Abstract
Efficient genetic modification of herpesviruses such as Kaposi's sarcoma-associated herpesvirus (KSHV) has come to rely on bacterial artificial chromosome (BAC) technology. In order to facilitate this approach, we generated a new KSHV BAC clone, called BAC16, derived from the rKSHV.219 virus, which stems from KSHV and Epstein-Barr virus-coinfected JSC1 primary effusion lymphoma (PEL) cells. Restriction enzyme and complete sequencing data demonstrate that the KSHV of JSC1 PEL cells showed a minimal level of sequence variation across the entire viral genome compared to the complete genomic sequence of other KSHV strains. BAC16 not only stably propagated in both Escherichia coli and mammalian cells without apparent genetic rearrangements, but also was capable of robustly producing infectious virions (∼5 × 107/ml). We also demonstrated the utility of BAC16 by generating deletion mutants of either the K3 or K5 genes, whose products are E3 ligases of the membrane-associated RING-CH (MARCH) family. While previous studies have shown that individual expression of either K3 or K5 results in efficient downregulation of the surface expression of major histocompatibility complex class I (MHC-I) molecules, we found that K5, but not K3, was the primary factor critical for the downregulation of MHC-I surface expression during KSHV lytic reactivation or following de novo infection. The data presented here demonstrate the utility of BAC16 for the generation and characterization of KSHV knockout and mutant recombinants and further emphasize the importance of functional analysis of viral genes in the context of the KSHV genome besides the study of individual gene expression.
INTRODUCTION
Kaposi's sarcoma-associated herpesvirus (KSHV) is the etiologic agent of Kaposi's sarcoma (KS), a vascular neoplasm of endothelial cell origin (9, 25, 46). KSHV infection is also associated with two rare lymphoproliferative disorders of B-cell origin: primary effusion lymphoma (PEL), and multicentric Castleman's disease (MCD) (9, 20, 70). KSHV-associated pathogenesis is facilitated by an immunocompromised state, particularly in the development of KS, which is the most common AIDS-associated malignancy (10). Compared to the general population, incidence rates of KS are more than 600-fold higher in people with HIV/AIDS (10, 15). KS development is tightly correlated with the level of KSHV-specific CD8 T cells (37). Moreover, both AIDS-related and iatrogenic (transplant-related) KS show tumor regression following immune restoration by highly active antiretroviral therapy (HAART) or cessation of immunosuppression, respectively (2, 7). Thus, cell-mediated immune responses are strongly implicated in the control of KSHV-driven pathogenesis.
Much progress has been made in the development of both cell culture and animal models to study KSHV (14, 32, 45, 48, 52, 71). However, reverse genetics approaches have not been utilized to their full potential to characterize KSHV gene function in these biologically relevant settings. Herpesvirus mutagenesis has come to rely on bacterial artificial chromosome (BAC) and “recombineering” technology to generate recombinant virus (69). As the basis for this technique, a BAC-containing clone of the complete viral genome has to be generated, enabling propagation of the viral genome in Escherichia coli and avoiding the need for cumbersome cloning techniques (47). The successful generation of such a clone often requires optimization to minimize genomic disruption by the BAC sequence and ensure accurate recapitulation of the viral life cycle in mammalian cells. Previously, a full-length BAC clone of the KSHV genome called BAC36 was generated, and it has been used to analyze the function of a number of viral genes (60, 75). However, recent evidence shows that BAC36 contains a duplication spanning a 9-kb region of the KSHV genome, including 6 complete KSHV open reading frames (ORFs): K5, K6, K7, ORF16, ORF17, ORF18, and part of ORF19 (72). This second copy is located within the terminal repeat (TR) region of the genome. A further complication is that the BAC vector backbone of BAC36 was engineered to integrate between ORF18 and ORF19 within the long unique region (LUR) of the KSHV genome. Indeed, a single copy of the BAC vector was detected between ORF18 and ORF19, but it is situated within the TR-localized copy of this genomic fragment and not within the LUR, as originally intended. This duplication, coupled with the location of the BAC vector within the unstable TR region, could explain why BAC36 has been observed to undergo frequent unwanted homologous recombination events that result in the deletion of large portions of the LUR (72).
KSHV can establish latent infection in host cells, during which a restricted gene expression program is thought to limit the availability of viral proteins for proteolytic degradation and entry into the major histocompatibility complex class I (MHC-I) pathway. In addition, the KSHV genome encodes an array of immune modulatory gene products that are thought to be particularly important during the more vulnerable stages of the viral life cycle, such as the period immediately following de novo infection or lytic reactivation (17, 40). Two such genes, coding for the K3 and K5 proteins, also known as modulator of immune recognition 1 and 2 (MIR1 and MIR2), respectively, are each able to selectively modulate the localization and/or stability of certain plasma membrane proteins as part of a strategy to interfere with a variety of host cell events, including cell-to-cell interactions, such as those involved in the mobilization of adaptive immune responses (38). For example, MHC-I molecules are targeted for endocytosis and degradation in the presence of either K3 or K5 (18, 28, 31). Interestingly, compared with K5, overexpressed K3 is much more efficient at reducing MHC-I surface expression in either BJAB B lymphoblastoid cells or primary endothelial cells of the human microvascular endothelial cell (HMVEC) line (31, 41). In addition, many K5-specific substrates have been identified, including proteins involved in costimulation, such as ICAM-I and B7-2 (19, 30).
Both K3 and K5 are homologues of the membrane-associated RING-CH (MARCH) E3 ubiquitin ligase family and share similar structural features: a cytosolic amino-terminal RING-CH domain, two transmembrane domains linked by a short stretch of amino acids, and a cytosolic carboxyl-terminal domain (3, 12, 21, 56). This two-transmembrane-type structure is also predicted for most other MARCH proteins, including mouse hepatitis virus 68 (MHV-68) K3 (mK3), RFHV-K3 (rfK3), and M153R, as well as cellular MARCH proteins 1, 2, 3, 4, 8, 9, and 11 (24, 29, 50). Interestingly, this family of proteins has also been shown to primarily target type I plasma membrane proteins involved in immune responses and share many of the same substrates (50). The RING-CH domain is critical for the ability of K3 and K5 to ubiquitinate cytosolic residues of substrates, while the carboxy-terminal region immediately distal to the second transmembrane domain is involved in the subsequent sorting and degradation of substrates (30, 31, 56).
Relatively less is known about the functional contribution of K3 and K5 in the context of KSHV infection. Both K3 and K5 transcripts are induced as part of the lytic replication gene expression program (27, 61). Although its mRNA is not evident within the virion particle, K5 can be transiently expressed shortly after de novo KSHV infection (5, 35, 74). In addition, the K5 gene is inducible by Notch signaling and can be expressed independently of RTA, the master regulator of lytic replication (13, 51). K3 expression occurs from multiple transcripts, including a constitutively expressed transcript in PEL cells as well as immediate-early and early transcripts (55, 61). Small interfering RNA (siRNA)-mediated knockdown of K5 following de novo KSHV infection of endothelial cells showed that K5 is important for KSHV-mediated downregulation of MHC-I and ICAM-I during this early stage of infection (1). However, the additional contribution of other KSHV genes is difficult to analyze without the use of complete gene knockout viruses. Moreover, the role of K3 or K5 in MHC-I or ICAM-I downregulation has not been examined during other stages of the KSHV life cycle. Given the complications associated with BAC36, a new KSHV BAC clone generated using an alternative cloning strategy could provide a better foundation for reverse genetics studies of KSHV genes such as those coding for K3 and K5.
In this study, we generated an rKSHV.219-derived BAC clone of the full-length KSHV genome, called BAC16. Our results indicate that BAC16 is stable in E. coli and can produce infectious viruses upon reconstitution in mammalian cells. To demonstrate its utility, we generated deletion mutants of the K3 or K5 gene. We also engineered revertant and point mutant viruses derived from each deletion. While wild-type (WT) BAC16 can efficiently remove MHC-I and ICAM-I from the cell surface following induction of lytic replication, deletion or point mutants of K5 completely lost this ability. Downregulation activity was restored in the K5 revertant virus. Surprisingly, deletion of K3 did not result in any detectable defect in KSHV-mediated reduction of MHC-I surface expression during lytic reactivation.
MATERIALS AND METHODS
Virus and cells.
Vero, Vero-rKSHV.219, HEK293A, iSLK, and iSLK-BAC16 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. iSLK cells were cultured in the presence of 1 μg/ml puromycin and 250 μg/ml G418. BAC16 and its derivatives were introduced into iSLK cells via Fugene HD transfection. In brief, BAC DNA was isolated from a 5-ml bacterial culture and resuspended in a 40 μl of distilled water. iSLK cells were seeded at 2 × 105 cells/well of a 6-well plate or ∼70% confluence. On the following day, the medium was changed to optiMEM 30 min prior to addition of the transfection complexes (fetal bovine serum [FBS] and penicillin-streptomycin were excluded from the medium). Transfection complexes were prepared by combining approximately 25% (10 μl) of the total miniprepped BAC DNA and 90 μl of optiMEM, followed by the addition of 5 μl of Fugene HD. After 10 min of incubation at room temperature, the complexes were added to the cells. Three hours after the transfection complexes were added, FBS was added to the optiMEM to a final concentration of 10%. On following day, transfected cells were trypsinized and transferred to 10-cm dishes and cultured in the presence of DMEM supplemented with 10% FBS and 1% penicillin-streptomycin but in the absence of other antibiotics. Two days after transfection, iSLK-BAC cell lines were established and maintained in the presence of 1 μg/ml puromycin, 250 μg/ml G418, and 1,200 μg/ml hygromycin B.
