Abstract
Open reading frame 45 (ORF45) of Kaposi's sarcoma-associated herpesvirus (KSHV) is an immediate-early and tegument protein that plays critical roles in antagonizing host antiviral responses. We have previously shown (Zhu et al, Proc. Natl. Acad. Sci. U. S. A., 99:5573–5578, 2002) that ORF45 suppresses activation of interferon regulatory factor 7 (IRF7), a crucial regulator of type I interferon gene expression, by blocking its virus-induced phosphorylation and nuclear accumulation. We report here further characterization of the mechanisms by which ORF45 inhibits IRF7 phosphorylation. In most cell types, IRF7 is phosphorylated and activated by IKKε and TBK1 after viral infection. We found that phosphorylation of IRF7 on Ser477 and Ser479 by IKKε or TBK1 is inhibited by ORF45. The inhibition is specific to IRF7 because phosphorylation of its close relative IRF3 is not affected by ORF45, implying that ORF45 does not inactivate the kinases directly. In fact, we found that ORF45 is phosphorylated efficiently on Ser41 and Ser162 by IKKε and TBK1. We demonstrated that ORF45 competes with the associated IRF7 and inhibits its phosphorylation by IKKε or TBK1 by acting as an alternative substrate.
INTRODUCTION
Type I interferons (IFNs) constitute the first line of host immune defense against viral infection. Their expression is tightly regulated through interferon regulatory factors (IRFs), particularly IRF3 and IRF7 (20, 45). Host cells sense invading viruses with pathogen recognition receptors, such as membrane-bound Toll-like receptors, cytosolic retinoic acid-inducible gene I (RIG-I)-like receptors, and others, which trigger a variety of cellular signaling pathways that converge on the activation of IRF3 and IRF7 (26, 43). Once activated, these IRFs bind to the promoters of target genes and induce expression of IFNs and IFN-stimulated genes (ISGs), which collectively lead to the establishment of antiviral states (21).
Activation of IRF3 and IRF7 depends on phosphorylation of their C-terminal serine residues upon viral infection. Phosphorylation of IRF3 and IRF7 causes conformation changes, homo- and/or heterodimerization among them, nuclear translocation, cooperation with cofactors, and ultimately activation of transcription of type I IFN genes (19, 21, 35, 37, 41). Although IRF7 and IRF3 are highly similar in their primary protein structures and modes of activation, they are not functionally redundant and differ in transcription profiles. IRF3 is ubiquitously expressed, but IRF7 is expressed at low levels in most cells (with the notable exception of the professional IFN-producing plasmacytoid dendritic cells), and its expression is upregulated by IFN and viral infection through a positive feedback loop (21, 49). Therefore, despite its low expression in most cell types, IRF7 plays crucial roles in regulation of type I IFN gene expression, as has been revealed by studies with IRF7−/− knockout mice (22). Its critical role in host antiviral immune defense is also reflected by the variety of viruses, including Kaposi's sarcoma-associated herpesvirus (KSHV), that encode proteins to counter the actions of IRF7 (5, 6, 25, 60, 66).
KSHV, also known as human herpesvirus 8 (HHV-8), is etiologically associated with a number of human cancers, such as Kaposi's sarcoma (KS), primary effusion lymphoma, and multicentric Castleman's disease (7, 16, 17). It has two alternative life cycles, latency and lytic replication. Latency is a dormant state during which only a few viral genes are expressed, whereas the lytic cycle leads to expression of a full panel of viral genes, assembly and release of progeny virus particles, and de novo infection of other cells (15, 17, 55). KSHV maintains latency in the majority of spindle cells in the KS lesion, whereas lytic replications occur in a small proportion of infected cells (56). Because viral infection in general comes under host immune surveillance, both phases of the KSHV lytic infectious cycle, primary de novo infection and reactivation from latency, would elicit host antiviral immune responses. Evasion of these responses is therefore crucial for persistent infection with KSHV.
To combat the host antiviral responses, KSHV has evolved elaborate mechanisms to counter the IFN-dependent antiviral defenses (11, 32, 46). We have previously shown that ORF45 of KSHV interacts with IRF7 and suppresses it activation (66). KSHV ORF45 is an immediate-early protein that is expressed very soon after viral lytic reactivation (65). It is phosphorylated and has a distinct subcellular localization (31, 69). ORF45 is also a tegument protein that is delivered into infected cells as a constituent of viral particles upon infection (64, 68, 69). Its unique temporal and spatial expression gives it a unique advantage in combating host antiviral responses during the very early stages of infection and allows it to suppress IRF7 activation, block the positive-feedback loop, and prevent expression of IFNs and ISGs and therefore establishment of antiviral states. Although several other viral factors, such as RTA (replication and transcription activator) and vIRFs (viral interferon regulatory factors), are also known to interfere with IFN induction or IFN-mediated signaling (30, 32, 46, 64), we demonstrated that wild-type KSHV induces little antiviral response, whereas the ORF45-null mutant triggers a greater response (68), suggesting that ORF45 is a potent factor in antagonizing host antiviral defenses.
To improve our understanding of the potency of ORF45 in antagonizing host antiviral responses, we defined the mechanisms by which ORF45 suppresses IRF7 activation. Previously, we had demonstrated that ORF45 blocks virus-mediated IRF7 phosphorylation, but the underlying mechanisms remained unclear (66). The virus-activated kinases responsible for phosphorylation of IRF7 and IRF3 have been identified as IKKε and TBK1 (8, 10, 13, 52). In the study we report here, we found that ORF45 inhibited phosphorylation of IRF7 but not IRF3 by these kinases. ORF45 apparently does not inactivate IKKε or TBK1 directly. In fact, it is phosphorylated by these two kinases. We have demonstrated that ORF45 is phosphorylated more efficiently than IRF7 by IKKε and TBK1 and that it inhibits IRF7 phosphorylation competitively by acting as an alternative substrate.
MATERIALS AND METHODS
Cells and reagents.
HEK293T cells and HEK293-TLR3 cells (kindly provided by Katherine Fitzgerald) were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, and necessary antibiotics. iSLK cells expressing doxycycline-inducible replication and transcription activator (RTA) (encoded by a key KSHV gene that is essential and sufficient for initiating the lytic replication program from latency) were cultured and maintained as previously described (40). BAC36-wt, a bacterial artificial chromosome clone that carries the entire KSHV genome (63), and BAC36-stop45, an ORF45-null mutant that carries a premature stop codon in the ORF45 coding region, were described previously (67). iSLK/BAC36 and iSLK/BAC36-stop45 were derived from iSLK cells carrying latent BAC36-wt or BAC36-stop45 genomes (27). The mouse anti-Flag, anti-HA, and anti-β-actin antibodies, 12-O-tetradecanoylphorbol-13-acetate (TPA), sodium butyrate, EZview red anti-Flag M2 and anti-HA affinity beads, 3×Flag peptides, and 3×HA peptides were purchased from Sigma. Rabbit anti-IRF7 and anti-IRF3 were purchased from Santa Cruz Biotech. Rabbit anti-IKKε and anti-pIRF3 (Ser396) were purchased from Cell Signaling. Mouse anti-pIRF7 (Ser477/479) was initially supplied by Rongtuan Lin and John Hiscott and later custom-ordered from BD Biosciences. Mouse anti-phospho-serine/threonine antibody (catalog no. 612548) was also purchased from BD Biosciences. Rabbit anti-TBK1 antibody was purchased from Millipore. Anti-ORF45 antibody was described previously (64). Rat anti-LANA antibody was purchased from Advanced Biotechnologies.
Plasmids.
Plasmid pCR3.1-ORF45 was described previously (66). Plasmids expressing Flag-IRF7, Flag-IRF3, and Flag-ORF45 were generated by subcloning of the coding sequence into pCMV-TAG3 (Stratagene). Plasmids pKH3-TBK1 and pKH3-IKKε were generated by cloning of the PCR-amplified cDNA fragments into pKH3 vector. Plasmids encoding glutathione S-transferase (GST) fusion proteins of ORF45 amino acids (aa) 1 to 115, aa 116 to 237, aa 238 to 332, and aa 333 to 407 and of IRF7 aa 452 to 503 were generated by insertion of PCR-amplified fragments into pGEX-5X (Pharmacia). Point mutations were generated with a QuikChange mutagenesis kit (Stratagene). To generate ORF45 or IRF7 short peptides fused to GST as kinase substrates, we annealed pairs of oligonucleotides (synthesized at Integrated DNA Technologies) and cloned them into pGEX-5X vector for protein expression in Escherichia coli. Some of the annealed oligonucleotides were also cloned into pEBG vector for expression in mammalian cells. The oligonucleotide sequences are as follows: ORF45-S41 sense, GATCCCACCTCCACCTCTTTCATCACTTCCAGGATTTGGCAGGC; ORF45-S41 antisense, TCGAGCCTGCCAAATCCTGGAAGTGATGAAAGAGGTGGAGGTGG; ORF45-S162 sense, GATCACGGCGGAGTCCGAGGCGTCCATGGGATGGGTTAGTCAGC; ORF45-S162 antisense, TCGAGCTGACTAACCCATCCCATGGACGCCTCGGACTCCGCCGT; ORF45-S41A sense, GATCCCACCTCCACCTCTTTCAGCACTTCCAGGATTTGGCAGGC; ORF45-S41A antisense, TCGAGCCTGCCAAATCCTGGAAGTGCTGAAAGAGGTGGAGGTGG; ORF45-S162A sense, GATCACGGCGGAGTCCGAGGCGGCCATGGGATGGGTTAGTCAGC; ORF45-S162A antisense, TCGAGCTGACTAACCCATCCCATGGCCGCCTCGGACTCCGCCGT; ORF45-S41/162 sense, GATCCCACCTCCACCTCTTTCATCACTTCCAGGATTTGGCAGGACGGCGGAGTCCGAGGCGTCCATGGGATGGGTTAGTCAGC; ORF45-S41/162 antisense, TCGAGCTGACTAACCCATCCCATGGACGCCTCGGACTCCGCCGTCCTGCCAAATCCTGGAAGTGATGAAAGAGGTGGAGGTGG; IRF7-S477/479 sense, GATCTCTTCCCTGGATAGCAGCAGCCTCAGCCTCTGCCTGTCCAGCGCCC; and IRF7-S477/479 antisense, TCGAGGGCGCTGGACAGGCAGAGGCTGAGGCTGCTGCTATCCAGGGAAGA. All clones were sequenced for assurance of accuracy.
