Telomerase adds simple-sequence repeats to the ends of linear chromosomes to counteract the loss of end sequence inherent in conventional DNA replication. Catalytic activity for repeat synthesis arises from the cooperation of the telomerase reverse transcriptase protein (TERT) and the template-containing telomerase RNA (TER). TERs vary widely in sequence and structure but share a set of motifs required for TERT binding and catalytic activity. Species-specific TER motifs play essential roles in RNP biogenesis, stability, trafficking, and regulation. Remarkably, the biogenesis pathways that generate mature TER differ between and even within phylogenetic groups. This review highlights the diversity of strategies for telomerase RNP biogenesis, RNP assembly, and telomere recruitment among ciliates, yeasts, and vertebrates and suggests common themes in these pathways and their regulation.
Keywords: ribonucleoprotein biogenesis, telomerase RNA, telomerase reverse transcriptase, telomerase holoenzyme, telomere maintenance
Abstract
Telomerase adds simple-sequence repeats to the ends of linear chromosomes to counteract the loss of end sequence inherent in conventional DNA replication. Catalytic activity for repeat synthesis results from the cooperation of the telomerase reverse transcriptase protein (TERT) and the template-containing telomerase RNA (TER). TERs vary widely in sequence and structure but share a set of motifs required for TERT binding and catalytic activity. Species-specific TER motifs play essential roles in RNP biogenesis, stability, trafficking, and regulation. Remarkably, the biogenesis pathways that generate mature TER differ across eukaryotes. Furthermore, the cellular processes that direct the assembly of a biologically functional telomerase holoenzyme and its engagement with telomeres are evolutionarily varied and regulated. This review highlights the diversity of strategies for telomerase RNP biogenesis, RNP assembly, and telomere recruitment among ciliates, yeasts, and vertebrates and suggests common themes in these pathways and their regulation.
INTRODUCTION
The evolution of linear chromosomes in the common ancestor of eukaryotes occurred despite several challenges posed by this mode of genome organization. First, conventional primer-requiring DNA polymerases cannot copy the 3′ end of a DNA template, leading to the progressive loss of end sequence with every round of DNA replication. Second, chromosome ends resemble double-stranded DNA breaks whose recognition and repair results in chromosome fusions. Third, DNA ends are vulnerable to the destructive action of exonucleases. A solution to these challenges was to form a protective cap structure from a tandem array of telomeric repeats with a net length of tens to thousands of base pairs (Gomes et al. 2010; Jain and Cooper 2010 O'Sullivan and Karlseder 2010). Telomeric DNA termini typically have a 3′ overhang, which can invade the duplex to form a loop that sequesters the single-stranded end. Single-stranded and double-stranded DNA-binding proteins coat the telomeric repeat DNA to assemble an end-capping structure (de Lange 2010; Stewart et al. 2012).
Telomere maintenance in most eukaryotes depends on new repeat synthesis by the specialized reverse transcriptase (RT) telomerase (Blackburn et al. 2006). Telomerase is a ribonucleoprotein (RNP) enzyme with two catalytically essential subunits: the telomerase reverse transcriptase protein (TERT) and the telomerase RNA (TER). TERT copies a template within TER to determine the repeat sequence, and outside of the active site additional domains of both subunits contribute to synthesis (Blackburn and Collins 2011). Many telomerase enzymes maintain productive binding to the substrate DNA during synthesis across the entire template (nucleotide addition processivity) and also during multiple rounds of template copying (repeat addition processivity, RAP). The processivity and fidelity of repeat synthesis vary across telomerases from different organisms and even telomerase from the same organism under different growth conditions (Chang et al. 2007; Collins 2009; Zhao et al. 2011). It is possible that some TERT proteins do not copy an internal template, perhaps reflected in the evolutionary loss of entire TERT domains (Blackburn and Collins 2011). Other organisms including Drosophila have lost TERT and TER entirely and instead use recombination- or transposition-based mechanisms for telomere maintenance (Pardue and DeBaryshe 2011).
In general, TERTs contain four domains: the telomerase “essential” N-terminal (TEN) domain, the telomerase RNA-binding domain (TRBD), the RT domain, and the C-terminal extension (CTE). The TEN domain traps single-stranded telomeric DNA and also interacts with TER (O'Connor et al. 2005; Jacobs et al. 2006; Robart and Collins 2011). The single-stranded DNA-handling function of the TEN domain likely promotes processive repeat synthesis by capturing the substrate and maintaining association with the single-stranded product (Collins 2011). The TRBD confers the specificity of interaction between TERT and TER (Lai et al. 2001; O'Connor et al. 2005). In the RT domain, motifs conserved in the evolutionarily related retrotransposon RTs form the active site, where aspartic acid residues coordinate the magnesium ions necessary for catalysis of dNTP addition (Lingner et al. 1997b; Belfort et al. 2011). In TERTs, this domain also positions the template and aligns the substrate 3′ end (Xie et al. 2010; Qi et al. 2012). The CTE may enhance nucleic acid association and/or otherwise contribute to RT domain function (Autexier and Lue 2006; Wyatt et al. 2010). A crystal structure of TERT from the flour beetle Tribolium castaneum, which lacks a TEN domain, revealed CTE contacts with the TRBD that create a protein ring around a model RNA–DNA hybrid (Gillis et al. 2008; Mitchell et al. 2010). It is possible that the CTE influences the telomerase catalytic cycle by affecting the conformation of this ring. Further insights will be greatly aided by high-resolution structure of TERT domain complexes with an endogenous TER.
