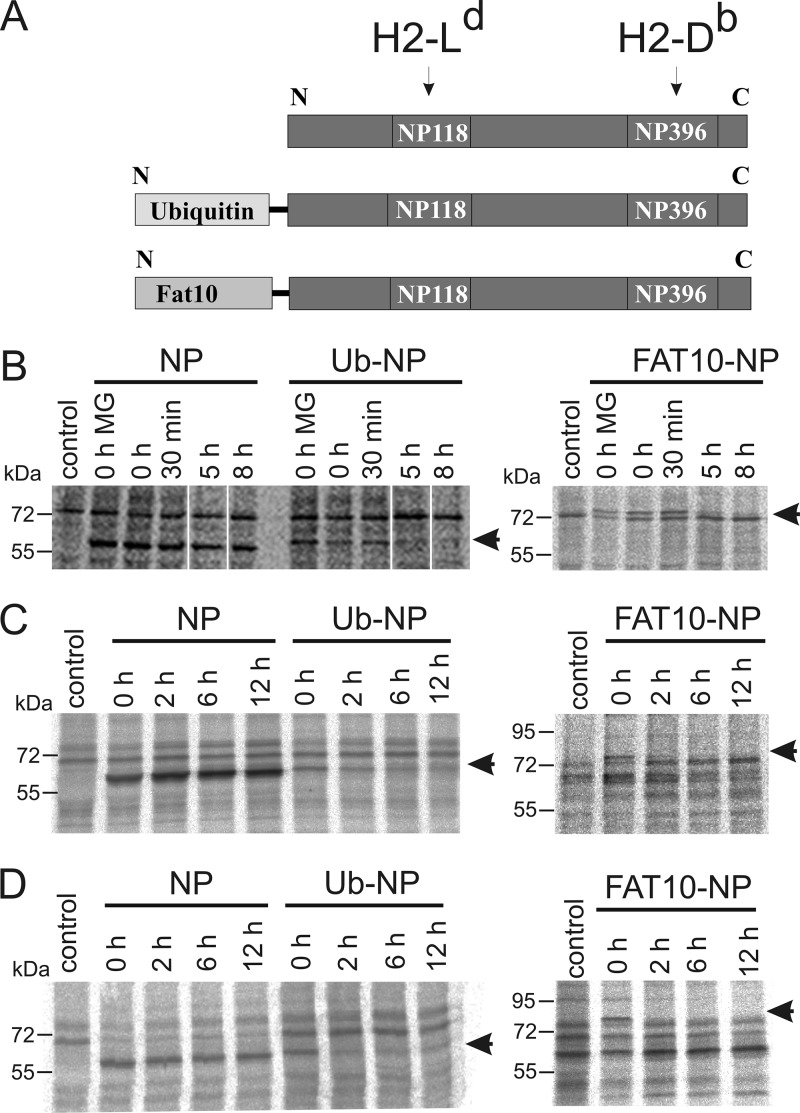
N-terminal fusion of the LCMV nucleoprotein with ubiquitin or the ubiquitin-like modifier FAT10 enhances its degradation rate.
Direct and cross-presentation are promoted by distinct properties of antigens, especially with respect to their degradation rate. To evaluate the impact of both pathways on vaccination, we generated three different constructs encoding LCMV NP variants which are degraded at different rates (Fig. 1A). The LCMV NP with its two immunodominant epitopes, NP118 (H-2Ld) and NP396 (H-2Db), was used either without any modification or as a fusion protein with ubiquitin or FAT10. The C-terminal GG motif from both ubiquitin and Fat10 was mutated to avoid cleavage by deconjugating enzymes, before fusion to the N terminus of the NP. The constructs were cloned into DNA vaccination vectors and recombinant VV, which were both applied in this study. To show the altered antigen stability of fusion proteins, we performed radioactive pulse-chase experiments after transient transfection of DNA vaccination vectors in HEK293 cells (Fig. 1B) or after infection with recombinant VV in murine cell lines (Fig. 1C and D). While no NP-specific immunoprecipitates could be detected in the nontransfected/infected control, bands of the expected sizes were found for NP (62 kDa), Ub-NP (70.5 kDa), and Fat10-NP (80 kDa) constructs in the transfected cells. While in all cases the NP alone was found to be stable during the 8- to 12-h time period analyzed, fusion with ubiquitin enhanced the degradation rate of the NP, leading to almost complete degradation with a half-life of less than 2 h. Interestingly, fusion to FAT10 also led to a destabilization of the NP, similar to what we have observed for the ubiquitin fusion. The enhanced degradation rates of NP fusion proteins in this experiment validate the NP constructs as adequate for a comparative study of antigen stability and its effect on antigen presentation.
Fig 1.
Characterization of protein expression constructs used for the analysis of antigen presentation after DNA immunization and infection with recombinant vaccinia virus (VV). (A) For both DNA and viral constructs, the lymphocytic choriomeningitis virus (LCMV) nucleoprotein (NP) was used either unmodified or cloned in a linear N-terminal fusion with either ubiquitin (Ub-NP) or FAT10 (FAT10-NP). Arrows indicate the two NP epitopes that were used in this study to measure antigen presentation, the H2-Ld-restricted NP118-126 (NP118) and the H2-Db-restricted NP396-404 (NP396). (B to D) Pulse-chase experiments to analyze the stability of different NP constructs. (B) HEK293 cells were transiently transfected; B8Db (C) and DC2.4 (D) cells were infected with recombinant VV encoding the indicated proteins. All cells were metabolically labeled with [35S]methionine. After the indicated time points, cells were lysed and NP was immunoprecipitated with the anti-NP antibody KL53. Samples were then separated by SDS-PAGE, dried, and imaged on a radio imager. MG, proteasome inhibitor MG132. Arrows indicate the size of NP (62 kDa), Ub-NP (70.5 kDa), and Fat10-NP (80 kDa). Representative results are shown.
Targeting the LCMV NP for rapid degradation increases direct presentation in vitro.