Immunoprecipitation (IP).
Forty-eight hours after transfection, HEK293T cells were washed with cold phosphate-buffered saline (PBS) and lysed with whole-cell lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% NP-40, 1 mM sodium orthovanadate [Na3VO4], 40 mM β-glycerophosphate, 1 mM sodium fluoride, 10% glycerol, 5 mM EDTA, 5 μg/ml of aprotinin, 5 μg/ml of leupeptin, 5 mM benzamidine, and 1 mM phenylmethylsulfonyl fluoride). Cell lysates were centrifuged at 10,000 × g for 10 min at 4°C and incubated with EZview red anti-Flag M2 or anti-HA beads for 4 h or overnight at 4°C. After washing with lysis buffer and then TBS (50 mM Tris-HCl [pH 7.4], 150 mM NaCl), proteins were eluted by incubation with 150 μg/ml 3×Flag peptide or 100 μg/ml 3×HA peptide in TBS for 1 h at 4°C and then analyzed by Western blotting.
Expression and preparation of recombinant proteins.
Full-length His-tagged ORF45 protein was expressed and purified from the baculovirus expression system as described previously (29, 66). GST fusion proteins were expressed and purified from bacteria. Briefly, E. coli BL21 cultures transformed with plasmids encoding GST or GST fusion proteins were induced with 200 μM isopropyl-β-d-thiogalactopyranoside for 3 h at room temperature. Cells were pelleted, washed, and resuspended in PBS. The cell suspension was sonicated, and Triton X-100 was added to a final concentration of 1%. After 30 min of incubation at 4°C with gentle agitation, cell debris was removed by centrifugation at 10,000 × g for 10 min. The supernatant was incubated with glutathione beads at 4°C overnight. After five washes with PBS, GST proteins were eluted with 10 mM reduced glutathione–50 mM Tris-HCl, pH 8.0. The eluates were dialyzed in buffer A150 (25 mM Tris-Cl [pH 7.5], 1 mM EDTA, 0.1% NP-40, 10% glycerol, and 150 mM NaCl) overnight at 4°C. The purity was assessed by Coomassie brilliant blue staining of proteins separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The protein concentration was determined with a bicinchoninic acid protein assay kit (Pierce). The purified GST proteins were divided into aliquots and stored at −80°C until use.
In vitro kinase assay.
HEK293T cells seeded into 100-mm dishes were transfected with 10 μg of HA-tagged IKKε or TBK1 expression plasmids by the calcium phosphate method. Two days after transfection, whole-cell lysates were prepared, and HA-IKKε or HA-TBK1 proteins were immunoprecipitated with 50 μl of EZview red affinity beads. After two washes with lysis buffer and three washes with TBS buffer, the immunoprecipitated beads were resuspended in 100 μl of TBS plus 1 mM phenylmethylsulfonyl fluoride, 1 mM Na3VO4, and 1× protease inhibitor cocktail (Roche). The in vitro kinase reaction was performed by incubation of 5 μl of IP complexes with 2.5 μg GST substrates in 25 μl of 1× kinase assay buffer (25 mM HEPES [pH 7.5], 50 mM NaCl, 20 mM β-glycerophosphate, 1 mM dithiothreitol, 20 mM MgCl, 1 mM Na3VO4, 1 μg/ml bovine serum albumin, 20 μM ATP, and 5 μCi [γ-32P]ATP). The reaction mixtures were kept at 30°C for 30 min, and reactions were stopped by the addition of 2× Laemmli sample buffer. After fractionation of samples by SDS-PAGE, the gel was dried and exposed to X-ray film or analyzed with a PhosphorImager. The immunoprecipitated HA-TBK1 and HA-IKKε proteins were processed for Western analysis with anti-HA antibody.
RNA interference (RNAi).
Small interfering RNAs (siRNAs) against human TBK1 or IKKε were purchased from Santa Cruz Biotech. Transfection of siRNAs was performed according to the manufacturer's protocols.
Luciferase reporter assays.
Luciferase assays were performed as previously described (33, 66). Briefly, HEK293T cells seeded in a 24-well plate were transfected by luciferase reporter and pRL-TK internal control plasmids with Lipofectamine 2000 transfection reagent (Invitrogen). Eight hours after transfection, each well was infected with 80 hemagglutinin units of Sendai viruses (purchased from Charles River). Dual-luciferase assays were performed 24 h after transfection. The relative luciferase activity was expressed in arbitrary units by normalization of firefly luciferase activity to Renilla luciferase activity. Data are the averages from three independent experiments, and error bars represent standard deviations.
Gel filtration chromatography.
The cells were harvested 2 days after transfection, washed with cold phosphate-buffered saline, resuspended in buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 30 mM NaF, 40 mM glycerophosphate, 5 mM EDTA, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride, 5 μg/ml aprotinin, 5 μg/ml leupeptin, and 5 μg/ml pepstatin A), and lysed by three rounds of freezing and thawing followed by 10 s of sonication in a microultrasonic cell disrupter. Cell lysates were clarified by ultracentrifugation at 100,000 × g for 1 h at 4°C. The supernatants were fractionated on a Superdex 200 HR 10/30 gel column as previously described (29).
RESULTS
KSHV ORF45 specifically inhibits IRF7 phosphorylation on Ser477 and Ser479 by IKKε and TBK1.
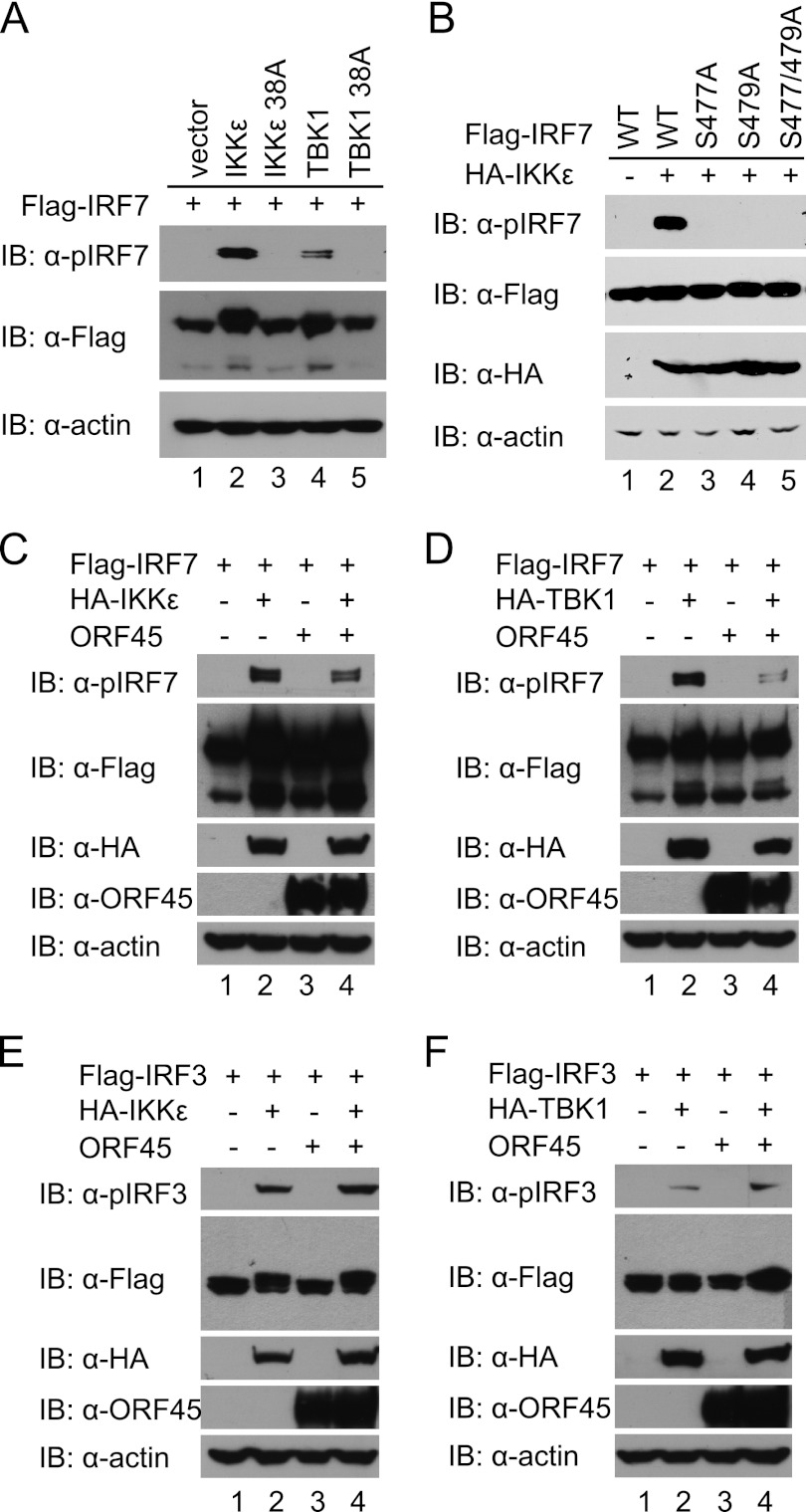
The latent IRF7 and IRF3 are activated by phosphorylation on the C-terminal serine/threonine residues by IKKε and TBK1 (8, 13, 52). Despite extensive efforts, the exact phosphorylation sites of human IRF3 by IKKε and TBK1 remain debatable, but substantial evidence supports Ser396 as the primary phosphorylation site (9, 12, 34, 39, 51). The sites of human IRF7 direct phosphorylation by IKKε and TBK1 were reported to be Ser477 and Ser479 (42). The results were confirmed by our own mutational and functional analyses (data not shown). We next used the phosphorylation-specific antibody anti-pIRF7 (Ser477/479) to examine IKKε- and TBK1-induced IRF7 phosphorylation in cells. We coexpressed IRF7 with IKKε or TBK1 in HEK293T cells and then used this antibody to detect the phosphorylated form of IRF7 by Western blotting. As shown in Fig. 1, a strong signal was detected in cells when the wild-type IRF7 was coexpressed with the wild-type IKKε or TBK1 but not in the kinase-dead mutants (Fig. 1A). Abolishment of the signal when either or both of Ser477 and Ser479 were changed to alanine attested to the specificity of this antibody (Fig. 1B). Importantly, coexpression of KSHV ORF45 inhibited the phosphorylation of IRF7 (Fig. 1C and D) but not that of IRF3 on Ser396 (Fig. 1E and F). These results suggest that ORF45 inhibits IKKε/TBK1-induced IRF7 phosphorylation on Ser477 and Ser479 in cells.
