Abstract
Polypeptide 2CATPase is one of the most thoroughly studied but least understood proteins in the life cycle of poliovirus. Within the protein, multiple functional domains important for uncoating, host cell membrane alterations, and RNA replication and encapsidation have previously been identified. In this study, charged to alanine-scanning mutagenesis was used to generate conditional-lethal mutations in hitherto-uncharacterized domains of the 2CATPase polypeptide, particularly those involved in morphogenesis. Adjacent or clustered charged amino acids (2 to 4), scattered along the 2CATPase coding sequence, were replaced with alanines. RNA transcripts of mutant poliovirus cDNA clones were transfected into HeLa cells. Subsequently, 10 lethal, 1 severely temperature-sensitive, 2 quasi-infectious, and 3 wild type-like mutants were identified. Using a luciferase-containing reporter virus, we demonstrated RNA replication defects in all lethal and quasi-infectious mutants. Temperature-sensitive mutants were defective in RNA replication only at the restricted temperatures. Furthermore, we characterized a quasi-infectious mutant (K6A/K7A) that produced a suppressor mutation (G1R) and a novel 2B∧2CATPase cleavage site (Q∧R). Surprisingly, this cleavage site mutation did not interfere with normal processing of the polyprotein. These mutants have led to the identification of several new sites within the 2CATPase polypeptide that are required for RNA replication. In addition, analysis of the suppressor mutants has revealed a new domain near the C terminus of 2CATPase that is involved in encapsidation, possibly achieved through interaction with an amino acid sequence between NTP binding motifs A and B of 2CATPase. Most importantly, the identification of suppressor mutations in both 2CATPase and the capsid domains (VP1 and VP3) of poliovirus has confirmed that an interaction between 2CATPase and capsid proteins is involved in viral morphogenesis.
INTRODUCTION
Poliovirus (PV) protein 2CATPase is a highly conserved nonstructural protein common to picornaviruses. Through use of genetic and drug inhibition studies, multiple functional domains have been identified in 2CATPase, but the exact role(s) of the protein in the viral life cycle remains elusive. Previous mutational studies of the conserved functional domains in PV 2CATPase have primarily yielded lethal or poor growth phenotypes due to defects in RNA replication and/or encapsidation. So far, only two sets of conditionally defective temperature-sensitive (ts) mutants, which are located near the C terminus of the protein, have been identified (22, 23, 32). Additional ts PV 2CATPase mutants would be particularly useful for identifying proteins/domains involved in the process of viral morphogenesis.
PV is a plus-strand RNA virus in the genus Enterovirus of the Picornaviridae family. The RNA genome is 7,500 nucleotides (nt) long and encodes a polyprotein that contains one structural (P1) and two nonstructural (P2, P3) domains (47) (Fig. 1A). The polyprotein is processed into functional precursors and mature viral proteins by viral proteinases 3Cpro/3CDpro and 2Apro (18, 43, 50). 2CATPase is a complex nonstructural protein, which contains a nucleoside triphosphate-binding motif (27) and displays ATPase activity in vitro (26, 36). This ATPase activity is inhibited by guanidine hydrochloride (GnHCl) (33), a well-known and potent inhibitor of PV RNA replication (11), and specific mutations in 2CATPase have been shown to confer GnHCl resistance or dependence (34). In infected cells, this 329-amino-acid protein complexes with other proteins to form viral RNA replication complexes on the surface of remodeled cytoplasmic vesicles (9). Genetic studies have implicated 2CATPase in a number of different functions in viral growth, including virus uncoating (22), host cell membrane rearrangements (2, 10, 37, 39), RNA binding and replication (3, 5, 6, 8, 21, 23, 30, 40–42), and encapsidation (25, 44, 46). The N terminus of 2CATPase, harboring an amphipathic helix (30), contains oligomerization domain (1) and RNA-binding domain (35) and anchors the protein to membranes (14) (Fig. 1B). Near its C terminus, 2CATPase contains a second amphipathic helix, also implicated in membrane binding (39), and a cysteine-rich region, which binds Zinc++ (32). The central and C-terminal domains of the polypeptide possess serpin (serine protease inhibitor) motifs and, indeed, 2CATPase can inhibit 3Cpro proteinase activity in vitro and in vivo (7). The protein also has the ability to oligomerize (1) and to interact with viral proteins 2B and 2BCATPase (12), 3A and 3AB (49), 3Cpro (7), and VP3 (25).
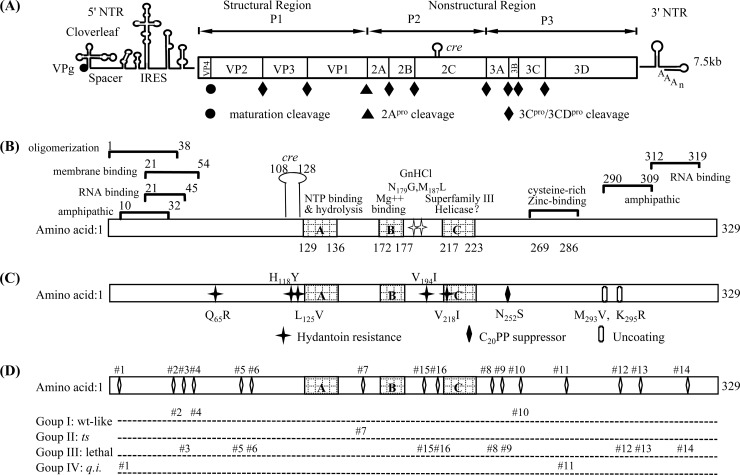
Fig 1.
Genomic structure of poliovirus, functional motifs, and grouping of the 16 alanine-scanning mutants of protein 2CATPase. (A) The PV RNA contains a long 5′ nontranslated region (5′ NTR), a single open reading frame, a short 3′ nontranslated region (3′ NTR), and a poly(A) tail. The proteinase cleavage sites and the P1, P2, and P3 domains of the polyprotein are shown. (B) Functional domains of the 2CATPase protein. (C) Locations of previously identified mutations in 2CATPase involved in encapsidation or uncoating. H118Y and V194I are newly discovered hydantoin-resistant mutations (Paul et al., unpublished). (D) Locations and grouping of the 16 alanine-scanning mutants of 2CATPase.
Based on sequence analyses, protein 2CATPase was classified as a member of superfamily III helicases (16) and it forms ring-like hexamers typical of various helicases (1). These helicases contain a small putative helicase domain, which has 3 conserved motifs, including the two classical ATP-binding motifs. In 2CATPase, the two sites common to other helicases are the A site (GxxxxGKS), which is involved in the binding of ATP, and the B site (DD), which interacts with Mg++. The third site, motif C, whose function is unknown, consists of an invariant N residue preceded by a stretch of moderately hydrophobic residues and is located downstream of motif B. However, numerous attempts to discover helicase activity associated with the 2CATPase protein have failed (36) (D. W. Kim and E. Wimmer, unpublished results). Downstream of motif C is residue N252, which is involved in an interaction with capsid protein VP3 of coxsackie A virus 20 (CAV20), in the context of a chimeric virus (C20PP) (25). A small RNA hairpin in the coding sequence of 2CATPase, a cis-acting replication element, Cre (nt 4444 to 4504) (15), is required for RNA synthesis as the template for the uridylylation of the terminal protein VPg (31).
The involvement of 2CATPase in PV encapsidation has been demonstrated previously through both drug inhibition studies and genetic experiments. Hydantoin, a drug that inhibits PV morphogenesis, yields resistant mutants that map to protein 2CATPase. The resistant mutations (44) (A. V. Paul et al., unpublished data) are located in the N-terminal and central portions of the 2CATPase coding region, and there is no clear relationship between these and other known motifs (Fig. 1C). Two sets of genetic experiments linked 2CATPase to encapsidation. The first involved a ts mutant with an insertion in 2CATPase (23). This mutant yielded suppressor mutations (M293V, K295R) in 2CATPase that proved to be cold sensitive in uncoating (22), an observation suggesting that 2CATPase has a role in determining unknown aspects of virion structure. In the second, we used a chimera containing the nonstructural proteins of PV and the capsid of the closely related CAV20 virus (C20PP). This chimera, whose proliferation was defective only in encapsidation, yielded a suppressor mutation either in 2CATPase or in capsid protein VP3 of CAV20, suggesting that the specificity of encapsidation is determined by an interaction between 2CATPase and the capsid (25).
Many mutants of 2CATPase have been studied in the past, but so far, only two sets of ts mutants have been identified. One of these displays cold sensitivity, as described above, and is involved in uncoating (22). The other leads to a ts defect in RNA replication (32). The best-known and most commonly used method for the generation of ts mutants is clustered charged to alanine mutagenesis, which was used successfully to produce ts mutants in RNA polymerase 3Dpol of poliovirus (13). This method was originally designed to study the role of surface charges in the functions of proteins, as summarized by Diamond and Kirkegaard (13). It has been suggested that the production of ts phenotypes by this method is related to the fact that charged residues (particularly clusters) in proteins frequently reside on solvent exposed surfaces. Disruption of these residues is believed to interfere with electrostatic or hydrogen bonding interactions between molecules or with the solvent.