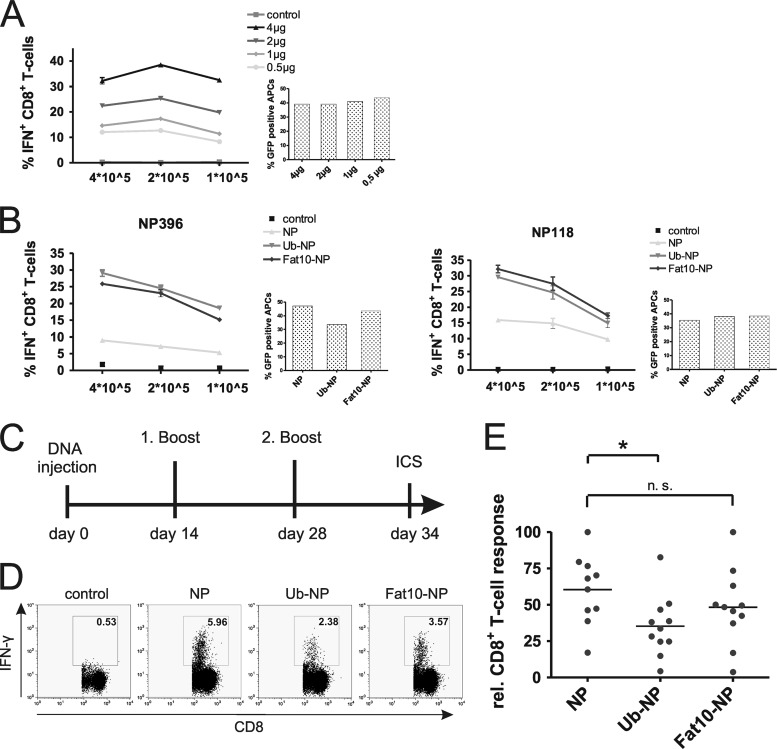
We first analyzed the impact of protein stability on direct presentation after transfection of DNA vaccine constructs in vitro. To detect possible differences between individual NP constructs, we first had to ascertain that our transfections would not lead to saturated antigen presentation with maximal CD8+ T-cell activation. Therefore, we titrated the amount of plasmid DNA transfected into the antigen-presenting cells (Fig. 2A). As expected, we observed increasing antigen presentation with the transfection of increasing amounts of NP-encoding plasmid. Although the best CD8+ T-cell activation was observed at concentrations of approximately 4 μg/well of a 6-well plate, we decided to use 1 μg plasmid DNA for all further transfections. This way we could exclude saturated presentation for individual constructs.
Fig 2.
Evaluation of DNA vaccines after transfection of lymphocytic choriomeningitis virus (LCMV) nucleoprotein (NP)-encoding constructs in vitro and immunization in vivo. (A) Titration of pCMV NP transfection. Indicated amounts of pCMV NP DNA were transfected into B8Db cells, and presentation of the epitope NP396 was analyzed by NP396-specific CD8+ T cells after 18 h. Antigen presentation was measured by intracellular cytokine staining (ICS). Cotransfection of a GFP-expressing plasmid served as a control. (B) Indicated numbers of B8-Db cells (H-2d/Db) transfected with DNA constructs were analyzed for NP396- and NP118-specific presentation, which was analyzed after 18 h by coincubation with a constant number of epitope-specific CD8+ T cells and ICS. GFP cotransfection served as a control. The displayed experiment shows representative results for two independent repeats. (C) Experimental setup of in vivo immunization. Mice were injected and electroporated intramuscularly with 100 μg DNA into the limbs on day 0. Homologous boosts were performed on days 14 and 28. On day 34, mice were sacrificed and splenocytes were restimulated with NP396-404 peptide. (D) Representative ICS results for nonimmunized mice (control) or mice immunized with the indicated DNA constructs. (E) Summary of DNA vaccination results from two independent experiments (n = 11). For better comparison of individual experiments relative CD8+ T cells, responses are shown (highest response of each individual experiment was set to 100%) and were determined using the following equation: relative response = (100/percent highest value of IFN-γ+ T cells) × percent IFN-γ+ T cells (sample). Statistical analysis was performed by unpaired Student's t test. *, P ≤ 0.05.
We analyzed the role of antigen stability for direct presentation in the mouse fibroblast cell line B8Db. DNA constructs were transfected for 18 h, and antigen presentation was analyzed using primary NP-specific CD8+ T-cell lines as a read-out system (Fig. 2B). Interestingly, NP396 as well as NP118 presentation was about 2- to 3-fold increased after transfection of fusion proteins compared to the stable wild-type NP, which only led to a basal presentation of epitopes. Plasmids encoding GFP were cotransfected in all samples to confirm equal transfection efficiencies by flow cytometry.
Rapid antigen degradation does not enhance immunogenicity of DNA vaccines.
In DNA vaccination, constructs encoding the antigen of interest are usually injected intramuscularly. Therefore, one can assume that the majority of cells transfected after DNA injection are muscle cells and cells of the connective tissue. However, the priming of T-cell responses can only be induced by professional APCs. Hence, either the number of APCs directly transfected during DNA vaccination must be large enough to stimulate immune responses or antigens have to be acquired exogenously and processed via cross-presentation.
To test whether targeting of antigens for rapid degradation can be an advantage for the induction of immune responses via DNA vaccination, we immunized mice with constructs expressing either the stable NP or fusion proteins of the NP and ubiquitin or FAT10 (Fig. 2C). After two booster injections, we analyzed the immune response in mice by intracellular cytokine staining (Fig. 2D). Interestingly, we found that the percentage of NP396-specific CD8+ T cells was significantly higher in mice immunized with the construct expressing the stable NP compared to ubiquitin-NP (Fig. 2E). Responses against the FAT10-NP fusion protein were also reduced compared to the stable NP, but differences were not significant. Therefore, our experiments clearly indicate that targeting the LCMV NP for rapid degradation is of no benefit for a DNA vaccination approach. In contrast, the long-lived protein is able to induce the strongest immune responses. These findings have to be taken into account when designing new DNA vaccines.
Enhanced direct presentation of LCMV NP-derived epitopes after infection with recombinant VVs expressing short-lived NP fusion proteins in vitro.
Recombinant VV was introduced not only as a potential vaccine against smallpox but also as a tool to initiate immune responses against various other immune targets (3). Since optimization of expression constructs is an important issue for successful vaccination strategies, we were interested in studying the role of antigen stability for the immune response of a recombinant antigen expressed by VV. Therefore, we generated recombinant VVs expressing long-lived LCMV NP or short-lived fusion proteins of the NP with either ubiquitin or Fat10 (Fig. 1).