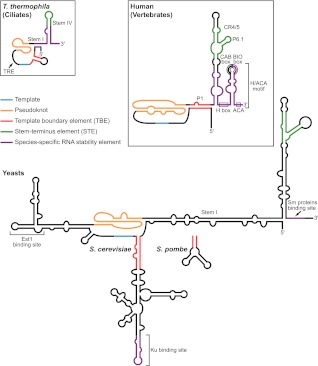
TERs vary more than TERTs in their folded domain organization, with disparate sequences and sizes ranging from about 150 nucleotides (nt) in ciliates to more than 10 times that length in some yeasts. However, functional assays suggest that several TER elements may be considered shared between ciliates, yeasts, and vertebrates, as illustrated in Figure 1 and reviewed extensively elsewhere, and therefore not individually referenced below (Theimer and Feigon 2006; Collins 2009; Blackburn and Collins 2011; Zhang et al. 2011; Podlevsky and Chen 2012). First is the single-stranded template, whose complement typically corresponds to about 1.5 repeats of the telomeric DNA sequence. Notable exceptions to this template length occur in yeasts with long and/or degenerate telomeric repeat sequences (Lue 2010). The 3′ region of the template aligns the DNA substrate, while the 5′ portion is copied. Adjacent to the template is a unique pseudoknot fold stabilized by triple-helix formation. Despite its structural conservation and importance for TER folding overall, the specific function of the TER pseudoknot remains an outstanding question. Based on studies to date, we suggest that a unifying model for pseudoknot function would be in positioning the template relative to the active site and other TER and TERT domains.
FIGURE 1.
Diagram of TER secondary structures highlighting functional motifs. The template, pseudoknot, TBE, and STE are common to ciliate, yeast, and vertebrate TERs. The STE is distal stem–loop IV in T. thermophila, conserved region 4/5 (CR4/5) in human, and a three-way helix junction in yeasts. Species-specific RNP stability elements recruit p65 in T. thermophila, H/ACA proteins in human, and Sm proteins in yeasts (also Ku in Saccharomyces). The binding sites for holoenzyme proteins that do not affect RNA stability, namely Est1 in yeasts and the CAB box-binding protein WDR79/TCAB1 in humans, are indicated. T. thermophila TER also contains a template recognition element (TRE) that contributes to template utilization.
The template-pseudoknot domain is typically closed by long-range base-pairing of sequence at or near the TER 5′ end (Fig. 1). This stem or a separate hairpin immediately 5′ of the template can provide a 5′ template boundary element (TBE) to prevent copying of nontemplate TER sequence. Separate from all of the other conserved TER elements described above is a “stem terminus” element (STE), comprised of either a terminal hairpin, a hairpin from a three-way junction, or a three-way junction alone (Fig. 1). The STE stimulates telomerase activity, at least in part, through direct interaction with the TERT TRBD. Because some STE sequence substitutions reduce activity without an evident change in TERT binding, the STE motif is likely to have an additional role in RNP folding and/or catalytic activation. One hypothesis for this additional role is an allosteric influence on TERT and/or TER domain conformation.
Cellular telomerase holoenzymes are multisubunit complexes that fractionate by gel filtration with an apparent mass of 500 kDa or more (Collins 2009). However, only TERT and TER are required to reconstitute telomerase catalytic activity on oligodeoxynucleotide substrates in vitro (Weinrich et al. 1997). Beyond the TER motifs that bind TERT and orchestrate the telomerase catalytic cycle, TERs possess additional motifs that recruit proteins involved in TER processing and RNP stability or localization in vivo. In addition, other telomerase holoenzyme proteins direct and regulate physical interactions with telomeric chromatin and functional engagement with the chromosome 3′ end. The identities of telomerase holoenzyme proteins and their exact mechanisms of function differ between organisms, but some of their biochemical roles bear striking similarities. This review aims to describe the unique and shared features of telomerases from ciliates, yeasts, and vertebrates with an emphasis on pathways of telomerase RNA biogenesis and RNP assembly. Telomerase physical recruitment to, elongation of, and regulation at telomeres are modulated by dynamic telomeric chromatin reviewed extensively elsewhere (Stern and Bryan 2008; Moser and Nakamura 2009; Schoeftner and Blasco 2009; de Lange 2010; Stewart et al. 2012). Most telomerase studies have involved Tetrahymena thermophila, Saccharomyces cerevisiae, Schizosaccharomyces pombe, mice, or cultured human cells. Less-intensively studied but important additional model systems include other ciliates, yeasts, vertebrates and also plants (Collins 1999; Teixeira and Gilson 2005; Watson and Riha 2010; Cifuentes-Rojas et al. 2011; Yu 2012).
CILIATE TELOMERASE
Ciliate telomerase RNA structure
Ciliate TERs are the smallest to be characterized to date. The model laboratory ciliate, T. thermophila, possesses a 159-nt TER (Greider and Blackburn 1989) that includes all of the conserved motifs summarized above (Fig. 1). The 9-nt template is flanked at its 5′ end by a TBE and at its 3′ end by a pseudoknot. These three motifs are enclosed by long-range base-pairing of stem I (Fig. 1). In addition to defining the template boundary, the TBE and its adjacent single-stranded regions also provide the high-affinity TERT-binding site (Autexier and Greider 1995; Lai et al. 2002). A recent single-molecule fluorescence resonance energy transfer study found that the T. thermophila TER pseudoknot remains stably folded throughout the catalytic cycle (Mihalusova et al. 2011). Remarkably, the pseudoknot can form even when some of the stem pairing is disrupted (Mihalusova et al. 2011), likely accounting for the modest impact of some, but not other, pseudoknot-destabilizing mutations on telomerase holoenzyme assembly and activity (Gilley and Blackburn 1999; Cunningham and Collins 2005).