Plasmids.
pBelo45 was constructed by replacing the fragment of pBeloBAC11 (NEB) with an EF1α-driven (pTracer; Invitrogen) green fluorescent protein-internal ribosome entry site-hygromycin (GFP-IRES-HYG) cassette (pL_UGIH; Signaling-Gateway) and KSHV genomic sequences positioned on either side of a unique PmeI site. KSHV genomic DNA on the left-hand side of the insertion site was PCR amplified from rKSHV.219 using the ORF57 primer set containing the indicated restriction enzyme sites for cloning purposes and a loxP site (Table 1). This PCR product was cloned into pSP72 via unique XhoI and EcoRI sites, resulting in pSP72A. Similarly, KSHV genomic DNA from the right-hand side of the insertion site was PCR amplified using the K9 primer set, and the product was introduced into pSP72A via PmeI and ClaI, resulting in pSP72B. The GFP-IRES-HYG cassette was PCR amplified from pL_UGIH using the GFP-HYG primer set. In order to place these coding sequences under the control of the EF1α promoter, the GFP-IRES-HYG PCR product was digested with SpeI and HpaI and ligated with SpeI- and PmeI-digested pTracer. The resulting plasmid, pTracer-GFP-IRES-HYG, contains the GFP-IRES-HYG coding sequence positioned downstream of the EF1α promoter. The EF1α-GFP-HYG-pA primer set was used to PCR amplify the complete selection cassette from pTracer-GFP-IRES-HYG. This PCR product was cloned into pSP72B using the unique ClaI and HpaI sites, resulting in pSP72C. The selection cassette and adjacent KSHV genomic targeting DNA were subcloned into pBeloBAC11, released from pSP72C by FspI and HpaI digestion, and ligated into pBeloBAC11 using the unique SrfI and HpaI sites, resulting in pBelo45. Proper orientation of all ligation products was verified by restriction enzyme digestion, and primer walking was used to completely sequence pBelo45. The sequence of the non-KSHV portion of the pBelo45 vector backbone is available in Fig. S1 in the supplemental material.
Table 1.
Primers and plasmids used in this study
| Primer name | Sequencea | Template (reference) | Resulting plasmid name(s) |
|---|---|---|---|
| Generation of pBelo45 | |||
| ORF57-1 | CCGCTCGAGXhoICGGTGCGCAFspIATAACTTCGTATAATGTATGCTATACGAAGTTATLoxPctggtggcggtctggtg | rKSHV.219 (68) | pSP72A |
| ORF57-2 | GAATTCEcoRICCGGCGCGCCGTTTAAACPmeIatgataattgacggtgagagcccccgc | ||
| K9-1 | AGCTTTGTTTAAACPmeIatggacccaggccaaagaccgaacccttttggggcgcc | rKSHV.219 (68) | pSP72B |
| K9-2 | ATCGATClaIgtggcacccaacatccattatggaaaaaccccgcgccaccttccgcc | ||
| GFP-HYG-1 | GGACTAGTSpeIGCCACCKozakatggtgagcaagggcgaggagct | pL_UGIH (Cell Signaling) | pTracer-GFP-IRES-HYG |
| GFP-HYG-2 | GTTAACHpaIctattcctttgccctcggacgagtgctggggcgt | ||
| EF1a-GFP-HYG-pA-1 | CCATCGATClaIATAACTTCGTATAATGTATGCTATACGAAGTTATLoxPgctccggtgcccgtcagtgggcagagcg | pTracer-GFP-IRES-HYG (this study) | pSP72C |
| EF1a-GFP-HYG-pA-2 | GTTAACHpaIcatagagcccaccgcatccccagcatgcctgctattgtcttcc | ||
| Generation of BAC16 mutants | |||
| Kan-in-K3-1 | ATCCTTCCACAGGGTTTGCCTGGGGGTGGCTATGGTTCCATGGGCGTGATTAGGAAACGTtagggataacagggtaatcgatttattc | pEPCMV-in (65) | pTracer-K3-in, pTracer-K3-RINGmut-in |
| Kan-in-K3-2 | tgctagccagtgttacaaccaattaacc | ||
| Kan-in-K5-1 | CCCTCTAGAXbaIACCGTTGTTTTTTGGATGATTTTTCCGCACCGGCTTTTTTGTGGGCGCGCAtagtgctagccagtgttacaaccaattaac | pEPCMV-in (65) | pTracer-K5-in, pTracer-K5-RINGmut-in |
| Kan-in-K5-2 | GGTTCTAGAXbaItagggataacagggtaatcgatttattc | ||
| DELK3-1 | GGGTTAATGCCATGTTTTATTGTGGGTTCTCTCTCAGGATAAGTATATAAGAGCACACTGaggatgacgacgataagtaggg | pEPkan-S (65) | BAC16-ΔK3, BAC16-ΔK3 ΔK5 |
| DELK3-2 | GGTAAACACCACCAACCACACAGTGTGCTCTTATATACTTATCCTGAGAGAGAACCCACAcaaccaattaaccaattctgattag | ||
| DELK5-1 | GGGCGTCACGTCACATATCTCTGTGCACCCAAGTGGTTGTCTCTGCAGCTGGGGTGGAAGaggatgacgacgataagtaggg | pEPkan-S (65) | BAC16-ΔK5, BAC16-ΔK3 ΔK5 |
| DELK5-2 | TCCCCTTTCCCTTTTTCAGACTTCCACCCCAGCTGCAGAGACAACCACTTGGGTGCACAGcaaccaattaaccaattctgattag | ||
| K3in-1 | CGAGGGTATAGGTAAACACCACCAACCACACAGTGTGCTCTTATATACTTtcaatggtgatggtgatgatgaccggtacgcgtag | pTracer-K3-in, pTracer-K3-RINGmut-in (this study) | BAC16-K3-RING-C→S, BAC16-K3rev |
| K3in-2 | CACTTGTTGCAGGGGTTAATGCCATGTTTTATTGTGGGTTCTCTCTCAGGATatggaagatgaggatgttcctgtctg | ||
| K5in-1 | GGTGCATAACACCCAGGGCGTCACGTCACATATCTCTGTGCACCCAAGTGGTTGTtcaatggtgatggtgatgatgaccggtac | pTracer-K5-in, pTracer-K5-RINGmut-in (this study) | BAC16-K5-RING-C→S, BAC16-K5rev |
| K5in-2 | CACTCTGCTCACCTCCCCTTTCCCTTTTTCAGACTTCCACCCCAGCTGCAGAGatggcgtctaaggacgtagaagagg |
Enzyme recognition sequences are underlined and followed by a subscript indicating the cognate enzyme/site name. Sequences homologous to the template are shown in lowercase. Sequences that are absent from BAC16 are shown in italics and for cloning purposes only. Sequence duplications introduced for “scarless” elimination of the KanR cassette are shown in boldface.
In order to introduce K3 or K5 coding sequences (either WT or RING-C→S versions) into the KSHV genome, we generated universal transfer constructs derived from plasmids harboring V5-His-tagged K3 or K5 coding sequences within the pTracer vector (39) using a strategy described previously (65). In brief, a positive selection marker and an adjacent I-SceI recognition site were cloned into a unique restriction site within the desired insertion sequence. A duplication of at least 40 bp immediately adjacent to the restriction site is included, enabling “scarless” removal of the Kan cassette following I-SceI-mediated cleavage. For our purposes, the Kan-in-K3 primer set was used to PCR amplify a DNA fragment containing a Kan selection cassette and an adjacent I-SceI site from pEPkan-S (65). This DNA fragment was introduced into either the pTracer K3 WT or RINGmut vector using blunt ligation via a unique EcoRV site occurring within the K3 coding sequence. A 60-bp duplication of the K3 coding sequence immediately downstream of the EcoRV site was included on the 5′ end of the Kan-in-K3-1 primer. An NheI digestion was used to identify clones with a correctly oriented ligation product such that the duplication flanks the intervening Kan and I-SceI sequences. A similar procedure was used to generate pTracer-K5-in and pTracer-K5-RING-C→S-in by utilizing a unique XbaI site within the parental pTracer-K5 WT and RING-C→S constructs.
219BAC construction.
219BAC virus was generated by spontaneous homologous recombination following transfection of pBelo45 linearized with PmeI into Vero cells stably carrying rKSHV.219 (68). Two days following transfection, recombinant BAC virus was selected using 400 μg/ml of hygromycin. To enrich for infectious 219-BAC, hygromycin-resistant cells were treated with 75 nM trichostatin A (TSA) and the virus-containing supernatant was used to infect naïve Vero cells and establish a new hygromycin-resistant cell line. A total of three rounds of serial propagation to naïve Vero cells were carried out to ensure that BAC clones recovered from Vero cells were infectious.
BAC DNA isolation and analysis.
Circular viral DNA was extracted from Vero cells using a genomic DNA isolation kit (Qiagen). One hundred nanograms of genomic DNA (gDNA) was used to electroporate E. coli DH10B (2.0 kV, 200 Ω, and 25 μF). BAC DNA was purified from chloramphenicol-resistant colonies using an alkaline lysis procedure followed by isopropanol precipitation. Purified BAC DNA was digested with KpnI, CpoI, or SbfI and separated on 0.8% agarose gels or using pulsed-field gel electrophoresis (PFGE) (CHEF-DR II; Bio-Rad) under the following conditions: 1% PFGE-grade agarose; 6 V/cm for 15 h; initial and final switch times of 1 and 5 s, respectively; and 14°C. For Southern blot hybridization, resolved BAC DNA fragments were transferred to a nylon membrane and hybridized with 32P-labeled probes. Complete sequencing of BAC16 and BAC25 was performed using a Solexa sequencer, and gaps were sequenced using the Sanger method.
Production of BAC16 virus stock.
BAC16 stocks were prepared from stable iSLK cells as previously described, but using a puromycin-resistant version of these cells in order to allow selection of hygromycin-resistant BAC16-containing cells (49). Briefly, BAC16 DNA was introduced into iSLK-puro cells and selected with 1,200 μg/ml of hygromycin B. Stable iSLK-BAC16 cells were induced in the presence of both doxycycline (1 μg/ml) and sodium butyrate (1 mM) and the absence of hygromycin, puromycin, and G418. Four days later, supernatant was collected and cleared of cells and debris by centrifugation (950 g for 10 min at 4°C) and filtration (0.45 μm). Virus particles were pelleted by ultracentrifugation (25,000 g for 3 h at 4°C) using an SW32 Ti rotor.