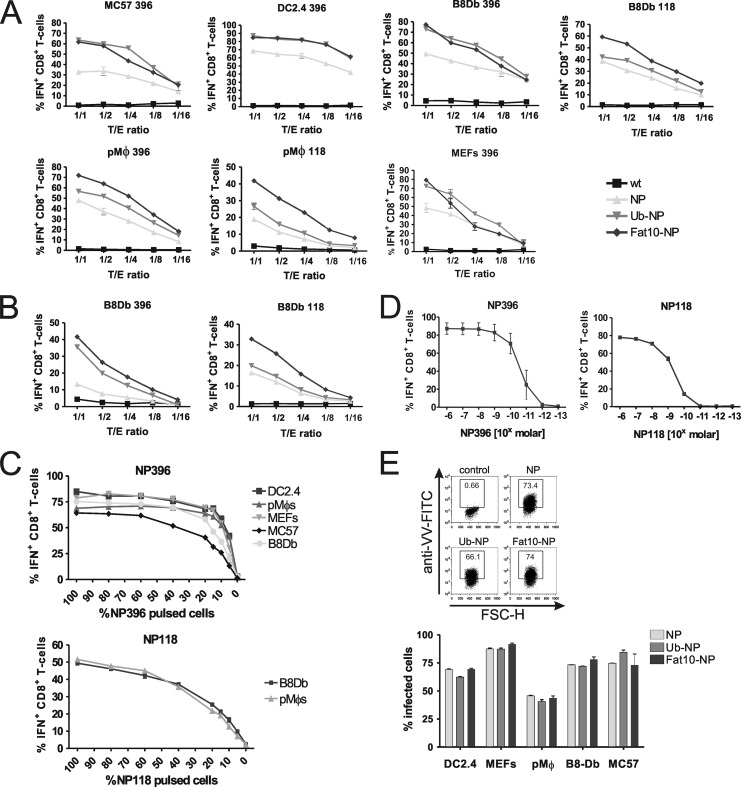
We wanted to investigate whether differences in antigen stability can also influence the direct presentation of antigens after VV infection. Therefore, we infected various cell types with NP-expressing recombinant vaccinia virus strains and detected the direct presentation of NP-derived epitopes by NP-specific CD8+ T-cell lines and intracellular cytokine staining for IFN-γ (Fig. 3A). Interestingly, and similar to our DNA vaccine experiments (Fig. 2B), we found the unstable forms of the NP to be presented with significantly higher efficiency than epitopes derived from the long-lived NP. This observation was true for both epitopes analyzed, the NP396 and the NP118, and it was valid both 4 (Fig. 3A) and 2 h (Fig. 3B) after infection, indicating that this was not a kinetic effect. Additionally, a direct comparison of the two NP fusion proteins Ub-NP and Fat10-NP reveals that Fat10-NP seems to present with the highest efficiency of all cell types and epitopes analyzed. Antigen presentation and activation of CD8+ T cells was not saturated, since differences were observed for all titrated target-to-effector (T/E) ratios.
Fig 3.
Direct presentation of NP epitopes in vitro after infection of the indicated cell lines with recombinant vaccinia viruses (VV) encoding either the long-lived lymphocytic choriomeningitis virus nucleoprotein (NP), short-lived fusion proteins of the NP with either ubiquitin (Ub-NP) or FAT10 (FAT10-NP), or wild-type vaccinia virus as a control. Four h (A) or 2 h (B) after infection, cells were washed with PBS and the indicated dilutions were incubated with 2 × 105 CD8+ T cells specific for the NP epitopes NP396 (H2-Db) and NP118 (H2-Ld). Activation of CD8+ T cells was analyzed by intracellular cytokine staining (ICS) for gamma interferon (IFN). Results are representative of two independent experiments. (C) Analysis of transfection/infection efficiency on CD8+ T-cell activation. Indicated cells were either left untreated or pulsed with NP peptides for 1 h at 37°C. Pulsed and nonpulsed cells were mixed in titrated ratios to a final concentration of 2 × 105 cells/well. CD8+ T cells were added in the same concentration, and their activation was analyzed by ICS. (D) Titration of peptide antigen sensitivity of CTL lines as used in panels A to C. Restimulated T cells lines with the indicated specificity for NP118 and NP396 were treated with the indicated concentrations of peptide for 3 h in the presence of brefeldin A and subsequently subjected to intracellular IFN-γ staining and flow-cytometric analysis. The experiment has been repeated twice with a similar outcome. (E) Intracellular staining for vaccinia virus. Indicated cells were infected with the indicated recombinant VV for 4 h. Afterwards cells were fixed and intracellularly stained with FITC-conjugated anti-vaccinia antibody. A representative flow cytometric analysis and a graph bar of two experiments performed with mean values and SEM are shown.
Small differences in infection rates might lead to changes in antigen presentation. To analyze the impact of such differences, we mixed different types of peptide-pulsed and nonpulsed cells at the indicated ratios and measured the activation of CD8+ T-cell lines (Fig. 3C). While there were strong differences in antigen presentation with increasing amounts of peptide-loaded cells in the low range, activation was almost saturated with around 50% peptide-loaded cells, and no major differences between cell types were recorded. Moreover, a titration of NP118 and NP396 peptides revealed that the NP118- and NP396-specific T-cell lines were of the expected sensitivity, with a detection threshold of 10−9 M for NP118 and 10−10 M for NP396 (Fig. 3D). As an additional control to show equal infection rates of recombinant VV for all cell types used, we performed intracellular vaccinia virus stainings and analysis of infected cells by flow cytometry. As indicated in the bar graph (Fig. 3E, bottom), the infection rates for NP, Ub-NP, and Fat10-NP were similar in all cell types tested. From these controls, we conclude that differences in antigen presentation seen for recombinant VV-infected cells were due to intrinsic properties of the antigens encoded.
Stable antigen shows the strongest CD8+ T-cell responses in mice after infection with recombinant vaccinia virus in vivo.
