Abstract
The hepatitis E virus (HEV) polyproline region (PPR) is an intrinsically unstructured region (IDR). This relaxed structure allows IDRs, which are implicated in the regulation of transcription and translation, to bind multiple ligands. Originally the nucleotide variability seen in the HEV PPR was assumed to be due to high rates of insertion and deletion. This study shows that the mutation rate is about the same in the PPR as in the rest of the nonstructural polyprotein. The difference between the PPR and the rest of the polyprotein is due to the higher tolerance of the PPR for substitutions at the first and second codon positions. With this higher promiscuity there is a shift in nucleotide occupation of these codons leading to translation of more cytosine residues: a shift that leads to more proline, alanine, serine, and threonine being encoded rather than histidine, phenylalanine, tryptophan, and tyrosine. This pattern of amino acid usage is typical of proline-rich IDRs. Increased usage of cytosine also leads to >22% of all amino acids in the PPR being prolines. Alignments of PPR sequences from HEV strains representing all genotypes indicate that all zoonotic isolates share an ancestor, and the carboxyl half of the PPR is more tolerant of mutations than the amino half. The evolution of HEV PPR, in contrast with that of the rest of the nonstructural polyprotein, is molded by pressures that lead toward increased proline usage with a corresponding decrease in the usage of aromatic amino acids, favoring formation of IDR structures.
INTRODUCTION
Hepatitis E virus (HEV) is a single-stranded, positive-sense RNA virus. The genome, which is 5′ capped and has a 3′ poly(A) tail, consists of three overlapping open reading frames (ORFs). The 5′-most ORF (ORF1) is encoded by nonstructural genes, the next 5′-most ORF (ORF3) is a phosphoprotein involved in viral regulation, and the 3′-most ORF (ORF2) is the viral capsid (1, 11). HEV causes both epidemic and sporadic jaundice (15, 21, 27). It is classified as belonging to four recognized mammalian genotypes (1–4, 21). Genotypes 1 and 2 infect only humans and are transmitted fecal-orally. Genotypes 3 and 4 infect several animals, including humans, swine, boar, deer, and mongooses (19). Besides these four genotypes there are additional mammalian HEV strains that have been isolated from rabbits (33), rats (14), and wild boars (24). The relationship between these more recently characterized strains and the recognized genotypes is still a matter of research and debate. Moreover, nonmammalian HEV strains have been found in chickens (13) and cutthroat trout (4).
The HEV nonstructural genes are most closely related to a group of viruses called the rubi-like viruses because of homology between the nonstructural genes of HEV and those of rubivirus (16). From the amino to the carboxyl terminus of the ORF1 polyprotein, these genes are the viral transferase, the Y domain, a papain-like cysteine protease, a region of unknown function, the polyproline region (PPR), the macro domain (also called the X domain), the helicase, and the RNA-directed RNA polymerase.
The PPR is also called the hypervariable region because it has higher genetic diversity than any other region in the genome (20, 23, 29). Koonin et al. (16) suggested that this region serves as a proline hinge. More recently it was determined that the region is intrinsically disordered and may regulate transcription and translation (23). Intrinsically disordered regions (IDRs) do not have stable tertiary structure (6, 9, 28). They have lower amino acid complexity, with a high proportion of polar and charged amino acids (Ala, Gly, Pro, and Ser), and a low content of bulky hydrophobic amino acids (Ile, Met, Phe, Trp, and Try) (7, 10). This disordered structure allows IDRs to assume several configurations thereby expediting the binding of this region to multiple ligands and facilitating its regulatory role (9, 10).
Because of the hypervariable sequence in the PPR some researchers avoid this region or exclude it from phylogenetic analysis of the ORF1 polyprotein (22), although it does contain phylogenetic information that has been used to genotype HEV strains (2, 3, 5, 18). The discovery of insertions and deletions in HEV genotype 3 PPR led to the assumption that the evolution of the PPR was too complex to model because of the difficulty of reconstructing its indel history (23). This assumption is questioned by data from the current study.
MATERIALS AND METHODS
Sequences from HEV genotypes 1, 3, and 4, avian HEV and rubivirus were examined (see Table S1 in the supplemental material). ORF1 sequences were split into two regions. The first region was the PPR. The second was the rest of the ORF1 polyprotein without the PPR (here, the nonpolyproline region [nPPR]). In genotypes 1, 3, and 4, the PPR was located using the conserved sequences that flank it (23). The flanking sequences for avian HEV were those obtained from the NCBI alignment for CDD:152960 (http://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi?ascbin=8&maxaln=10&seltype=2&uid=152960). In rubivirus, the PPR was estimated to be situated between amino acid 702 and amino acid 813 from a plot of the Shannon entropy for the nonstructural genes using the longest continuous region for which the entropy value was >0.1 (23).
Sequences were aligned in Clustal X (version 2.1) (17) and adjusted manually to optimize the alignment of purines and pyrimidines.
Sequences were segregated by codon position, and nucleotide counts were done using a Perl script. The number of segregating sites, nucleotide diversity, transition/transversion bias, and codon usage were calculated in Mega5 (version 5.05) (25). Shannon entropy was calculated by codon position in BioEdit (version 7.0.5.3) (12). Nucleotide sequence alignments were done initially in Clustal X2 (version 2.1) (17) and modified with manual adjustments.
Bayesian estimation of the mean substitution rate and the relative substitution rates at each codon position in the PPR and the nPPR were calculated using BEAST (version 1.6.1) (8). A general time-reversal substitution model was used with estimated base frequencies. A site-heterogeneity model with invariant sites and four gamma categories was used with codon positions segregated into three partitions by position. Substitution-rate parameters, the rate-heterogeneity model, and base frequencies were unlinked across codon positions. A relaxed, uncorrelated, lognormal, molecular clock was used. A constant-size tree prior was used, with the initial tree generated by unweighted-pair group method using average linkages (UPGMA). This series of analyses was conducted on HEV genotype 1, 3, and 4 sequences and rubiviruses because of the number of sequences available but not on avian HEV because of the limited number of sequences available. Because of the indels observed in subgenotypes 3a, 3e, and 3f (23), the insertions in 3a sequences were removed from an alignment of genotype 3 sequences. Additionally, because of the 27-amino-acid insertion seen in some 3f sequences all other sequences were aligned against the 3f repeat closer to the carboxyl terminus of the nPPR, and the 27 amino acids closer to the amino terminus were deleted from all 3f sequences containing the insertion.