Fig 1.
KSHV ORF45 specifically inhibits phosphorylation of IRF7 on Ser477 and Ser479 by IKKε or TBK1. (A) Phosphorylation of human IRF7 on Ser477 and Ser479 by IKKε and TBK1. HEK293T cells were transfected with 1 μg of Flag-tagged IRF7 and 100 ng of HA-IKKε/TBK1 or their kinase-dead mutant (S38A) plasmids. Cell lysates were prepared at 48 h after transfection and immunoblotted with anti-pIRF7 (Ser477/479) antibody. (B) Specific recognition of IRF7 phosphorylation on Ser477 and Ser479 by the anti-pIRF7 (Ser477/479) antibody. HEK293T cells were transfected with 100 ng HA-IKKε plus 1 μg of Flag-tagged expression plasmids encoding wild-type IRF7 or the S477A, S479A, or S477/479A mutant. At 48 h after transfection, cell lysates were prepared and immunoblotted with antibodies as indicated. (C and D) ORF45 inhibits phosphorylation of IRF7 by IKKε and TBK1. HEK293T cells were transfected with 1 μg of Flag-IRF7 together with 100 ng of HA-IKKε (C) or HA-TBK1 (D) expression plasmid in the presence or absence of 2 μg of pCR3.1-ORF45. At 48 h after transfection, cell lysates were prepared and analyzed by Western blotting with antibodies as indicated. (E and F) ORF45 does not inhibit phosphorylation of IRF3 by IKKε or TBK1. HEK293T cells were transfected with 1 μg Flag-IRF3 together with 100 ng of HA-IKKε (E) or HA-TBK1 (F) expression plasmids in the presence or absence of 2 μg of pCR3.1-ORF45. Cell lysates were prepared and analyzed by Western blotting with the specified antibodies.
KSHV ORF45 does not interfere with interactions between IKKε/TBK1 kinases and their substrates.
Having confirmed that KSHV ORF45 inhibits IRF7 phosphorylation by IKKε and TBK1, we next wished to define the underlying mechanisms. We envision three possible scenarios: (i) ORF45 directly inhibits the kinase activity of IKKε and TBK1; (ii) ORF45 interferes with interactions between the kinases and substrates; (iii) ORF45 inhibits IRF7 phosphorylation specifically. Because ORF45 did not inhibit IKKε/TBK1-induced phosphorylation of IRF3 but inhibited that of IRF7 under the same conditions (compare Fig. 1E and F to Fig. 1C and D), the first scenario seemed unlikely.
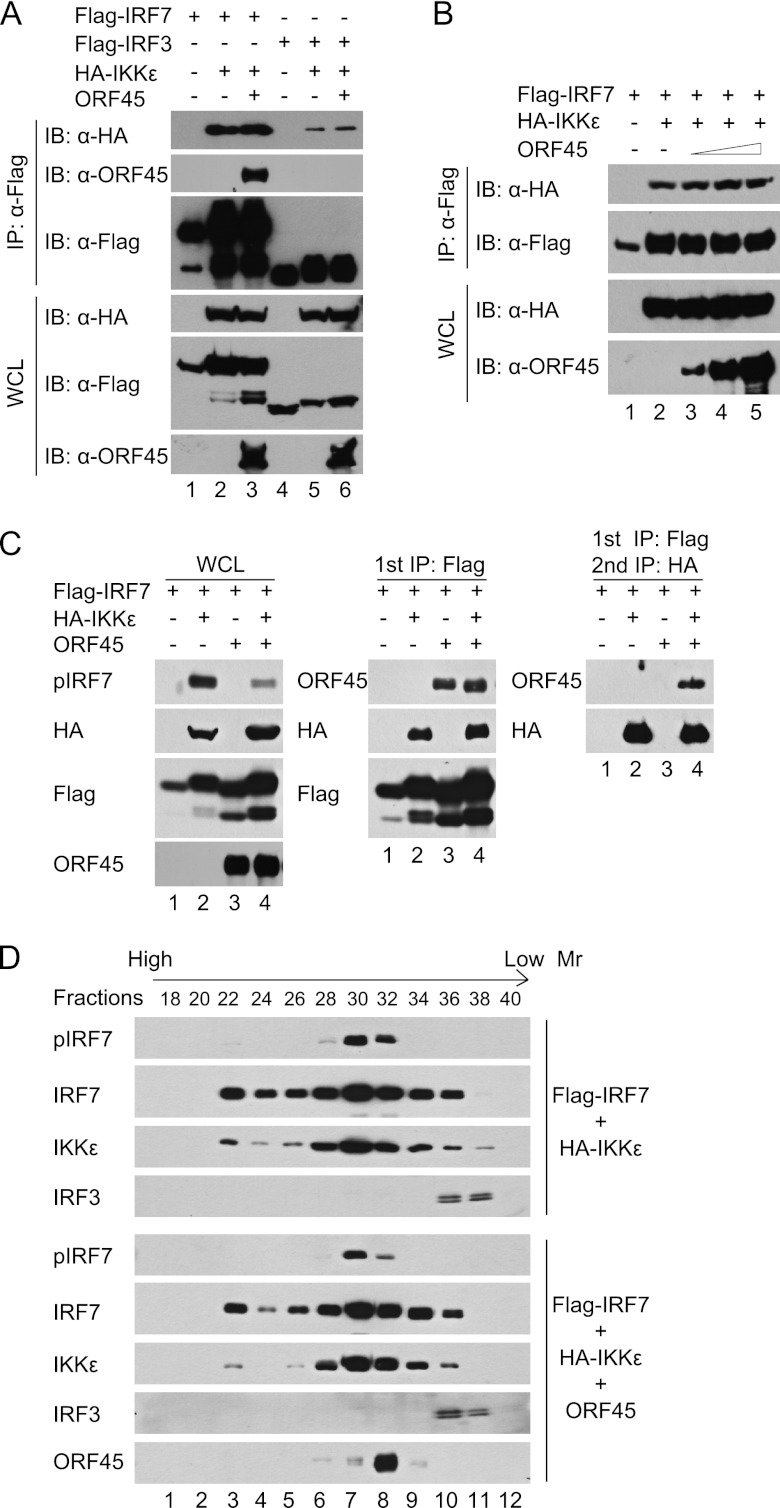
Next, to determine whether ORF45 interferes with interactions between the kinases and substrates, we expressed Flag-IRF7 or Flag-IRF3 in HEK293T cells with HA-IKKε in the presence or absence of ORF45, immunoprecipitated the cell lysates with anti-Flag affinity resins, and then analyzed the immunocomplexes by Western blotting. As shown in Fig. 2A, both IRF7 and IRF3 bound to IKKε, IRF7 appeared to bind better than IRF3, ORF45 specifically interacted with IRF7 but not IRF3, and ORF45 had little effect on the interactions between IKKε and either IRF. When ORF45 was expressed at higher levels, the interaction between IKKε and IRF7 remained unaffected (Fig. 2B). Similar results were obtained with TBK1 (data not shown). These results suggest that ORF45 does not interfere with the interactions between the kinases and their substrates.
Fig 2.
KSHV ORF45 does not interfere with interaction between the IKKε/TBK1 kinases and their substrates. (A and B) ORF45 does not interfere with the association between IRF7 and IKKε. Plasmids expressing Flag-IRF7 (5 μg), Flag-IRF3 (5 μg), HA-IKKε (1 μg), and ORF45 [5 μg (A) or 2, 5, and 10 μg (B)] were cotransfected into HEK293T cells in different combinations as indicated. At 48 h after transfection, cell lysates were prepared and subjected to immunoprecipitation with anti-Flag M2 affinity beads. The input whole-cell lysates (WCL) and immunoprecipitation (IP) complexes were analyzed by immunoblotting with anti-Flag, anti-HA, and anti-ORF45 antibodies. (C) ORF45, IRF7, and IKKε associate with each other in the same complexes in cells. HEK293T cells were transfected with Flag-IRF7-, HA-IKKε-, and ORF45-expressing plasmids in different combinations as indicated. Cell lysates were immunoprecipitated with anti-Flag M2 affinity beads and eluted with 3×Flag peptides. The eluates were then immunoprecipitated with anti-HA affinity beads and eluted with 3×HA peptides. Cell lysates and eluates were analyzed by immunoblotting with the specified antibodies. (D) ORF45 cofractionates with IRF7 and IKKε but not with IRF3. Lysates of HEK293T cells expressing Flag-IRF7 and HA-IKKε in the presence or absence of ORF45 were fractionated on a Superdex 200 HR 10/30 column. Fractions were analyzed by immunoblotting with the indicated antibodies.
If ORF45 does not prevent IKKε/TBK1 from interacting with IRF7, they all may associate with one another, because IRF7 interacts with both ORF45 (66) and IKKε/TBK1 (Fig. 2A). Indeed, when the lysates of cells expressing Flag-IRF7, HA-IKKε, and ORF45 were sequentially immunoprecipitated with anti-Flag and then with anti-HA affinity resins, ORF45 was detected not only in the eluates of the first anti-Flag IP but also in the second anti-HA IP complexes, indicating that all three proteins can reside in the same complexes. Similar results were obtained with TBK1 (data not shown).