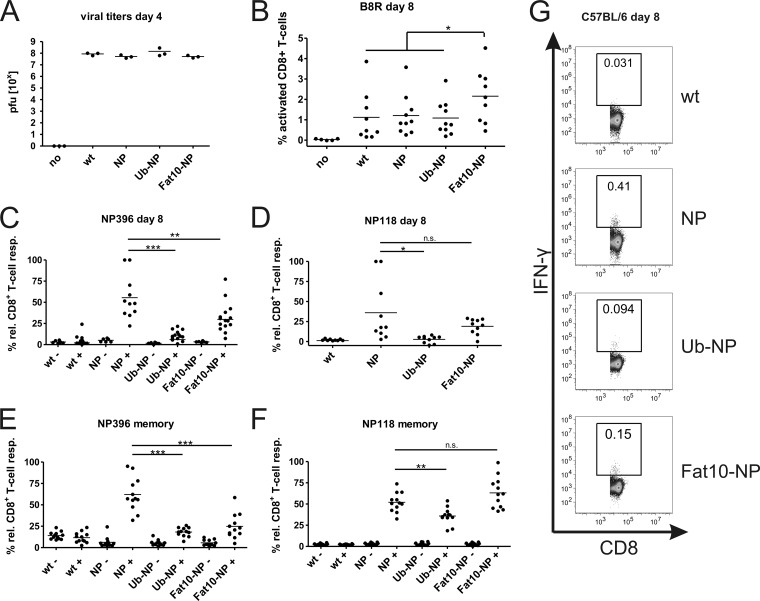
After analyzing direct presentation in vitro, we were interested in studying the role of antigen stability after VV infection in vivo. In an initial experiment, we compared the viral titers of wild-type vaccinia virus and the different recombinant VV on day 4 after infection to ensure that possible differences in immune induction were not due to alterations between individual virus clones (Fig. 4A). While no virus could be detected in ovaries of uninfected mice, comparable titers of virus were found in mice infected with wild-type and recombinant VVs. No significant difference could be calculated between individual virus strains. CD8+ T-cell responses against vaccinia virus in C57BL/6 mice are dominated by the B8R protein-derived epitope B8R20-27. As an additional control, we compared the B8R-specific immune responses in mice immunized with either wild-type VV or different rVV strains (Fig. 4B). Since the only difference between individual viruses is the introduction of NP and NP derivatives, the B8R-specific response should be similar in mice infected with all viruses. In fact, there was no significant difference in the B8R-specific immune response on day 7 after infection with wild-type VV and the recombinant VV clones NP and Ub-NP and only slightly higher numbers of B8R-specifc CD8+ T cells in mice immunized with the rVV expressing Fat10-NP. When we analyzed the activation of NP-specific CD8+ T cells, we observed the highest induction in mice injected with the recombinant VV expressing the long-lived form of the NP (Fig. 4C and D). In contrast to this, there was hardly any response detectable in mice immunized with the virus expressing the short-lived Ub-NP. A similar result was obtained when analyzing memory mice 3 weeks after VV infection (Fig. 4E and F). Since there was no difference in viral propagation and induction of B8R-specific immune responses between mice infected with rVV-NP, rVV-Ub-NP, and wild-type VV (Fig. 4B), we conclude from these experiments that the stable antigen is superior in inducing CD8+ T-cell responses after vaccinia virus infection in this system. However, an intermediate phenotype was observed in the CTL responses after infection with recombinant VV expressing Fat10-NP (Fig. 4C and G). CD8+ T-cell responses were significantly stronger than those of rVV Ub-NP, but they failed to reach the level of NP396-specific responses elicited by rVV NP, whereas the memory responses to NP118 in BALB/c mice elicited by infection with rVV NP and rVV FAT10-NP were equally strong (Fig. 4F).
Fig 4.
Analysis of CD8+ T-cell responses in mice immunized with different recombinant vaccinia viruses (VV). (A) Comparison of viral titers in ovaries of C57BL/6 mice on day 4 after infection to wild-type vaccinia virus, recombinant VV expressing either the long-lived lymphocytic choriomeningitis virus (LCMV) nucleoprotein (NP), the short-lived fusion proteins with ubiquitin (Ub-NP) or FAT10 (FAT10-NP), or with PBS as a control (no). Results are indicated in PFU per one pair of ovaries. (B to F) Spleens of infected and control mice (no) were removed on day 8 after infection, and splenocytes were restimulated in vitro with the VV epitope B8R (B) or the LCMV epitope NP396 (C) or NP118 (D). (E and F) Mice were infected as described above, and splenocytes were restimulated 3 weeks after infection (memory response) with LCMV epitope NP396 (E) or NP118 (F). After 5 h, CD8+ T-cell activation was analyzed by intracellular cytokine staining (ICS) for gamma interferon, and samples were analyzed by flow cytometry. (G) Representative ICS stainings for an experiment as described in panel C. Dot plots shown in panels C to F show the percentages of IFN-γ-positive cells of all CD8+ lymphocytes; each dot represents the results from one mouse analyzed. Combined results from three independent experiments are shown. For better comparison of individual experiments, relative CD8+ T-cell responses are shown (% rel. CD8+ T-cell resp.). The highest response of each individual experiment was set to 100%. Relative response = (100/percent IFN-γ+ T cells [highest value]) × percent IFN-γ+ T cells (sample). Statistical analysis was performed by using an unpaired Student's t test with Welch's correction if variances were significantly different: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
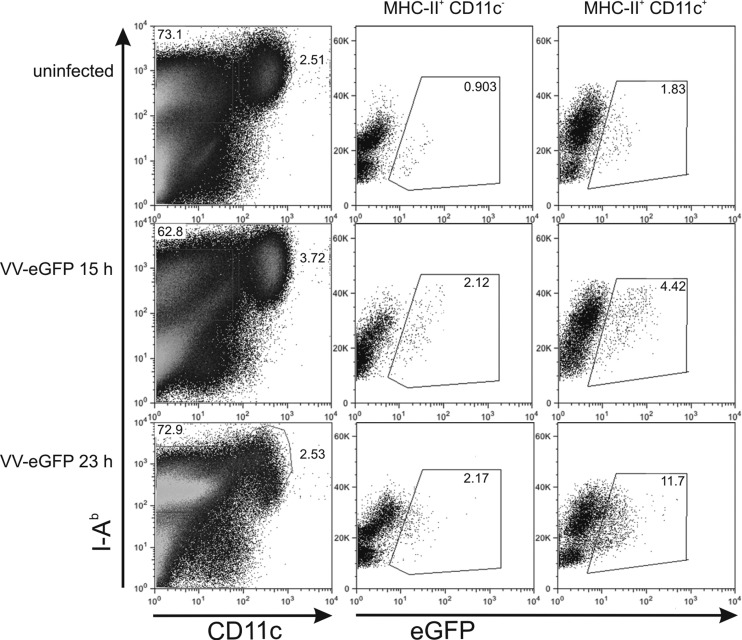
Finally, we investigated whether the striking difference between short-lived and long-lived variants of NP with respect to NP118 and NP396 antigen presentation in vitro compared to CD8+ T-cell priming in vivo was due to the inability of the used rVV strain to infect DC in mice. C57BL/6 mice were infected with rVV expressing GFP (rVV-GFP), which resulted in GFP expression in MHC-II+ CD11c− cells and MHC-II+ CD11c+ DCs in the spleen 15 and 23 h after infection, strongly suggesting that rVV-GFP had infected DCs in vivo (Fig. 5). Therefore, we conclude that the inferior induction of NP-specific CTL responses by rapidly degraded variants of NP compared to stable NP in vivo cannot be attributed to the inability of the used rVV strain to directly infect CD11c+ dendritic cells.