RESULTS
Comparison of codon degeneracy pattern between PPR and the nPPR.
An examination of a variety of codon properties shows that the expected levels of substitution by codon position are maintained because of codon degeneracy (30) in the nPPR (Table 1). The second position is the most conserved position in the codons followed by the first position, and the third position is the least conserved (Table 1, conserved). This pattern is also reflected by the number of segregating sites per codon position, S. The lowest conservation at the third codon position is seen in genotypes 3 and 4, which may be due to the wider host range seen in these genotypes compared with the other viruses in Table 1 (19). Nucleotide diversity also reflects codon degeneracy, with more nucleotide divergence seen in the third codon position, and, as with position conservation and S, genotypes 3 and 4 exhibit the highest nucleotide divergence at the third codon position. The values for the first and second codon positions are more similar among all the viruses examined. The data for the third codon position in avian HEV suggest that it is intermediate between genotypes 3 and 4 and between genotype 1 and rubivirus, suggesting further that avian HEV has a wider host range than that seen in genotype 1 and rubivirus but not as wide as that seen in genotypes 3 and 4.
Table 1.
Codon values for the nPPR and PPR
| Genotype | Parametera | Valueb |
|||||
|---|---|---|---|---|---|---|---|
| nPPR |
PPR |
||||||
| 1 | 2 | 3 | 1 | 2 | 3 | ||
| Genotype 1 | Conserved | 0.90 | 0.93 | 0.38 | 0.63 | 0.52 | 0.34 |
| S | 171 | 122 | 1010 | 26 | 34 | 47 | |
| π | 0.023 | 0.013 | 0.18 | 0.10 | 0.12 | 0.18 | |
| Mean Hx | 0.037 | 0.022 | 0.28 | 0.15 | 0.19 | 0.27 | |
| μ | 0.30 | 0.15 | 2.55 | 0.76 | 0.88 | 1.36 | |
| Genotype 3 | Conserved | 0.82 | 0.91 | 0.060 | 0.11 | 0.20 | 0.043 |
| S | 409 | 270 | 1533 | 104 | 102 | 115 | |
| π | 0.048 | 0.014 | 0.38 | 0.29 | 0.27 | 0.42 | |
| Mean Hx | 0.077 | 0.027 | 0.61 | 0.50 | 0.45 | 0.71 | |
| μ | 0.25 | 0.048 | 2.70 | 0.82 | 0.64 | 1.54 | |
| Genotype 4 | Conserved | 0.81 | 0.90 | 0.058 | 0.18 | 0.16 | 0.00 |
| S | 324 | 189 | 1532 | 70 | 73 | 85 | |
| π | 0.038 | 0.010 | 0.36 | 0.28 | 0.28 | 0.48 | |
| Mean Hx | 0.062 | 0.020 | 0.57 | 0.46 | 0.47 | 0.80 | |
| μ | 0.25 | 0.044 | 2.70 | 0.78 | 0.74 | 1.48 | |
| Avian | Conserved | 0.90 | 0.97 | 0.31 | 0.53 | 0.60 | 0.13 |
| S | 147 | 43 | 998 | 41 | 35 | 77 | |
| π | 0.053 | 0.016 | 0.38 | 0.26 | 0.23 | 0.52 | |
| Mean Hx | 0.062 | 0.018 | 0.46 | 0.32 | 0.28 | 0.63 | |
| Rubivirus | Conserved | 0.92 | 0.96 | 0.40 | 0.61 | 0.62 | 0.25 |
| S | 161 | 84 | 1197 | 44 | 43 | 84 | |
| π | 0.017 | 0.0060 | 0.14 | 0.10 | 0.078 | 0.18 | |
| Mean Hx | 0.027 | 0.011 | 0.22 | 0.17 | 0.14 | 0.31 | |
| μ | 0.28 | 0.10 | 2.62 | 0.70 | 0.77 | 1.53 | |
Conserved, fraction of conserved nucleotides; S, number of segregating sites; π, nucleotide diversity; Mean Hx, mean Shannon entropy; μ, relative mutation rate.
Numbers above columns represent codon positions.
Lower nucleotide conservation of the nPPR with higher numbers of segregating sites, and increased nucleotide diversity and entropy, implies a higher tolerance for nucleotide substitutions and thus a higher rate of substitution. A review of the corresponding data for the PPR shows that the first and second codon positions are more tolerant of substitutions than in the nPPR, but the bias toward higher substitution rate at the third codon position compared with the first and second codon positions is still maintained, although not at the levels seen in the nPPR (Table 1). Further, the lower levels of substitution seen at the second codon position versus the first in the nPPR are not as pronounced in the PPR. This leveling of values among all three codon positions in the PPR is observed across all the variables analyzed. The higher nucleotide diversity seen in the nPPR in genotypes 3 and 4 is also seen in the PPR but is not as pronounced.