To provide further evidence that ORF45, IRF7, and IKKε/TBK1 associate with each other in cells, we fractionated the lysates of cells overexpressing IRF7, IKKε, and ORF45. As shown in Fig. 2D, IRF7 cofractionated with IKKε in a broad range of fractions (fractions 22 to 36), whereas IRF3 was detected in a narrow range of fractions with lower molecular weights (fractions 36 to 38). Noticeably, ORF45 was detected in fractions 28 to 34, in which both IRF7 and IKKε were present but IRF3 were absent, suggesting that ORF45, IRF7, and IKKε associate with each other in complexes distinct from those containing IRF3. Furthermore, the levels of pIRF7 in fractions of lysates of ORF45-expressing cells were overall lower than those in the corresponding fractions of lysates of control cells expressing no ORF45. The effect was most obvious in fraction 32, in which the amount of ORF45 protein peaked. In sum, these results suggest that ORF45 does not disrupt the association between IRF7 and IKKε/TBK1; rather, they associate with each other in the same complexes.
KSHV ORF45 is phosphorylated by IKKε and TBK1 in vitro.
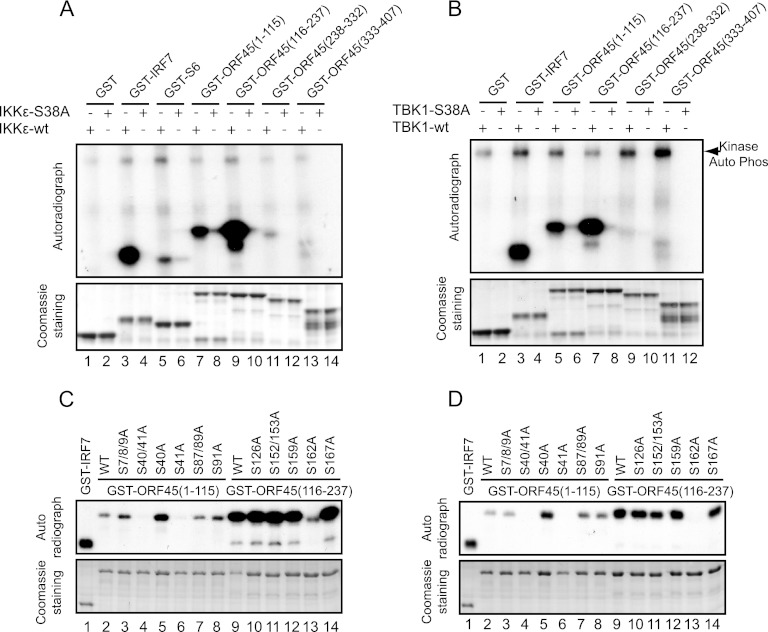
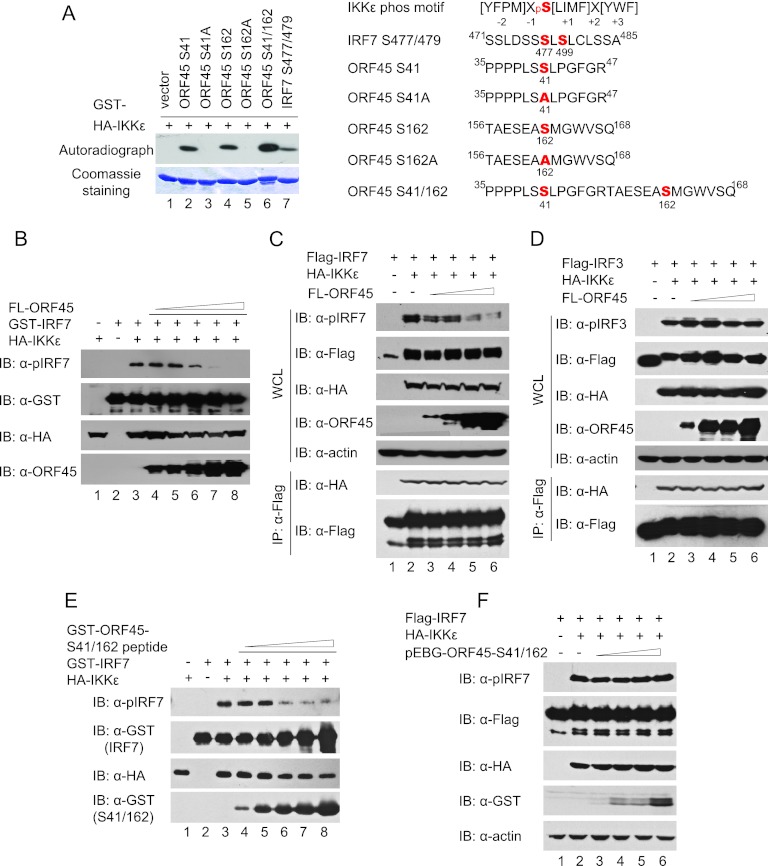
The full-length ORF45 protein purified from the baculovirus expression system can be self-phosphorylated in vitro (data not shown), presumably because of the tightly associated kinases such as the p90 ribosomal S6 kinase, as we recently reported (28, 29). When ORF45 protein was incubated with IKKε or TBK1, its phosphorylation was further increased (data not shown), suggesting that ORF45 is phosphorylated by these kinases. Because the full-length ORF45 was not expressed well in bacteria, we next divided ORF45 into four portions, aa 1 to 115, aa 116 to 237, aa 238 to 332, and aa 333 to 407, and expressed and purified each as a GST-tagged fusion protein in E. coli. In vitro kinase assays revealed that the first two fragments, aa 1 to 115 and aa 116 to 237, were phosphorylated efficiently by IKKε and TBK1 but not by the kinase-dead mutant (Fig. 3A and B). We further mapped the phosphorylation sites in these two fragments by introducing a series of serine-to-alanine mutations and used the GST fusion protein mutants as substrates in kinase assays. We found that replacement of Ser41 with alanine in the fragment from aa 1 to 115 abolished the phosphorylation, whereas mutation at other serine residues had minimal effect (Fig. 3C and D). Similarly, replacement of Ser162 with alanine in the fragment from aa 116 to 237 also abolished the phosphorylation (Fig. 3C and D). These results indicate that KSHV ORF45 is phosphorylated by IKKε and TBK1 on Ser41 and Ser162.
Fig 3.
KSHV ORF45 is phosphorylated by IKKε or TBK1 on Ser41 and Ser162. (A and B) KSHV ORF45 is phosphorylated by IKKε and TBK1. Four ORF45 fragments, GST-ORF45 (aa 1 to 115), GST-ORF45 (aa 116 to 237), GST-ORF45 (aa 238 to 332), and GST-ORF45 (aa 333 to 407), as well as GST-IRF7 (aa 452 to 503) and control GST-S6 and GST proteins were purified from Escherichia coli and used for in vitro kinase assays with IKKε (A) or TBK1 (B). (C and D) Identification of the primary sites of ORF45 phosphorylated by IKKε or TBK1. A series of serine-to-alanine mutant proteins of GST-ORF45 (aa 1 to 115) and GST-ORF45 (aa 116 to 237) were expressed and purified from E. coli. Equal amounts of these proteins were subjected to in vitro kinase assays with IKKε (C) or TBK1 (D).
KSHV ORF45 is phosphorylated by IKKε and TBK1 in cells.
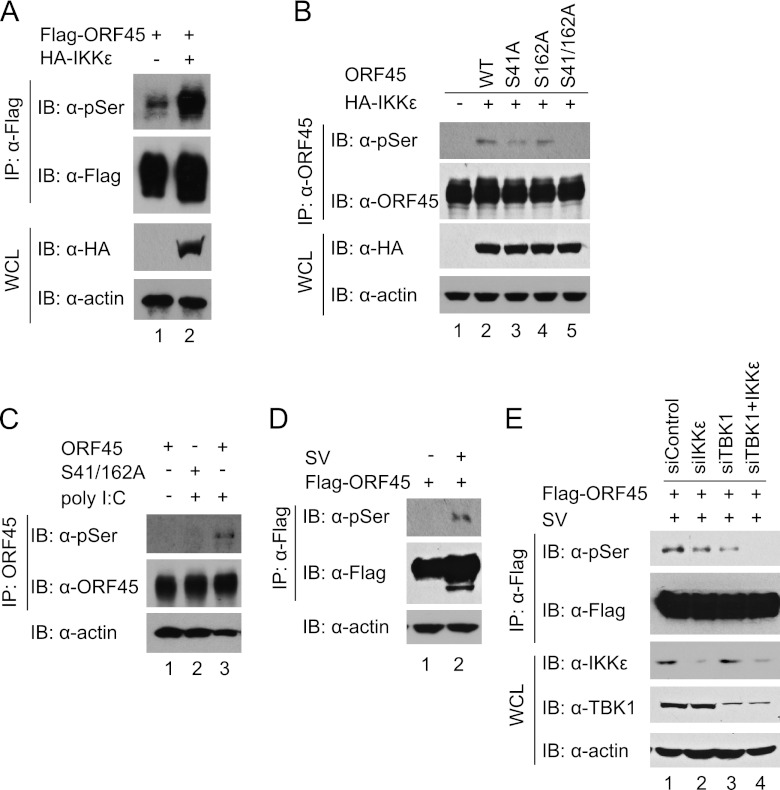
Having identified the phosphorylation sites of ORF45 by IKKε and TBK1, we further determined whether ORF45 is phosphorylated by these kinases in cells. Because a phosphorylation-specific antibody to ORF45 is not available, we used an anti-phospho-serine/threonine antibody instead to detect gross phosphorylation of serine and threonine residues of ORF45. Basal phosphorylation of ORF45 was detected in HEK293T cells (Fig. 4A); the level of ORF45 phosphorylation was significantly increased by coexpression of IKKε (Fig. 4A) or TBK1 (data not shown). The IKKε-induced ORF45 phosphorylation was reduced significantly by mutation of both Ser41 and Ser162 but was reduced only slightly by mutation of either residue (Fig. 4B), suggesting that both these two residues are the primary phosphorylation sites. The phosphorylation of ORF45 was increased by TLR3 ligand poly(I·C) (Fig. 4C) and by Sendai virus infection (Fig. 4D). Furthermore, the phosphorylation of ORF45 was reduced by knockdown of both TBK1 and IKKε by siRNAs (Fig. 4E). These experiments suggest that ORF45 is phosphorylated on Ser41 and Ser162 by IKKε and TBK1 in cells.