Fig 5.
Infection of dendritic cells with rVV-GFP in vivo. C57BL/6 mice were injected i.p. with 3 × 106 PFU rVV-eGFP. At 15 or 23 h after infection, mice were euthanized and splenocytes were stained with anti-I-Ab (MHC-II) and anti-CD11c antibodies. MHC-II+ CD11c− and MHC-II+ CD11c+ cells were gated as indicated, and eGFP fluorescence was assessed by flow cytometry. The experiment has been reproduced twice with similar outcomes.
DISCUSSION
The development of novel vaccines against infectious diseases and cancer is still a challenging field of biomedical research. Immunogenicity, biological safety, and cost efficiency are key words often discussed in connection with the requirements of novel vaccines. However, biological safety and high immunogenicity are especially important parameters, and they are often mutually exclusive. In this report, we studied the molecular requirements of antigen stability for two kinds of vaccines that are promising candidates for future immunotherapy and vaccination.
DNA vaccines combine many advantages as tools in clinical immunology (8). Current approaches that intend to enhance DNA-mediated immune responses include various delivery systems (e.g., electroporation and gene gun), coadministration of cytokines, or other proinflammatory molecules (26). Although CD4+ T-cell responses can be induced by DNA vaccination, the majority of studies concentrate on the initiation of CD8+ T-cell responses, since these cells are important for clearing infected or malignant cells. There is an ongoing debate on the molecular mechanisms involved in the initial priming of CD8+ T cells after DNA vaccination. Of special interest are the cell types involved in antigen priming and whether direct or cross-presentation accounts for antigen-specific responses. Initial studies in the 1990s have clearly identified DCs as the major APC type in charge of priming CD8+ T cells after DNA vaccination (24). Because the vast majority of antigen after intramuscular injection of DNA is produced by myocytes (15), it was questioned whether direct presentation by the relatively low number of transfected APCs could account for the overall CD8+ T-cell priming, as documented in some reports (1, 35). For cross-presentation, antigens expressed by myocytes and other tissue-specific cells would have to be transferred to APCs that can internalize exogenous antigens and present peptides in the context of MHC class I. Indeed, such a transfer of antigens from myocytes to APCs was formally shown and associated with antigen cross-presentation (17). The understanding of the antigen presentation pathway after DNA immunization is important to further enhance its immunogenicity.
In this study, we investigated the role of antigen stability on the efficiency of DNA vaccination. By N-terminal fusion of the LCMV NP to either ubiquitin or Fat10, we were able to convert the long-lived nature of the NP into rapidly degraded derivatives that resemble the viral protein in all parameters (e.g., epitopes and expression profile) except its half-life (Fig. 1). Rapidly degraded proteins and DRiPs were found to be the major sources of the direct MHC class I antigen presentation pathway (5, 51). In accordance with these findings, one would assume that proteasomal targeting of an antigen can lead to enhanced direct presentation. By transfecting mouse fibroblasts, we could show that ubiquitin-NP and Fat10-NP fusion proteins are more efficiently presented in vitro than the construct expressing the long-lived wild-type NP (Fig. 2). This experiment argues against a DRiP-only hypothesis in direct MHC class I presentation, because additional degradation of normally stable proteins seems to enhance the peptide load.
The dependence of direct presentation on rapid protein degradation, in combination with the finding that cross-presentation of the LCMV NP entirely depends on the stable antigen (7, 16), allows an interpretation of our findings concerning the general mechanism of antigen presentation in DNA vaccination in vivo. If direct presentation by APCs would be the major mechanism, proteasomal targeting of the antigen should enhance CD8+ T-cell activation, as seen in vitro. Vice versa, the long-lived wild-type protein should give the best results if a response is based on cross-presentation. Interestingly, we found the strongest immune activation in mice that were immunized with the long-lived wild-type NP. The responses after injection of ubiquitin-NP were significantly reduced. This finding contradicts an earlier study with the opposite results using the same NP constructs (38). However, in that study the authors could only indirectly show the NP-specific immune responses by analysis of viral clearance after an LCMV infection that was performed weeks after immunization. This rather indirect method was required because the low intrinsic immunogenicity of DNA constructs did not allow direct ex vivo analyses. We circumvented this difficulty by using a novel electroporation device as a tool to enhance immunogenicity, as also demonstrated by other recent studies (8). Thus, we were able to analyze CD8+ T-cell responses directly ex vivo, which makes our results more reliable and allows a different interpretation of the data: cross-presentation of DNA-encoded antigens has to make a major contribution to the overall immune response to DNA vaccines. Otherwise, we could not explain why short-lived NP fusion proteins show enhanced antigen presentation in vitro but reduced responses in vivo. Our data are in accordance with a similar study showing the requirement of the stable NP of influenza A virus in DNA tattoo vaccination in order to induce effective immune responses (11). Other lines of evidence indicate cross-presentation of DNA-encoded antigens after DNA immunization. In a cellular transfer model, it was shown that donor-restricted immune responses were still initiated in mice if APCs were injected as long as 21 days after DNA immunization (14). It is therefore likely that antigens expressed by other cell types were acquired and immune responses induced via cross-presentation. Strong evidence for a role of cross-presentation also comes from two studies showing efficient CD8+ T-cell priming in mice that were immunized with DNA constructs expressing the antigen under a tissue-specific promoter not active in APCs (13, 29). Assuming tight promoter regulation, these studies show that DNA-mediated immune responses can be entirely independent of direct presentation. Since our experiments did not show total inhibition of immune responses after targeting of the LCMV NP for rapid degradation, we conclude from our experiments that cross-presentation as well as direct presentation contribute to the overall priming of CD8+ T cells after DNA immunization. This idea fits well with recent studies showing a redundant function of direct and cross-presentation in similar approaches (36, 48). Depending on the nature and dose of antigen, both pathways seem to be possible (25).