The relative substitution rates seen in genotype 4 and rubivirus confirm this tolerance for substitutions (Table 1). The relative rates of substitution in the nPPR for genotype 4 and rubivirus, respectively, are 61 and 25 times higher in the third codon position than in the second codon, and the relative substitution rates are 5.7 and 2.7 times higher at the first codon position than the second (Table 1, μ). However, in the PPR for genotype 4 and rubivirus, the relative substitution rates are twice as high at the third codon position than at the second, and the rates at the first and second codon positions are about equal. The relative substitution rates for genotype 1 are similar to those for rubivirus, and the genotype 3 rates are similar to those for genotype 4 (Table 1). The mean substitution rates as calculated in BEAST using a relaxed, uncorrelated lognormal clock for the nPPR are 1.6 × 10−3 and 5.7 × 10−4 for genotype 4 and rubivirus, respectively, and those for the PPR are 3.7 × 10−3 and 1.1 × 10−3 for genotype 4 and rubivirus, respectively. These results indicate that the overall substitution rate is about twice higher in the PPR than the nPPR. However, if the relative substitution rate by codon position is taken into account, the estimated substitution rate for the third codon position is about the same in both regions of the ORF1 polyprotein (1.4 × 10−3 versus 1.8 × 10−3 for genotype 4 and 5.0 × 10−4 versus 5.4 × 10−4 for rubivirus). These data suggest that the difference between the nPPR and the PPR is not due to a difference in rate of mutation but in higher promiscuity at the first and second codon positions in the PPR.
Preponderance of Pro in PPR.
Codon usage in the nPPR and the PPR shows there is also a difference in codon usage between them (Table 2). The most frequently used codons in the PPR are used at higher rates than in the nPPR. This is probably due to lower amino acid complexity in the PPR (23). The difference is also seen with those codons used the least. The PPR has more codons that are not used than does the nPPR, and the frequency of occurrence is lower for the least-used codons in the PPR (Table 2). Another difference is the higher usage of codons with C at the second codon position in the PPR. That leads to a higher content of Pro, Ala, Ser, and Thr. The bias toward Pro is further increased by the preference for codons with C in the first codon position, while the nPPR shows a preference for G in this position. The preference for C in the PPR is so high that the most highly used codon in avian HEV, genotypes 3 and 4, is the Pro codon, and >22% of the PPR codons in all the viruses examined encode Pro. Among the least used codons in the PPR are codons encoding His, Phe, Trp, and Tyr (Table 2). This pattern of codon usage is what would be expected for intrinsically disordered proline-rich regions (9, 10).
Table 2.
Codon usage in the nPPR and PPRa
| Genotype and frequency | nPPR |
PPR |
||
|---|---|---|---|---|
| Codon (aa) | Frequency | Codon (aa) | Frequency | |
| Genotype 1 | ||||
| Most frequent | GCC(A) | 5.31 | GCC(A) | 12.59 |
| GAG(E) | 4.29 | CCU(P) | 9.93 | |
| GGC(G) | 3.75 | UCU(S) | 6.15 | |
| GCU(A) | 3.37 | CCC(P) | 6.15 | |
| CGC(R) | 3.07 | CCG(P) | 5.87 | |
| ACC(T) | 3.04 | ACC(T) | 3.92 | |
| CUC(L) | 2.95 | GAG(E) | 3.92 | |
| CAG(Q) | 2.90 | GCU(A) | 3.64 | |
| GUU(V) | 2.65 | AUA(I) | 2.94 | |
| GUC(V) | 2.37 | GCG(A) | 2.94 | |
| GAU(D) | 2.37 | GAU(D) | 2.52 | |
| UUU(F) | 2.35 | AGU(S) | 2.38 | |
| Least frequent | ACG(T) | 0.68 | CGA(R) | 0.14 |
| UCA(S) | 0.67 | UCG(S) | 0.00 | |
| AGU(S) | 0.62 | UAC(Y) | 0.00 | |
| AUA(I) | 0.53 | AAU(N) | 0.00 | |
| GUA(V) | 0.49 | AAA(K) | 0.00 | |
| AGC(S) | 0.45 | AAG(K) | 0.00 | |
| AAA(K) | 0.44 | UGU(C) | 0.00 | |
| CAA(Q) | 0.41 | UGC(C) | 0.00 | |
| UUA(L) | 0.35 | UGG(W) | 0.00 | |
| AGA(R) | 0.24 | CGC(R) | 0.00 | |
| CGA(R) | 0.20 | AGA(R) | 0.00 | |
| GGA(G) | 0.19 | GGA(G) | 0.00 | |
| Genotype 3 | ||||