In ciliate TERs, terminal stem IV and its loop comprise the STE (Fig. 1). Nucleotides within loop IV contribute to TERT binding in a manner important for RNP conformational stability in vitro and biological stability in vivo (Stone et al. 2007; Robart et al. 2010). T. thermophila TER also has another motif immediately 3′ of the template termed the template recognition element (TRE) (Fig. 1), which contributes to efficient copying through mid-template positions and improves the in vitro use of oligonucleotide templates physically separate from the rest of TER (Miller and Collins 2002; Cunningham and Collins 2005). Curiously, cells expressing a TER with TRE sequence substitutions have longer than normal telomeres, perhaps linked to altered enzyme pausing during repeat synthesis (Cunningham and Collins 2005).
Ciliate telomerase RNP maturation and TERT–TER assembly
Ciliate telomerase RNP assembly begins with TER transcription by RNA polymerase III (Greider and Blackburn 1989). Mature TER retains the 3′ uridine-rich termination sequence of the primary transcript (Fig. 2). In T. thermophila the TER 3′ polyuridine tail along with stems I and IV provides a binding site for the telomerase-specific p65 protein (Fig. 2). This La-family telomerase holoenzyme protein is required for TERT and TER accumulation in vivo (Witkin and Collins 2004; O'Connor and Collins 2006). Here, we will use the designation “RNP catalytic core” to describe the physiologically assembled minimal catalytically active RNP (for example, the p65–TER–TERT ternary complex), which includes subunits essential for TER folding and stability in vivo but not for optimized TERT + TER minimal RNP reconstitution in vitro. The interaction of p65 with TER stabilizes a kink in stem IV necessary for tight TERT binding (Stone et al. 2007; Singh et al. 2012). Binding of p65 also induces additional conformational changes in stem–loop IV and beyond that promote TERT assembly and catalytic activity (Prathapam et al. 2005; Richards et al. 2006; Berman et al. 2010; Akiyama et al. 2012; Singh et al. 2012).
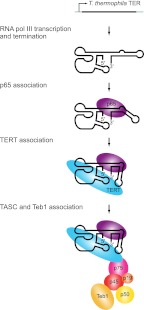
FIGURE 2.
T. thermophila telomerase RNP biogenesis. T. thermophila TER is transcribed by RNA polymerase III. The binding of p65 stabilizes a kink in stem IV that optimally positions loop IV for TERT binding. The p65-TER–TERT ternary complex then interacts with a complex of p75, p50, p45, and p19 that recruits the single-stranded telomeric DNA-binding protein Teb1.
Bridging the ciliate RNP catalytic core to telomere substrates
The T. thermophila telomerase holoenzyme has been purified to homogeneity, enabling extensive subunit characterization. Beyond the RNP catalytic core, five associated proteins designated p19, p45, p50, p75, and Teb1 (Fig. 2) are required for telomere maintenance in vivo (Witkin and Collins 2004; Witkin et al. 2007; Min and Collins 2009). Four of these five proteins (p19, p45, p50, and p75) contribute to the telomere adaptor subcomplex (TASC) that bridges the RNP catalytic core to Teb1 (Min and Collins 2009; Eckert and Collins 2012). Teb1 joining generates the complete telomerase holoenzyme, which in T. thermophila retains a remarkably stable grip on product DNA and thus supports high RAP in vitro (Greider 1991).
Teb1 binds directly and with high affinity to single-stranded telomeric repeat DNA (Min and Collins 2009, 2010; Zeng et al. 2011). Although Teb1 has an oligonucleotide/oligosaccharide (OB) fold architecture homologous to the large subunit of Replication Protein A (RPA), it is telomerase-specific in its physical interactions and sequence-specific in its recognition of tandem T2G4 repeats. Teb1 stimulation of RAP in vitro results in part from activating protein–protein interaction(s) and in part from its binding to nascent product DNA (Min and Collins 2010), which could be important in vivo independent of elongation RAP per se. Because the Teb1 and p50 subunits are extensively proteolyzed and/or substoichiometric in the pool of holoenzyme purified from cell extract (Min and Collins 2009), a heterologous system for reconstitution of recombinant holoenzyme will be required to fully understand the roles of TASC and Teb1 in telomere elongation. T. thermophila telomerase physical recruitment to and/or functional activation at telomeres may also involve the telomere-associated Pat1 protein, whose depletion causes gradual telomere shortening without compromised end-protection (Linger et al. 2011).
YEAST TELOMERASE
Yeast telomerase RNA structure
Yeast telomerase RNAs are relatively large (1000 nt or more) with a template, TBE, and pseudoknot brought together by long-range base-pairing of stem I (Fig. 1). In the S. cerevisiae TER, TLC1, a high-affinity binding site for the TERT protein Est2 is within this conserved region (Livengood et al. 2002; Zappulla et al. 2005). Most of yeast TER length derives from three long stems, or “arms,” extending from the conserved central region (Fig. 1). One of these arms is positioned immediately 5′ of the template and provides a TBE (Tzfati et al. 2000; Seto et al. 2003; Box et al. 2008b). In addition, in Saccharomyces but not other yeast species, the terminal stem–loop of this arm forms a binding site for the Ku heterodimer (Fig. 1), which enhances in vivo RNP accumulation, nuclear localization, and telomere recruitment (Peterson et al. 2001; Stellwagen et al. 2003; Fisher et al. 2004; Gallardo et al. 2008; Mozdy et al. 2008). The second RNA arm occurs between the template and pseudoknot and contains a binding site for the regulatory protein Est1 (Fig. 1), which together with another holoenzyme subunit, Est3, allows telomere elongation in vivo (see below and for review Osterhage and Friedman 2009; DeZwaan and Freeman 2010). A similar motif in the S. pombe TER, TER1, interacts with an Est1 ortholog (Webb and Zakian 2012), but no Est3 ortholog is known to exist in this species.