Mutagenesis of BAC16 in GS1783.
To modify the KSHV genome, BAC16 was introduced into the GS1783 E. coli strain (a kind gift from Greg Smith) by electroporation (0.1-cm cuvette, 1.8 kV, 200 Ω, 25 μF). Gene deletions were introduced as previously described (64). Briefly, PCR amplification was used to generate a linear DNA fragment containing a kanamycin resistance expression cassette, an I-SceI restriction enzyme site, and flanking sequences derived from KSHV genomic DNA, each of which includes an ∼40-bp copy of a duplication shown in boldface in Table 1. This fragment was then electroporated into GS1783 cells harboring BAC16 and transiently expressing gam, bet, and exo. These three proteins are required for homologous recombination of linear DNA fragments with a target sequence and can be expressed in a temperature-inducible manner from the lambda Red operon engineered within the endogenous GS1783 chromosome. Integration of the Kanr/I-SceI cassette was verified by PCR and restriction enzyme digestion of the purified BAC DNA. The GS1783 strain is also equipped with an arabinose-inducible gene encoding the I-SceI enzyme. Upon treatment with 1% l-arabinose, the integrated Kanr/I-SceI cassette is cleaved, resulting in a transiently linearized BAC16. A second Red-mediated recombination between the duplicated sequences results in recircularization of the BAC DNA and “scarless” loss of the Kanr/I-SceI cassette. Kanamycin-sensitive colonies were screened via replica plating. The amino acid coding sequences of the K3 or K5 gene from BAC16-WT were used along with the DELK3 and DELK5 primer sets, respectively, to yield BAC16-ΔK3 and BAC16ΔK5 (Table 1). Similarly, the K5 coding sequence was removed from BAC16-ΔK3 to generate BAC16-ΔK3ΔK5 lacking both coding sequences. The K3in primer set was used to amplify from the K3 and Kan portions of the pTracer-K3in and pTracer-k3-RING-C→S-in plasmids. These PCR products were introduced into BAC16-ΔK3 to yield revertant or RING-C→S mutant BAC16. Similarly, the K5in primer set was used to generate BAC16-K5rev and BAC16-K5-RING-C→S.
Quantification of infectious virus and KSHV DNA levels in cells.
Various amounts of cell-free virus supernatant were diluted in fresh medium (1-ml final volume/well) and used to inoculate 293A cells that were seeded at approximately 2 × 105cells/well in 12-well plates 12 h prior to infection. Following inoculation, plates were immediately centrifuged (2,000 × g for 45 min at 30°C) and then placed back into the CO2 incubator. One hour later, the inoculum was removed and replaced with fresh medium. Cells were collected 24 h later and washed once with cold phosphate-buffered saline (PBS). The percentage of GFP-positive cells was determined using the CantoII fluorescence-activated cell sorter (FACS) (BD Bioscience, San Jose, CA). Infectious units (IU) are expressed as the number of GFP-positive cells in each well at the time of analysis. Purification of genomic DNA from infected cells and the quantification of KSHV DNA level were described previously in reference 66. KSHV ORF11-specfic primers were used to quantify the amount of KSHV DNA in BAC16-transfected stable iSLK cell lines. The KSHV DNA amount was normalized to the amount of purified host cellular DNA.
Immunoblotting.
iSLK-BAC16 stable cell lines were harvested with radioimmunoprecipitation assay (RIPA) buffer supplemented with complete protease inhibitor cocktail (Roche). For immunoblotting, 30 μg of each protein sample was resolved by SDS-PAGE and transferred onto a polyvinylidene difluoride membrane (Bio-Rad). The membranes were blocked using 5% nonfat milk and then probed with antibodies diluted in PBS-Tween (PBS-T). The following primary antibodies were used: rabbit anti-K2 (ABI 13-214-050), rabbit anti-K3 (16), mouse anti-K8 (Abcam ab36617), rat anti-LANA (ABI 11-007), and mouse anti-β-tubulin (Santa Cruz SC-5274). Mouse anti-RTA and mouse anti-K5 antibodies were kind gifts from Koichi Yamanishi (Osaka University, Japan). Secondary horseradish peroxidase-conjugated anti-mouse and anti-rabbit antibodies were purchased from Cell Signaling.
Flow cytometry.
293A and iSLK cells were detached from the plate with 0.25% trypsin at the indicated time points postinfection for 293A or post-doxycycline treatment for iSLK and washed once with PBS. All subsequent steps were carried out at 4°C. Approximately 5 × 105 cells per sample were incubated in ice-cold FACS blocking buffer (PBS with 3% FBS) for 20 min. Cells were incubated in antibody solution (PBS plus antibody) for 20 min. MHC-I surface expression was probed using mouse anti-HLA-A, -B, and -C (BD Pharmingen, no. 555551) and either goat anti-mouse IgG-allophycocyanin (APC) (BD 550826) or goat anti-mouse IgG-APC-Cy7 (Biolegend, no. 405316). ICAM-I was probed using mouse anti-CD54 (BD, no. 559771). Isotype control samples were stained with anti-mouse IgG1, κ-APC (BD 555751) (ICAM-I), or anti-mouse IgG1, κ, and goat anti-mouse IgG-APC-Cy7 (Biolegend 405316). Relative surface staining of ∼20,000 cells/sample was quantified on a CantoII FACS. Data were analyzed using FlowJo v.6.4.7.
Nucleotide sequence accession number.
The sequence of BAC16, not including the BAC insertion, has been submitted to GenBank (accession no. GQ994935).
RESULTS
Generation of an rKSHV.219-derived BAC clone.
To facilitate the study of KSHV gene function, we constructed an rKSHV.219-derived, infectious BAC clone of the full-length KSHV genome (Fig. 1A). As previously reported, rKSHV.219 was generated by the insertion of an RFP-GFP-Puror cassette at nucleotide (nt) 83527 (GQ994935), which lies within an intergenic region of the KSHV derived from JSC1 PEL cells (68). In order to introduce BAC vector elements into the rKSHV.219 genome, we replaced the RFP-GFP-Puror cassette with a loxP-flanked mini-F-GFP-Hygror vector. For this purpose, we generated a modified pBeloBAC11 plasmid, pBelo45, which includes KSHV genomic sequences immediately upstream and downstream of nt 83527 (GQ994935) (Fig. 1A) (see Materials and Methods).
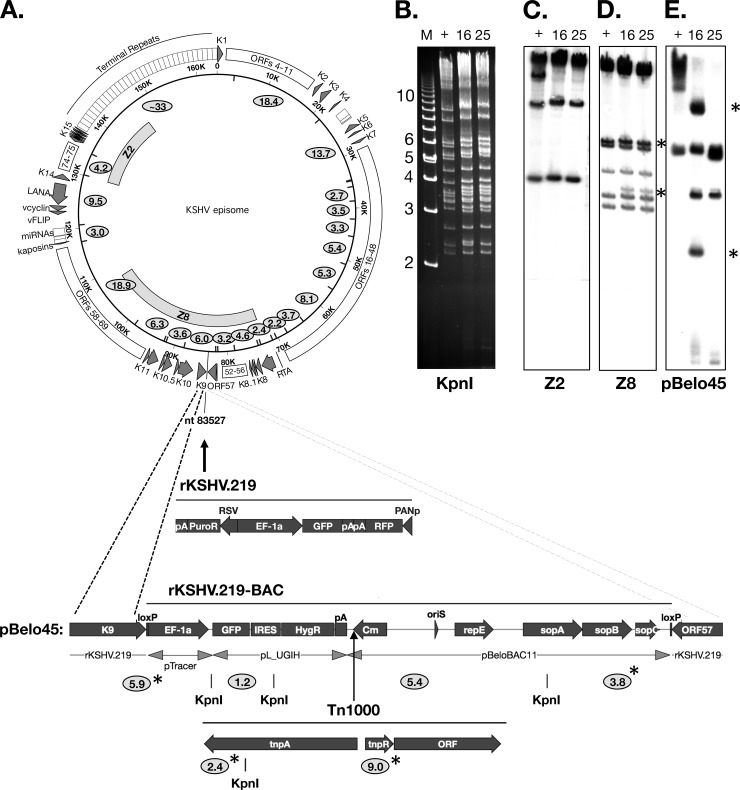
Fig 1.
Construction and analysis of rKSHV.219-derived BAC clones. (A) Schematic diagram of the KSHV genome, the rKSHV.219 and 219-BAC insertion site (GQ994935), the RFP-GFP-Puror cassette, and the pBelo45 targeting construct containing the BAC vector, GFP-Hygror cassette, flanking loxP sites, and flanking sequences for homologous recombination (dashed lines). A majority of 219BAC clones also contain a Tn1000 insertion immediately following the TAA codon of the cam gene, as depicted. KpnI recognition sites are indicated by inward tick marks or below the diagrammed sequence features. KpnI fragment sizes (in kilobase pairs) are indicated as ovals and are based on the GK18 sequence (NC_009333) and the pBelo45 sequence. Fragment sizes marked with an asterisk result from the depicted integration events. (B) Gel electrophoresis of KpnI-digested BAC DNA. Two different clones of 219BAC, BAC16, and BAC25, were analyzed. BAC36 DNA was used as a positive control (+) (75). M, 1-kb marker. (C to E) Southern blot hybridization using a 32P-labeled probe from Z2 (C), Z8 (D), and pBelo45 (E) to detect KSHV and 219BAC-specific sequences. The 2.4- and 9.0-kb fragments in panel E are the result of a Tn1000 insertion.
Following linearization at a unique PmeI site engineered between the homology arms, pBelo45 was transfected into Vero cells stably harboring rKSHV.219. Recombinant BAC virus was selected in the presence of hygromycin B and the absence of puromycin. In order to enrich for infectious, full-length rKSHV.219-BAC, hygromycin-resistant cells were treated with a histone deacetylase (HDAC) inhibitor, trichostatin A (TSA), to induce lytic replication. The virus-containing supernatants were used to infect naïve Vero cells and establish a new hygromycin-resistant cell line. Two additional rounds of serial propagation to naïve Vero cells resulted in the loss of TSA-induced RFP expression, presumably due to enrichment of hygromycin-resistant 219BAC virus. Circular KSHV DNA was extracted from the final round of infected, hygromycin-selected Vero cells and then introduced into E. coli DH10B by electroporation.