| Most frequent | GAG(E) | 4.42 | CCC(P) | 9.05 |
| GCC(A) | 4.35 | CCU(P) | 7.15 | |
| GCU(A) | 3.22 | CCA(P) | 7.04 | |
| GGC(G) | 3.21 | CCG(P) | 6.59 | |
| CAG(Q) | 2.76 | GCC(A) | 4.58 | |
| GUU(V) | 2.67 | UCU(S) | 4.13 | |
| CUU(L) | 2.48 | GCU(A) | 3.91 | |
| UUU(F) | 2.47 | GAG(E) | 3.35 | |
| CCU(P) | 2.43 | AGU(S) | 2.79 | |
| GAU(D) | 2.41 | UCC(S) | 2.57 | |
| GUG(V) | 2.21 | ACC(T) | 2.46 | |
| CGU(R) | 2.16 | ACA(T) | 2.46 | |
| Least frequent | UCA(S) | 0.87 | AUG(M) | 0.22 |
| CUA(L) | 0.86 | AAU(N) | 0.22 | |
| AGG(R) | 0.77 | AAA(K) | 0.22 | |
| AAA(K) | 0.74 | UAC(Y) | 0.11 | |
| UCG(S) | 0.73 | CAU(H) | 0.11 | |
| AGU(S) | 0.67 | CAA(Q) | 0.11 | |
| CAA(Q) | 0.61 | UGU(C) | 0.11 | |
| AGC(S) | 0.60 | CGG(R) | 0.11 | |
| CGA(R) | 0.52 | AGA(R) | 0.11 | |
| GUA(V) | 0.46 | CAC(H) | 0.00 | |
| GGA(G) | 0.44 | UGC(C) | 0.00 | |
| AGA(R) | 0.39 | CGA(R) | 0.00 | |
| Genotype 4 | ||||
| Most frequent | GAG(E) | 4.40 | CCC(P) | 7.13 |
| GCC(A) | 3.73 | CCA(P) | 6.42 | |
| GCU(A) | 3.29 | CCU(P) | 5.83 | |
| CUU(L) | 3.08 | CCG(P) | 4.88 | |
| GGC(G) | 3.02 | GCU(A) | 4.76 | |
| GUU(V) | 2.87 | UCU(S) | 4.40 | |
| CAG(Q) | 2.75 | GUG(V) | 3.92 | |
| UUU(F) | 2.66 | GAU(D) | 3.45 | |
| GAU(D) | 2.57 | GCC(A) | 3.33 | |
| CCU(P) | 2.41 | GUU(V) | 3.21 | |
| GGU(G) | 2.25 | GAG(E) | 2.85 | |
| GUC(V) | 2.21 | GGC(G) | 2.62 | |
| Least frequent | UCG(S) | 0.88 | ACG(T) | 0.24 |
| ACG(T) | 0.84 | CAU(H) | 0.24 | |
| CUA(L) | 0.83 | AAA(K) | 0.24 | |
| AGC(S) | 0.76 | GGA(G) | 0.24 | |
| AGG(R) | 0.75 | AAC(N) | 0.12 | |
| GUA(V) | 0.63 | AAG(K) | 0.12 | |
| CGA(R) | 0.62 | UAU(Y) | 0.00 | |
| AGU(S) | 0.60 | UAC(Y) | 0.00 | |
| UUA(L) | 0.57 | UGG(W) | 0.00 | |
| CAA(Q) | 0.57 | CGA(R) | 0.00 | |
| GGA(G) | 0.36 | AGA(R) | 0.00 | |
| AGA(R) | 0.17 | AGG(R) | 0.00 | |
| Avian | ||||
| Most frequent | GAG(E) | 3.66 | CCG(P) | 6.86 |
| GUG(V) | 3.61 | CCA(P) | 5.26 | |
| GCC(A) | 3.41 | CCU(P) | 5.03 | |
| CAG(Q) | 3.20 | CCC(P) | 4.58 | |
| GAU(D) | 3.19 | GCA(A) | 4.58 | |
| GUU(V) | 3.06 | GAG(E) | 3.89 | |
| GGG(G) | 2.53 | GGU(G) | 3.66 | |
| GAC(D) | 2.51 | GGC(G) | 3.66 | |
| UUG(L) | 2.41 | CAG(Q) | 3.43 | |
| GCU(A) | 2.38 | GCU(A) | 3.20 | |
| GCG(A) | 2.34 | GCC(A) | 3.20 | |
| GGC(G) | 2.31 | CGC(R) | 2.75 | |
| Least frequent | AGG(R) | 0.84 | AAG(K) | 0.46 |
| CUC(L) | 0.83 | UGU(C) | 0.46 | |
| AUC(I) | 0.75 | AGG(R) | 0.46 | |
| UCC(S) | 0.75 | GUG(V) | 0.23 | |
| UCG(S) | 0.72 | ACA(T) | 0.23 | |
| CAA(Q) | 0.71 | UGC(C) | 0.23 | |
| CGA(R) | 0.69 | AGU(S) | 0.23 | |
| AGC(S) | 0.68 | AGA(R) | 0.23 | |
| AGU(S) | 0.66 | UUU(F) | 0.00 | |
| UCA(S) | 0.64 | UAU(Y) | 0.00 | |
| GGA(G) | 0.33 | UAC(Y) | 0.00 | |
| AGA(R) | 0.28 | UGG(W) | 0.00 | |
| Rubivirus | ||||
| Most frequent | GCC(A) | 7.80 | CCC(P) | 10.38 |
| CGC(R) | 6.58 | CCG(P) | 10.21 | |
| GGC(G) | 4.90 | GCC(A) | 8.15 | |
| CUC(L) | 4.55 | GCG(A) | 7.79 | |
| GAG(E) | 4.52 | GAC(D) | 7.70 | |
| GAC(D) | 4.48 | CGC(R) | 6.45 | |
| GCG(A) | 4.37 | GGC(G) | 4.12 | |
| CCC(P) | 3.78 | CCA(P) | 3.49 | |
| ACC(T) | 3.22 | AGC(S) | 2.78 | |
| GUC(V) | 3.13 | GCA(A) | 2.51 | |
| UGC(C) | 2.99 | GUC(V) | 2.42 | |
| CAC(H) | 2.80 | GAG(E) | 2.33 | |
| Least frequent | UAU(Y) | 0.34 | CAU(H) | 0.09 |
| ACA(T) | 0.33 | UUU(F) | 0.00 | |
| UUU(F) | 0.31 | UUC(F) | 0.00 | |
| AGG(R) | 0.30 | UUA(L) | 0.00 | |
| GGA(G) | 0.29 | AUA(I) | 0.00 | |
| UCU(S) | 0.26 | AUG(M) | 0.00 | |
| UCA(S) | 0.25 | UAU(Y) | 0.00 | |
| UGU(C) | 0.24 | AAA(K) | 0.00 | |
| AGA(R) | 0.24 | UGU(C) | 0.00 | |
| CUA(L) | 0.23 | UGG(W) | 0.00 | |
| UUA(L) | 0.13 | AGA(R) | 0.00 | |
| AUA(I) | 0.12 | GGA(G) | 0.00 | |
The table lists codons, the amino acid (aa) encoded by the codon (in parentheses), and frequency of use (as a percentage). Codons are listed from most to least frequent. The space in the table separates the 12 most frequently and the 12 least frequently used codons for each genotype. Termination codons have been removed from the table.