The third RNA arm is the extremely long stem I, extended by a hairpin formed by fold-back of the 3′ end (Fig. 1). This 3′ terminal hairpin contains the STE as a three-way junction motif that stimulates catalytic activity and may contact TERT in Kluyveromyces lactis and likely other yeasts (Brown et al. 2007). A single-stranded uridine-rich motif near the 3′ end of yeast TERs binds to Sm proteins (Fig. 1), which contribute to the maturation and stability of the RNA (Seto et al. 1999; Tang et al. 2012). Studies of TLC1 have demonstrated that the RNA arms can be truncated substantially and still support activity in vitro or prevent senescence in vivo (Zappulla et al. 2005), giving rise to the impression of TER as a conformationally flexible scaffold for protein assembly (Zappulla and Cech 2006). However, minimized TLC1 accumulates to levels much lower than wild type, maintains short telomeres, and reduces fitness (Zappulla et al. 2005).
Studies of yeast TERs have provided insights into the function of the pseudoknot. In K. lactis TER, some pseudoknot sequence substitutions prevent copying of the full template (Tzfati et al. 2003). Another study used fragments of TLC1 assembled with Est2 in rabbit reticulocyte lysate to determine that disruption of the pseudoknot triple helix reduced telomerase activity but not Est2 binding (Qiao and Cech 2008). A role for the pseudoknot in substrate DNA positioning was proposed based on the crosslinking of pseudoknot nucleotides within a minimal recombinant RNA alone to the 3′ end of a telomeric DNA oligonucleotide (Qiao and Cech 2008).
Yeast telomerase RNP maturation and TERT–TER assembly
Studies of yeast telomerase RNA biogenesis have focused on S. cerevisiae TLC1 and more recently also on S. pombe TER1. Both are independently transcribed by RNA polymerase II and accumulate primarily as unpolyadenylated species (Figs. 3, 4) with a minor fraction (5%–10%) occurring in a polyadenylated form (Chapon et al. 1997; Leonardi et al. 2008). Telomerase holoenzyme contains the unpolyadenylated form (Bosoy et al. 2003; Leonardi et al. 2008).
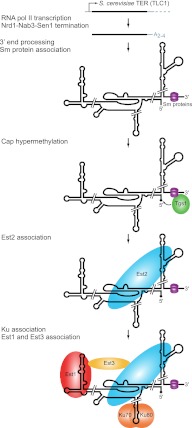
FIGURE 3.
S. cerevisiae telomerase RNP biogenesis. S. cerevisiae TLC1 is transcribed by RNA polymerase II, and transcription is terminated by the Nrd1–Nab3–Sen1 pathway. The 3′ end is processed by the nuclear exosome and TRAMP complex, leaving a short adenosine-rich tail that is ultimately removed. Sm protein binding near the 3′ end stabilizes the RNA and recruits the cap hypermethylase Tgs1, which modifies the 5′ cap to TMG, indicated by a diamond. The TERT subunit Est2 binds directly to TLC1. The Ku heterodimer recognizes a distinct binding site on TLC1 to promote RNP accumulation and nuclear import. The regulatory subunit Est1 can bind directly to TLC1 and with Est3 stimulates telomerase function at telomeres.
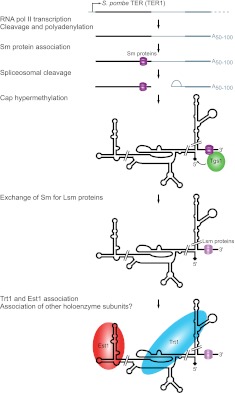
FIGURE 4.
S. pombe telomerase RNP biogenesis. S. pombe TER1 is transcribed by RNA polymerase II as a precursor that includes a downstream intron and exon. TER1 secondary structure has yet to be experimentally validated; it is illustrated here as similar to other yeast TERs. Sm proteins assemble on the precursor near the mature TER1 3′ end. Spliceosomal cleavage at the 5′ splice site releases mature TER1, which escapes ligation to the downstream exon. Sm proteins then recruit the cap hypermethylase Tgs1, which modifies the 5′ cap to TMG, indicated by a diamond. Sm proteins are then replaced by Lsm proteins that protect the 3′ end from degradation, followed by assembly of Est1 and the TERT subunit Trt1.
Although previously suggested to be a precursor, recent results suggest that the polyadenylated form of TLC1 is not an intermediate in the accumulation of the unpolyadenylated mature form (Noël et al. 2012). The 3′ end of functional TLC1 is produced by the Nrd1–Nab3–Sen1 pathway (Fig. 3), which also matures the transcripts of small nuclear (sn) and small nucleolar (sno) RNAs in yeast (Kuehner et al. 2011; Noël et al. 2012). In this pathway, specific patterns of RNA polymerase II C-terminal domain phosphorylation recruit the RNA-binding proteins Nrd1 and Nab3, which recognize sequences in the precursor transcript downstream from the mature RNA 3′ end. They interact with the Sen1 helicase, which is thought to terminate transcription by unwinding the RNA–DNA hybrid in the RNA polymerase active site. The TRAMP (Trf4, Air2, Mtr4) complex then adds a short polyadenosine tail to the 3′ end (Fig. 3), which is ultimately removed by the nuclear exosome (Jamonnak et al. 2011; Kuehner et al. 2011).