Genetic analysis of candidate clones.
Restriction endonuclease and Southern blotting analyses were used to identify bacterial clones harboring a pBelo45 cassette that had correctly recombined with the full-length KSHV genome. Of the 32 total clones analyzed, BAC16 and BAC25 were among 14 clones that appeared to harbor a complete KSHV genome. Digestion of BAC16 and BAC25 with KpnI revealed KSHV DNA fragments of various sizes, which matched well with the predicted digestion pattern based on published genomic sequence data (NC_009333) and migrated comparably to most of the fragments from BAC36, a previously characterized full-length BAC clone of KSHV (Fig. 1A and B). Southern blot hybridization with a labeled Z2 probe spanning the right-hand side of the KSHV genome detected the expected fragment sizes of 4.2-kb and 9.5-kb as well as a high-molecular-weight fragment predicted to contain the TR region. An extra fragment of ∼13 kb detected in BAC36 likely correlates with the presence of a duplicated sequence that was recently shown to be present in the TR region (72). Fragments of 5.9-kb and 3.8-kb predicted to result from proper BAC integration within the KSHV genome were evident in both BAC16 and BAC25 and also detectable by Southern blot analysis using either a KSHV-specific probe (Z8) or a pBelo45-specific probe (Fig. 1D and E). In addition, a fragment of 1.2 kb derived exclusively from the BAC cassette was apparent in both BAC16 and BAC25 clones and detected only by the pBelo45 probe (data not shown). However, a 5.4-kb fragment, also consisting exclusively of BAC vector DNA, while detected in BAC25 was absent in BAC16, and instead, fragments of ∼2.4 kb and ∼9.0 kb appeared only in BAC16 (Fig. 1E). Sequencing of the ∼9.0-kb fragment and direct sequencing of the BAC16 vector backbone revealed that the additional DNA fragments resulted from insertion of a Tn1000 transposon between the chloramphenicol and hygromicin resistance genes (Fig. 1A) (data not shown).
Stable propagation of BAC16 in E. coli DH10B.
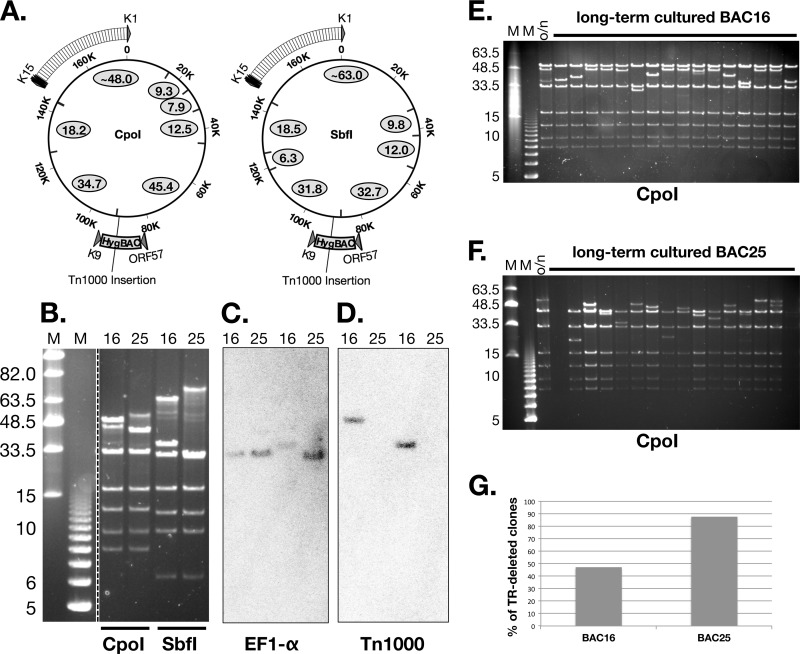
Besides BAC25, only one additional clone showed the predicted KpnI restriction pattern for 219BAC lacking a Tn1000 (data not shown). On the other hand, most clones appeared identical to BAC16, with the addition of KpnI fragments corresponding to a Tn1000 insertion located at the diagramed position (Fig. 1A). Given the pervasiveness of the Tn1000 sequence among the clones that we analyzed, we decided to perform additional Southern blot analysis to determine if Tn1000 had inserted elsewhere in either the BAC16 or BAC25 genomes. BAC16 and BAC25 DNAs were digested with either CpoI or SbfI, followed by Southern blot hybridization of the resolved fragments with a Tn1000-specific probe. Indeed, Tn1000 was only detected at the expected site of BAC16 and was completely absent from BAC25 (Fig. 2A, B, C, and D). Next, we compared the stability of BAC16 and BAC25 after serial passage in DH10B, an E. coli strain carrying a recA mutation. Following 5 days of daily passaging in liquid culture, single colonies were recovered on LB agar plates and purified BAC DNA was analyzed by CpoI digestion (Fig. 2E and F). DNA fragments derived exclusively from the long unique region (LUR) did not undergo significant changes compared to single-passage BAC DNA. However, as seen in two representative gels, the ∼48-kb and ∼54-kb fragments containing the TR region from BAC16 and BAC25, respectively, were significantly shorter in several long-term-cultured clones (Fig. 2E and F). Although instability of the TR region was observed in both BAC16 and BAC25, we found that only 15 out of 32 long-term-cultured clones from BAC16 had a shortened TR-containing fragment compared to 28 out of 32 such clones from BAC25 (Fig. 2G). Given that TR deletions were observed less frequently in long-term-cultured BAC16 clones, we chose to use BAC16 for further characterization and mutagenesis.
Fig 2.
Stability of BAC16 and BAC25. (A) Schematic of CpoI and SbfI recognition sites (inward tick marks). Predicted fragment sizes (in kb) are depicted as ovals for 219BAC lacking Tn1000. As the Tn1000 sequence does not contain any CpoI or SbfI sites, the 6.0-kb Tn1000 insertion within BAC16, the 45.4-kb (CpoI) and 31.8-kb (SbfI) fragment sizes are predicted to be 51.4 kb and 37.8 kb, respectively. (B) Gel electrophoresis of CpoI- or SbfI-digested BAC16 or BAC25 DNA. M, midrange PFGE or 1-kb size marker. (C and D) Southern blot hybridization with an EF1-α-specific probe (C) or a Tn1000-specific probe (D). (E and F) E. coli DH10B harboring BAC16 (E) or BAC25 (F) was passaged daily in liquid culture for a total of 5 days. DNAs isolated from single colonies derived from this long-term culture were analyzed by CpoI digestion and gel electrophoresis. BAC16 and BAC25 DNAs from overnight (o/n) cultures were analyzed in parallel. Panel G shows the percentage of TR-fragment-deleted clones (among a total of 32 clones) analyzed from each long-term culture.
Complete sequencing analysis of the KSHV BAC16.
BAC16 was completely sequenced via the Solexa sequencing method, and the sequence was deposited into GenBank. Importantly, sequence reads were represented equally across the LUR portion of BAC16, indicating a lack of a large-scale duplication. In previous work, several loci of the KSHV genome were sequenced via PCR amplification directly from JSC-1 cells, including the ORF26 region, ORF75-E, gB, gH, gL, gM, gN, K8.1, and ORF68 (53, 58). Comparison of these locus sequences with the BAC16 sequence showed 100% identity, with the exception of a synonymous point mutation at A242 of ORF75 (data not shown). This indicates that the genomic sequence of JSC-1-derived KSHV had not undergone any significant changes during its propagation as rKSHV.219 and BAC16 and that this sequence likely reflects the actual JSC-1 viral genome sequence. Moreover, with the exception of the K1 sequence, we found very low levels of sequence variations across the entire BAC16 genome compared to other complete genomic sequences of KSHV (BCBL-1, HQ404500; BC-1, U75698; and KS-derived, U93872, NC_009993).
Production of high-titer BAC-derived virus stock.
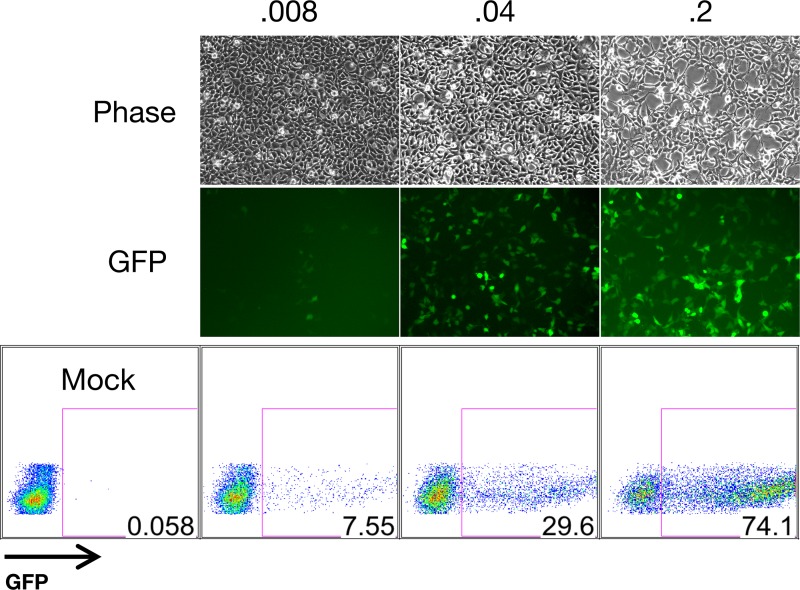
Inefficient production of cell-free infectious KSHV has limited the range of experimental systems utilized by researchers in the field. The combined use of exogenous RTA expression and sodium butyrate treatment was shown to synergistically activate the lytic cycle, yielding substantial quantities of rKSHV.219 from Vero cells (68). Furthermore, the use of spin infection is capable of increasing the infection levels approximately up to 70-fold (68, 73). Recently, the production of infectious cell-free KSHV was further refined by Myoung et al., who reported the utility of newly generated doxycycline-inducible RTA cell lines (called iVero and iSLK) for production of high-titer virus stocks, including BAC-derived stocks (49). Indeed, we were also able to produce such a virus stock with a titer of ∼5 × 107 from iSLK cells carrying WT BAC16 based on GFP-positive cells (see Materials and Methods). Using different volumes of virus stock, we were able to infect 293A cells and obtain various percentages of GFP-positive cells (Fig. 3). In addition, telomerase-immortalized endothelial (TIVE) cells and HMVECs, which are less susceptible to KSHV infection, were successfully infected with BAC16 (data not shown). Thus, BAC16 appears suitable for experiments requiring a high virus titer.