The distribution of nucleotides by codon position in the nPPR and PPR shows that specific changes lead to the shift in codon usage. Table 3 shows that there is a significant GC bias at codon positions 1 and 3 (P < 0.07) but not at position 2 (P > 0.5) of the nPPR. However, in the PPR the GC bias is seen in positions 1 and 2 (P < 0.001) but not at position 3 (P > 0.2). The nucleotide preference by codon position in the nPPR is for G at position 1, C at position 2, and a pyrimidine at position 3. In the PPR at position 1 this preference is for G in rubivirus and genotypes 1 and 4 but for C in genotype 3 and avian HEV; at position 2 for these viruses, the preference is for C, and at position 3, the preference is for a pyrimidine except for rubivirus, where it is for C. The greatest nucleotide bias is seen in the second codon position of the PPR, where C is preferred at significantly higher levels than any of the other three bases (P < 0.0001). A comparison of the second codon position between the PPR and the nPPR shows that although C is the preferred nucleotide, the nucleotide fraction of C is almost twice as large in the PPR, the other three nucleotides exhibiting decreases of 18% to 69% except for G in avian HEV, which shows almost no change. These differences indicate that although there is not much change in nucleotide preferences at codon position 3 in the PPR, there is an increase in the fractional content of C at positions 1 and 2, with the greatest shift in nucleotide preference occupying position 2 thus leading to a preference for Pro in the PPR.
Table 3.
Nucleotide fraction by codon positiona
| Genotype | Amino acid | Value |
|||||
|---|---|---|---|---|---|---|---|
| nPPR |
PPR |
||||||
| 1 | 2 | 3 | 1 | 2 | 3 | ||
| Genotype 1 | A | 19.31 | 24.09 | 8.07 | 16.11 | 14.00 | 9.68 |
| T | 18.41 | 27.33 | 29.90 | 11.71 | 15.14 | 33.10 | |
| C | 26.89 | 28.67 | 36.85 | 34.24 | 59.24 | 33.10 | |
| G | 35.38 | 19.92 | 25.17 | 37.94 | 11.62 | 24.12 | |
| Genotype 3 | A | 19.48 | 24.98 | 12.14 | 17.58 | 12.02 | 15.93 |
| T | 18.78 | 27.02 | 32.37 | 15.56 | 16.67 | 30.94 | |
| C | 26.54 | 27.67 | 29.60 | 36.71 | 58.16 | 28.96 | |
| G | 35.20 | 20.34 | 25.89 | 30.15 | 13.15 | 24.17 | |
| Genotype 4 | A | 19.32 | 24.77 | 12.28 | 10.78 | 15.01 | 18.67 |
| T | 18.53 | 27.10 | 33.67 | 17.32 | 21.31 | 32.04 | |
| C | 26.70 | 27.86 | 28.24 | 34.61 | 52.38 | 26.86 | |
| G | 35.45 | 20.27 | 25.81 | 37.30 | 11.31 | 22.43 | |
| Avian | A | 20.44 | 25.97 | 11.24 | 13.73 | 19.45 | 21.28 |
| T | 17.35 | 28.01 | 26.00 | 9.15 | 16.48 | 26.54 | |
| C | 25.14 | 25.29 | 34.54 | 39.13 | 43.25 | 24.94 | |
| G | 37.06 | 20.73 | 28.22 | 37.99 | 20.82 | 27.23 | |
| Rubivirus | A | 15.58 | 22.87 | 6.64 | 11.80 | 17.97 | 10.39 |
| T | 12.84 | 22.78 | 10.41 | 6.28 | 7.06 | 9.50 | |
| C | 31.02 | 29.54 | 53.96 | 40.25 | 54.68 | 49.42 | |
| G | 40.56 | 24.82 | 29.00 | 41.66 | 20.29 | 30.69 | |
Integers above columns represent codon positions. All other values are percentages.
Transitions more common than transversions in both PPR and the nPPR.
The estimated transition/transversion bias for these viruses ranges from 6.2 to 11 in the nPPR and from 3.2 to 17 in the PPR. This bias suggests that transitional mutations are much more favored among these viruses than are transversions. One explanation is that transitions are less likely to result in the generation of stop codons (31), and transversions result in more diverse amino acid substitutions and significantly different chemical composition (31, 32). Given the high transition/transversion bias it might be possible to discern evolutionary patterns in the PPRs of these viruses.
Ancestry differences.