The processing pathway that generates mature S. pombe TER1 is surprisingly different from that used by S. cerevisiae TLC1 (Fig. 4). The 3′ extended, polyadenylated form of TER1 does appear to serve as the precursor for mature TER1 3′ end formation (Box et al. 2008a). An intron following the exon encoding mature TER1 is recognized by the spliceosome, and cleavage occurs at the 5′ splice site without ligation (Fig. 4), an outcome potentially favored by a weak 3′ splice-site sequence and the atypical length of sequence between this site and the branch point (Box et al. 2008a). The same mechanism may generate the 3′ end of Candida TERs, which exhibit a similar conservation of 5′ splice site and branch point sequences (Gunisova et al. 2009).
TLC1 and TER1, like snRNAs, assemble with Sm proteins and acquire a 2,2,7-trimethylguanosine (TMG) cap (Figs. 3, 4). The Sm proteins SmB, SmD1, SmD2, SmD3, SmE, SmF, and SmG form a heteroheptameric ring that binds a single-stranded uridine-rich region at the 3′ ends of TLC1 and TER1, promoting RNP accumulation and also TER cap hypermethylation via a protein–protein interaction with the TMG synthase Tgs1 (Seto et al. 1999; Leonardi et al. 2008; Tang et al. 2012). Kluyveromyces and Candida TERs conserve this sequence element and thus likely also Sm protein interactions (Gunisova et al. 2009). In S. pombe but not S. cerevisiae, the Sm ring on TER is replaced by a related Lsm2–8 complex that promotes the association of TER1 with the S. pombe TERT protein, Trt1 (Fig. 4), and protects the mature TER1 3′ end from exonuclease activity (Tang et al. 2012).
Studies of yeast TER subcellular trafficking have primarily investigated S. cerevisiae TLC1. TLC1 cap hypermethylation occurs in the nucleolus (Mouaikel et al. 2002). TMG-capped TLC1 is then exported to the cytoplasm via the Crm1 export complex and is later reimported via Mtr10 and/or Kap122 importins (Ferrezuelo et al. 2002; Gallardo et al. 2008). In S phase, TLC1 can be detected at a few clustered telomeres (Gallardo et al. 2011). The findings that all three telomerase holoenzyme Est proteins are required for endogenous TLC1 nuclear localization, and that overexpressed Est1 or Est2 localizes to the nucleus, suggest the possibility that these proteins assemble on TLC1 in the cytoplasm and facilitate its nuclear reentry (Teixeira et al. 2002; Gallardo et al. 2008). However, unlike the case for T. thermophila TER, the biological accumulation of yeast TERs does not require TER assembly with TERT (Lingner et al. 1997a).
Bridging the yeast RNP catalytic core to telomere substrates
S. cerevisiae Est1 and Est3 are required for telomere elongation in vivo but not for RNP catalytic core activity in vitro (Lingner et al. 1997a). It was thus proposed that these proteins physically recruit the RNP to telomeres or subsequently activate it for telomere elongation. In addition to binding TLC1 (Fig. 3), S. cerevisiae Est1 interacts with the single-stranded telomeric DNA-binding protein Cdc13, which alternatively can form a telomere-associated RPA-like complex with Stn1 and Ten1 designated CST (Qi and Zakian 2000; Grandin et al. 2001; Pennock et al. 2001; Gao et al. 2007; Wu and Zakian 2011). Many studies have characterized the interaction of Est1 and Cdc13 as a physical bridge of the telomerase RNP catalytic core to a telomere substrate, as thoroughly reviewed elsewhere (Osterhage and Friedman 2009; DeZwaan and Freeman 2010). Although Est2 is detected at telomeres by chromatin immunoprecipitation (ChIP) throughout the cell cycle, Est1 is not telomere-associated during G1 when it is degraded by the proteasome (Taggart et al. 2002; Osterhage et al. 2006). These and other findings have led to the suggestion that in addition to physically bridging a telomerase RNP to a telomere, Est1 also stimulates S. cerevisiae telomerase catalytic activity as part of its biological function (DeZwaan and Freeman 2010).
Recent studies have begun to define the biochemical role(s) of Est3 in yeast telomerase holoenzymes. Candida albicans Est3 stimulates in vitro elongation of some oligonucleotide primers, and Saccharomyces castelli or S. cerevisiae Est3 stimulates catalytic activity overall (Hsu et al. 2007; Lee et al. 2010; Talley et al. 2011). Est3 interacts directly with the Est2 TEN domain in S. cerevisiae (Fig. 3) and S. castelli (Lee et al. 2010; Talley et al. 2011). A study of recombinant Est3 from yeast lacking a known Est1 demonstrated crosslinking to single-stranded telomeric DNA dependent on Est3 interaction with the Est2 TEN domain (Yen et al. 2011). Incorporation of Est3 into the S. cerevisiae or C. albicans telomerase holoenzyme has a variably reported dependence on Est1 (Osterhage et al. 2006; Hsu et al. 2007; Lee et al. 2010; Tuzon et al. 2011).