Compared to DNA vaccines, recombinant VV vaccines have the advantage of showing significantly higher transfection/infection efficiency that leads to the expression and presentation of recombinant antigens. Similar to the situation described above, the extent to which direct and/or cross-priming contributes to the initial CD8+ T-cell activation after infection with VV has not been elucidated definitively. We generated recombinant VV strains that either express the long-lived wild-type form of the LCMV NP or short-lived fusion proteins of NP with ubiquitin or Fat10. Similar to our DNA data, we found enhanced direct presentation in vitro if cells were infected with recombinant VV strains expressing the short-lived fusion proteins (Fig. 3). Due to efficient infection of VV, we could analyze various murine cell types that all showed similar results. Interestingly, cells infected with recombinant VV expressing Fat10-NP resulted in the highest level of CD8+ T-cell activation in a number of cell lines. This is in line with the rapid degradation kinetic of Fat10-NP (Fig. 1). The fact that destabilization of antigens can lead to increased antigen presentation in vitro was already shown in an earlier publication by reducing the half-life of an influenza virus NP via the N-end rule (46).
Interestingly, our studies indicated different antigen preferences for CD8+ T-cell priming in vivo. The recombinant VV strain expressing the long-lived wild-type form of the NP was found to be most potent in inducing CD8+ T cells in primary as well as memory responses (Fig. 4). In contrast to this, the short-lived ubiquitin-NP induced only weak responses. These findings are especially striking because there was no difference in viral replication in vivo and in responses to the VV epitope B8R. Moreover, we could not exclude that the vaccinia virus strain used in this study infected dendritic cells, as infection of mice with rVV-GFP led to prominent GFP expression, especially of MHC-II+ CD11c+ DCs but also of MHC-II+ CD11c− cells (Fig. 5). This result strongly suggests that DCs were directly infected with rVV-GFP, because an endocytic uptake of GFP+ cells would lead to rapid degradation of GFP in endolysosomes of DC and quenching of the acid-sensitive fluorescence of GFP. It appears that a direct infection of DC with VV and synthesis of rVV-encoded proteins in them nevertheless does not lead to a superior priming of NP118- and NP396-specific T cells in vivo by rapidly degraded Ub-NP or FAT10-NP. It is possible that rVV-infected DC contribute little to T-cell priming, because VV lyses cells shortly after infection, which may hinder T-cell priming, which is known to rely on a long-lasting DC–T-cell interaction in vivo. Alternatively, VV replication may limit the priming potential of DC by downregulating factors needed for T-cell stimulation, like cytokines or costimulatory receptors.
In line with our data on DNA vaccination, our results clearly favor a dominant, if not exclusive, role of cross-presentation for MHC class-restricted immune responses after VV infection. Interestingly, a very similar study with equal results was performed using a recombinant VV expressing either the long-lived tyrosinase or a short-lived ubiquitin fusion (18). Also in this study, the wild-type protein was much more potent in inducing immune responses than its short-lived derivative. In a different report, coexpression of inhibitory proteins of cytomegalovirus that block direct presentation reduced responses to a recombinant antigen (6). Therefore, the authors argued that cross-presentation of viral antigens is a mechanism to overcome viral evasion strategies in case direct presentation is inhibited. Rapid degradation of antigen was also shown to inhibit CD8+ T-cell responses against other viruses, like vesicular stomatitis virus, Sendai virus (28), and others (43).
There are very few studies that are in favor of direct presentation as the dominant mechanism in VV infection. Surprisingly, proteasomal targeting of HIV antigens was shown to increase CD8+ T-cell induction (45), which is in contrast to the previously mentioned studies and our results. Other reports showed an importance of direct presentation (6, 42). A study using the HIV protein Gag showed increased presentation of unstable antigen in vitro, but differences between long- and short-lived antigens in vivo have not been detected (49). The reason for differences between our and the aforementioned studies are unclear but might be due to HIV-specific antigen properties. Overall, we conclude that antigen cross-presentation is an important mechanism to induce CD8+ T-cell responses after VV infection, an idea that is in line with studies on other cytopathic viruses (41). There are ideas to increase immunogenicity of recombinant vaccines by expression of minigenes (2, 50). However, we cannot support this idea with our data. Additionally, recombinant VVs expressing minigenes of CD8+ T-cell epitopes were shown to fail in inducing immune responses via cross-presentation in vivo, while direct presentation in vitro was efficient (33). The few exceptions that appear in the literature most probably are due to special properties of the antigen used and should not be overestimated compared to the increasing number of reports in favor of cross-presentation.
Finally, it has to be discussed to what extent Fat10 fusion to the LCMV NP alters its potential in antigen presentation and CD8+ T-cell priming. In our in vitro transfection experiments, we detected better direct presentation for the Fat10-NP construct than the fusion protein ubiquitin-NP. Considering this fact and the rapid degradation rate of Fat10-NP, one could assume a loss of immunogenicity in DNA vaccination and recombinant VV infection that is similar to what we observed for ubiquitin-NP. However, in all of our in vivo experiments, the Fat10-NP constructs showed an intermediate phenotype. In both systems, the NP-specific responses of Fat10-NP were stronger than those of the virus expressing ubiquitin-NP. Memory responses in BALB/c mice showed the highest level of T-cell activation for the Fat10-NP-expressing VV. This observation cannot be explained yet. However, although viral replication was similar for the Fat10-NP-expressing strain, we observed significantly higher responses to the vaccinia epitope B8R. Therefore, the enhanced presentation could be due to effects not related to the Fat10 construct but to uncharacterized differences in the viral strains.
Apart from this difference, one could speculate that Fat10-mediated degradation leads to a different pathway from those occurring after modification with ubiquitin. It was shown in numerous publications that not only stable proteins but also stabilized peptides in association with cellular factors like heat shock proteins (HSPs) can be an efficient source of antigen for cross-presentation (10). Here, we speculate that intermediates of Fat10-NP but not ubiquitin-NP are stabilized to represent an antigen source for cross-presentation. This way, the IFN-γ-inducible Fat10 would allow the stabilization of antigens during inflammation that would normally be prone to ubiquitinylation and degraded into small peptides that have no access to the cross-presentation pathway. However, we have no evidence for such a pathway, the discovery of which could provide novel insights into the biology of Fat10 to further distinguish its function from that of ubiquitin.