Fig 3.
Infectious BAC16 virus production. A BAC16 virus stock was generated as described in Materials and Methods. Four 15-cm plates containing iSLK cells harboring BAC16 were seeded at ∼70% confluence and treated with 1 μg/ml of doxycycline and 1 mM sodium butyrate for 96 h. Approximately 40 ml of virus-containing supernatants were concentrated via centrifugation using a SW32 rotor. Virus pellets were then resuspended in the residual medium (∼300 μl). Then 293A cells were infected with 0.008, 0.04, or 0.2 μl of this BAC16 virus stock and analyzed by fluorescence microscopy and flow cytometry at 24 h postinfection.
Generation of K3- and K5-deleted recombinant KSHV.
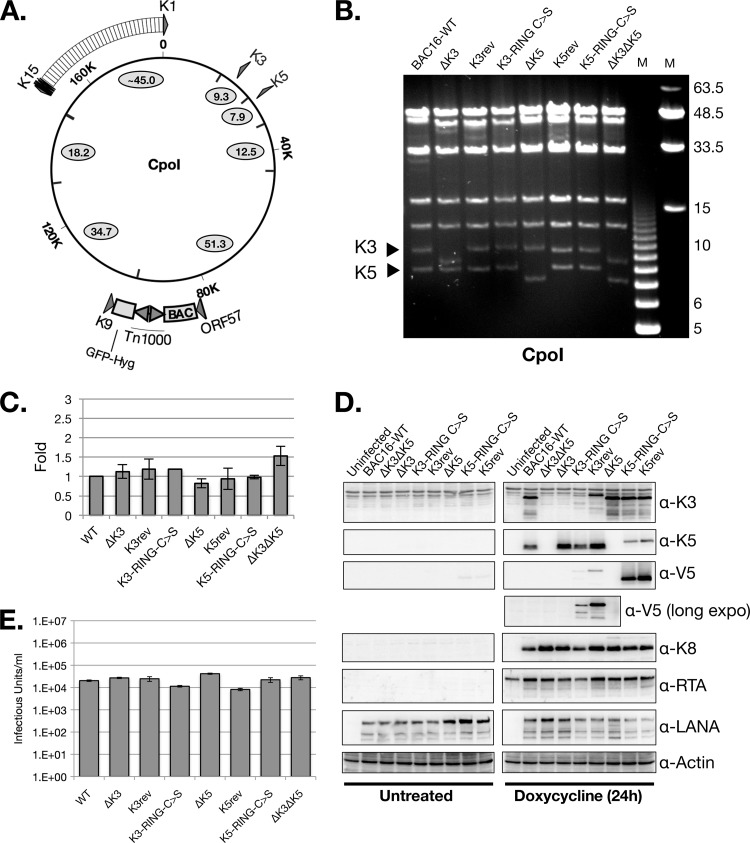
In order to demonstrate the feasibility of BAC16 for the study of KSHV gene function, we generated a set of K3 and K5 deletion mutants. The entire coding sequence of either the K3 or K5 gene was removed from BAC16 using “scarless” mutagenesis (64, 65). Then, V5-His-tagged K3 and K5 coding sequences were inserted into their endogenous loci within the ΔK3 and ΔK5 genomes, respectively, to generate tagged revertant viruses. Likewise, a catalytically dead RING-C→S mutant coding sequence of either K3 or K5 was introduced into the ΔK3 and ΔK5 BACs, respectively. Each mutant genome was analyzed by PCR, restriction enzyme digestion, and direct sequencing analysis (Fig. 4A and B) (data not shown). The K3 and K5 genes reside on CpoI fragments of 9.3 and 7.9 kb, respectively (Fig. 4A). These fragments were predictably reduced to 8.3 kb and 7.1 kb following the targeted deletion of either the K3 or K5 gene, respectively, and restored to their WT sizes in the V5-His-tagged BACs: K3rev, K3-RING-C→S, K5rev, and K5-RING-C→S (Fig. 4B). Importantly, CpoI fragments of other parts of the BAC16 genome did not show any detectable size changes following genetic manipulation (Fig. 4B).
Fig 4.
Characterization of WT and mutant BAC16 virus. (A) Schematic diagram of CpoI digestion of BAC16 (including Tn1000 sequence) (B) Pulsed-field gel analysis of CpoI-digested WT and mutant BAC16 DNAs. Triangles denote the 9.3-kb and 7.9-kb fragments predicted to harbor the K3 and K5 coding sequences, respectively. (C) iSLK cells stably transfected with WT or mutant BAC16 were analyzed by qPCR to determine relative DNA copy number. (D) The same set of cells was analyzed by Western blot analysis using the indicated antibodies (α-K3, α-K5, etc). long expo, long exposure. (E) Infectious units were quantified from viral supernatants harvested from the indicated iSLK-BAC16 cell lines following 4 days of treatment with both doxycycline and sodium butyrate (see Materials and Methods). Note, the viral supernatants from this experiment were not concentrated by centrifugation and thus had a lower titer than those in Fig. 3.
Characterization of BAC-derived WT and mutant viruses.
WT, null, point mutant, and revertant viruses were reconstituted in iSLK cells, which contain a doxycycline-inducible RTA expression system stably integrated in the cellular chromosome (49). Upon doxycycline treatment, RTA expression is sufficient to initiate the lytic replication phase that results in the induction of K3 and K5 gene expression and culminates in the release of infectious virion particles from the cells. GFP expression from BAC16 appeared stable, and full-length BAC genomes were readily recovered from infected cells, indicating stable propagation of BAC16 and BAC16 mutants in eukaryotic cells (data not shown). Following the hygromycin selection of BAC16-transfected cells, comparable amounts of viral DNA were found in iSLK cells harboring the different KSHV-BACs (Fig. 4C). Viral gene expression was analyzed by Western blotting in the absence and presence of doxycycline treatment (Fig. 4D). As expected, the deletion mutants showed no detectable protein expression from their respective deleted genes. Moreover, the expression level of K3 did not appear to be affected by the absence of K5 and vice versa. Protein expression was restored in the respective revertant and enzymatically dead RING-C→S mutant BACs, albeit at lower levels. LANA or K8 protein levels were comparable in all cell lines. As expected, since iSLK cells have been engineered to express RTA in a doxycycline-inducible manner via an expression cassette integrated within the cellular DNA, RTA expression was detected in all the doxycycline-treated iSLK cells, including uninfected cells (49).
Next, we determined whether the absence of K3 or K5 had an effect on the efficiency of infectious virus production from iSLK cells. Following 2 days of doxycycline treatment, serial dilutions of cell-free supernatants was transferred to 293A cells and infectious virus was quantified by FACS analysis of GFP expression at 24 h postinfection (Fig. 4E). The levels of infectious viruses were comparable between the WT and derivative viruses, indicating that neither K3 deletion nor K5 deletion significantly affects virus production. This is in contrast to published data that showed that the siRNA-mediated depletion of K5 in KSHV-infected HeLa cells significantly reduced infectious virus release (44). The reason for this discrepancy is unknown but perhaps reflects differences in tetherin expression levels between HeLa and iSLK cells as it was shown previously that knockdown of the tetherin in K5-depleted cells restores infectious virus release from HeLa cells (44).
K5 but not K3 is required for KSHV-mediated reduction of MHC-I surface expression during viral reactivation in iSLK cells.
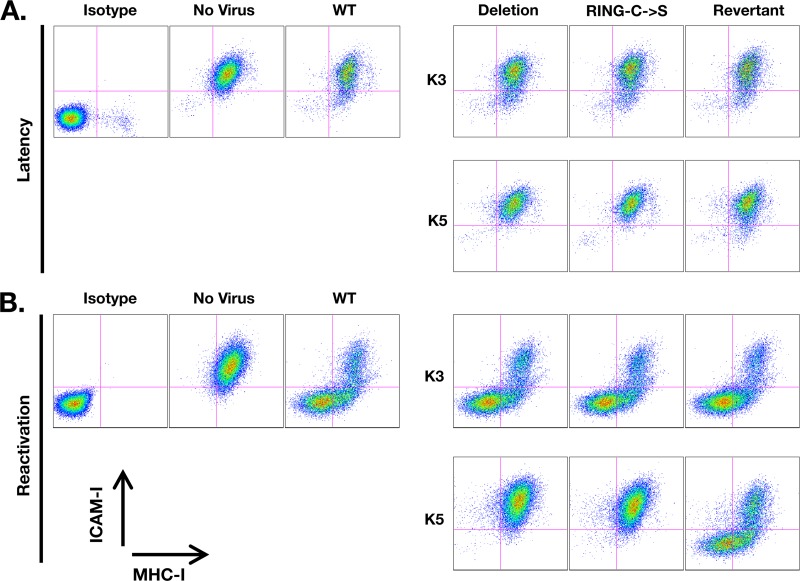
MHC-I and ICAM-I surface expression levels were compared between iSLK cells harboring WT, ΔK3, ΔK5, K3rev, K5rev, K3-RING-C→S, K5-RING-C→C, and ΔK3 ΔK5 BAC16 viruses (Fig. 5A and B). Without treatment, there was no significant difference in the levels of MHC-I surface expression of iSLK cells harboring WT and recombinant BAC16 compared to that of naïve iSLK cells. However, ICAM-I surface expression was slightly reduced in small percentages of WT BAC16 cells relative to uninfected iSLK cells (Fig. 5A). This phenotype was dependent on the expression of WT K5 as a deletion mutant of K5 lacked the ability to downregulate ICAM-I. This phenotype was restored in cells harboring the K5 revertant but not in those carrying the K5-RING-C→S mutant, confirming the previous reports that an intact RING-CH domain is required for K5-mediated downregulation of ICAM-I (30). Furthermore, this indicates that K5 is functionally active in a small percentage of cells under nonlytic replicating conditions. Finally, our data confirm the previous report that K5 downregulates ICAM-I more efficiently than MHC-I (1).