Fig 4.
KSHV ORF45 is phosphorylated by IKKε or TBK1 in cells. (A) Phosphorylation of ORF45 is increased by IKKε. HEK293T cells were transfected with 5 μg of Flag-ORF45 in the absence or presence of 500 ng of HA-IKKε expression vectors. At 48 h after transfection, cell lysates were prepared and immunoprecipitated with anti-Flag M2 affinity beads. The IP complexes were immunoblotted with anti-phospho-serine/threonine antibody for detection of gross phosphorylation of ORF45. The input WCL and IP complexes were also analyzed with the indicated antibodies as controls. (B) Phosphorylation of ORF45 by IKKε is reduced by mutations of Ser41 and Ser162. HEK293T cells were transfected with 5 μg of wild-type ORF45 or ORF45-S41A, ORF45-S162A, or ORF45-S41/162A in the absence or presence of 500 ng of HA-IKKε expression plasmids. At 48 h after transfection, cell lysates were prepared and immunoprecipitated with anti-ORF45 antibody. The input WCL and IP complexes were analyzed by immunoblotting with anti-phospho-serine/threonine and other antibodies as indicated. (C) Phosphorylation of ORF45 induced by TLR3 ligand poly(I·C) is reduced by mutation of Ser41 and Ser162. HEK293-TLR3 cells were transfected with 5 μg of wild-type ORF45 or ORF45-S41/162A mutant expression vectors. At 48 h after transfection, the cells were treated with poly(I·C) for 30 min. Cell lysates were prepared, immunoprecipitated, and analyzed as described above. (D) Phosphorylation of ORF45 is increased by Sendai virus infection. HEK293T cells were transfected with 5 μg of Flag-ORF45 expression vector. At 24 h after transfection, the cells were treated with Sendai virus for 24 h. Cell lysates were prepared, immunoprecipitated, and analyzed as described above. (E) Phosphorylation of ORF45 is reduced by knockdown of TBK1 and IKKε by siRNAs. HEK293T cells were transfected with commercially validated siRNAs against TBK1 and IKKε or the accompanying control. The cells were further transfected with Flag-ORF45 expression plasmid 24 h later. Phosphorylation of ORF45 was analyzed as described above.
KSHV ORF45 competitively inhibits phosphorylation of IRF7 by IKKε and TBK1.
Because ORF45 neither affected the kinase activity of IKKε or TBK1 nor interfered with the general interaction between the kinases and substrates but was efficiently phosphorylated by IKKε and TBK1, we speculated that ORF45 could compete with IRF7 as an alternative substrate to inhibit its phosphorylation specifically. Indeed, phosphorylation of ORF45 fragments by IKKε or TBK1 seems more efficient than that of IRF7 (Fig. 3). To compare these substrates directly, we made GST-tagged short peptides containing the phosphorylation sites of IRF7 (Ser477/479) or ORF45 (Ser41, Ser162, and Ser41/Ser162) and the corresponding mutations in which the sites were replaced by alanine and used them in kinase assays. The results indicated that the short peptides of ORF45 are phosphorylated more efficiently than is IRF7 (Fig. 5A, compare lanes 2, 4, and 6 to lane 7).
Fig 5.
KSHV ORF45 competitively inhibits IRF7 phosphorylation by IKKε or TBK1. (A) ORF45 is phosphorylated more efficiently than IRF7 by IKKε and TBK1. Short GST-fused peptides containing the phosphorylation sites of IRF7 (Ser477/479), ORF45 Ser41, Ser162, and chimeric Ser41/Ser162, and the corresponding S41A and S162A mutants were expressed and purified from E. coli. Equal amounts of the proteins were subjected to in vitro kinase assays with HA-IKKε as described above (left). The sequences of the peptides were aligned to the consensus substrate phosphorylation motif of IKKε identified by Hutti et al. (24) (right). (B) ORF45 competitively inhibits phosphorylation of IRF7 by IKKε and TBK1 in vitro. GST-IRF7 (aa 283 to 503) proteins were mixed with increasing amounts of full-length (FL) ORF45 proteins in reactions for in vitro kinase assays. The assays were performed as described for Fig. 4 except that IRF7 substrate phosphorylation was detected by Western blotting with anti-pIRF7(Ser477/479) antibody. (C) ORF45 competitively inhibits phosphorylation of IRF7 by IKKε/TBK1 in cells. Plasmids expressing Flag-IRF7 (1 μg), IKKε (100 ng), and increasing amounts of FL-ORF45 (100 ng, 500 ng, 2 μg, and 4 μg) were transfected into HEK293T cells. At 48 h after transfection, cell lysates were prepared, immunoprecipitated, and analyzed by Western blotting with antibodies as indicated. (D) ORF45 does not inhibit phosphorylation of IRF3 by IKKε/TBK1 in cells. Plasmids expressing Flag-IRF7 (1 μg), IKKε (100 ng), and increasing amounts of FL-ORF45 (100 ng, 500 ng, 2 μg, and 4 μg) were transfected into HEK293T cells. At 48 h after transfection, cell lysates were prepared, immunoprecipitated, and analyzed by Western blotting with antibodies as indicated. (E) A short GST-fused peptide containing ORF45 phosphorylation sites (GST-ORF45-S41/162) competitively inhibits phosphorylation of IRF7 by IKKε/TBK1 in vitro. GST-IRF7 (aa 283 to 503) proteins were mixed with increasing amounts of GST-ORF45-S41/162 peptide (shown in panel A) in reactions for in vitro kinase assays. The assays were performed as described for panel B. (F) GST-ORF45-S41/162 peptide does not inhibit phosphorylation of IRF7 by IKKε/TBK1 in cells. Plasmids expressing Flag-IRF7 (1 μg), IKKε (100 ng), and increasing amounts of mammalian GST expression vector pEBG-ORF45-S41/162 (100 ng, 500 ng, 2 μg, and 4 μg) were transfected into HEK293T cells. At 48 h after transfection, cell lysates were prepared and analyzed by Western blotting with antibodies as indicated. Identical results were obtained when IKKε was replaced with TBK1 in these experiments and thus were not shown.
Next we determined whether ORF45 competitively inhibited IRF7 phosphorylation when they were mixed together. When increasing amounts of full-length ORF45 protein were mixed with GST-IRF7 (aa 283 to 503) and IKKε in in vitro kinase assays, phosphorylation of IRF7 on Ser477 and Ser479 as detected by anti-pIRF7 (Ser477/479) was inhibited by ORF45 in a dose-dependent manner (Fig. 5B). Similarly, expression of full-length ORF45 also inhibited IKKε-induced IRF7 phosphorylation on Ser477/479 in cells in a dose-dependent manner (Fig. 5C) but, as expected, had no effect on IRF3 phosphorylation (Fig. 5D). In similar assays, although a GST fusion peptide that contains both Ser41 and Ser162 (ORF45 Ser41/162; depicted in Fig. 5A) inhibited IRF7 phosphorylation at higher concentrations (Fig. 5E), expression of the same GST fusion peptide had no effect on IRF7 phosphorylation in cells (Fig. 5F), suggesting that inhibition of IRF7 phosphorylation in cells requires the interaction between ORF45 and IRF7. Together, these results suggest that KSHV ORF45 inhibits phosphorylation of IRF7 by IKKε and TBK1 as a more efficient substrate and the optimal inhibition requires the interactions between ORF45 and IRF7.
The ORF45 phosphorylation sites Ser41 and Ser162 are critical for inhibition of IRF7 phosphorylation by IKKε and TBK1.