The results presented in this report provide strong evidence that stable antigens should be preferentially used to achieve robust immune responses after DNA vaccination and infection with recombinant VV. Our results are in accordance with other studies showing the importance of cross-presentation in these systems and should be taken into account for the development of novel vaccination strategies that aim to improve immunogenicity.
ACKNOWLEDGMENTS
We thank Peter Öhlschläger for providing the electroporation device (in collaboration with Ichor Medical Systems, San Diego, CA) and Gerardo Alvarez for assistance with DNA immunization. Michael Basler is acknowledged for help with recombinant vaccinia viruses.
This work was supported by International Graduate School IRTG1331 and the German Research Foundation (DFG), grant number GR 1517/10-1.
Footnotes
Published ahead of print 3 July 2012
REFERENCES
- 1. Akbari O, et al. 1999. DNA vaccination: transfection and activation of dendritic cells as key events for immunity. J. Exp. Med. 189:169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. An LL, Whitton JL. 1997. A multivalent minigene vaccine, containing B-cell, cytotoxic T-lymphocyte, and Th epitopes from several microbes, induces appropriate responses in vivo and confers protection against more than one pathogen. J. Virol. 71:2292–2302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Basler M, Groettrup M. 2007. No essential role for tripeptidyl peptidase II for the processing of LCMV-derived T cell epitopes. Eur. J. Immunol. 37:896–904 [DOI] [PubMed] [Google Scholar]
- 4. Basler M, Youhnovski N, van den Broek M, Przybylski M, Groettrup M. 2004. Immunoproteasomes down-regulate presentation of a subdominant T cell epitope from lymphocytic choriomeningitis virus. J. Immunol. 173:3925–3934 [DOI] [PubMed] [Google Scholar]
- 5. Basta S, Alatery A. 2007. The cross-priming pathway: a portrait of an intricate immune system. Scand. J. Immunol. 65:311–319 [DOI] [PubMed] [Google Scholar]
- 6. Basta S, Chen WS, Bennink JR, Yewdell JW. 2002. Inhibitory effects of cytomegalovirus proteins US2 and US11 point to contributions from direct priming and cross-priming in induction of vaccinia virus-specific CD8(+) T cells. J. Immunol. 168:5403–5408 [DOI] [PubMed] [Google Scholar]
- 7. Basta S, Stoessel R, Basler M, van den Broek M, Groettrup M. 2005. Cross-presentation of the long-lived lymphocytic choriomeningitis virus nucleoprotein does not require neosynthesis and is enhanced via heat shock proteins. J. Immunol. 175:796–805 [DOI] [PubMed] [Google Scholar]
- 8. Belakova J, Horynova M, Krupka M, Weigl E, Raska M. 2007. DNA vaccines: are they still just a powerful tool for the future? Arch. Immunol. Ther. Exp. 55:387–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bevan MJ. 1976. Cross-priming for a secondary cytotoxic response to minor H antigens with H-2 congenic cells which do not crossreact in cytotoxic assay. J. Exp. Med. 143:1283–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Binder RJ. 2006. Heat shock protein vaccines: from bench to bedside. Int. Rev. Immunol. 25:353–375 [DOI] [PubMed] [Google Scholar]
- 11. Bins AD, Wolkers MC, van den Boom MD, Haanen JB, Schumacher TN. 2007. In vivo antigen stability affects DNA vaccine immunogenicity. J. Immunol. 179:2126–2133 [DOI] [PubMed] [Google Scholar]
- 12. Carvalho JA, Rodgers J, Atouguia J, Prazeres DM, Monteiro GA. 2010. DNA vaccines: a rational design against parasitic diseases. Expert Rev. Vaccines 9:175–191 [DOI] [PubMed] [Google Scholar]
- 13. Cho JH, Youn JW, Sung YC. 2001. Cross-priming as a predominant mechanism for inducing CD8(+) T cell responses in gene gun DNA immunization. J. Immunol. 167:5549–5557 [DOI] [PubMed] [Google Scholar]
- 14. Doe B, Selby M, Barnett S, Baenziger J, Walker CM. 1996. Induction of cytotoxic T lymphocytes by intramuscular immunization with plasmid DNA is facilitated by bone marrow-derived cells. Proc. Natl. Acad. Sci. U. S. A. 93:8578–8583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Donnelly JJ, Liu MA, Ulmer JB. 2000. Antigen presentation and DNA vaccines. Am. J. Respir. Crit. Care Med. 162:S190–S193 [DOI] [PubMed] [Google Scholar]
- 16. Donohue KB, et al. 2006. Cross-priming utilizes antigen not available to the direct presentation pathway. Immunology 119:63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fu TM, et al. 1997. Priming of cytotoxic T lymphocytes by DNA vaccines: requirement for professional antigen presenting cells and evidence for antigen transfer from myocytes. Mol. Med. 3:362–371 [PMC free article] [PubMed] [Google Scholar]
- 18. Gasteiger G, Kastenmuller W, Ljapoci R, Sutter G, Drexler I. 2007. Cross-priming of cytotoxic T cells dictates antigen requisites for modified vaccinia virus Ankara vector vaccines. J. Virol. 81:11925–11936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Graham FL, Smiley J, Russell WC, Nairn R. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 36:59–74 [DOI] [PubMed] [Google Scholar]
- 20. Groettrup M, Khan S, Schwarz K, Schmidtke G. 2001. Interferon-γ inducible exchanges of 20S proteasome active site subunits: why? Biochimie 83:367–372 [DOI] [PubMed] [Google Scholar]
- 21. Groettrup M, Pelzer C, Schmidtke G, Hofmann K. 2008. Activating the ubiquitin family: UBA6 challenges the field. Trends Biochem. Sci. 33:230–237 [DOI] [PubMed] [Google Scholar]
- 22. Groettrup M, et al. 1995. The interferon-γ-inducible 11S regulator (PA28) and the LMP2/LMP7 subunits govern the peptide production by the 20S proteasome in vitro. J. Biol. Chem. 270:23808–23815 [DOI] [PubMed] [Google Scholar]
- 23. Groettrup M, et al. 2001. Structural plasticity of the proteasome and its function in antigen processing. Crit. Rev. Immunol. 21:339–359 [PubMed] [Google Scholar]