In this study, we have used the clustered charged to alanine mutagenesis method and generated 16 mutants of PV 2CATPase that contain a minimum of two adjacent or closely positioned charged to alanine changes. The viability of the mutants was tested at different temperatures (33°C, 37°C, and 39.5°C), and the growth phenotypes of the resulting viruses, if any, were analyzed at the three temperatures. We also searched for and identified suppressor mutants, which are particularly useful in the identification of viral proteins that might interact with 2CATPase.
We have identified 10 lethal, 1 ts, and 2 quasi-infectious (q.i.) mutants, all of which were defective in RNA replication to some extent. Only three mutants possessed wild-type (Wt)-like growth phenotypes. Interestingly, we obtained suppressor mutations not only in protein 2CATPase but also in capsid proteins VP3 and VP1—the first demonstration of genetic suppression of a 2CATPase defect by capsid proteins in the background of the PV genome. The data presented here reinforce our previous conclusion that an interaction between 2CATPase and the capsid proteins is required for viral encapsidation. Our results also suggest the existence of a domain near the C terminus of the 2CATPase polypeptide that is involved in viral morphogenesis, possibly achieved through interaction with an amino acid sequence between NTP binding motifs A and B of 2CATPase.
MATERIALS AND METHODS
Cells.
HeLa R19 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) (Life Technology) supplemented with 10% bovine calf serum (BCS), 100 units of penicillin, and 100 mg/ml of streptomycin. Transfection or passages of HeLa cells were carried out with 2% BCS.
Plasmids.
pT7PVM contains a full-length infectious cDNA of PV1 (M). The pGEM-T vector was obtained from Promega. pR-PPP is an infectious Renilla luciferase (R-Luc) reporter virus construct in which Renilla luciferase (311 amino acids [aa]) is expressed as an N-terminal fusion to the PV polyprotein (25). pFPP is a reporter replicon in which the P1 capsid was replaced by a Firefly luciferase (550-aa) coding sequence.
T vector-based site-directed mutagenesis.
Site-directed mutagenesis was used to obtain the desired mutations. In each mutation, clustered positively charged lysine (K) and arginine (R) or negatively charged aspartic acid (D) and glutamic acid (E) residues (2 to 4) in the PV 2CATPase protein were replaced with neutral alanine by changing the corresponding codons. First, a Wt 2CATPase nucleotide sequence flanking the SnaBI and HpaI restriction sites in pT7PVM was amplified by PCR and ligated into the pGEM-T vector, which was used as the template for site-directed mutagenesis in the 2CATPase coding sequence. Then, 16 pairs of oligonucleotide primers were designed that contained the alanine-scanning mutations and introduced into the pGEM-T vector that contained the Wt 2CATPase sequence, using a Stratagene QuikChange site-directed mutagenesis kit according to the instruction manual. The mutated sites and the corresponding codon changes are summarized in Table 1. After sequencing analysis, the designed 2CATPase mutations were then subcloned from the pGEM-T vector into pT7PVM or the Renilla luciferase reporter virus (R-PPP) or F-Luc replicon using restriction sites SnaBI and XhoI (mutants 1 to 6) or XhoI and HpaI (mutants 7 to 16).
Table 1.
List of the 16 clustered charged-to-alanine mutants of 2CATPase and the corresponding amino acid and nucleotide changes
| Mutant (mutations) | Wild-type codons | Alanine codonsa |
|---|---|---|
| 1 (K6A/K7A) | AAG AAG | gcG gcG |
| 2 (K33A/E34A/K35A) | AAG GAG AAA | gcG GcG gcA |
| 3 (R41A/D42A/K43A) | AGA GAT AAG | gcA GcT gcG |
| 4 (K49A/R51A) | AAA CTT AGA | gcA CTT gcA |
| 5 (K88A/R89A) | AAG AGG | gcG gcG |
| 6 (E97A/K99A/R100A) | GAA GCC AAA AGA | GcA GCC gcA gcA |
| 7 (E148A/R149A/E150A) | GAA AGA GAA | GcA gcA GcA |
| 8 (R240A/R241A) | AGG CGC | gcG gcg |
| 9 (D245A/D247A) | GAC ATG GAC | GcC ATG GcC |
| 10 (R256A/D257A) | AGA GAT | gcA GcT |
| 11 (K279A/R280A) | AAG AGA | gcG gcA |
| 12 (D294A/K295A) | GAC AAA | GcC gcA |
| 13 (R298A/R300A) | AGA GTT AGA | gcA GTT gcA |
| 14 (E313A/R314A/R316A/R317A) | GAG AGA AAC AGA AGA | GcG gcA AAC gcA gcA |
| 15 (D183A/D186A/K188A) | GAT GGT GCG GAC ATG AAG | GcT GGT GCG GcC ATG gcG |
| 16 (E207A/E208A/K209A) | GAG GAG AAA | GcG GcG gcA |
Lowercase letters indicate the nucleotide changes.
RNA transcription, transfection, and in vitro translation.
Wt and mutant plasmids of pT7PVM were linearized at a unique EcoRI restriction site and used as templates for in vitro RNA synthesis using T7 RNA polymerase. RNA transcripts (3 to 10 μg) were transfected into 35-mm-diameter HeLa R19 cell monolayers by the DEAE-dextran method as described previously (45) and incubated at 33°C, 37°C, and 39.5°C. Three days posttransfection or at the time of full cytopathic effect (CPE), viruses, if any, were harvested. Full CPE was defined as the point where 90% to 95% of the cells displayed CPE. Lysates of cells transfected with mutants lacking CPE were inoculated into fresh 35-mm-diameter HeLa R19 cell monolayers for up to four subsequent serial passages. To assess the virus titers (PFU/ml) and plaque phenotypes of the viable alanine-scanning mutants, any samples displaying full CPE at 33°C were then subjected to plaque assays at 33°C, 37°C, and 39.5°C. The identity of the viruses was confirmed or determined by reverse transcription-PCR (RT-PCR)/sequencing analysis.
HeLa cell S10 cytoplasmic extracts were prepared and in vitro RNA translations were performed with these cytoplasmic extracts at 34°C, as previously described (28).
Plaque assays.
Plaque assays were performed on HeLa R19 monolayers using 0.6% tragacanth gum. After 72 h of incubation at 33°C or 48 h of incubation at 37°C or 39.5°C, the viral plaques were developed by 1% crystal violet staining (25).
RT-PCR and sequencing analysis of viral RNAs isolated from purified plaques.
Single plaques were isolated from the plaque assay plates before staining and amplified by one passage at the same temperature in fresh 35-mm-diameter HeLa R19 monolayers. Total RNA was extracted from 200 μl lysates with 1 ml TRIzol reagent (Invitrogen) and reverse transcribed into cDNA using SuperScript III reverse transcriptase (Invitrogen). The PCR products generated using an Expand Long Template PCR system (Roche) were purified and sequenced.
Luciferase assays.
Dishes (35-mm diameter) of HeLa R19 monolayer cells were transfected with 5 μg of R-PPP RNA transcripts (linearized with PvuI for pR-PPP) and were incubated at 33°C, 37°C, and 39.5°C as needed in standard tissue culture medium (DMEM) with 2% BCS in the presence and absence of 2 mM GnHCl. Luciferase activity was determined in the lysates of cells harvested 16 h posttransfection. Cell lysates (20 μl) were mixed with 20 μl of Renilla luciferase (R-Luc) assay reagent (Promega luciferase assay system; catalog no. E2810), and R-Luc activity was measured in an OPTOCOMP I luminometer (MGM Instruments, Inc.). Cell lysates (250 μl) from transfections in the absence of GnHCl were used to infect HeLa R19 cells in the presence and absence of 2 mM GnHCl, and luciferase activity was determined in the lysates of cells harvested 8 h postinfection. For the measurement of firefly luciferase (F-Luc) (linearized with DraI for pFPP), the F-Luc substrate (Promega luciferase assay system; catalog no. E1501) was used. Cells were harvested at time points corresponding to the greatest F-Luc signal achievable at each temperature after transfection: 10 h at 33°C, 8 h at 37°C, and 6 h at 39.5°C (19).
RESULTS
Since in vivo PV RNA replication is tightly linked to translation and encapsidation is tightly linked to RNA replication (4, 48), it is difficult to design experiments that clearly distinguish between defects in replication and defects in encapsidation. We define encapsidation as the formation of mature virus particles capable of initiating an infectious cycle when added to fresh HeLa cells. Since poliovirus does not have an active mechanism of egress from HeLa cells, we always assayed encapsidated viruses by freeze-thawing of the host cells after transfection or infection, e.g., total virus is harvested from inside and outside the cells (note that full CPE eventually leads to a destruction of the host cells). It should be noted that all enteroviruses follow a strict rule: active translation is necessary in cis for RNA replication, and active RNA replication is necessary in cis for encapsidation (48). Thus, a lack of RNA synthesis results in complete absence of encapsidation. However, even efficient translation does not guarantee efficient RNA replication, and efficient RNA replication does not guarantee efficient encapsidation.
In this study, we subjected the PV 2CATPase protein to clustered charged to alanine mutagenesis, a method that has previously been shown to preferentially yield ts phenotypes in the poliovirus RNA polymerase 3Dpol (13). Such mutants might exhibit significant differences in the temperature sensitivity of the protein's various functions and facilitate studies of encapsidation separate from replication.
(i) Construction of 16 clustered charged to alanine-scanning mutants of PV 2CATPase.