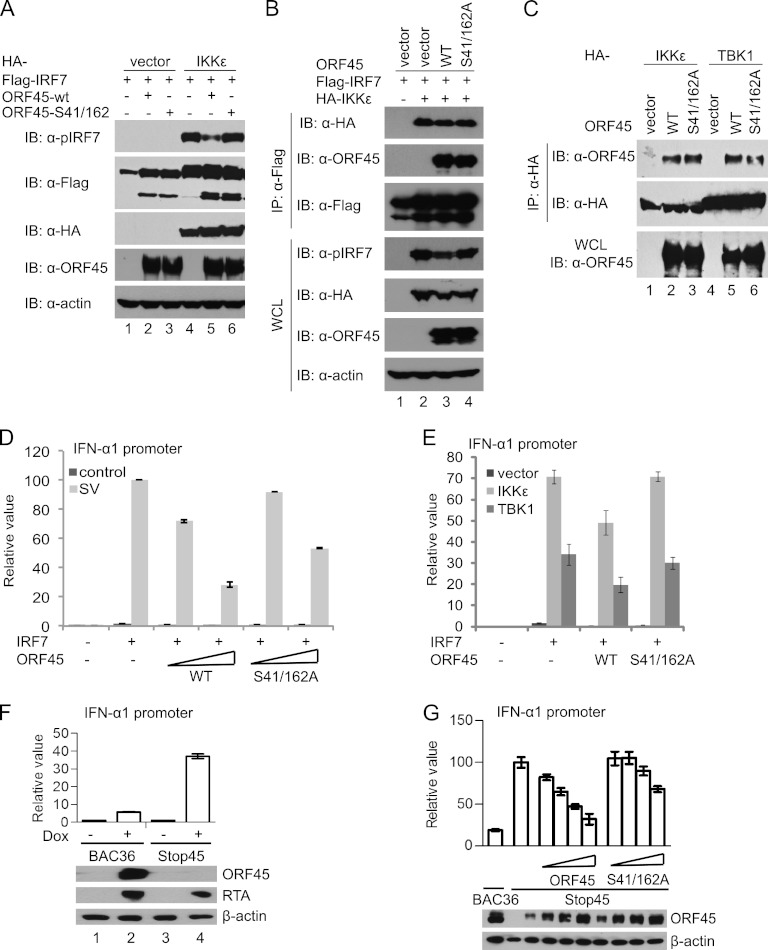
Finally, to determine whether mutation of the phosphorylation sites in ORF45 affects its inhibition of IRF7 phosphorylation, we transfected Flag-IRF7, HA-IKKε, or HA-TBK1 and ORF45 or its phosphorylation site mutant ORF45-S41/162A into HEK293T cells in different combinations. Phosphorylation of IRF7 was determined by Western blotting with the phosphorylation-specific antibody. As shown in Fig. 6A, the wild-type ORF45 inhibited IRF7 phosphorylation by IKKε (Fig. 6A) and by TBK1 (data not shown), but the phosphorylation site mutant ORF45-S41/162A lost its inhibition ability. The mutant nevertheless interacted with IRF7 as well as the wild-type ORF45, and the mutation had no apparent effect on IKKε/IRF7 interaction (Fig. 6B). The mutant also apparently bound to IKKε/TBK1 as efficiently as did the wild-type ORF45 (Fig. 6C). However, luciferase reporter assays revealed that the ORF45-S41/162A mutant inhibited the transactivation activity of IRF7 less effectively than did the wild-type ORF45 (Fig. 6D and E), suggesting a critical role for Ser41 and Ser162 of ORF45 in the inhibition of IRF7 phosphorylation by IKKε and TBK1. To determine the role of endogenous ORF45 in inhibition of IRF7 during KSHV lytic infection, we wished to assay IRF7 activity in cells infected by the wild-type or the ORF45-null mutant viruses. We introduced the wild-type BAC36-wt and the ORF45-null BAC36-stop45 BAC DNAs into iSLK cells and established stable cell lines (27, 63, 67). iSLK cells express doxycycline-inducible RTA and therefore support robust lytic replication of KSHV upon treatment with doxycycline (40). Because the anti-pIRF7 antibody was not sensitive enough to detect the endogenous active IRF7, a method to assay IRF7-specific activity in cells was still lacking. We instead used reporter assays to measure IRF7 transactivation activity. We transfected IRF7 and IFN-α1 reporter plasmids into these cells and then treated the cells with doxycycline to initiate KSHV lytic replication. As shown in Fig. 6F, IRF7 transactivation activity in the BAC36-wt infected cells was only moderately increased by doxycycline treatment, but the increase was significantly higher in cells infected by the ORF45-null mutant, suggesting that ORF45 is required for inhibition of IRF7 during KSHV lytic replication. To determine the role of Ser41 and Ser162 in the inhibition of IRF7, we transfected the iSLK/BAC36-stop45 cells with IRF7 reporter constructs in the presence of increasing amounts of the wild-type ORF45 or the ORF45-S41/162A mutant plasmid. As shown in Fig. 6G, the wild-type ORF45 inhibited IRF7 in a dose-dependent manner, whereas the ORF45-S41/162A mutant had a diminished effect under similar conditions. The critical role of Ser41 and Ser162 of ORF45 in the inhibition of IRF7 phosphorylation by IKKε and TBK1 suggests that ORF45, although it does not interfere with the interaction of the kinases to substrate IRF7, inhibits phosphorylation of IRF7 by acting as an alternative substrate.
Fig 6.
Ser41 and Ser162 of KSHV ORF45 contribute to inhibition of IRF7 phosphorylation and transactivation activity. (A) Ser41 and Ser162 of KSHV ORF45 are required for inhibition of IKKε- or TBK1-induced IRF7 phosphorylation. HEK293T cells were transfected with 1 μg of Flag-IRF7, 100 ng of HA-IKKε, and 2 μg of wild-type ORF45 or ORF45-S41/162A mutant expression plasmids. At 48 h after transfection, cell lysates were prepared and analyzed by Western blotting with the indicated antibodies. Similar results were obtained with TBK1 (data not shown). (B) Mutation of Ser41 and Ser162 of ORF45 does not affect IRF7/ORF45 or IRF7/IKKε interaction. HEK293T cells were transfected with 5 μg of Flag-IRF7, 500 ng of HA-IKKε, and 10 μg of wild-type ORF45 or ORF45-S41/162A mutant expression vectors. At 48 h after transfection, cell lysates were prepared and immunoprecipitated with anti-Flag M2 affinity beads. The input WCL and IP complexes were analyzed by immunoblotting with antibodies as indicated. (C) Mutation of both Ser41 and Ser162 of ORF45 does not affect interactions between ORF45 and IKKε or TBK1. HEK293T cells were transfected with IKKε (500 ng) or TBK1 (500 ng) with wild-type ORF45 or ORF45-S41/162A mutant expression vectors. At 48 h after transfection, cell lysates were prepared and immunoprecipitated with anti-Flag M2 affinity beads. The input WCL and IP complexes were analyzed by immunoblotting with antibodies as indicated. (D) Mutation of both Ser41 and Ser162 of ORF45 impairs the inhibition of virus-induced IRF7 transactivation activity. HEK293T cells were transfected with IFN-α1 promoter reporter, pRL-TK, IRF7, and increasing amounts of ORF45 expression plasmids as indicated. Eight hours after transfection, each well was infected with 80 hemagglutinin units of Sendai viruses (SV). At 24 h after transfection, cell lysates were prepared and used for dual-luciferase assays. The relative luciferase activity was expressed in arbitrary units by normalization of firefly luciferase activity to Renilla luciferase activity. Values are averages ± standard deviations from three experiments. (E) Mutation of both Ser41 and Ser162 of ORF45 impairs the inhibition of IKKε/TBK1-induced IRF7 transactivation activity. HEK293T cells were transfected with the IFN-α1 promoter reporter, IRF7, and 60 ng ORF45 with or without 2 ng IKKε expression plasmids. At 24 h after transfection, cell lysates were prepared and used for dual-luciferase assays. (F) ORF45 contributes to inhibition of IRF7 during KSHV lytic replication. The iSLK/BAC36-wt and iSLK/BAC-stop45 cells seeded in 24-well plates were transfected with IFN-α1 promoter reporter, pRL-TK, and IRF7 plasmids using the Fugene 6 reagent. Six hours after transfection, cells were treated with 2 μg/ml doxycycline (Dox) and 1 mM sodium butyrate. At 24 h after transfection, cell lysates were prepared and used for dual-luciferase assays and for Western blot analysis. (G) Ser41 and Ser162 are required for inhibition of IRF7 by ORF45 during KSHV lytic replication. The iSLK/BAC36-wt and iSLK/BAC-stop45 cells were transfected with IFN-α1 promoter reporter, pRL-TK, IRF7, and increasing amounts of the wild-type ORF45 or the S41/162A mutant plasmids as indicated. Six hours after transfection, the cells were treated and assayed as described above.
DISCUSSION
Phosphorylation on serine residues in the C-terminal regulatory region is the most critical step in the activation of latent IRF7 that leads to subsequent expression of type I IFNs (35, 37). In most cell types (all except the plasmacytoid dendritic cells), IKKε and TBK1 are the kinases responsible for phosphorylation of IRF7 and IRF3 after viral infection, regardless of the types of pathogen recognition receptors are involved (8, 10, 13, 38, 41, 52). We therefore sought to determine whether and how ORF45 inhibits IRF7 phosphorylation by IKKε and TBK1. Because our in vivo and in vitro assays confirmed that phosphorylation of IRF7 on Ser477 and Ser479 by these kinases was inhibited by KSHV ORF45, that ORF45 seemed not to inactivate IKKε or TBK1 directly, and that ORF45 is phosphorylated efficiently on Ser41 and Ser162 by IKKε and TBK1, we conclude that KSHV ORF45 inhibits IRF7 phosphorylation by IKKε or TBK1 by acting as an alternative substrate.
Because of the critical roles of IKKε and TBK1 in regulation of IFN expression, they are common targets of a variety of viruses seeking to evade host antiviral responses. Activation of IKKε and TBK1 is regulated by their associations with cellular proteins, such as suppressor of IKKε, TRAF family member-associated NF-κB activator (TANK), similar to NAP1 TBK1 adaptor (SINTBAD), cylindromatosis (CYLD), and DEAD box RNA helicase (DDX3X) (14, 18, 23, 47, 54, 62). The interactions are exploited by a number of viruses as strategies for inhibition of activation of IKKε and TBK1. For example, the M protein of severe acute respiratory syndrome coronavirus and the Gn protein of pathogenic hantavirus interfere with the formation of a complex between TNF receptor-associated factor 3 and IKKε/TBK1 (2, 53); K7 of vaccinia virus and the polymerase of hepatitis B virus interfere with IKKε/TBK1 by attacking DEAD box RNA helicase (50, 59, 61). The above strategies are probably not used by ORF45, however, because it does not directly affect the kinase activities of IKKε and TBK1. Other viral proteins, like ORF45, are found to be phosphorylated by IKKε/TBK1, including the P protein of Borna disease virus (58), VP35 of Ebola virus (44), and the V proteins of several paramyxoviruses (36). Although competition as an alternative substrate was proposed as the underlying mechanism, the exact sites of phosphorylation by IKKε/TBK1 on these proteins have not been identified.