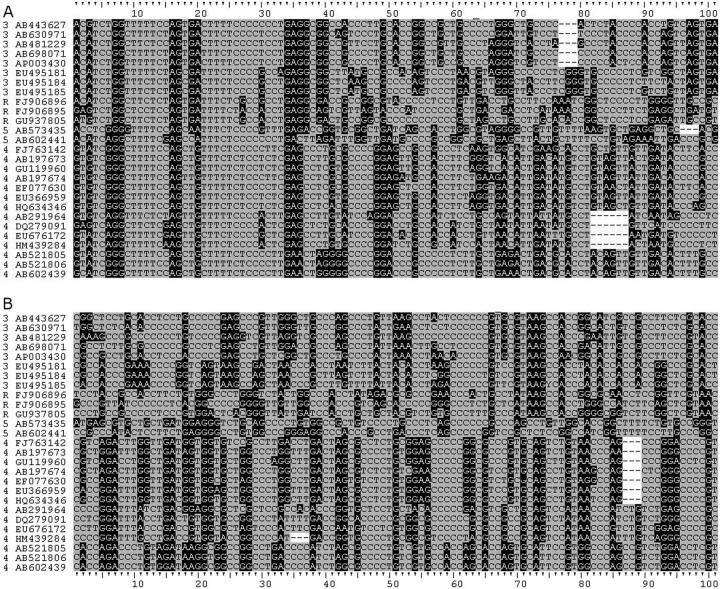
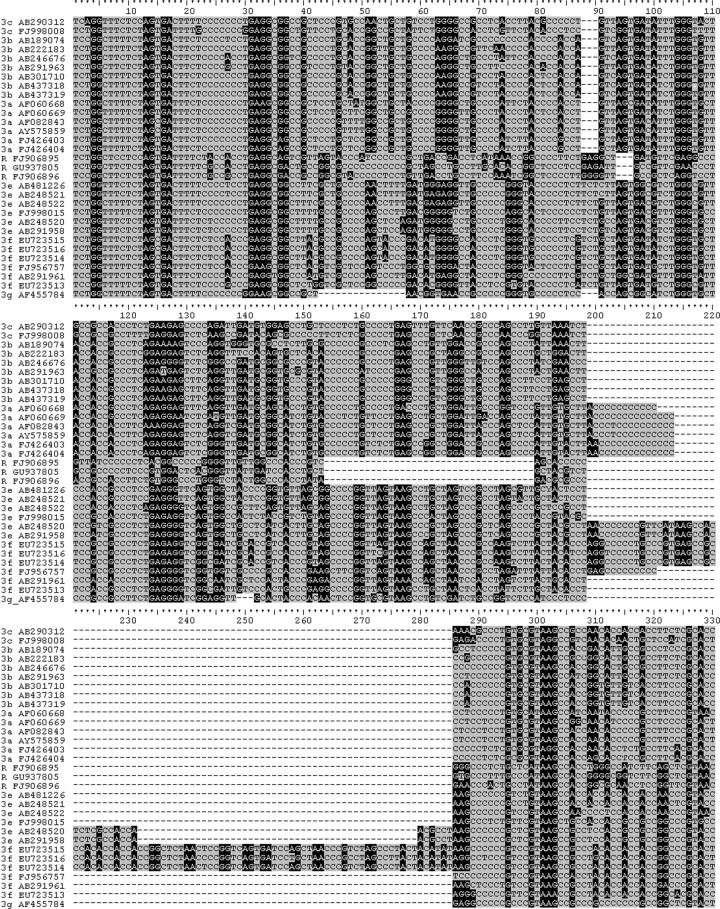
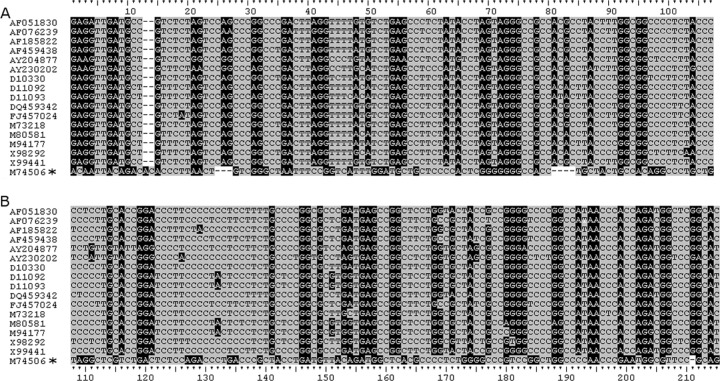
Examining the amino half of the PPR of zoonotic HEVs shows that there is homology among them, suggestive of descent from a common ancestor. As expected from phylogenetic trees, genotypes 4 and the Japanese wild boar sequences exhibit more similarity, and genotype 3 and the Chinese rabbit sequences are more similar to each other at the amino end of the PPR (Fig. 1). In the carboxyl end of the PPR, this clustering is still evident; however, the similarity seen among all of these sequences at the amino end of the PPR is less evident at the carboxyl end, suggesting that the amino half of the PPR may not tolerate mutations as well as the carboxyl half. Further examination of the genotype 3 sequences and the Chinese rabbit sequences shows similarities and differences between sequences. Like the 3a, 3e, and 3f sequences, the rabbit sequences have a deletion in the carboxyl half of the PPR although not where insertions and deletions in 3a, 3e, and 3f occur (Fig. 2). The PPR sequence alone is not enough to determine whether or not the rabbit sequences belong to a separate genotype from genotype 3. An examination of genotypes 1 and 2 shows that their amino ends are not similar to those from the zoonotic HEVs (Fig. 3A), and unlike the situation in the zoonotic sequences, the amino ends of the PPR in genotypes 1 and 2 are not similar enough to suggest descent from an ancestor common to the two of them or to the zoonotic HEVs, due perhaps to the low numbers of sequences (there being only one sequence from genotype 2). Some similarity is seen when out-of-frame shifts are allowed, implying the existence of an anthroponotic ancestor. Like the zoonotic HEVs, genotypes 1 and 2 are less similar at the carboxyl end of the PPR than the amino end (Fig. 3B), suggesting that the carboxyl end of the PPR is more susceptible to substitution.
Fig 1.
Alignment of zoonotic HEV PPRs. Sequences were aligned in Clustal X2 and adjusted manually. Purines, black background with white text; pyrimidines, gray background with black text; dashes represent gaps. Sequence IDs are prefixed with their genotype: 3, genotype 3; 4, genotype 4; 5, Japanese wild boar sequences; R, Chinese rabbit sequences. (A) Sequences aligned from nucleotide position 2121. (B) Sequences aligned from the carboxyl-terminal conserved sequence. A ruled guide is attached to each alignment.
Fig 2.
Alignment of complete PPR from genotype 3 and rabbit sequences. Alignment of zoonotic HEV PPRs. Sequences were aligned in Clustal X2 and adjusted manually. Sequences are prefixed with their genotype and subgenotype or R for the rabbit sequences. See the Fig. 1 legend for background shading. A continuous ruled guide is attached to the alignment for reference.
Fig 3.
Alignment of anthroponotic HEV PPRs. Sequences were aligned in Clustal X2 and adjusted manually. All sequences belong to genotype 1 except for M74506, marked with an asterisk, which is genotype 2. See the Fig. 1 legend for background shading. (A) Sequences aligned from nucleotide position 2121. (B) Sequences aligned from nucleotide 2334 (M80581). The ruled guide is continuous across the entire alignment.