With the aim of discovering novel functional domains in the 2CATPase protein that are involved in encapsidation, we designed and constructed 16 clustered charged to alanine-scanning mutants spreading over the entire length of the polypeptide. We targeted clusters (2 to 4) of adjacent or nearby charged amino acids for substitution with alanine residues (Fig. 1D; Table 1).
(ii) All 16 2CATPase mutants exhibited normal translation and processing of the PV polyprotein.
To examine the possibility that the mutants exhibited defects in translation or polyprotein processing, we translated the Wt and all of the 16 mutant RNA transcripts in vitro in HeLa cell extracts (28). After 8 h of incubation at 34°C, samples of the reactions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). As shown in Fig. 2, all of the mutants exhibited normal translation and polyprotein processing profiles. Some of the 2CATPase-related polypeptides (P2, 2BCATPase, and 2CATPase) had aberrant migration patterns (Table 2; Fig. 2), presumably due to changes in the charge of the protein resulting from substitutions of charged residues with alanine.
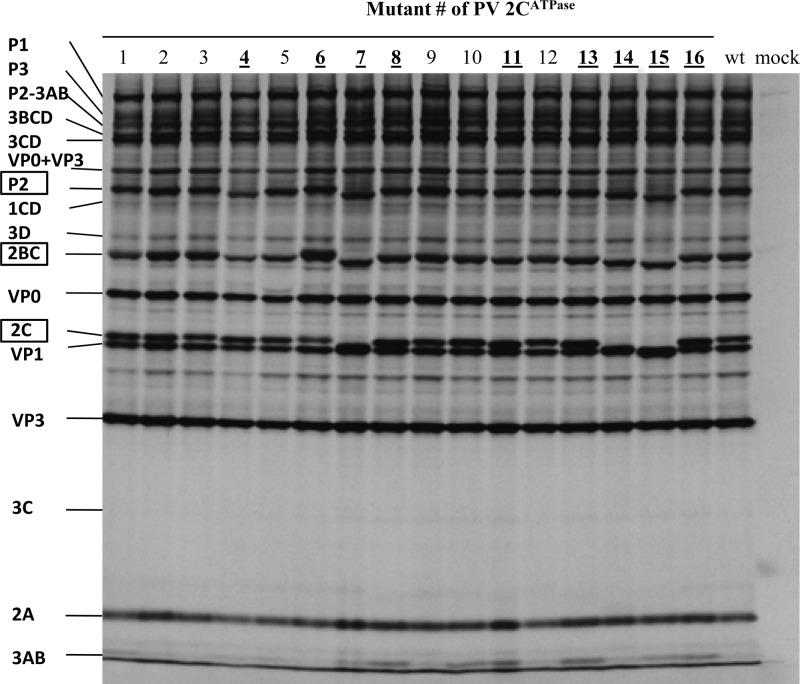
Fig 2.
In vitro translation of Wt and mutant RNA transcripts. RNA transcripts (200 ng) derived from the Wt and 16 alanine-scanning mutant constructs were translated in vitro in HeLa cell extracts at 34°C for 8 h, as described in Materials and Methods. Positions of the precursor and mature proteins are indicated, and the 2CATPase-related proteins are boxed. Those mutants that exhibited a shift in the migration of their 2CATPase-related proteins (P2, 2BC, and 2CATPase) are bolded and underlined.
Table 2.
Summary of the growth phenotypes of 16 alanine-scanning mutants of 2Ca
| Group (no. of mutants with indicated phenotype) | Alanine-scanning mutant (mutations) | Time of full CPE at indicated temp (°C)b |
Translation and polyprotein processing | P2/2BC/2C migration on SDS-PAGE gel | Replication | Reversion(s)/suppressor mutation(s) | Defect(s) | ||
|---|---|---|---|---|---|---|---|---|---|
| 33 | 37 | 39.5 | |||||||
| I (3) Wt like | 2 (K33A/E34A/K35A) | Tf | Tf | Tf | + | + | + | − | None |
| 4 (K49A/R51A) | Tf | Tf | Tf | + | Shift | + | − | None | |
| 10 (R256A/D257A) | Tf | Tf | Tf | + | + | + | − | None | |
| II (1) ts | 7 (E148A/R149A/E150A) | Tf | None or passage 1 | None | + | Shift | ts | 2C | ts, replication |
| III (10) lethal | 3 (R41A/D42A/K43A) | None | None | None | + | + | − | − | Replication |
| 5 (K88A/R89A) | None | None | None | + | + | − | − | Replication | |
| 6 (E97A/K99A/R100A) | None | None | None | + | Shift | − | − | Replication | |
| 8 (R240A/R241A) | None | None | None | + | Shift | − | − | Replication | |
| 9 (D245A/D247A) | None | None | None | + | + | − | − | Replication | |
| 12 (D294A/K295A) | None | None | None | + | + | − | − | Replication | |
| 13 (R298A/R300A) | None | None | None | + | Shift | − | − | Replication | |
| 14 (E313A/R314A/R316A/R317A) | None | None | None | + | Shift | − | − | Replication | |
| 15 (D183A/D186A/K188A) | None | None | None | + | Shift | − | − | Replication | |
| 16 (E207A/E208A/K209A) | None | None | None | + | Shift | − | − | Replication | |
| IV (2) q.i. | 1 (K6A/K7A) | Passage 2 | Passage 1 | Passage 1 | + | + | q.i. | 2C | Replication |
| 11 (K279A/R280A) | Passage 1 | Passage 1 | None or passage 1 | + | Shift | q.i. | 2C, VP3, VP1 | Replication and Encapsidation | |
ts, temperature sensitive; q.i., quasi-infectious.
Tf, full CPE at transfection; None, no CPE; None or passage 1, no CPE or full CPE at passage 1.
(iii) Analysis of the growth phenotypes of the 16 2CATPase mutants and their division into four groups.
To examine the growth properties of the mutants at different temperatures, Wt and mutant RNA transcripts were transfected into HeLa R19 cells and incubated at 33°C, 37°C, and 39.5°C for up to 72 h or until full CPE developed. Lysates of mutants producing no CPE on transfection were subjected to up to 4 serial blind passages at the same temperatures. The time required for full CPE to develop was recorded (Table 2), and the virus titers and plaque phenotypes of each viable mutant were determined at all three temperatures. Based on their proliferation phenotypes, the 16 mutants were divided into four groups: I, the wild-type-like group (three mutants); II, the temperature-sensitive (ts) group (one mutant); III, the lethal group (nine mutants); and IV, the quasi-infectious (q.i.) group (two mutants) (Fig. 1D).
Group I mutants (mutants 2, 4, and 10) possessed Wt-like growth phenotypes, producing full CPE after transfection (33°C, 37°C, and 39.5°C) (Table 2) and Wt-like virus titers and plaque sizes (data not shown). They were not further characterized. However, mutant 2 was slightly ts at 39.5°C, with 1-to-2-log-lower virus production compared to the Wt or to the other two mutants in this group.
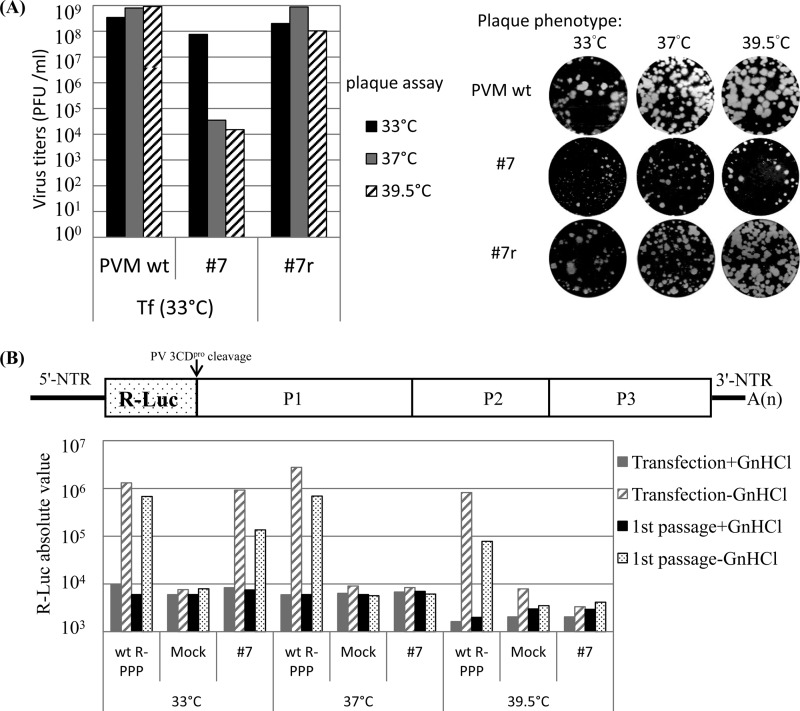
Group II presented only one (severely) temperature-sensitive mutant (mutant 7). Transfection of HeLa cells with RNA transcripts of mutant 7 at 33°C resulted in full CPE after 48 h, yielding virus titers that were similar to that of Wt virus. The plaque size, however, was greatly reduced even at the permissive temperature (Fig. 3A). Progeny viruses of mutant 7 isolated at 33°C were ts at 37°C and 39.5°C (Fig. 3A). When RNA transcripts of mutant 7 were transfected at 37°C, sometimes CPE developed after the first passage. However, attempts to isolate viable variants failed when transfection was performed at 39.5°C, even after four blind passages (Table 2).