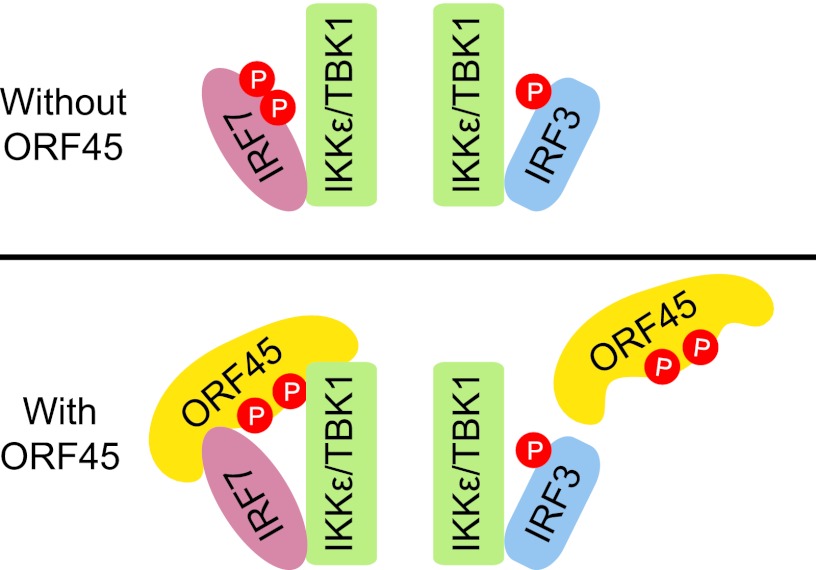
Recently, the consensus phosphorylation sites of IKKε substrates were identified as [YFPM]XpS[LIMF]X[YWF], in which the hydrophobic +1 position seems to be the most critical (Fig. 6A) (24). Both Ser477 and Ser479 (DSSpSLpSLCLS) of IRF7 are followed by an L at +1, but an aromatic residue at +3 or −2 was absent in both cases. Therefore, neither of the serine residues is predicted to be optimal for phosphorylation by IKKε/TBK1. Inspection of the sequence of ORF45 revealed that Ser41 (PLSpSLPGF) is followed at +1 by L and Ser162 (SEApSMGWV) by M; an F is at the +4 position of Ser41, and a W is at +3 of Ser162. These features may explain why ORF45 behaves as a better substrate in the in vitro kinases assays. Although it acts as a better substrate, ORF45 inhibits IRF7 phosphorylation specifically and has little effect on IRF3 phosphorylation in cells. The specificity of inhibition is intriguing but conceivably could be explained by the specific interaction between ORF45 and IRF7. Our data suggest that ORF45 interacts with IRF7 but not IRF3 and that ORF45, IRF7, and IKKε/TBK1 associate with each other in the same complexes (Fig. 2). As a result, the IRF7-bound ORF45 can compete with IRF7 for phosphorylation by these kinases because ORF45 is a more efficient substrate than IRF7. In contrast to IRF7, IRF3 does not interact with ORF45 (Fig. 2). Therefore, phosphorylation of IRF3 by IKKε/TBK1 is not affected by ORF45 in cells because ORF45 does not inactivate these kinases (Fig. 7).
Fig 7.
Schematic diagram of ORF45-mediated specific inhibition of IRF7 phosphorylation by IKKε/TBK1. ORF45 interacts with IRF7 but not with IRF3. The IRF7-bound ORF45 competes with IRF7 for phosphorylation by IKKε/TBK1. Because IRF3 does not interact with ORF45, its phosphorylation by IKKε/TBK1 is not affected.
Activation of a kinase does not always result in phosphorylation of all of its substrates. For example, IRF3 is activated in only a subset of signaling pathways that activate TBK1. A recent study demonstrated that the stimulator of IFN genes (STING) acts as a scaffold to recruit both TBK1 and IRF3 and therefore to specify IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway (57). Because of their critical roles in cellular signaling transduction, the scaffold proteins can be conceivably exploited by pathogens to alter protein-protein interactions, consequently reshaping the cellular signaling output to facilitate their survival and propagation. ORF45 appears to act as a scaffold by interacting with both IRF7 and IKKε/TBK1. Because ORF45 is a more efficient substrate than IRF7 and the phosphorylation sites (Ser41 and Ser 162) of ORF45 might be more accessible to the kinases than those of IRF7, ORF45 inhibits phosphorylation of the associated IRF7. ORF45 is known to bind to the internal inhibitory domain (aa 283 to 466) of IRF7. Our structural modeling of IRF7 indicates that the inhibitory domain of IRF7 forms a tight compact hydrophobic core that probably opens up when the C-terminal region is phosphorylated by IKKε/TBK1 (48). Binding of ORF45 may cause structural constriction of IRF7 and may also interfere with access of the IRF7 phosphorylation residues (Ser477 and Ser479) to IKKε/TBK1 but not affect the physical interactions of IRF7 with the kinases. Further structural analyses of IRF7 and its complex with ORF45 will help to resolve the mechanistic details. In addition to interactions with IRF7 and IKKε/TBK1, ORF45 also interacts with a number of viral and cellular proteins (including RSK and ERK) through defined motifs, further supporting the notion that ORF45 acts as a molecular scaffold and therefore alters the protein interactome during KSHV lytic replication (28, 29).
Unique among viral proteins that interfere with IRF3/IRF7 phosphorylation, ORF45 selectively inhibits IRF7 phosphorylation but spares the IKKε/TBK1 kinases themselves. IKKε and TBK1 are expected to have other substrates that may or may not be directly linked to innate immune responses. In fact, both IKKε and TBK1 have been identified as products of tumor genes that have transforming potential and are overrepresented in certain cancers (1, 3, 4). Because the overall activities of these kinases are not affected, the oncogenic potential of IKKε and TBK1 may not be inhibited by ORF45. The lytic cycle is well known to contribute to viral pathogenesis and tumorigenesis. Therefore, the unique mode of leaky inhibition of IKKε/TBK1 by ORF45 may have important implications in KSHV pathogenesis.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant R01DE016680 to F.Z. and NIH R01CA86839 to Y.Y.
We are grateful to Katherine Fitzgerald for providing HEK293-TLR3 cells, to Rongtuan Lin and John Hiscott for providing anti-pIRF7 (Ser477/479) antibody, and to Jinjong Myoung and Don Ganem for providing the iSLK cells. We thank members of the Zhu laboratory for critical reading of the manuscript and for helpful discussions. We also thank Anne B. Thistle at the Florida State University for excellent editorial assistance.
Footnotes
Published ahead of print 11 July 2012
REFERENCES
- 1. Adli M, Baldwin AS. 2006. IKK-i/IKKepsilon controls constitutive, cancer cell-associated NF-kappaB activity via regulation of Ser-536 p65/RelA phosphorylation. J. Biol. Chem. 281:26976–26984 [DOI] [PubMed] [Google Scholar]
- 2. Alff PJ, Sen N, Gorbunova E, Gavrilovskaya IN, Mackow ER. 2008. The NY-1 hantavirus Gn cytoplasmic tail coprecipitates TRAF3 and inhibits cellular interferon responses by disrupting TBK1-TRAF3 complex formation. J. Virol. 82:9115–9122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barbie DA, et al. 2009. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462:108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boehm JS, et al. 2007. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell 129:1065–1079 [DOI] [PubMed] [Google Scholar]
- 5. Buettner N, et al. 2010. Thogoto virus ML protein is a potent inhibitor of the interferon regulatory factor-7 transcription factor. J. Gen. Virol. 91:220–227 [DOI] [PubMed] [Google Scholar]
- 6. Chang TH, et al. 2009. Ebola Zaire virus blocks type I interferon production by exploiting the host SUMO modification machinery. PLoS Pathog. 5:e1000493 doi:10.1371/journal.ppat.1000493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chang Y, et al. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865–1869 [DOI] [PubMed] [Google Scholar]
- 8. Chau TL, et al. 2008. Are the IKKs and IKK-related kinases TBK1 and IKK-epsilon similarly activated? Trends Biochem. Sci. 33:171–180 [DOI] [PubMed] [Google Scholar]
- 9. Chen W, et al. 2008. Contribution of Ser386 and Ser396 to activation of interferon regulatory factor 3. J. Mol. Biol. 379:251–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Clement JF, Meloche S, Servant MJ. 2008. The IKK-related kinases: from innate immunity to oncogenesis. Cell Res. 18:889–899 [DOI] [PubMed] [Google Scholar]
- 11. Coscoy L. 2007. Immune evasion by Kaposi's sarcoma-associated herpesvirus. Nat. Rev. Immunol. 7:391–401 [DOI] [PubMed] [Google Scholar]
- 12. Dragan AI, Hargreaves VV, Makeyeva EN, Privalov PL. 2007. Mechanisms of activation of interferon regulator factor 3: the role of C-terminal domain phosphorylation in IRF-3 dimerization and DNA binding. Nucleic Acids Res. 35:3525–3534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fitzgerald KA, et al. 2003. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4:491–496 [DOI] [PubMed] [Google Scholar]
- 14. Friedman CS, et al. 2008. The tumour suppressor CYLD is a negative regulator of RIG-I-mediated antiviral response. EMBO Rep. 9:930–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ganem D. 2007. Kaposi's sarcoma-associated herpesvirus, p 2847–2888 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 5th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 16. Ganem D. 2010. KSHV and the pathogenesis of Kaposi sarcoma: listening to human biology and medicine. J. Clin. Invest. 120:939–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Greene W, et al. 2007. Molecular biology of KSHV in relation to AIDS-associated oncogenesis. Cancer Treat. Res. 133:69–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guo B, Cheng G. 2007. Modulation of the interferon antiviral response by the TBK1/IKKi adaptor protein TANK. J. Biol. Chem. 282:11817–11826 [DOI] [PubMed] [Google Scholar]
- 19. Hiscott J. 2007. Triggering the innate antiviral response through IRF-3 activation. J. Biol. Chem. 282:15325–15329 [DOI] [PubMed] [Google Scholar]
- 20. Honda K, Takaoka A, Taniguchi T. 2006. Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity 25:349–360 [DOI] [PubMed] [Google Scholar]
- 21. Honda K, Taniguchi T. 2006. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat. Rev. Immunol. 6:644–658 [DOI] [PubMed] [Google Scholar]
- 22. Honda K, et al. 2005. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434:772–777 [DOI] [PubMed] [Google Scholar]
- 23. Huang J, et al. 2005. SIKE is an IKK epsilon/TBK1-associated suppressor of TLR3- and virus-triggered IRF-3 activation pathways. EMBO J. 24:4018–4028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hutti JE, et al. 2009. Phosphorylation of the tumor suppressor CYLD by the breast cancer oncogene IKKepsilon promotes cell transformation. Mol. Cell 34:461–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Joo CH, et al. 2007. Inhibition of interferon regulatory factor 7 (IRF7)-mediated interferon signal transduction by the Kaposi's sarcoma-associated herpesvirus viral IRF homolog vIRF3. J. Virol. 81:8282–8292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kawai T, Akira S. 2006. Innate immune recognition of viral infection. Nat. Immunol. 7:131–137 [DOI] [PubMed] [Google Scholar]