Fig 5.
Flow cytometry analysis of ICAM-I and MHC-I surface expression of iSLK-BAC16 cell lines during latency (A) or reactivation after 24 h of doxycycline treatment (B). Each iSLK-BAC16 stable cell line sample was stained with both MHC-I (W6/32) and ICAM-I antibodies. iSLK cells lacking KSHV were also stained with the indicated antibodies or with isotype control antibodies (see Materials and Methods). Live cells were gated via forward scatter (FSC) and side scatter (SSC) profiling and analyzed for GFP, APC (ICAM-I), and APC-Cy7 (MHC-I) fluorescence. All BAC16 cell lines were ∼100% GFP positive (data not shown).
Upon doxycycline treatment, we found that both MHC-I and ICAM-I were efficiently removed from the cell surface in the majority of WT BAC16 stable cells (Fig. 5B). K5-deficient viruses showed the complete abrogation of downregulation of both ICAM-I and MHC-I surface expression compared to WT (Fig. 5B). A K5 revertant virus was nearly as effective as the WT at reducing MHC-I and ICAM-I surface expression. However, a virus engineered to express the K5 gene harboring point mutations (C→S) in the RING domain was not able to restore KSHV-mediated reduction of MHC-I or ICAM-I surface expression (Fig. 5B). On the other hand, deletion of the K3 gene showed no significant defect in MHC-I downregulation, nor did the K3 RING-C→S virus (Fig. 5B). Thus, K5 appears to be the major viral protein required for MHC-I downregulation during KSHV reactivation in iSLK cells.
K5, but not K3, is required for the reduction of MHC-I surface expression following de novo infection.
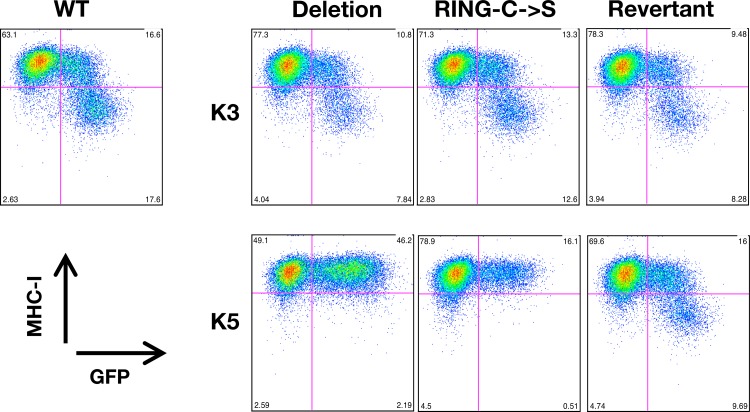
Previous work has demonstrated a critical role of K5 in MHC-I and ICAM-I downregulation following de novo infection of human umbilical vein endothelial cells (HUVECs) by using siRNA-mediated knockdown of K5 (1). However, due to the imperfect efficiency of siRNA approaches, it remains unclear whether other KSHV genes can also modulate surface expression of MHC-I molecules during this stage of infection. Thus, we measured MHC-I surface expression following infection of 293A cells with WT, ΔK3, ΔK5, K3rev, K5rev, K3-RING-C→S, and K5-RING-C→S viruses (Fig. 6). 293A cells infected with WT BAC16 showed a significant population of cells with reduced MHC-I surface expression. On the other hand, the ΔK5 virus showed a complete lack of KSHV-mediated reduction of MHC-I at 36 h postinfection of 293A cells, indicating that K5 is the primary gene that appears to play a role in MHC-I downregulation at this stage of infection (Fig. 6). Moreover, this ability could be restored in the K5 revertant virus, confirming that loss of MHC-I downregulation in the deletion virus was indeed due to the lack of the K5 gene (Fig. 6). In contrast, BAC16-ΔK3 could downregulate MHC-I surface expression comparably to WT BAC16 following de novo infection (Fig. 6). This indicates that K5, but not K3, is required for the reduction of MHC-I surface expression following de novo infection.
Fig 6.
MHC-I surface expressions of 293A cells upon infection with WT or mutant BAC16 virus. At 36 h postinfection with WT or mutant BAC16 virus (multiplicity of infection [MOI] of 0.5), GFP-positive 293A cells were gated and examined for MHC-I surface expression.
DISCUSSION
In this study, we report the construction of a new infectious recombinant KSHV bacmid called BAC16 and demonstrate its utility in studying gene function during the course of KSHV infection. Previous reverse genetics studies have relied on BAC36 for the genetic manipulation of KSHV. However, recent evidence suggests that this clone may present unnecessary complications for reverse genetics studies, especially those involving genes that reside within the duplicated region of the BAC36 genome, which may be a reason for a low level of viral production (72). Besides lacking genomic duplications or other rearrangements of the LUR, BAC16 differs from BAC36 in several aspects. First, the BAC vector and selection cassette were inserted within the intergenic region between ORF57 and K9, whereas the BAC insertion of BAC36 is located at a PmeI site between ORF18 and ORF19 (75). A BAC cassette was inserted into an analogous location within the rhesus monkey rhadinovirus (RRV) genome (between ORF57 and R9), resulting in its successful cloning (23). Second, BAC16 is a clone of rKSHV.219, a JSC-1-derived recombinant virus, while BAC36 is derived from BCBL-1 cells. While both BAC16 and BAC36 harbor the predominant (P) allele of K15, their sequences belong to different subtypes: C3 and A3, respectively (53, 76). Subtype sequence variation can be found throughout the genome and is particularly evident within two variable regions of the K1 coding sequence (76). However, the consequence of this sequence variation is unclear. The pBelo45 construct may prove useful for the cloning and characterization of other strains of KSHV. Finally, BAC16 was constructed using a GFP-IRES-HYG cassette rather than the dual promoters used with BAC36. Based on FACS analysis, GFP expression was stably maintained in close to 100% of hygromycin-resistant iSLK-BAC16 stable cell lines for the duration of their propagation (∼3 months) (data not shown). In contrast, GFP-negative, hygromycin-resistant cells have been frequently observed in BAC36 stable cell lines (11).
One peculiar aspect of this study is the unexpected presence of Tn1000 in the majority of clones we analyzed. Furthermore, the Tn1000 insertion sites were identical in all of the Tn1000-containing clones: the site is located adjacent to the pA signal for the BAC vector cam and Hygror genes, an AT-rich locus. Although the target sequence specificity of Tn1000 has not been well studied, related transposons such as Tn3 and Tn1456 show a strong preference for insertion into AT-rich target sequences (34, 67). We also observed enhanced stability of BAC16, a Tn1000-containing clone, compared to the BAC25, which does not contain a Tn1000 insertion. Interestingly, the presence of Tn1000 in a pBR322-derived plasmid has been shown to stabilize that plasmid (6). It is possible that the presence of Tn1000 in BAC16 may contribute to its stability compared to BAC25. Finally, we did not observe any unwanted transposition events in BAC16, even after long-term culturing.
K3 and K5 have been well characterized as posttranslational regulators of several plasma membrane proteins, notably MHC-I. In addition, several other KSHV gene products have been implicated in the potential modulation of MHC-I gene expression at the transcription level, including a global suppressor of cellular gene expression, shutoff and exonuclease (SOX), encoded by ORF37; a p300 transcription modulator, viral interferon regulatory factor-1 (vIRF-1); and an NF-κB inducer, viral FLICE inhibitory protein (vFLIP) (26, 36). Our data suggest that only one of these genes, coding for K5, is critical for downregulating MHC-I surface expression during KSHV reactivation in endothelial cells or de novo infection of 293A cells. Without K5, MHC-I surface expression levels in KSHV-reactivating cells and de novo-infected 293A cells were identical to that of uninfected cells. This is in contrast to another gammaherpesvirus, MHV-68, which encodes one homologue of the KSHV K3 and K5 proteins, called mK3 (8). While MHC-I surface expression was significantly increased in ΔmK3-infected cells compared with WT-infected cells, it was not restored to the level found uninfected cells, suggesting that an MHV-68 gene or genes other than mK3 also reduce MHC-I surface expression (59).
Overexpression of either K3 or K5 in KSHV-negative cells has revealed a variety of substrates; K3 has been shown to target all HLA allotypes, CD1d, IFNGR1, PECAM (CD31), and ALCAM (CD166), while K5 substrates include HLA-A and -B, CD1d, ICAM-I, B7-2, IFNGR1, MICA/B, AICL, PECAM, ALCAM, and BST-2 (tetherin) (4, 18, 19, 27, 28, 30, 31, 39, 41, 42, 44, 57, 62). More recent evidence suggests a broad role for K5 in various aspects of cellular physiology and homeostasis, including remodeling of endothelial cell junctions through downregulation of VE-cadherins, modulation of iron import and export via HFE degradation, inhibition of BMPRII signaling, and increase of monocyte metabolism and proliferation via modulation of the localization-dependent activity of certain receptor tyrosine kinases (22, 33, 43, 54). Interestingly, the latter function can be mediated by K5 mutants lacking an intact RING-CH domain, suggesting E3 ligase activity is not required for certain K5 functions. Analysis of these additional functions using the BAC16 mutant K3 and K5 viruses may provide a better understanding of how these functions are important for the KSHV life cycle.