S. pombe telomerase holoenzyme assembly differs from that of S. cerevisiae in Est protein association (Fig. 4). Reciprocally with some other characterized yeasts, S. pombe has Est1, but not an identified Est3 (Beernink et al. 2003). S. pombe Est1 has been proposed to recruit telomerase to telomeres and stimulate its catalytic activity through interactions with TER1 (Fig. 4) and the telomere-associated protein Ccq1 (Tomita and Cooper 2008; Moser et al. 2011; Webb and Zakian 2012). This role of Est1 is restricted to S phase by cell-cycle-regulated phosphorylation of Ccq1, which is essential for Est1 binding (Moser et al. 2011; Yamazaki et al. 2012). Curiously, unlike the case for S. cerevisiae TLC1, removing the S. pombe TER1 binding site for Est1 does not preclude the maintenance of short but stable telomere lengths (Webb and Zakian 2012).
The Ku heterodimer is best known for its role in the recognition and repair of double-stranded DNA breaks by nonhomologous end-joining, but it also has roles at normal telomeres (Fisher and Zakian 2005). Interestingly, in S. cerevisiae, Ku also interacts directly with TLC1 (Fig. 3; Peterson et al. 2001; Stellwagen et al. 2003). ChIP studies suggest that Ku is required for telomerase association with telomeres during G1 (Fisher et al. 2004). In part, this observation could reflect the role of Ku in nuclear import of TLC1 RNPs (Gallardo et al. 2008). Some models suggest that Ku could act as a bridge by simultaneously binding TLC1 and telomeric DNA, but in vitro Ku binds DNA and RNA in a mutually exclusive manner (Pfingsten et al. 2012). Ku is not genetically essential in S. cerevisiae and is not associated with telomerase in other yeasts, suggesting that it may be a recent evolutionary addition to Saccharomyces telomerases.
HUMAN TELOMERASE
Human telomerase RNA structure
The mature 451-nt human TER, designated hTR in its original identification (Feng et al. 1995), is intermediate in length between ciliate and yeast TERs (Fig. 1). Phylogenetic comparison of vertebrate TER sequences revealed several conserved regions (CRs), including the template and pseudoknot (Chen et al. 2000; Zhang et al. 2011). In most vertebrate TERs, the template and pseudoknot are enclosed by the P1 stem, which also contributes the TBE (Fig. 1). However, P1 is absent from rodent TERs, in which the template is only ∼2 nt from the RNA 5′ end (Hinkley et al. 1998; Chen et al. 2000). CR4/5, and in specific hTR stem–loop P6.1 and the three-way helical junction at its base (Fig. 1), are the activity-stimulating STE (Chen et al. 2002; Robart and Collins 2010). Physically separate template-pseudoknot and CR4/5 RNAs can reconstitute activity with human TERT expressed in rabbit reticulocyte lysate (Mitchell and Collins 2000). Also, hTR lacking a template can elongate a DNA primer hybridized to an oligonucleotide RNA template (Qi et al. 2012). In assays of this trans template human telomerase activity, deletion of the pseudoknot reduces but does not eliminate catalytic activity (Qi et al. 2012), demonstrating that the pseudoknot is not strictly required for active-site use.
In addition to the conserved TER motifs described above, hTR contains vertebrate-specific TER motifs required for mature RNA biogenesis, RNP assembly, subcellular trafficking, and regulation in vivo. The 5′ end of hTR contains several guanosine (G) tracts that increase mature RNA accumulation, likely by folding as a G-quadruplex that is subsequently resolved by the helicase DHX36 (Lattmann et al. 2011; Sexton and Collins 2011). This region is missing from rodent TERs, consistent with the observation that G-quadruplex formation is stimulatory but not essential for hTR biogenesis (Sexton and Collins 2011). The 3′ half of vertebrate TERs adopts a fold shared with the H/ACA family of RNAs (Fig. 1) consisting of two hairpins connected by a single-stranded “hinge” region (H box) and followed by a single-stranded tail with an ACA (Mitchell et al. 1999; Chen et al. 2000). Eukaryotic H/ACA RNAs generally function as guides for site-specific RNA pseudouridylation, using pockets in the hairpin stems to hybridize to sequences flanking the target uridine(s). H/ACA snoRNAs target ribosomal RNA, while small Cajal body (sca) RNAs target snRNAs (Kiss et al. 2010). In the mature telomerase RNP, each H/ACA RNA hairpin assembles with a set of four proteins: the pseudouridylase dyskerin, NOP10, NHP2, and GAR1 (Fig. 5; Collins 2008; Egan and Collins 2010). As in a canonical H/ACA RNA, the hTR H/ACA motif is required for hTR accumulation in vivo as a biologically stable RNP (Mitchell et al. 1999; Mitchell and Collins 2000; Fu and Collins 2003). However, as no putative target sequence complementary to either of the hTR H/ACA hairpin pockets has been identified, telomerase apparently lacks canonical H/ACA RNP function.
FIGURE 5.
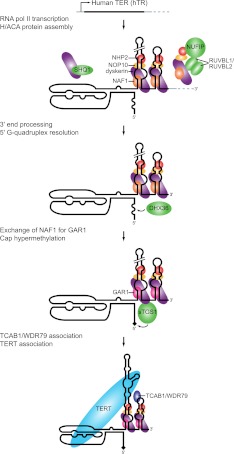
Human telomerase RNP biogenesis. The human TER, hTR, is transcribed by RNA polymerase II as a precursor that cotranscriptionally assembles with the H/ACA protein heterotrimer of dyskerin, NHP2, and NOP10 bound to the H/ACA RNP assembly chaperone NAF1. This process is aided by the dyskerin chaperone SHQ1 and a complex of NUFIP and the helicases RUVBL1 and RUVBL2. NUFIP interacts with NHP2, and the RUVBL1/RUVBL2 heterodimer interacts with dyskerin to promote H/ACA RNP assembly. The hTR precursor transcript is then processed at its 3′ end by an unknown mechanism. The G-quadruplex structure that can form near the 5′ end, represented by a wavy line, is resolved by the helicase DHX36. Then, NAF1 is exchanged for the mature H/ACA RNP protein GAR1, and sTGS1 modifies the 5′ cap to TMG, indicated by a diamond. TCAB1 and TERT bind to hTR in the active telomerase holoenzyme.