Fig 3.
Mutant 7 is severely temperature sensitive and exhibits a RNA replication defect at the nonpermissive temperatures. (A) Growth phenotypes of mutant 7 (E148A/R149A/E150A) and its partial revertant variant 7r (E148A/R149A). The titers of lysates of the Wt and mutant 7 from the 33°C transfections (Tf) and of plaque-purified variant 7r were determined at 33°C, 37°C, and 39.5°C by plaque assay. Virus titers are shown on the left and plaque sizes on the right of the panel. For mutant 7, plaque pictures were taken from different dilutions (10−5, 10−2, and 10−2) at 33°C, 37°C, and 39.5°C, respectively. (B) Analysis of RNA replication phenotype of mutant 7. The genome structure of the R-Luc reporter virus (R-PPP) is shown on top of the panel. RNA replication and encapsidation by the Wt and mutant 7 were assayed at different temperatures using the luciferase reporter virus. Transfection of RNA transcripts and luciferase assays were performed as described in Materials and Methods. R-PPP, Renilla luciferase gene fused to three domains of the PV polyprotein; Mock, no viral RNA added.
Group III mutants (10 constructs) exhibited lethal growth phenotypes at all three temperatures: 33°C, 37°C, and 39.5°C. No progeny viruses could be isolated, even after four blind serial passages (Table 2).
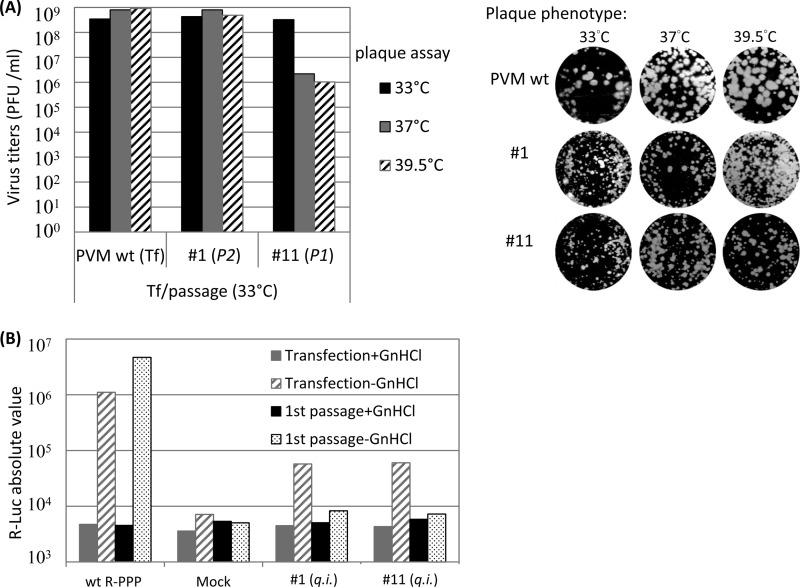
Group IV mutants were quasi-infectious, and their genetic analyses yielded the most interesting results. As the designation “quasi-infectious” (q.i.) implies, these mutants produced progeny viruses only after the evolution of genetic variants, in our case, suppressor mutations. The phenotypes of the suppressor mutants of mutants 1 and 11, however, were very different.
To enlarge the possibilities for the emergence of suppressor mutations, we passaged mutants 1 and 11 either once (passage 1) or twice (passage 2) at 33°C, 37°C, or 39.5°C until full CPE was observed (Table 2). Indeed, different kinds of suppressor mutations of mutants 1 and mutants 11 were identified, as discussed below. Interestingly, one of the suppressor mutants of mutant 11 was ts (Fig. 4A).
Fig 4.
Quasi-infectious mutants 1 and 11 are defective in RNA replication. (A) Growth phenotypes of mutants 1 and 11. At the time of full CPE (passage 2 with mutant 11; passage 1 with mutant 1), the transfection lysates at 33°C were collected and the virus titers were determined by plaque assay at 33°C, 37°C, and 39.5°C. Virus titers are illustrated on the left side and plaque sizes on the right side of the panel. For mutants, plaque pictures are taken from different dilutions at different temperatures as follows: for mutant 1, 10−5, 10−6, and 10−5 at 33°C, 37°C, and 39.5°C, respectively; and for mutant 11, 10−5, 10−3, and 10−3 at 33°C, 37°C, and 39.5°C, respectively. (B) HeLa R19 cells were transfected with 5 μg of Wt or mutant Renilla luciferase reporter virus RNA transcripts in both the presence and absence of 2 mM GnHCl, and then they were subjected to passage once at 33°C on HeLa cells. Luciferase activities were determined as described in Materials and Methods. Mock, no viral RNA added.
(iv) Analysis of RNA replication and encapsidation phenotypes of the defective 2CATPase mutants using Renilla reporter polioviruses (R-PPP).
To further characterize RNA replication and encapsidation phenotypes, we applied the strategy of the use of Renilla luciferase (R-Luc) reporter viruses (R-PPP) (25) in which the R-Luc coding sequence is fused to the N terminus of the PV polyprotein (PPP, indicating the three domains of the PV polyprotein). During translation of R-PPP transcripts or genomic RNA, the R-Luc reporter polypeptide is cleaved from the polyprotein by 3CDpro (Fig. 3B) (25), which, in turn, signals the extent of viral RNA translation or replication. RNA transcripts of Wt pR-PPP or mutant pR-PPPs were transfected into HeLa cells, in both the absence and presence of guanidine HCl (GnHCl), a strong inhibitor of PV RNA synthesis (34). Luciferase activity was measured at 16 h posttransfection. In the presence of GnHCl, the R-Luc signal represents translation of the input RNA, whereas in the absence of the drug, the signal is a measure of viral RNA replication (25). To test for encapsidation, cell lysates from transfections in the absence of GnHCl were subjected to passage to fresh cells. These cells were then incubated, again in both the absence and presence of GnHCl, followed by measurement of luciferase signal at 8 h postinfection. Encapsidation was inferred from the detection of luciferase activity in the lysates of the cells that were subjected to passage (25).
The genomes of all 13 group II to IV mutants were analyzed by the R-PPP strategy. Both replication and encapsidation of ts mutant 7 (group II) at 33°C were slightly less efficient than that of the Wt (Fig. 3B). However, at 37°C and 39.5°C, both processes were completely inhibited, indicating that the defect of mutant 7 in proliferation is likely related to RNA replication. All group III mutants were found to be defective in RNA replication and, therefore, also in encapsidation (data not shown), whereas q.i. mutants 1 and 11 (group IV) exhibited a less severe replication phenotype at 33°C (Fig. 4B).
(v) A partial reversion rescued the ts phenotype of mutant 7.
When HeLa cells were transfected with transcripts of mutant 7 at 33°C, spontaneously formed variants capable of growing at 39.5°C were found in the 33°C lysate. The strategy for analyzing mutants and variants was to plaque assay and propagate the 33°C-transfection lysates at both 33°C and 39.5°C (Fig. 3A). Following reverse transcription and PCR (RT-PCR) of the total RNA, PCR fragments of full-length PV genomes propagated either at 33°C or at 39.5°C (selecting the revertant/suppressor mutants of ts mutant 7) were sequenced. As expected, only the mutant 7 genomes (E148A/R149A/E150A) but no revertants or suppressors of mutant 7 were detected in 33°C-propagated lysates (due to the overwhelming excess of ts mutant 7 genotypes in the original inoculum). At 39.5°C, however, only the unique variant 7r (E148A/R149A), which contained an A150E reversion, could replicate and be expanded. This partial revertant, variant 7r (E148A/R149A), exhibited a wt-like growth phenotype when assayed at all three temperatures (Fig. 3A). This result suggests that the presence of an alanine residue at position 150 was the primary cause of the ts phenotype of mutant 7. This was confirmed by constructing an E150A 2CATPase mutant of Wt PV that indeed exhibited a ts phenotype similar to that of mutant 7 (data not shown).
(vi) Suppressor mutations of q.i. mutant 1 generated a novel Q∧R cleavage site at 2B∧2CATPase.
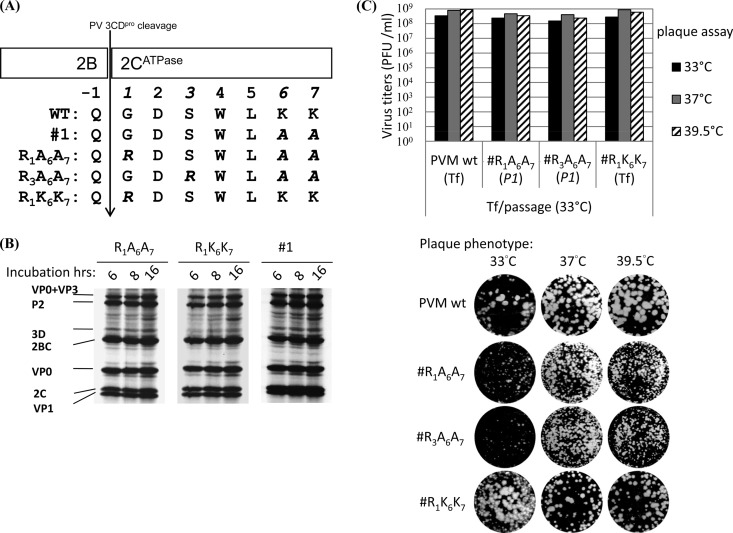
Mutant 1 (K6A/K7A) yielded two types of suppressor mutations (G1R and S3R) at the N terminus of 2CATPase during the first passage on HeLa cells at 37°C. Both of these mutants had substituted a neutral residue (G1 or S3) upstream of the original mutated sites (K6A/K7A) for a positively charged arginine (R) residue, while retaining the original (K6A/K7A) mutations (Fig. 5A). Interestingly, the G1R suppressor mutation generated an unusual Q∧R 2B∧2CATPase proteinase cleavage site for 3CDpro (Q∧G in the Wt; Fig. 5A). To confirm that the G1R and S3R mutations were responsible for rescuing the q.i. phenotype of the original mutant 1, we introduced these changes individually into the mutant 1 background in pT7PVM, yielding two variants, R1A6A7 and R3A6A7 (Fig. 5A). In addition, to examine the effect of the G1R mutation alone, we constructed a R1K6K7 mutant (Fig. 5A).