- 24. Gurunathan S, Klinman DM, Seder RA. 2000. DNA vaccines: immunology, application, and optimization. Annu. Rev. Immunol. 18:927–974 [DOI] [PubMed] [Google Scholar]
- 25. Heath WR, et al. 2004. Cross-presentation, dendritic cell subsets, and the generation of immunity to cellular antigens. Immunol. Rev. 199:9–26 [DOI] [PubMed] [Google Scholar]
- 26. Kutzler MA, Weiner DB. 2004. Developing DNA vaccines that call to dendritic cells. J. Clin. Investig. 114:1241–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu MA. 2011. DNA vaccines: an historical perspective and view to the future. Immunol. Rev. 239:62–84 [DOI] [PubMed] [Google Scholar]
- 28. Lizee G, et al. 2003. Control of dendritic cell cross-presentation by the major histocompatibility complex class I cytoplasmic domain. Nat. Immunol. 4:1065–1073 [DOI] [PubMed] [Google Scholar]
- 29. Loirat D, et al. 1999. Muscle-specific expression of hepatitis B surface antigen: no effect on DNA-raised immune responses. Virology 260:74–83 [DOI] [PubMed] [Google Scholar]
- 30. Mackett M, Smith GL, Moss B. 1982. Vaccinia virus: a selectable eukaryotic cloning and expression vector. Proc. Natl. Acad. Sci. U. S. A. 79:7415–7419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Moss B. 2011. Smallpox vaccines: targets of protective immunity. Immunol. Rev. 239:8–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Moss B, Elroy-Stein O, Mizukami T, Alexander WA, Fuerst TR. 1990. Product review. New mammalian expression vectors. Nature 348:91–92 [DOI] [PubMed] [Google Scholar]
- 33. Norbury CC, et al. 2004. CD8(+)T cell cross-priming via transfer of proteasome substrates. Science 304:1318–1321 [DOI] [PubMed] [Google Scholar]
- 34. Pavelic V, Matter MS, Mumprecht S, Breyer I, Ochsenbein AF. 2009. CTL induction by cross-priming is restricted to immunodominant epitopes. Eur. J. Immunol. 39:704–716 [DOI] [PubMed] [Google Scholar]
- 35. Porgador A, et al. 1998. Predominant role for directly transfected dendritic cells in antigen presentation to CD8+ T cells after gene gun immunization. J. Exp. Med. 188:1075–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Prasad SA, Norbury CC, Chen W, Bennink JR, Yewdell JW. 2001. Cutting edge: recombinant adenoviruses induce CD8 T cell responses to an inserted protein whose expression is limited to nonimmune cells. J. Immunol. 166:4809–4812 [DOI] [PubMed] [Google Scholar]
- 37. Ramirez MC, Sigal LJ. 2004. The multiple routes of MHC-I cross-presentation. Trends Microbiol. 12:204–207 [DOI] [PubMed] [Google Scholar]
- 38. Rodriguez F, Zhang J, Whitton JL. 1997. DNA immunization: ubiquitination of a viral protein enhances cytotoxic T-lymphocyte induction and antiviral protection but abrogates antibody induction. J. Virol. 71:8497–8503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schulz M, et al. 1989. Major histocompatibility complex-dependent T cell epitopes of lymphocytic choriomeningitis virus nucleoprotein and their protective capacity against viral disease. Eur. J. Immunol. 19:1657–1667 [DOI] [PubMed] [Google Scholar]
- 40. Schwarz K, et al. 2000. Overexpression of the proteasome subunits LMP2, LMP7, and MECL-1 but not PA28α/β enhances the presentation of an immunodominant lymphocytic choriomeningitis virus T cell epitope. J. Immunol. 165:768–778 [DOI] [PubMed] [Google Scholar]
- 41. Shen LJ, Rock KL. 2004. Cellular protein is the source of cross-priming antigen in vivo. Proc. Natl. Acad. Sci. U. S. A. 101:3035–3040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shen X, Wong SB, Buck CB, Zhang J, Siliciano RF. 2002. Direct priming and cross-priming contribute differentially to the induction of CD8+ CTL following exposure to vaccinia virus via different routes. J. Immunol. 169:4222–4229 [DOI] [PubMed] [Google Scholar]
- 43. Sigal LJ, Crotty S, Andino R, Rock KL. 1999. Cytotoxic T-cell immunity to virus-infected non-haematopoietic cells requires presentation of exogenous antigen. Nature 398:77–80 [DOI] [PubMed] [Google Scholar]
- 44. Talavera A, Rodriguez JM. 1992. Isolation and handling of recombinant vaccinia viruses. Methods Mol. Biol. 8:235–248 [DOI] [PubMed] [Google Scholar]
- 45. Tobery TW, Siliciano RF. 1997. Targeting of HIV-1 antigens for rapid intracellular degradation enhances cytotoxic T lymphocyte (CTL) recognition and the induction of de novo CTL responses in vivo after immunization. J. Exp. Med. 185:909–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Townsend A, et al. 1988. Defective presentation to class I-restricted cytotoxic T lymphocytes in vaccinia-infected cells is overcome by enhanced degradation of antigen. J. Exp. Med. 168:1211–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ulmer JB, Otten GR. 2000. Priming of CTL responses by DNA vaccines: direct transfection of antigen presenting cells versus cross-priming. Dev. Biol. 104:9–14 [PubMed] [Google Scholar]
- 48. Wolkers MC, Stoetter G, VythDreese FA, Schumacher TNM. 2001. Redundancy of direct priming and cross-priming in tumor-specific CD8(+) T cell responses. J. Immunol. 167:3577–3584 [DOI] [PubMed] [Google Scholar]
- 49. Wong SB, Buck CB, Shen X, Siliciano RF. 2004. An evaluation of enforced rapid proteasomal degradation as a means of enhancing vaccine-induced CTL responses. J. Immunol. 173:3073–3083 [DOI] [PubMed] [Google Scholar]
- 50. Xiang R, et al. 2000. An autologous oral DNA vaccine protects against murine melanoma. Proc. Natl. Acad. Sci. U. S. A. 97:5492–5497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yewdell JW, Anton LC, Bennink JR. 1996. Defective ribosomal products (DRiPs): a major source of antigenic peptides for MHC class I molecules. J. Immunol. 157:1823–1826 [PubMed] [Google Scholar]