DISCUSSION
Because of the indels seen in the HEV PPR genotype 3 (Fig. 2), it was assumed that much of the hypervariability seen in the PPR is due to insertions and deletions (23). The current study shows instead that much of the variability seen in the PPR is due to higher rates of nucleotide substitution at the first and second codon positions in the PPR.
Although the PPR is hypervariable, this hypervariability is not due to a higher substitution rate in the PPR compared to the nPPR. The same substitution rate appears to be operational in both regions (Table 1). The difference is that fewer mutations in the first and second codon positions are lethal in the PPR. Most likely this higher promiscuity, seen at the first and second codon positions in the PPR, is due to its intrinsically disordered structure. The lack of a well-defined tertiary protein structure means that substitutions in the first and second codon positions, which are more likely to result in nonsynonymous amino acid switches, are allowed more often than in the nPPR, where a tertiary structure must be maintained constitutively for proper function. However, the PPR does have constraints, as suggested by the higher usage of structure-breaking Pro codons (6, 7). The bias toward transitional substitutions may be because these substitutions are less prone than transversional substitutions to generate stop codons and because transversions lead to more diverse amino acid substitutions and significantly different chemical composition in the resultant peptide (31, 32).
Codon usage in the nPPR and the PPR shows there is a shift toward using C at the first and second codon positions in the PPR (Table 2). This is due to a shift away from using A and T at these positions and a reduction in the use of G at the second codon position (Table 3). Although the usage of G at the first codon position does not change much, the usage of C increases with the decreases in A and T (Table 3). The shift at codon position 2 is even more dramatic: from about equal usage of all nucleotides at the second codon position in the nPPR to C occurring at >50% of the second codon positions in the PPR (Table 3). This in turn results in a shift toward high usages of Pro, Ala, Ser, and Thr in the PPR, so marked that the most frequently used codon in genotypes 3 and 4 and avian HEV is Pro (Table 2). Even in genotype 1 and rubivirus, >22% of all codons in this region are Pro codons (Table 2). The decrease in A/T usage leads to a decrease in His, Phe, Trp, and Tyr. These are the patterns of amino acid usage typical of IDRs (7, 28). The decrease in A at the first codon position and A and G at the second codon position of the PPR means that transversional substitutions have occurred; these transversions appear to be more common among genotypes and subgenotypes (Fig. 1, 2, and 3).
Although the first and second codon positions are more promiscuous in the PPR than the nPPR, alignments of zoonotic HEVs suggest that this promiscuity is greater in the carboxyl half of the PPR than in the amino half (Fig. 1). The carboxyl half of the PPR is also where most of the recognized indel activity occurs in the PPR (Fig. 2). This difference suggests that the carboxyl half of the PPR is more mutable than the amino end, and the carboxyl half of the PPR may be more involved in binding multiple ligands (23).
Evolution is more easily traced in the nPPR because of the tertiary structural constraints required by the nonstructural genes for them to function properly. In contrast, because of the higher promiscuity toward substitutions and the lack of intrinsic structure or active-site amino acids, it is much more difficult to trace evolution in the PPR alone. However, an alignment of zoonotic HEVs shows that there is a similarity in purine/pyrimidine (transitional substitution) banding in the amino half of the PPR, suggesting that these isolates share an ancestor (Fig. 1). This commonality is not seen in the carboxyl half of these PPR sequences, perhaps due to higher mutability in that domain. An alignment of the PPR for the anthroponotic genotypes 1 and 2 does not exhibit an easily recognized similarity of purine/pyrimidine banding, perhaps because only one example of the genotype 2 PPR sequence exists; nonetheless, out-of frame shifting of the alignment implies a common ancestor (Fig. 3).
The similarity of sequence (Fig. 3) and lower nucleotide diversity (Table 1) seen in genotype 1 suggest that less substitution occurs in genotype 1 than in genotypes 3 or 4. This could be because the zoonotic HEVs have a wider host range, and higher nucleotide diversity is required for adaptation of these strains to their hosts. Another explanation is that modern genotype 1 is actually composed of a subset of subgenotypes from a genotype 1 ancestor. Paleoepidemiological research indicates that epidemic HEV was more common in Australia, North America, and Europe in the 18th and 19th centuries than today (26). An analysis of the evolution of HEV suggests further that genotype 1 went through an evolutionary bottleneck about 80 to 90 years ago (22). Improvements in sanitation in developed countries from the early 20th century could have forced genotype 1 through an evolutionary bottleneck that led to the extinction of genotype 1 in Australia, North America, and Europe, with the only surviving subtypes of genotype 1 being found in developing countries. More isolates of genotypes 1 and 2 are needed to better define the evolution of these genotypes and of the PPR in mammalian HEVs.
The hypervariability seen in the HEV PPR appears to be due to increased rates of substitution in the PPR compared to the nPPR, but the impetus for this hypervariability is increased promiscuity toward substitution at the first and second codon positions in the PPR. In conjunction with this promiscuity is a shift in nucleotide usage toward increased usage of C such that Pro codons are among the most favored in the PPR, and the decreased usage of A and T results in decreased use of His, Phe, Trp, and Typ codons. This shift leads to a region with a high number of structure-breaking Pro residues and few aromatic residues, thereby accounting for the proline richness seen in IDRs.
Supplementary Material
ACKNOWLEDGMENTS
I thank Chong-Gee Teo for discussions and review of this paper, and I acknowledge the helpful suggestions of reviewers from CDC and the journal.
The findings and conclusions in this article are those of the author and do not necessarily represent the views of the Centers for Disease Control and Prevention/Agency for Toxic Substances and Disease Registry.