Fig 5.
Quasi-infectious mutant 1 yielded suppressor mutants harboring a novel Q∧R3CDpro protease cleavage site at 2B∧2CATPase. (A) Location of suppressor mutations G1R and S3R of 2CATPase alanine-scanning mutant 1. The amino acid sequence around the 2B∧2CATPase cleavage site of the Wt, mutant 1, R1A6A7 (G1R), and R3A6A7 (S3R) suppressor mutants is shown. The G1R mutation was introduced into the Wt PV background (R1K6K7), which is also included in the panel. A vertical arrow shows the 3CDpro cleavage site at 2B∧2CATPase. The last amino acid in 2B is indicated by −1 and the amino acids of 2CATPase by 1 to 7. (B) In vitro translation assays of mutant 1 (Wt-like) and of two reconstructed suppressor mutants, R1A6A7 and R1K6K7. Translations were carried out in HeLa cell extracts in vitro at 34°C for 6 h, 8 h, and 16 h (see Materials and Methods). Only the upper portion of the gel (VP1 and up) is shown. (C) Growth phenotypes of the Wt and of all three derivatives of mutant 1. RNA transcripts were transfected into HeLa cells, and the lysates were collected at the time of full CPE. At 33°C, the R1A6A7 mutant yielded full CPE only at passage 1. The titers of the lysates were determined by plaque assay at 33°C, 37°C, and 39.5°C. The virus titers are shown at the top and the plaque phenotypes at the bottom of the panel.
We first investigated the processing efficiency of the novel Q∧R cleavage site by translating in vitro the RNA transcripts of the reconstructed R1A6A7 and R1K6K7 variants at different time points. Normal polyprotein translation and processing and, especially, normal production of 2CATPase were observed (Fig. 5B). This suggested that the G1R suppressor mutation confers a normal translation and protein processing pattern to both the R1A6A7 (G1R mutation in the context of mutant 1) and R1K6K7 (G1R mutation in the context of Wt PV) mutants, an observation indicating that Q∧R can be efficiently cleaved by 3CDpro. Suppressor variant R3A6A7 also showed normal in vitro translation and polyprotein processing (data not shown). We also tested the growth phenotypes of these three variants. The R1A6A7 and R3A6A7 reconstructed suppressor variants yielded progeny on HeLa cells at 33°C during the first passage (Fig. 5C). The virus titers were comparable to that of the Wt at all 3 temperatures tested (33°C, 37°C, and 39.5°C), but the plaque size was small (Fig. 5C). The reconstructed R1K6K7 mutant, however, yielded full CPE immediately after transfection, and both virus titer and plaque size were normal (Fig. 5C). All three reconstructed mutants showed full CPE upon transfection at 37°C and 39.5°C (data not shown).
(vii) Suppressor mutations of q.i. mutant 11 revealed genetic suppression of 2CATPase encapsidation defects by capsid proteins VP1 and VP3.
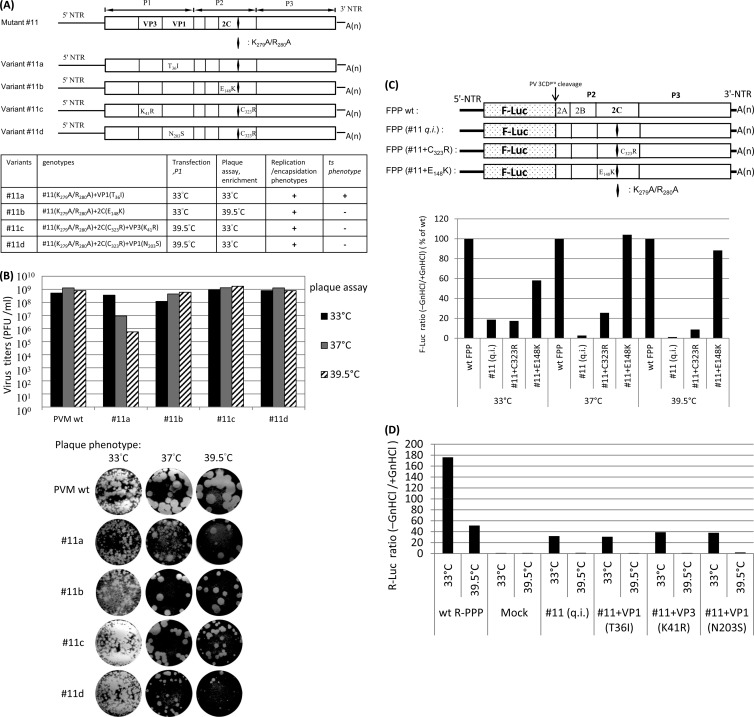
Mutant 11 (K279A/R280A), just like mutant 1, was quasi-infectious, and thus, progeny viruses isolated from HeLa cell lysates were always found to be genetic variants thereof. Direct revertants of the mutations at positions 279 and/or 280 of mutant 11, however, were not observed, presumably because of the large number (four) of nucleotides changed. To extend the opportunity of finding suppressor mutations, we carried out transfections, passage, plaque assays, and enrichment of variants at a combination of the two different temperatures 33°C and 39.5°C (Fig. 6A, bottom panel). Application of different temperatures to a mutant is expected to maximize the number of emerging suppressor mutations. Accordingly, RNA transcripts of mutant 11 were transfected into HeLa cells either at 33°C or at 39.5°C and, after development of full CPE (passage 1), emerging viruses were plaque purified and expanded, again either at 33°C or at 39.5°C (Fig. 6A, bottom panel). The full-length genomes of four isolated variants (variants 11a, 11b, 11c, and 11d) were then sequenced. Note that all four suppressor variants of mutant 11 still carried the original mutant 11 mutation K279A/R280A, as indicated by the diamond in Fig. 6A.
Fig 6.
Growth phenotypes of four suppressor variants of mutant 11. (A) Four suppressor variants of mutant 11, all of which retain the original mutant 11 mutation at K279A/R280A, as indicated by the diamond in the schematic genomes. The genomic structures of mutant 11 and its four suppressor variants are shown on the top. The locations of suppressor mutations in 2CATPase (either C323R or E148K) and/or capsid proteins VP3 (K41R) and VP1 (T36I or N203S) are indicated. The genotypes and the selection temperatures and phenotypes of the four suppressor variants are summarized at the bottom. (B) Growth phenotypes of the four suppressor variants of mutant 11. Plaques were picked and amplified at the temperatures indicated in panel A, and the titers of the lysates were determined by plaque assay at 33°C, 37°C, and 39.5°C and incubated for 72 h. Note that plaque pictures of variant 11 at 37°C and 39.5°C were taken from a different dilution (10−4) instead of 10−7 for the Wt and other variants. (C) The genomic structures of F-Luc replicons (FPP) of the Wt and mutant 11 and its suppressor variants, FPP (mutant 11 plus C323R) and FPP (mutant 11 plus E148K), which carry only single suppressor mutations in 2CATPase (either C323R or E148K) in addition to the original mutant 11 mutation at K279A/R280A, are shown at the top. RNA replication of these constructs was assayed at 33°C, 37°C, and 39.5°C in the absence and presence of GnHCl (see Materials and Methods). The ratio of F-Luc levels observed in the absence and presence of GnHCl is plotted; at each temperature, the ratio of the Wt is taken as 100%. (D) Capsid suppressor mutations do not rescue the RNA replication defect of mutant 11. HeLa R19 cells were transfected with 5 μg of Renilla luciferase reporter virus RNA transcripts of the Wt or mutant 11 and its variants with capsid suppressor mutations at 33°C or 39.5°C in both the presence and absence of 2 mM GnHCl. Luciferase activities were determined as described in Materials and Methods. The ratio of R-Luc levels observed in the absence and presence of GnHCl is plotted. Mock, no viral RNA added.
The results of these experiments are summarized in Table 2 and Fig. 6. The most intriguing variant identified was variant 11a, which was isolated by transfection, first passage, and plaque purification at 33°C. Variant 11a contained a single suppressor mutation in capsid protein VP1 (T36I) (Fig. 6A). Although this variant was found to be ts at 37°C and 39.5°C, it proliferated at 33°C to titers equivalent to that of Wt PV (Fig. 6B), an observation demonstrating that the defect of mutant 11 was fully rescued at this temperature by “cross-talk” of an as-yet-unknown character between VP1 and 2CATPase.