Individual expression of either K3 or K5 is sufficient to reduce MHC-I surface expression, and this effect is more dramatic in cells overexpressing K3 compared to K5 (18, 31). However, the ability of KSHV to reduce MHC-I surface expression in iSLK cells was not affected by the absence of K3. Moreover, deletion of the K3 gene from the ΔK5 virus did not produce any changes in MHC-I surface expression compared to single deletion of the K5 gene (data not shown). Our data suggest that K3 does not appear to significantly contribute to KSHV-mediated reduction of MHC-I surface expression during reactivation in SLK cells. Interestingly, we observed a lower level of K3 protein compared with that of K5, as assessed by Western blotting of the V5-tagged revertant viruses. However, additional work is needed to clarify whether this is due to a generally low level of K3 expression in all reactivating cells or whether K3 expression is limited to a very small fraction of these cells. If the latter is true, the K3-expressing cell population may be too small to detect a significant population of MHC-I-reduced cells by flow cytometry but nonetheless show detectable K3 expression via Western blotting.
Another possibility is the presence of a KSHV-specific factor that can posttranslationally inactivate K3. Thus, the regulatory mechanism or mechanisms that determine whether K3 is expressed or active may be the key to understanding its role in the context of KSHV infection. In this study, we examined the K3 function in endothelial cells. However, K3 may be active or expressed more robustly in other cell types, such as the B cells, the other major target cell of KSHV and the major latent reservoir of KSHV. In addition, our study was limited to 12, 24, and 48 h postreactivation and 36 h postinfection (Fig. 5 and 6) (data not shown). Although we did not detect any K3-specific functionality using K3-deficient viruses, K3 may still play a role in MHC-I downregulation during other periods of the KSHV life cycle. The broader substrate range of K5 includes activating (MICA/B and AICL) and costimulatory molecules (ICAM-1 and B7-2) involved in NK cell activation, but spares HLA-C and HLA-E (19, 62, 63). On the other hand, indiscriminant targeting of HLA allotypes by K3, including the NK cell inhibitory ligands of killer cell immunoglobulin-like receptors (KIRs), HLA-C and HLA-E, causes slightly increased susceptibility to NK cell lysis in K3-expressing cells (30). Thus, K3 expression may be detrimental for cells, especially in the absence of K5, suggesting a need for the tightly controlled gene expression regulation of K3.
The data presented here and in the accompanying article by Toth et al. (66a) demonstrate the utility of BAC16 for the generation and characterization of KSHV knockout and mutant viruses. The KSHV genome encodes several gene products with seemingly redundant function. A stable BAC provides a basis for the targeted mutagenesis procedures necessary to tease apart these overlapping functions and characterize viral gene function in biologically relevant settings. Like the K3 and K5 genes, other KSHV immune evasion genes have been characterized extensively in overexpression systems. Understanding how these and other genes contribute to the establishment of persistent infection and pathogenesis requires a reverse genetics approach wherein the effects of viral genetic deficiencies are evaluated in a relevant infection model. Such approaches will hopefully lead to a better understanding of KSHV pathogenesis.
Supplementary Material
ACKNOWLEDGMENTS
This work was partly supported by CA082057, CA31363, CA115284, DE019085, AI073099, the Hastings Foundation, DFG SFB796 (A.E.), BaCaTec (A.E. and J.U.J.), and the Fletcher Jones Foundation (J.U.J.).
We thank Koichi Yamanishi, Patrick Moore, and Greg Smith for reagents. Finally, we thank all of J.J.'s lab members for discussions.
Footnotes
Published ahead of print 27 June 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Adang LA, Tomescu C, Law WK, Kedes DH. 2007. Intracellular Kaposi's sarcoma-associated herpesvirus load determines early loss of immune synapse components. J. Virol. 81:5079–5090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barozzi P, et al. 2008. Changes in the immune responses against human herpesvirus-8 in the disease course of posttransplant Kaposi sarcoma. Transplantation 86:738–744 [DOI] [PubMed] [Google Scholar]
- 3. Bartee E, Mansouri M, Hovey Nerenberg BT, Gouveia K, Fruh K. 2004. Downregulation of major histocompatibility complex class I by human ubiquitin ligases related to viral immune evasion proteins. J. Virol. 78:1109–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bartee E, McCormack A, Fruh K. 2006. Quantitative membrane proteomics reveals new cellular targets of viral immune modulators. PLoS Pathog. 2:e107 doi:10.1371/journal.ppat.0020107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bechtel J, Grundhoff A, Ganem D. 2005. RNAs in the virion of Kaposi's sarcoma-associated herpesvirus. J. Virol. 79:10138–10146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bellani M, Shlain V, Nudel C, Sanchez Rivas C. 1997. Tn1000 (gamma delta) insertions stabilize pBR322-derived recombinant plasmids in Escherichia coli. Biotechnol. Lett. 19:1–5 [Google Scholar]
- 7. Bihl F, et al. 2007. Kaposi's sarcoma-associated herpesvirus-specific immune reconstitution and antiviral effect of combined HAART/chemotherapy in HIV clade C-infected individuals with Kaposi's sarcoma. AIDS 21:1245–1252 [DOI] [PubMed] [Google Scholar]
- 8. Boname JM, Stevenson PG. 2001. MHC class I ubiquitination by a viral PHD/LAP finger protein. Immunity 15:627–636 [DOI] [PubMed] [Google Scholar]
- 9. Boshoff C, et al. 1995. Kaposi's sarcoma-associated herpesvirus infects endothelial and spindle cells. Nat. Med. 1:1274–1278 [DOI] [PubMed] [Google Scholar]
- 10. Boshoff C, Weiss R. 2002. AIDS-related malignancies. Nat. Rev. Cancer 2:373–382 [DOI] [PubMed] [Google Scholar]
- 11. Budt M, Hristozova T, Hille G, Berger K, Brune W. 2011. Construction of a lytically replicating Kaposi's sarcoma-associated herpesvirus. J. Virol. 85:10415–10420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cadwell K, Coscoy L. 2005. Ubiquitination on nonlysine residues by a viral E3 ubiquitin ligase. Science 309:127–130 [DOI] [PubMed] [Google Scholar]
- 13. Chang H, Dittmer DP, Shin YC, Hong Y, Jung JU. 2005. Role of Notch signal transduction in Kaposi's sarcoma-associated herpesvirus gene expression. J. Virol. 79:14371–14382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chang H, et al. 2009. Non-human primate model of Kaposi's sarcoma-associated herpesvirus infection. PLoS Pathog. 5:e1000606 doi:10.1371/journal.ppat.1000606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chaturvedi AK, Mbulaiteye SM, Engels EA. 2008. Underestimation of relative risks by standardized incidence ratios for AIDS-related cancers. Ann. Epidemiol. 18:230–234 [DOI] [PubMed] [Google Scholar]
- 16. Ciufo DM, et al. 2001. Spindle cell conversion by Kaposi's sarcoma-associated herpesvirus: formation of colonies and plaques with mixed lytic and latent gene expression in infected primary dermal microvascular endothelial cell cultures. J. Virol. 75:5614–5626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Coscoy L. 2007. Immune evasion by Kaposi's sarcoma-associated herpesvirus. Nat. Rev. Immunol. 7:391–401 [DOI] [PubMed] [Google Scholar]
- 18. Coscoy L, Ganem D. 2000. Kaposi's sarcoma-associated herpesvirus encodes two proteins that block cell surface display of MHC class I chains by enhancing their endocytosis. Proc. Natl. Acad. Sci. U. S. A. 97:8051–8056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Coscoy L, Ganem D. 2001. A viral protein that selectively downregulates ICAM-1 and B7-2 and modulates T cell costimulation. J. Clin. Invest. 107:1599–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Du MQ, Bacon CM, Isaacson PG. 2007. Kaposi sarcoma-associated herpesvirus/human herpesvirus 8 and lymphoproliferative disorders. J. Clin. Pathol. 60:1350–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Duncan LM, et al. 2006. Lysine-63-linked ubiquitination is required for endolysosomal degradation of class I molecules. EMBO J. 25:1635–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Durrington HJ, et al. 2010. Identification of a lysosomal pathway regulating degradation of the bone morphogenetic protein receptor type II. J. Biol. Chem. 285:37641–37649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Estep RD, Powers MF, Yen BK, Li H, Wong SW. 2007. Construction of an infectious rhesus rhadinovirus bacterial artificial chromosome for the analysis of Kaposi's sarcoma-associated herpesvirus-related disease development. J. Virol. 81:2957–2969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fruh K, Bartee E, Gouveia K, Mansouri M. 2002. Immune evasion by a novel family of viral PHD/LAP-finger proteins of gamma-2 herpesviruses and poxviruses. Virus Res. 88:55–69 [DOI] [PubMed] [Google Scholar]
- 25. Ganem D. 2010. KSHV and the pathogenesis of Kaposi sarcoma: listening to human biology and medicine. J. Clin. Invest. 120:939–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Glaunsinger B, Ganem D. 2004. Lytic KSHV infection inhibits host gene expression by accelerating global mRNA turnover. Mol. Cell 13:713–723 [DOI] [PubMed] [Google Scholar]
- 27. Haque M, et al. 2000. Identification and analysis of the K5 gene of Kaposi's sarcoma-associated herpesvirus. J. Virol. 74:2867–2875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Haque M, et al. 2001. Major histocompatibility complex class I molecules are down-regulated at the cell surface by the K5 protein encoded by Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8. J. Gen. Virol. 82:1175–1180 [DOI] [PubMed] [Google Scholar]
- 29. Harris S, Lang SM, Means RE. 2010. Characterization of the rhesus fibromatosis herpesvirus MARCH family member rfK3. Virology 398:214–223 [DOI] [PubMed] [Google Scholar]
- 30. Ishido S, et al. 2000. Inhibition of natural killer cell-mediated cytotoxicity by Kaposi's sarcoma-associated herpesvirus K5 protein. Immunity 13:365–374 [DOI] [PubMed] [Google Scholar]