Footnotes
Published ahead of print 18 July 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Ahmad I, Holla RP, Jameel S. 2011. Molecular virology of hepatitis E virus. Virus Res. 161:48–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arankalle VA, Chobe LP, Chadha MS. 2006. Type-IV Indian swine HEV infects rhesus monkeys. J. Viral Hepat. 13:742–745 [DOI] [PubMed] [Google Scholar]
- 3. Arankalle VA, Paranjape S, Emerson SU, Purcell RH, Walimbe AM. 1999. Phylogenetic analysis of hepatitis E virus isolates from India (1976–1993). J. Gen. Virol. 80:1691–1700 [DOI] [PubMed] [Google Scholar]
- 4. Batts W, Yun S, Hedrick R, Winton J. 2011. A novel member of the family Hepeviridae from cutthroat trout (Oncorhynchus clarkii). Virus Res. 158:116–123 [DOI] [PubMed] [Google Scholar]
- 5. Chatterjee R, et al. 1997. African strains of hepatitis E virus that are distinct from Asian strains. J. Med. Virol. 53:139–144 [PubMed] [Google Scholar]
- 6. Dosztányi Z, Chen J, Dunker K, Simon I, Tompa P. 2006. Disorder and sequence repeats in hub proteins and their implications for network evolution. J. Proteome Res. 5:2985–2995 [DOI] [PubMed] [Google Scholar]
- 7. Dosztányi Z, Csizmok V, Tompa P, Simon I. 2005. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J. Mol. Biol. 347:827–839 [DOI] [PubMed] [Google Scholar]
- 8. Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dunker AK, et al. 2008. The unfoldomics decade: an update on intrinsically disordered proteins. BMC Genomics 9(suppl. 2):S1 doi:10.1186/1471-2164-9-S2-S1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dyson HJ, Wright PE. 2005. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6:197–208 [DOI] [PubMed] [Google Scholar]
- 11. Emerson SU, et al. 2004. Hepevirus, p 853–857 In Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA. (ed), Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses Elsevier/Academic Press, London, United Kingdom [Google Scholar]
- 12. Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95–98 [Google Scholar]
- 13. Haqshenas G, Shivaprasad HL, Woolcock PR, Read DH, Meng XJ. 2001. Genetic identification and characterization of a novel virus related to human hepatitis E virus from chickens with hepatitis-splenomegaly syndrome in the United States. J. Gen. Virol. 82:2449–2462 [DOI] [PubMed] [Google Scholar]
- 14. Johne R, et al. 2010. Detection of a novel hepatitis E-like virus in faeces of wild rats using a nested broad-spectrum RT-PCR. J. Gen. Virol. 91:750–758 [DOI] [PubMed] [Google Scholar]
- 15. Khuroo MS. 2011. Discovery of hepatitis E: the epidemic non-A, non-B hepatitis 30 years down the memory lane. Virus Res. 161:3–14 [DOI] [PubMed] [Google Scholar]
- 16. Koonin EV, et al. 1992. Computer-assisted assignment of functional domains in the nonstructural polyprotein of hepatitis E virus: delineation of an additional group of positive-strand RNA plant and animal viruses. Proc. Natl. Acad. Sci. U. S. A. 89:8259–8263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Larkin MA, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 18. Legrand-Abravanel F, et al. 2009. Hepatitis E virus genotype 3 diversity, France. Emerg. Infect. Dis. 15:110–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Meng XJ. 2011. From barnyard to food table: the omnipresence of hepatitis E virus and risk for zoonotic infection and food safety. Virus Res. 161:23–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pudupakam RS, et al. 2009. Deletions of the hypervariable region (HVR) in open reading frame 1 of hepatitis E virus do not abolish virus infectivity: evidence for attenuation of HVR deletion mutants in vivo. J. Virol. 83:384–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Purcell RH, Emerson SU. 2008. Hepatitis E: an emerging awareness of an old disease. J. Hepatol. 48:494–503 [DOI] [PubMed] [Google Scholar]
- 22. Purdy MA, Khudyakov YE. 2010. Evolutionary history and population dynamics of hepatitis E virus. PLoS One 5:e14376 doi:10.1371/journal.pone.0014376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Purdy MA, Lara J, Khudyakov YE. 2012. The hepatitis E virus polyproline region is involved in viral adaptation. PLoS One 7:e35974 doi:10.1371/journal.pone.0035974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Takahashi M, Nishizawa T, Sato H, Sato Y, Jirintai Nagashima S, Okamoto H. 2011. Analysis of the full-length genome of a hepatitis E virus isolate obtained from a wild boar in Japan that is classifiable into a novel genotype. J. Gen. Virol. 92:902–908 [DOI] [PubMed] [Google Scholar]
- 25. Tamura K, et al. 2011. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Teo C-G. 2012. Fatal outbreaks of jaundice in pregnancy and the epidemic history of hepatitis E. Epidemiol. Infect. 140:767–787 [DOI] [PubMed] [Google Scholar]
- 27. Teshale EH, Hu DJ, Holmberg SD. 2010. The two faces of hepatitis E virus. Clin. Infect. Dis. 51:328–334 [DOI] [PubMed] [Google Scholar]
- 28. Tsai C-J, Ma B, Sham YY, Kumar S, Nussinov R. 2001. Structured disorder and conformational selection. Proteins 44:418–427 [DOI] [PubMed] [Google Scholar]
- 29. Tsarev SA, et al. 1992. Characterization of a prototype strain of hepatitis E virus. Proc. Natl. Acad. Sci. U. S. A. 89:559–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Watson JD, et al. 2007. Molecular biology of the gene, 6th ed Benjamin-Cummings, San Francisco, CA [Google Scholar]
- 31. Wong TS, Roccatano D, Schwaneberg U. 2007. Are transversion mutations better? A Mutagenesis Assistant Program analysis on P450 BM-3 heme domain. Biotechnol. J. 2:133–142 [DOI] [PubMed] [Google Scholar]
- 32. Wong TS, Roccatano D, Schwaneberg U. 2007. Challenges of the genetic code for exploring sequence space in directed protein evolution. Biocatal. Biotransformation 25:229–241 [Google Scholar]
- 33. Zhao C, et al. 2009. A novel genotype of hepatitis E virus prevalent among farmed rabbits in China. J. Med. Virol. 81:1371–1379 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.