Variant 11b, isolated after transfection and first passage at 33°C and plaque purification at 39.5°C, carried a single suppressor mutation in 2CATPase (E148K) (Fig. 6A). E148K fully rescued the growth phenotype of the parental mutant 11 at 37°C and 39.5°C, although the rescue was less robust at 33°C (Fig. 6B). Just as with mutant 11a, a single suppressor mutation in mutant 11b rescued the q.i. phenotype of mutant 11, although the former was a capsid VP1 (T36I) suppressor mutation whereas the latter was a 2CATPase (E148K) suppressor mutation.
Two other interesting suppressor mutants, variants 11c and 11d, were isolated after transfection and first passage at 39.5°C and plaque purified at 33°C (Fig. 6A). Both variants carried two suppressor mutations. They shared the same suppressor mutation in 2CATPase (C323R), but variant 11c had an additional mutation in VP3 (K41R) whereas variant 11d had an additional mutation in VP1 (N203S).
We were interested to test which of these suppressor mutations rescued RNA replication. Of particular interest were the suppressor mutations E148K of mutant 11b and C323R of mutant 11c/d, both mapping to 2CATPase. The experiments were carried out with an F-Luc replicon (F-PP) in which the capsid-coding region of the PV polyprotein was replaced by the F-Luc coding sequence (Fig. 6C) (24). This F-PP reporter replicon eliminates any effect of capsids and allows study of the effect of the mutations directly on RNA replication.
The replicon of FPP (mutant 11) could replicate at 33°C, although less robustly than the Wt (Fig. 6C), whereas at higher temperatures replication was negligible. The VP1 (T36I) capsid suppressor mutation did not improve replication of mutant 11, as shown by the R-PPP reporter virus data (Fig. 6D). However, suppressor mutant 11a produced, remarkably, virus yields similar to that of Wt PV at 33°C (Fig. 6B). These data indicate that at 33°C, a capsid mutation corresponding to VP1 (T36I) rescued the encapsidation defect of mutant 11.
F-PP (mutant 11 plus E148K) replicated well at 37°C and 39.5°C but not as well at 33°C, an observation indicating rescue of RNA replication of mutant 11 by the single suppressor mutation in 2CATPase (E148K). Since suppressor variant 11b grew well at 37°C and 39.5°C, with virus yields comparable to that of the Wt (Fig. 6B), it appears that rescue of the encapsidation defect of mutant 11 at these temperatures was achieved by 2CATPase (E148K). We propose that there is an interaction between the C-terminal domain of 2CATPase and an amino acid sequence between NTP binding motifs A and B of the protein.
The C323R mutation in variants 11c and 11d, on the other hand, had no effect at 33°C and only partially rescued RNA replication at 37°C and to a lesser extent at 39.5°C, as shown by the FPP data (mutant 11 plus C323R) in Fig. 6C. Indeed, mutation C323R was never observed in isolation but rather was always accompanied by a capsid mutation in either VP3 (mutant 11c) or VP1 (mutant 11d). Just as C323R itself was unable to rescue encapsidation, the capsid mutations in mutant 11c or 11d alone could rescue neither replication (Fig. 6D) nor subsequent encapsidation (data not shown) of mutant 11. Excitingly, the double suppressor mutants 11c and 11d exhibited growth properties similar to the those of the Wt virus since they proliferated at all three temperatures (Fig. 6B). These data strongly suggest that one of the two capsid mutations, VP1 (N203S) or VP3 (K41R), in cooperation with C323R in 2CATPase, rescued encapsidation of mutant 11. These genetic data strongly support the conclusion that 2CATPase acts in conjunction with capsid polypeptides during PV morphogenesis.
(viii) Conservation of the charged amino acids in C-cluster enterovirus 2CATPase sequences covaries with the debilitating effect of alanine mutations.
The more conserved the original charged amino acid at the sites of alanine scanning among human C-cluster enteroviruses, the more debilitated the PV 2CATPase alanine-scanning mutants. Except for mutant 4, the growth phenotypes of the 2CATPase alanine-scanning mutants described here covary with the extent of conservation of the corresponding charged amino acid sequence among the human C-cluster enteroviruses. The more conserved the cluster of charged amino acids, the more debilitated the alanine-scanning mutants (Table 3). All mutants with low conservation (4/4) of the charged amino acid were viable. The majority of mutants with high conservation (10/12) of the charged amino acid were nonviable.
Table 3.
Amino acid sequence conservation at the sites of alanine scanning of PV 2CATPase mutants among human C cluster enterovirusesa
| Group (no. of mutants with indicated phenotype) | Mutant and amino acids mutated | Amino acid sequence conservationb | Variation(s) of amino acids among human C cluster enteroviruses |
|---|---|---|---|
| I (3) Wt like | 2 K33/E34/K35 | Low | K33R/E34(N/T/C)/K35R |
| 4 K49/R51 | High | Only one R51K | |
| 10 R256/D257 | Low | R256(V/I/T)/D257(K/R) | |
| II (1) ts | 7 E148/R149/E150 | Low | E148K/R149(K/Q)/E150A |
| III (10) lethal | 3 R41/D42/K43 | High | R41K |
| 5 K88/R89 | High | K88R/R89K | |
| 6 E97/K99/R100 | High | K99R | |
| 8 R240/R241 | High | None | |
| 9 D245/D247 | High | D247E | |
| 12 D294/K295 | High | Only one K295R | |
| 13 R298/R300 | High | None | |
| 14 E313/R314/R316/R317 | High | R314K | |
| 15 D183/D186/K188 | High | None | |
| 16 E207/E208/K209 | High | None | |
| IV (2) q.i. | 1 K6/K7 | High | None |
| 11 K279/R280 | Low | K279S/R280(K/T) | |
| Suppressor mutations | G1 | High | Only one G1S |
| S3 | High | Only one S3G | |
| E148 | Low | E148K in CAV1/19/22 | |
| C323 | High | None |
Numbering of amino acid residues is according to the PVM sequence.
Low, amino acids varied in electronic charge; High, amino acids, some of which may have amino acid changes from K to R, R to K, D to E, G to S, or S to G as listed above, were 100% conserved in electronic charge.
DISCUSSION
The aim of this work was to identify and characterize novel functional domains of the PV protein 2CATPase and particularly ones involved in the process of encapsidation. Encapsidation is the last and a crucial step of the virus's life cycle. Successful assembly not only provides newly synthesized viral genomes with a protective shell but also ensures proper uncoating, which is required for attachment to and penetration into subsequent host cells to initiate the next round of viral infection. In addition, capsid proteins are among the major determinants of tropism due to their ability to bind to specific receptors for cell entry (29). Previous studies by both drug inhibition and genetic experiments have identified various sites in the 2CATPase polypeptide important for encapsidation (Fig. 1C). Hydantoin-resistant mutants in 2CATPase (Q65R, L125V, V218I [44], H118Y, and V194I [Paul et al., unpublished]) map to regions at or near A and C motifs of the NTP-binding domain and near the N terminus of the protein. Moreover, genetic studies have generated uncoating defect mutations (M293V, K295R) near the C terminus of the polypeptide (22). Finally, an extensive genetic study of enterovirus chimeras coupled with biochemical experiments has provided strong evidence that encapsidation of enteroviruses does not depend on an RNA packaging signal but rather on an interaction between the capsid protein(s) and the nonstructural protein 2CATPase (25). Specifically, genetic studies of the C-cluster coxsackie virus 20/poliovirus chimera (C20PP) implicated residue N252 (downstream of box C) of poliovirus 2CATPase as an essential contact point with CAV20 VP3 for morphogenesis (25).
In order to identify novel functional domains of PV 2CATPase, in particular, ones involved in encapsidation, we have carried out clustered charged to alanine-scanning mutagenesis of residues primarily located in heretofore-uncharacterized regions of the polypeptide. We were particularly interested in generating a conditional lethal mutant (e.g., a ts mutant) that would facilitate the dissection of individual steps in morphogenesis. Surprisingly, of the 16 charged to alanine mutants, only one (6%) exhibited a severely ts growth phenotype, a yield of conditional lethal mutants significantly lower than that observed in similar studies with PV RNA polymerase (35%) (13). This result is likely related to the multifunctional nature of the 2CATPase polypeptide. If we use a less strict standard of definition of the ts phenotype, mutant 2 is also ts, which amounts to a 12% ts growth phenotype. Most of the mutants were nonviable (63%) or q.i. (13%). All mutants but the three Wt-like ones possessed severe defects in RNA replication, an observation indicating that charged residues in the protein are very important for this function.