Within the hTR H/ACA 3′ hairpin loop, two additional motifs have been defined: the CAB box and the BIO box (Fig. 1). A CAB box is present in each hairpin loop of H/ACA scaRNAs and binds TCAB1/WDR79 (Fig. 5) to promote RNP concentration in Cajal bodies (Richard et al. 2003; Jády et al. 2004; Tycowski et al. 2009; Venteicher et al. 2009). In contrast, the BIO box is not shared by other H/ACA RNAs. Consistent with an hTR-specific requirement for BIO box function, the BIO box cooperates with other hTR-specific 3′ hairpin stem elements to promote mature RNP accumulation in vivo (Fu and Collins 2003; Egan and Collins 2012). BIO box mutant hTR precursor does not escape the site of transcription and is not 3′ end processed (Theimer et al. 2007).
Human telomerase RNP maturation
The hTR precursor is transcribed by RNA polymerase II and exonucleolytically processed at its 3′ end to the boundary of the H/ACA motif (Fig. 5). The presence of a downstream polyadenylation signal or U1 snRNA 3′ box/transcription terminator antagonizes hTR accumulation (Fu and Collins 2003), leaving the actual mechanism of nascent transcript termination and/or endonucleolytic cleavage an open question. The accumulation of mature hTR increases if a self-cleaving ribozyme producing a 2′,3′-cyclic phosphate is inserted immediately downstream from the processed 3′ end (Egan and Collins 2012), suggesting that precursor degradation by 3′ to 5′ exonuclease activity limits hTR accumulation. Unlike all other human H/ACA RNAs, 5′ to 3′ processing of the hTR precursor to the 5′ boundary of the H/ACA domain must be suppressed to retain the template and pseudoknot (Fig. 1).
Many general H/ACA RNP assembly factors are essential for hTR biogenesis (Fig. 5). A preformed H/ACA protein complex of NHP2, NOP10, dyskerin, and the H/ACA RNP assembly factor NAF1 is cotranscriptionally loaded on each hairpin of the nascent H/ACA RNA (Wang and Meier 2004; Darzacq et al. 2006; Richard et al. 2006). While most pseudouridine-guide H/ACA RNAs require the presence of both H/ACA motif hairpins to cooperatively bind the NHP2-NOP10-dyskerin-NAF1 assembly scaffold, hTR exhibits strongly enhanced H/ACA RNP assembly directed by its H/ACA domain 3′ hairpin and dependent on the hTR-specific BIO box (Egan and Collins 2012). The dyskerin chaperone SHQ1 is proposed to function as an RNA mimic, preventing nonspecific RNA binding by dyskerin prior to its assembly into an H/ACA protein core heterotrimer capable of specific H/ACA RNA hairpin binding (Walbott et al. 2011). H/ACA RNP assembly is also aided by a complex of NUFIP, which binds NHP2, and the helicases RUVBL1 and RUVBL2, which bind dyskerin (Boulon et al. 2008; Venteicher et al. 2008). Cellular depletion of any of these chaperone activities reduces mature hTR accumulation in vivo (Hoareau-Aveilla et al. 2006; Fu and Collins 2007; Boulon et al. 2008; Venteicher et al. 2008; Grozdanov et al. 2009).
After transcription, assembly with H/ACA core proteins, and RNA processing (Fig. 5), hTR is routed through Cajal bodies by the transport factors PHAX and Nopp140 (Yang et al. 2000; Boulon et al. 2004). There, the short form of TGS1 (sTGS1) gives hTR its TMG cap (Jády et al. 2004; Fu and Collins 2006; Girard et al. 2008). Cajal bodies are also sites of RNP remodeling, likely including the exchange of the RNP assembly factor NAF1 for the mature RNP component GAR1 (Darzacq et al. 2006). In a manner physically and functionally separable from these steps of RNP biogenesis, the mature RNP also concentrates in Cajal bodies by direct CAB box association with TCAB1/WDR79 (Tycowski et al. 2009; Venteicher et al. 2009). Cellular depletion of TCAB1/WDR79 does not affect hTR accumulation but does reduce RNP association with Cajal bodies and telomeres and results in telomere shortening (Venteicher et al. 2009; Zhong et al. 2011). TCAB1/WDR79 associates with overexpressed hTR partly independent of the CAB box, and, unlike the CAB box, is essential for telomere elongation (Cristofari et al. 2007; Fu and Collins 2007; Egan and Collins 2010; Stern et al. 2012). The significance of TCAB1/WDR79 for telomere maintenance is highlighted by the discovery of TCAB1/WDR79 gene mutations that cause dyskeratosis congenita (Zhong et al. 2011), a human disease of telomerase deficiency (Armanios 2009; Savage and Bertuch 2010).