- 31. Ishido S, Wang C, Lee BS, Cohen GB, Jung JU. 2000. Downregulation of major histocompatibility complex class I molecules by Kaposi's sarcoma-associated herpesvirus K3 and K5 proteins. J. Virol. 74:5300–5309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jones T, et al. 2012. Direct and efficient cellular transformation of primary rat mesenchymal precursor cells by KSHV. J. Clin. Invest. 122:1076–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Karki R, Lang SM, Means RE. 2011. The MARCH family E3 ubiquitin ligase K5 alters monocyte metabolism and proliferation through receptor tyrosine kinase modulation. PloS Pathog. 7:e1001331 doi:10.1371/journal.ppat.1001331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Karlovsky P, Vaskova M. 1986. Hot spot for Tn1000 insertions in cloned repressor gene of the L phage. Plasmid 16:219–221 [DOI] [PubMed] [Google Scholar]
- 35. Krishnan HH, et al. 2004. Concurrent expression of latent and a limited number of lytic genes with immune modulation and antiapoptotic function by Kaposi's sarcoma-associated herpesvirus early during infection of primary endothelial and fibroblast cells and subsequent decline of lytic gene expression. J. Virol. 78:3601–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lagos D, et al. 2007. Kaposi sarcoma herpesvirus-encoded vFLIP and vIRF1 regulate antigen presentation in lymphatic endothelial cells. Blood 109:1550–1558 [DOI] [PubMed] [Google Scholar]
- 37. Lambert M, et al. 2006. Differences in the frequency and function of HHV8-specific CD8 T cells between asymptomatic HHV8 infection and Kaposi sarcoma. Blood 108:3871–3880 [DOI] [PubMed] [Google Scholar]
- 38. Lehner PJ, Hoer S, Dodd R, Duncan LM. 2005. Downregulation of cell surface receptors by the K3 family of viral and cellular ubiquitin E3 ligases. Immunol. Rev. 207:112–125 [DOI] [PubMed] [Google Scholar]
- 39. Li Q, Means R, Lang S, Jung JU. 2007. Downregulation of gamma interferon receptor 1 by Kaposi's sarcoma-associated herpesvirus K3 and K5. J. Virol. 81:2117–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liang C, Lee JS, Jung JU. 2008. Immune evasion in Kaposi's sarcoma-associated herpes virus associated oncogenesis. Semin. Cancer Biol. 18:423–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Manes TD, Hoer S, Muller WA, Lehner PJ, Pober JS. 2010. Kaposi's sarcoma-associated herpesvirus K3 and K5 proteins block distinct steps in transendothelial migration of effector memory CD4+ T cells by targeting different endothelial proteins. J. Immunol. 184:5186–5192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mansouri M, et al. 2006. Kaposi sarcoma herpesvirus K5 removes CD31/PECAM from endothelial cells. Blood 108:1932–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mansouri M, Rose PP, Moses AV, Fruh K. 2008. Remodeling of endothelial adherens junctions by Kaposi's sarcoma-associated herpesvirus. J. Virol. 82:9615–9628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mansouri M, et al. 2009. Molecular mechanism of BST2/tetherin downregulation by K5/MIR2 of Kaposi's sarcoma-associated herpesvirus. J. Virol. 83:9672–9681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Matthews NC, Goodier MR, Robey RC, Bower M, Gotch FM. 2011. Killing of Kaposi's sarcoma-associated herpesvirus-infected fibroblasts during latent infection by activated natural killer cells. Eur. J. Immunol. 41:1958–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mesri EA, Cesarman E, Boshoff C. 2010. Kaposi's sarcoma and its associated herpesvirus. Nat. Rev. Cancer 10:707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Messerle M, Crnkovic I, Hammerschmidt W, Ziegler H, Koszinowski UH. 1997. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. U. S. A. 94:14759–14763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mutlu AD, et al. 2007. In vivo-restricted and reversible malignancy induced by human herpesvirus-8 KSHV: a cell and animal model of virally induced Kaposi's sarcoma. Cancer Cell 11:245–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Myoung J, Ganem D. 2011. Generation of a doxycycline-inducible KSHV producer cell line of endothelial origin: maintenance of tight latency with efficient reactivation upon induction. J. Virol. Methods 174:12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nathan JA, Lehner PJ. 2009. The trafficking and regulation of membrane receptors by the RING-CH ubiquitin E3 ligases. Exp. Cell Res. 315:1593–1600 [DOI] [PubMed] [Google Scholar]
- 51. Okuno T, et al. 2002. Activation of human herpesvirus 8 open reading frame K5 independent of ORF50 expression. Virus Res. 90:77–89 [DOI] [PubMed] [Google Scholar]
- 52. Parsons CH, et al. 2006. KSHV targets multiple leukocyte lineages during long-term productive infection in NOD/SCID mice. J. Clin. Invest. 116:1963–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Poole LJ, et al. 1999. Comparison of genetic variability at multiple loci across the genomes of the major subtypes of Kaposi's sarcoma-associated herpesvirus reveals evidence for recombination and for two distinct types of open reading frame K15 alleles at the right-hand end. J. Virol. 73:6646–6660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rhodes DA, Boyle LH, Boname JM, Lehner PJ, Trowsdale J. 2010. Ubiquitination of lysine-331 by Kaposi's sarcoma-associated herpesvirus protein K5 targets HFE for lysosomal degradation. Proc. Natl. Acad. Sci. U. S. A. 107:16240–16245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rimessi P, et al. 2001. Transcription pattern of human herpesvirus 8 open reading frame K3 in primary effusion lymphoma and Kaposi's sarcoma. J. Virol. 75:7161–7174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sanchez DJ, Coscoy L, Ganem D. 2002. Functional organization of MIR2, a novel viral regulator of selective endocytosis. J. Biol. Chem. 277:6124–6130 [DOI] [PubMed] [Google Scholar]
- 57. Sanchez DJ, Gumperz JE, Ganem D. 2005. Regulation of CD1d expression and function by a herpesvirus infection. J. Clin. Invest. 115:1369–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shin YC, et al. 2010. Glycoprotein gene sequence variation in rhesus monkey rhadinovirus. Virology 400:175–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Stevenson PG, et al. 2002. K3-mediated evasion of CD8(+) T cells aids amplification of a latent gamma-herpesvirus. Nat. Immunol. 3:733–740 [DOI] [PubMed] [Google Scholar]
- 60. Sturzl M, et al. 2009. The contribution of systems biology and reverse genetics to the understanding of Kaposi's sarcoma-associated herpesvirus pathogenesis in endothelial cells. Thromb. Haemost. 102:1117–1134 [DOI] [PubMed] [Google Scholar]
- 61. Taylor JL, Bennett HN, Snyder BA, Moore PS, Chang Y. 2005. Transcriptional analysis of latent and inducible Kaposi's sarcoma-associated herpesvirus transcripts in the K4 to K7 region. J. Virol. 79:15099–15106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Thomas M, et al. 2008. Down-regulation of NKG2D and NKp80 ligands by Kaposi's sarcoma-associated herpesvirus K5 protects against NK cell cytotoxicity. Proc. Natl. Acad. Sci. U. S. A. 105:1656–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Thomas M, Wills M, Lehner PJ. 2008. Natural killer cell evasion by an E3 ubiquitin ligase from Kaposi's sarcoma-associated herpesvirus. Biochem. Soc. Trans. 36:459–463 [DOI] [PubMed] [Google Scholar]
- 64. Tischer BK, Smith GA, Osterrieder N. 2010. En passant mutagenesis: a two step markerless red recombination system. Methods Mol. Biol. 634:421–430 [DOI] [PubMed] [Google Scholar]
- 65. Tischer BK, von Einem J, Kaufer B, Osterrieder N. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40:191–197 [DOI] [PubMed] [Google Scholar]
- 66. Toth Z, et al. 2010. Epigenetic analysis of KSHV latent and lytic genomes. PloS Pathog. 6:e1001013 doi:10.1371/journal.ppat.1001013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66a. Toth Z, et al. 2012. Negative elongation factor-mediated suppression of RNA polymerase II elongation of Kaposi's sarcoma-associated herpesvirus lytic gene expression. J. Virol. 86:9696–9707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tu CP, Cohen SN. 1980. Translocation specificity of the Tn3 element: characterization of sites of multiple insertions. Cell 19:151–160 [DOI] [PubMed] [Google Scholar]
- 68. Vieira J, O'Hearn PM. 2004. Use of the red fluorescent protein as a marker of Kaposi's sarcoma-associated herpesvirus lytic gene expression. Virology 325:225–240 [DOI] [PubMed] [Google Scholar]
- 69. Warden C, Tang QY, Zhu H. 2011. Herpesvirus BACs: past, present, and future. J. Biomed. Biotechnol. 2011:124595 doi:10.1155/2011/124595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wen KW, Damania B. 2010. Kaposi sarcoma-associated herpesvirus (KSHV): molecular biology and oncogenesis. Cancer Lett. 289:140–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wu W, et al. 2006. KSHV/HHV-8 infection of human hematopoietic progenitor (CD34+) cells: persistence of infection during hematopoiesis in vitro and in vivo. Blood 108:141–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yakushko Y, et al. 2011. Kaposi's sarcoma-associated herpesvirus bacterial artificial chromosome contains a duplication of a long unique-region fragment within the terminal repeat region. J. Virol. 85:4612–4617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Yoo SM, et al. 2008. Centrifugal enhancement of Kaposi's sarcoma-associated virus infection of human endothelial cells in vitro. J. Virol. Methods 154:160–166 [DOI] [PubMed] [Google Scholar]
- 74. Yoo SM, Zhou FC, Ye FC, Pan HY, Gao SJ. 2005. Early and sustained expression of latent and host modulating genes in coordinated transcriptional program of KSHV productive primary infection of human primary endothelial cells. Virology 343:47–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhou FC, et al. 2002. Efficient infection by a recombinant Kaposi's sarcoma-associated herpesvirus cloned in a bacterial artificial chromosome: application for genetic analysis. J. Virol. 76:6185–6196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zong JC, Metroka C, Reitz MS, Nicholas J, Hayward GS. 1997. Strain variability among Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genomes: evidence that a large cohort of United States AIDS patients may have been infected by a single common isolate. J. Virol. 71:2505–2511 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.