The current study has unveiled several new sites in PV 2CATPase essential for RNA replication. Since RNA replication is a prerequisite for encapsidation, we cannot rule out the possibility that these sites may also be involved in encapsidation. Of the 10 lethal mutations conferring RNA replication defects, only one (mutation 3) is located within a domain of known function (membrane binding) (Fig. 1B and D) whereas other lethal mutations are located upstream of motif A (mutations 5 and 6), between motifs B and C, or downstream of motif C (mutations 8 and 9) of the NTP-binding domain (27). The remaining three lethal mutations (mutations 12, 13, and 14) are located downstream of the cysteine-rich Zn++-binding domain near the C terminus of the polypeptide. Surprisingly, two (mutations 2 and 4) of the three mutations resulting in Wt-like growth phenotypes are located near the N terminus of the protein, where several functional domains overlap (Fig. 1B and D). In fact, mutant 2 exhibited a mild ts phenotype at 39.5°C, with 1-to-2-log-lower virus production compared to the Wt or the other two group I mutants (data not shown), though this was not investigated further. The third Wt-like mutant (mutant 10) (R256A/D257A) was the most robust mutant, with a growth phenotype identical to that of Wt PV (data not shown). Mutant 10 carries changes downstream of motif C (Fig. 1B and D) near N252, the site in 2CATPase known to interact with VP3 in the C20PP chimeric virus (25). Interestingly, the N252 residue itself can be mutated to Gly, Ser, or Ala without affecting PV morphogenesis (data not shown). These data suggest that a region downstream of motif C in PV 2CATPase near R252, R256, and R257 is tolerant to certain genetic changes. The same region downstream of motif C (S255 and A263) has previously been shown also to tolerate linker insertions (23).
In vitro translation in HeLa cell extracts (28) indicated that none of the alanine mutations of PV 2CATPase interfere with protein synthesis and proteolytic processing (Fig. 2). However, one of the suppressor mutations identified in this study provided novel insight into proteolytic processing of PV. The suppressor mutation G1R of mutant 1 (K6A/K7A) generated a unique 3Cpro/3CDpro proteinase cleavage site at 2B∧2CATPase. In contrast to most picornaviruses, polioviruses are “purists” in that all of their natural 3Cpro/3CDpro cleavage sites are Q∧G; thus, the generation of the Q∧R site was surprising. The only naturally occurring Q∧R site among picornavirus polyproteins exists in the hepatitis A virus (HAV) polyprotein between proteins 3Cpro and 3Dpol (38). Our in vitro translation assays demonstrated that processing between 2B and 2CATPase at the Q∧R cleavage site in the Q∧R suppressor polyproteins was just as efficient as at the original Q∧G site (Fig. 5B). It should be noted that the G-to-R mutation involves only a single nucleotide change instead of the two required for the A-to-R substitution (Table 1). This likely explains why the virus regenerated a positively charged residue at position 1 rather than reverting the nucleotides of the A codons at positions 6 or 7.
The other suppressor mutations revealed two novel functional domains in the 2CATPase polypeptide, which differ from those domains identified by the lethal mutations. These mutations, arising in the 2CATPase protein, always regenerated a charged amino acid that was originally eliminated by the charged to alanine substitution. These were either at the same location as (A150E) or at locations that differed from (E148K, C323R) that of the original residue change. The first domain that we propose to play a role in 2CATPase function(s) is located in the amino acid sequence between the A and B motifs of the NTP-binding domain, involving A150E and E148K. A150E is a partial revertant of ts mutant 7 (E148A/R149A/E150A), and E148K is a suppressor mutation of mutant 11 (K279A/K280A). The presence of a suppressor mutation (E148K) at this location suggests a functional interaction between the C terminus of the 2CATPase polypeptide and the amino acid sequence between motifs A and B of the NTP-binding domain (27), within either the 2CATPase protein or 2CATPase oligomers, which may contribute to the signature of different viruses. The second domain maps to the original mutated site (K279A/R280A) of mutant 11. These mutations are localized close to the C terminus of the 2CATPase polypeptide within the Zn++-binding domain that consists of five cysteines (C269, C272, C281, C282, and C286) residues and one histidine (H273) residue. It should be noted that residues K279A/R280A of mutant 11 are located just upstream of M293 and K295, which were previously shown to be important for uncoating and, presumably, for virion structure. Based on our results, we propose that the domain containing residues K279/R280 is involved in encapsidation.
Interestingly, the growth phenotypes of all but one (mutant 4) of our 2CATPase alanine-scanning mutants correlate with the extent of conservation of the charged amino acid sequence among human C cluster enteroviruses (Table 3). We thus predict that if we were to make the same alanine mutations at the corresponding highly conserved sites of related enteroviruses, the majority, if not all, of the mutants would exhibit lethal phenotypes. Interestingly, one suppressor variant, 11b (E148K plus K279A/R280A), mimics the electronic charges of three wild-type CAVs (CAV1/CAV19/CAV22) (K148 plus S279/T280) at aa 148 (positive charged), 279 (noncharged), and 280 (noncharged). This strongly supports our proposal that there is an interaction between the C-terminal domain of the 2CATPase protein and the amino acid sequence between NTP binding motifs A and B of the protein. This interaction might be either within the 2CATPase protein or between 2CATPase oligomers. Such an interaction of 2CATPase in PV may extend to other members of the Enterovirus genus.
The most important conclusion derived from our work is that both VP3 and VP1 communicate with 2CATPase, most likely with the C-terminal domain of the protein, and that this interaction is required for encapsidation. This conclusion is strongly supported by the emergence of suppressor variants 11a, 11c, and 11d of mutant 11 (Fig. 6). These suppressor variants were acquired by subjecting original mutant 11 to different selection pressures with a combination of low and high temperatures during transfection and passage and during plaque purification and enrichment of variants, a simple but powerful design. From the properties of the progeny viruses, we are able to explain why different variants were selected under different growth conditions. When selected at 33°C, mutant 11 was able to replicate its RNA but was unable to produce progeny viruses because of an encapsidation defect. This encapsidation defect can be corrected by capsid mutation VP1 (T36I), which evolves as a suppressor at 33°C. In addition, 2CATPase mutation E148K overcomes both the replication defect and the encapsidation defect of mutant 11. When selection is carried out at 39.5°C, the RNA replication defect of mutant 11 can be partially rescued by 2CATPase mutation C323R. It then needs an additional capsid mutation such as VP1 (N203S) or VP3 (K41R) to correct the defect in encapsidation. In either case, these genetic data strongly support the conclusion not only that 2CATPase is involved in encapsidation but also that it does so in conjunction with capsid polypeptides VP1 or VP3 to facilitate PV morphogenesis. Note that a stimulating influence of the capsid proteins on RNA replication has never been observed, a statement supported by numerous constructs reported in the literature.
However, we cannot at present determine whether VP1 or VP3 binds directly to 2CATPase or whether mutations in these capsid proteins structurally rearrange higher-order capsid precursors that can then directly bind to 2CATPase in encapsidation (25). The capsid precursor P1 of PV is processed and assembled to form protomers (VP0, VP3, VP1). Available evidence suggests that the protomer does not dissociate into the individual cleavage products in the cytoplasm of the infected cell but, rather, that it pentamerizes to (VP0,VP3,VP1)5—the building block of the immature capsid [(VP0,VP3,VP1)5]12—as discussed by Liu et al. (25). In the future, it will be interesting to determine whether or not there is a direct physical interaction between VP1 and 2CATPase and at which stage of an intermediate(s) 2CATPase associates with capsid proteins. Interestingly, the VP1 and VP3 suppressor mutations involved the exchange of a polar residue to one with increased hydrophobicity (for VP1, T36I, or N203S) or of one charged residue to another (for VP3, K41R). One of the VP1 residues (T36) is located near the N terminus of the VP1 protein, which was previously shown to be involved both in RNA release from the infecting virion and in the interaction with or insertion of the progeny RNA into capsid proteins during morphogenesis (20). On the basis of the study of two small N-terminal deletions of VP1 of poliovirus, Kirkegaard and colleagues have concluded that RNA packaging and RNA release are genetically linked but can be mutated separately in different VP1 alleles. Whether an interaction between VP1 and 2CATPase is required for these processes remains to be determined. Our results clearly demonstrate, however, that the selection of revertants or suppressor mutants generated from clustered charged amino acid to alanine mutagenesis is a useful method of identifying viral protein-interacting components of the replication-encapsidation complex.
ACKNOWLEDGMENTS
We thank Steffen Mueller for the Renilla reporter virus clone and for discussions and Jiang Yin for valuable information on picornavirus polyprotein processing.
C.S. was a Visiting Scholar from the University of Edinburgh, United Kingdom. This work was supported by a grant from the NIH (R37AI015122).