- 27. Kuang E, Fu B, Liang Q, Myoung J, Zhu F. 2011. Phosphorylation of eukaryotic translation initiation factor 4B (EIF4B) by open reading frame 45/p90 ribosomal S6 kinase (ORF45/RSK) signaling axis facilitates protein translation during Kaposi sarcoma-associated herpesvirus (KSHV) lytic replication. J. Biol. Chem. 286:41171–41182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kuang E, Tang Q, Maul GG, Zhu F. 2008. Activation of p90 ribosomal S6 kinase by ORF45 of Kaposi's sarcoma-associated herpesvirus and its role in viral lytic replication. J. Virol. 82:1838–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kuang E, Wu F, Zhu F. 2009. Mechanism of sustained activation of ribosomal S6 kinase (RSK) and ERK by Kaposi sarcoma-associated herpesvirus ORF45: multiprotein complexes retain active phosphorylated ERK and RSK and protect them from dephosphorylation. J. Biol. Chem. 284:13958–13968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee HR, Kim MH, Lee JS, Liang C, Jung JU. 2009. Viral interferon regulatory factors. J. Interferon Cytokine Res. 29:621–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li X, Zhu F. 2009. Identification of the nuclear export and adjacent nuclear localization signals for ORF45 of Kaposi's sarcoma-associated herpesvirus. J. Virol. 83:2531–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liang C, Lee JS, Jung JU. 2008. Immune evasion in Kaposi's sarcoma-associated herpes virus associated oncogenesis. Semin. Cancer Biol. 18:423–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liang Q, Deng H, Sun CW, Townes TM, Zhu F. 2011. Negative regulation of IRF7 activation by activating transcription factor 4 suggests a cross-regulation between the IFN responses and the cellular integrated stress responses. J. Immunol. 186:1001–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lin R, Heylbroeck C, Pitha PM, Hiscott J. 1998. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell. Biol. 18:2986–2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lin R, Mamane Y, Hiscott J. 2000. Multiple regulatory domains control IRF-7 activity in response to virus infection. J. Biol. Chem. 275:34320–34327 [DOI] [PubMed] [Google Scholar]
- 36. Lu LL, Puri M, Horvath CM, Sen GC. 2008. Select paramyxoviral V proteins inhibit IRF3 activation by acting as alternative substrates for inhibitor of kappaB kinase epsilon (IKKe)/TBK1. J. Biol. Chem. 283:14269–14276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Marie I, Smith E, Prakash A, Levy DE. 2000. Phosphorylation-induced dimerization of interferon regulatory factor 7 unmasks DNA binding and a bipartite transactivation domain. Mol. Cell. Biol. 20:8803–8814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Matsui K, et al. 2006. Cutting edge: role of TANK-binding kinase 1 and inducible IkappaB kinase in IFN responses against viruses in innate immune cells. J. Immunol. 177:5785–5789 [DOI] [PubMed] [Google Scholar]
- 39. Mori M, et al. 2004. Identification of Ser-386 of interferon regulatory factor 3 as critical target for inducible phosphorylation that determines activation. J. Biol. Chem. 279:9698–9702 [DOI] [PubMed] [Google Scholar]
- 40. Myoung J, Ganem D. 2011. Generation of a doxycycline-inducible KSHV producer cell line of endothelial origin: maintenance of tight latency with efficient reactivation upon induction. J. Virol. Methods 174:12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ning S, Pagano JS, Barber GN. 2011. IRF7: activation, regulation, modification and function. Genes Immun. 12:399–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Paz S, et al. 2006. Induction of IRF-3 and IRF-7 phosphorylation following activation of the RIG-I pathway. Cell Mol. Biol. (Noisy-le-grand) 52:17–28 [PubMed] [Google Scholar]
- 43. Pichlmair A, Reis e Sousa C. 2007. Innate recognition of viruses. Immunity 27:370–383 [DOI] [PubMed] [Google Scholar]
- 44. Prins KC, Cardenas WB, Basler CF. 2009. Ebola virus protein VP35 impairs the function of interferon regulatory factor-activating kinases IKKepsilon and TBK-1. J. Virol. 83:3069–3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Randall RE, Goodbourn S. 2008. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 89:1–47 [DOI] [PubMed] [Google Scholar]
- 46. Rezaee SA, Cunningham C, Davison AJ, Blackbourn DJ. 2006. Kaposi's sarcoma-associated herpesvirus immune modulation: an overview. J. Gen. Virol. 87:1781–1804 [DOI] [PubMed] [Google Scholar]
- 47. Ryzhakov G, Randow F. 2007. SINTBAD, a novel component of innate antiviral immunity, shares a TBK1-binding domain with NAP1 and TANK. EMBO J. 26:3180–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sathish N, Zhu FX, Golub EE, Liang Q, Yuan Y. 2011. Mechanisms of autoinhibition of IRF-7 and a probable model for inactivation of IRF-7 by Kaposi's sarcoma-associated herpesvirus protein ORF45. J. Biol. Chem. 286:746–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sato M, et al. 2000. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity 13:539–548 [DOI] [PubMed] [Google Scholar]
- 50. Schroder M, Baran M, Bowie AG. 2008. Viral targeting of DEAD box protein 3 reveals its role in TBK1/IKKepsilon-mediated IRF activation. EMBO J. 27:2147–2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Servant MJ, Grandvaux N, tenOever BR, Duguay D, Lin R, Hiscott J. 2003. Identification of the minimal phosphoacceptor site required for in vivo activation of interferon regulatory factor 3 in response to virus and double-stranded RNA. J. Biol. Chem. 278:9441–9447 [DOI] [PubMed] [Google Scholar]
- 52. Sharma S, tenOever BR, Grandvaux N, Zhou GP, Lin R, Hiscott J. 2003. Triggering the interferon antiviral response through an IKK-related pathway. Science 300:1148–1151 [DOI] [PubMed] [Google Scholar]
- 53. Siu KL, et al. 2009. Severe acute respiratory syndrome coronavirus M protein inhibits type I interferon production by impeding the formation of TRAF3 · TANK · TBK1/IKKε complex. J. Biol. Chem. 284:16202–16209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Soulat D, et al. 2008. The DEAD-box helicase DDX3X is a critical component of the TANK-binding kinase 1-dependent innate immune response. EMBO J. 27:2135–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Speck SH, Ganem D. 2010. Viral latency and its regulation: lessons from the gamma-herpesviruses. Cell Host Microbe 8:100–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Staskus KA, et al. 1997. Kaposi's sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J. Virol. 71:715–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tanaka Y, Chen ZJ. 2012. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci. Signal. 5:ra20 doi:10.1126/scisignal.2002521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Unterstab G, et al. 2005. Viral targeting of the interferon-β-inducing Traf family member-associated NF-κB activator (TANK)-binding kinase-1. Proc. Natl. Acad. Sci. U. S. A. 102:13640–13645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang H, Ryu WS. 2010. Hepatitis B virus polymerase blocks pattern recognition receptor signaling via interaction with DDX3: implications for immune evasion. PLoS Pathog. 6:e1000986 doi:10.1371/journal.ppat.1000986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wu L, et al. 2009. Epstein-Barr virus LF2: an antagonist to type I interferon. J. Virol. 83:1140–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yu S, et al. 2010. Hepatitis B virus polymerase inhibits RIG-I- and Toll-like receptor 3-mediated beta interferon induction in human hepatocytes through interference with interferon regulatory factor 3 activation and dampening of the interaction between TBK1/IKKepsilon and DDX3. J. Gen. Virol. 91:2080–2090 [DOI] [PubMed] [Google Scholar]
- 62. Zhang M, et al. 2008. Regulation of IkappaB kinase-related kinases and antiviral responses by tumor suppressor CYLD. J. Biol. Chem. 283:18621–18626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhou FC, et al. 2002. Efficient infection by a recombinant Kaposi's sarcoma-associated herpesvirus cloned in a bacterial artificial chromosome: application for genetic analysis. J. Virol. 76:6185–6196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhu FX, Chong JM, Wu L, Yuan Y. 2005. Virion proteins of Kaposi's sarcoma-associated herpesvirus. J. Virol. 79:800–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhu FX, Cusano T, Yuan Y. 1999. Identification of the immediate-early transcripts of Kaposi's sarcoma-associated herpesvirus. J. Virol. 73:5556–5567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhu FX, King SM, Smith EJ, Levy DE, Yuan Y. 2002. A Kaposi's sarcoma-associated herpesviral protein inhibits virus-mediated induction of type I interferon by blocking IRF-7 phosphorylation and nuclear accumulation. Proc. Natl. Acad. Sci. U. S. A. 99:5573–5578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhu FX, Li X, Zhou F, Gao SJ, Yuan Y. 2006. Functional characterization of Kaposi's sarcoma-associated herpesvirus ORF45 by bacterial artificial chromosome-based mutagenesis. J. Virol. 80:12187–12196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhu FX, Sathish N, Yuan Y. 2010. Antagonism of host antiviral responses by Kaposi's sarcoma-associated herpesvirus tegument protein ORF45. PLoS One 5:e10573 doi:10.1371/journal.pone.0010573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhu FX, Yuan Y. 2003. The ORF45 protein of Kaposi's sarcoma-associated herpesvirus is associated with purified virions. J. Virol. 77:4221–4230 [DOI] [PMC free article] [PubMed] [Google Scholar]