Footnotes
Published ahead of print 3 July 2012
REFERENCES
- 1. Adams P, Kandiah E, Effantin G, Steven AC, Ehrenfeld E. 2009. Poliovirus 2C protein forms homo-oligomeric structures required for ATPase activity. J. Biol. Chem. 284:22012–22021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aldabe R, Carrasco L. 1995. Induction of membrane proliferation by poliovirus proteins 2C and 2BC. Biochem. Biophys. Res. Commun. 206:64–76 [DOI] [PubMed] [Google Scholar]
- 3. Baltera RF, Jr, Tershak DR. 1989. Guanidine-resistant mutants of poliovirus have distinct mutations in peptide 2C. J. Virol. 63:4441–4444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baltimore D. 1969. Biochemistry of viruses. Marcel Dekker, Inc., New York, NY [Google Scholar]
- 5. Banerjee R, Echeverri A, Dasgupta A. 1997. Poliovirus-encoded 2C polypeptide specifically binds to the 3′-terminal sequences of viral negative-strand RNA. J. Virol. 71:9570–9578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Banerjee R, Tsai W, Kim W, Dasgupta A. 2001. Interaction of poliovirus-encoded 2C/2BC polypeptides with the 3′ terminus negative-strand cloverleaf requires an intact stem-loop b. Virology 280:41–51 [DOI] [PubMed] [Google Scholar]
- 7. Banerjee R, Weidman MK, Echeverri A, Kundu P, Dasgupta A. 2004. Regulation of poliovirus 3C protease by the 2C polypeptide. J. Virol. 78:9243–9256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Barton DJ, Flanegan JB. 1997. Synchronous replication of poliovirus RNA: initiation of negative-strand RNA synthesis requires the guanidine-inhibited activity of protein 2C. J. Virol. 71:8482–8489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bienz K, Egger D, Pasamontes L. 1987. Association of polioviral proteins of the P2 genomic region with the viral replication complex and virus-induced membrane synthesis as visualized by electron microscopic immunocytochemistry and autoradiography. Virology 160:220–226 [DOI] [PubMed] [Google Scholar]
- 10. Cho MW, Teterina N, Egger D, Bienz K, Ehrenfeld E. 1994. Membrane rearrangement and vesicle induction by recombinant poliovirus 2C and 2BC in human cells. Virology 202:129–145 [DOI] [PubMed] [Google Scholar]
- 11. Crowther D, Melnick JL. 1961. Studies of the inhibitory action of guanidine on poliovirus multiplication in cell cultures. Virology 15:65–74 [DOI] [PubMed] [Google Scholar]
- 12. Cuconati A, Xiang W, Lahser F, Pfister T, Wimmer E. 1998. A protein linkage map of the P2 nonstructural proteins of poliovirus. J. Virol. 72:1297–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Diamond SE, Kirkegaard K. 1994. Clustered charged-to-alanine mutagenesis of poliovirus RNA-dependent RNA polymerase yields multiple temperature-sensitive mutants defective in RNA synthesis. J. Virol. 68:863–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Echeverri A, Banerjee R, Dasgupta A. 1998. Amino-terminal region of poliovirus 2C protein is sufficient for membrane binding. Virus Res. 54:217–223 [DOI] [PubMed] [Google Scholar]
- 15. Goodfellow I, et al. 2000. Identification of a cis-acting replication element within the poliovirus coding region. J. Virol. 74:4590–4600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gorbalenya AE, Koonin EV. 1993. Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 3:419–429 [Google Scholar]
- 17. Reference deleted.
- 18. Hanecak R, Semler BL, Anderson CW, Wimmer E. 1982. Proteolytic processing of poliovirus polypeptides: antibodies to polypeptide P3-7c inhibit cleavage at glutamine-glycine pairs. Proc. Natl. Acad. Sci. U. S. A. 79:3973–3977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jahan N, Wimmer E, Mueller S. 2011. A host-specific, temperature-sensitive translation defect determines the attenuation phenotype of a human rhinovirus/poliovirus chimera, PV1(RIPO). J. Virol. 85:7225–7235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kirkegaard K. 1990. Mutations in VP1 of poliovirus specifically affect both encapsidation and release of viral RNA. J. Virol. 64:195–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kirkegaard K, Baltimore D. 1986. The mechanism of RNA recombination in poliovirus. Cell 47:433–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li JP, Baltimore D. 1990. An intragenic revertant of a poliovirus 2C mutant has an uncoating defect. J. Virol. 64:1102–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li JP, Baltimore D. 1988. Isolation of poliovirus 2C mutants defective in viral RNA synthesis. J. Virol. 62:4016–4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li X, Lu HH, Mueller S, Wimmer E. 2001. The C-terminal residues of poliovirus proteinase 2A(pro) are critical for viral RNA replication but not for cis[hyphen] or trans-proteolytic cleavage. J. Gen. Virol. 82:397–408 [DOI] [PubMed] [Google Scholar]
- 25. Liu Y, et al. 2010. Direct interaction between two viral proteins, the nonstructural protein 2C and the capsid protein VP3, is required for enterovirus morphogenesis. PLoS Pathog. 6:e1001066 doi:10.1371/journal.ppat.1001066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mirzayan C, Wimmer E. 1994. Biochemical studies on poliovirus polypeptide 2C: evidence for ATPase activity. Virology 199:176–187 [DOI] [PubMed] [Google Scholar]
- 27. Mirzayan C, Wimmer E. 1992. Genetic analysis of an NTP-binding motif in poliovirus polypeptide 2C. Virology 189:547–555 [DOI] [PubMed] [Google Scholar]
- 28. Molla A, Paul AV, Wimmer E. 1991. Cell-free, de novo synthesis of poliovirus. Science 254:1647–1651 [DOI] [PubMed] [Google Scholar]
- 29. Mueller S, Wimmer E, Cello J. 2005. Poliovirus and poliomyelitis: a tale of guts, brains, and an accidental event. Virus Res. 111:175–193 [DOI] [PubMed] [Google Scholar]
- 30. Paul AV, Molla A, Wimmer E. 1994. Studies of a putative amphipathic helix in the N-terminus of poliovirus protein 2C. Virology 199:188–199 [DOI] [PubMed] [Google Scholar]
- 31. Paul AV, Rieder E, Kim DW, van Boom JH, Wimmer E. 2000. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J. Virol. 74:10359–10370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pfister T, Jones KW, Wimmer E. 2000. A cysteine-rich motif in poliovirus protein 2C(ATPase) is involved in RNA replication and binds zinc in vitro. J. Virol. 74:334–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pfister T, Wimmer E. 1999. Characterization of the nucleoside triphosphatase activity of poliovirus protein 2C reveals a mechanism by which guanidine inhibits poliovirus replication. J. Biol. Chem. 274:6992–7001 [DOI] [PubMed] [Google Scholar]
- 34. Pincus SE, Diamond DC, Emini EA, Wimmer E. 1986. Guanidine-selected mutants of poliovirus: mapping of point mutations to polypeptide 2C. J. Virol. 57:638–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rodríguez PL, Carrasco L. 1995. Poliovirus protein 2C contains two regions involved in RNA binding activity. J. Biol. Chem. 270:10105–10112 [DOI] [PubMed] [Google Scholar]
- 36. Rodríguez PL, Carrasco L. 1993. Poliovirus protein 2C has ATPase and GTPase activities. J. Biol. Chem. 268:8105–8110 [PubMed] [Google Scholar]
- 37. Suhy DA, Giddings TH, Jr, Kirkegaard K. 2000. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 74:8953–8965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tesar M, et al. 1994. Expression of hepatitis A virus precursor protein P3 in vivo and in vitro: polyprotein processing of the 3CD cleavage site. Virology 198:524–533 [DOI] [PubMed] [Google Scholar]
- 39. Teterina NL, Gorbalenya AE, Egger D, Bienz K, Ehrenfeld E. 1997. Poliovirus 2C protein determinants of membrane binding and rearrangements in mammalian cells. J. Virol. 71:8962–8972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Teterina NL, Kean KM, Gorbalenya AE, Agol VI, Girard M. 1992. Analysis of the functional significance of amino acid residues in the putative NTP-binding pattern of the poliovirus 2C protein. J. Gen. Virol. 73(Pt. 8):1977–1986 [DOI] [PubMed] [Google Scholar]
- 41. Teterina NL, et al. 2006. Evidence for functional protein interactions required for poliovirus RNA replication. J. Virol. 80:5327–5337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tolskaya EA, et al. 1994. Genetic studies on the poliovirus 2C protein, an NTPase. A plausible mechanism of guanidine effect on the 2C function and evidence for the importance of 2C oligomerization. J. Mol. Biol. 236:1310–1323 [DOI] [PubMed] [Google Scholar]
- 43. Toyoda H, et al. 1986. A second virus-encoded proteinase involved in proteolytic processing of poliovirus polyprotein. Cell 45:761–770 [DOI] [PubMed] [Google Scholar]
- 44. Vance LM, Moscufo N, Chow M, Heinz BA. 1997. Poliovirus 2C region functions during encapsidation of viral RNA. J. Virol. 71:8759–8765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. van der Werf S, Bradley J, Wimmer E, Studier FW, Dunn JJ. 1986. Synthesis of infectious poliovirus RNA by purified T7 RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 83:2330–2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Verlinden Y, Cuconati A, Wimmer E, Rombaut B. 2000. The antiviral compound 5-(3,4-dichlorophenyl) methylhydantoin inhibits the post-synthetic cleavages and the assembly of poliovirus in a cell-free system. Antiviral Res. 48:61–69 [DOI] [PubMed] [Google Scholar]
- 47. Wimmer E, Hellen CUT, Cao XM. 1993. Genetics of poliovirus. Annu. Rev. Genet. 27:353–436 [DOI] [PubMed] [Google Scholar]
- 48. Wimmer E, Paul AV. 2010. The making of a picornavirus genome, p 33–55 In Ehrenfeld E, Domingo E, Roos RP. (ed), The picornaviruses. ASM Press, Washington, DC [Google Scholar]
- 49. Yin J, Liu Y, Wimmer E, Paul AV. 2007. Complete protein linkage map between the P2 and P3 non-structural proteins of poliovirus. J. Gen. Virol. 88:2259–2267 [DOI] [PubMed] [Google Scholar]
- 50. Ypma-Wong MF, Dewalt PG, Johnson VH, Lamb JG, Semler BL. 1988. Protein 3CD is the major poliovirus proteinase responsible for cleavage of the P1 capsid precursor. Virology 166:265–270 [DOI] [PubMed] [Google Scholar]