Human TERT–hTR assembly
TERT is not required for hTR biogenesis or biological accumulation, although it can have a stimulatory effect on the latter when overexpressed (Yi et al. 1999). The TERT TRBD binds the CR4/5 region of hTR, and both the TRBD and TEN domain also bind the template-pseudoknot region (Lai et al. 2001; Robart and Collins 2011). Curiously, at their endogenous levels, for most of the cell cycle hTR concentrates in Cajal bodies, while human TERT concentrates in other nuclear foci (Tomlinson et al. 2006). During S phase, human TERT concentrates in nucleoli and hTR-containing Cajal bodies move to the nucleolar periphery. Only hTR and TERT sites of colocalization, associated with Cajal bodies, are observed to also colocalize with telomeres (Tomlinson et al. 2006). Expression of TERT is required for detectable hTR telomere localization (Tomlinson et al. 2008). Telomeres are not clustered in human cells (Ludérus et al. 1996), and unlike in S. cerevisiae, telomerase appears to associate with most telomeres during a single S phase (Jády et al. 2006; Tomlinson et al. 2006; Zhao et al. 2011). How nuclear subdomains sequester and/or coordinate hTR and TERT complexes remains an open question (Collins 2008). Also open is the possibility of multiple states of telomerase–telomere interaction, for example, distinguishing initial recruitment from productive elongation or accounting for in vivo differences in elongation RAP (Zhao et al. 2011).
As described above for the initial biogenesis of a biologically stable hTR RNP, the subsequent assembly of hTR with TERT is a highly chaperoned process. TERT assembly with hTR is proposed to rely on the protein-folding chaperone HSP90, because the HSP90 inhibitor geldanamycin reduces the production of active human telomerase RNP and induces proteasome-mediated degradation of TERT (Holt et al. 1999; Forsythe et al. 2001; Kim et al. 2005). Expression of a dominant-negative form of the snRNP assembly factor SMN disrupts the localization of TERT in vivo and its assembly with hTR in vitro, suggesting that SMN could play a role in human telomerase RNP catalytic core assembly (Bachand et al. 2002). The DNA-dependent protein kinase-interacting protein KIP could also promote telomerase holoenzyme assembly, since its overexpression increases telomerase activity and telomere length without affecting the levels of TERT mRNA or hTR (Lee et al. 2004). Furthermore, holoenzyme assembly may be regulated by hEST1A/SMG6, an ortholog of yeast Est1 that interacts with hTR and TERT and whose overexpression decreases telomere length (Reichenbach et al. 2003; Snow et al. 2003; Redon et al. 2007).
Bridging the human RNP catalytic core to telomere substrates
Human telomerase recruitment to telomeres requires the TIN2 and TPP1 telomere proteins (Abreu et al. 2010; Tejera et al. 2010), which are anchored to the double-stranded region of the telomere through TRF1 and TRF2 (de Lange 2010; Stewart et al. 2012). The TPP1 OB fold is proposed to interact directly with the TERT TEN domain (Xin et al. 2007; Zaug et al. 2010). TPP1 is also linked to single-stranded telomeric DNA through POT1. A POT1–TPP1 complex stimulates the RAP of a minimal recombinant RNP assembled in rabbit reticulocyte lysate by maintaining association with the product DNA and aiding template translocation (Wang et al. 2007; Latrick and Cech 2010). However, POT1 is not required for telomerase recruitment to telomeres when assayed by ChIP or fluorescence in situ hybridization (Abreu et al. 2010), suggesting that its recruitment role does not require POT1-mediated linkage to single-stranded telomeric DNA.
The human Ku heterodimer has also been suggested to interact with hTR and/or TERT (Chai et al. 2002; Ting et al. 2005), but the in vivo functional significance of the interactions reported to date has not been established. Human Ku70 interacts with TRF2 to localize Ku to telomeres, where it appears to prevent telomere loss (Hsu et al. 1999; Song et al. 2000; Wang et al. 2009). Thus, it is possible that Ku has a role in recruiting human telomerase to telomeres distinct from the roles of Ku in yeast.
COMMON THEMES IN TELOMERASE BIOGENESIS AND REGULATION
Despite remarkably rapid divergence in TER structure and cellular pathways of telomerase RNP maturation, holoenzyme assembly, and recruitment to telomeres, some general features are shared among ciliates, yeasts, and vertebrates. TERs in all three phylogenetic groups recruit proteins prior to TERT binding that remain part of the active holoenzyme and are required for RNA stability in vivo: p65 in ciliates, Sm and Lsm proteins in yeasts, and H/ACA proteins in vertebrates. TERs also contain structurally related template-pseudoknot and STE motifs to mediate their interactions with the TERT TRBD, although the relative affinities of these interactions differ between species. The specificity of TERT–TER interaction in vivo may have remained dependent on the existence of at least two distinct TER–TERT binding interfaces, separated in TER secondary structure but brought together in tertiary structure by prior steps of RNP assembly in vivo. Mechanisms of holoenzyme recruitment to and activation at telomeres also potentially exhibit similarities across phylogenetic groups. The biochemical and regulatory activities of Teb1 and TASC in T. thermophila could parallel the roles of Cdc13, Est1, and Est3 in yeasts and TIN2-bound TPP1 in vertebrates. Future work will be required to further define the precise biological roles of all of the telomerase holoenzyme proteins. Studies of diverse organisms have revealed a great deal of general insight about telomerase biogenesis and regulation, yet much still remains to be understood about the cellular pathways that produce this enzyme and control its activity.
ACKNOWLEDGMENTS
We thank Mary Couvillion for critical reading of this manuscript and members of the Collins lab for helpful discussions. This effort was supported by NIH research grants HL079585 and GM054198 to K.C. and NIH training grant CA009041.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.034629.112.
REFERENCES
- Abreu E, Aritonovska E, Reichenbach P, Cristofari G, Culp B, Terns RM, Lingner J, Terns MP 2010. TIN2-tethered TPP1 recruits human telomerase to telomeres in vivo. Mol Cell Biol 30: 2971–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama BM, Loper J, Najarro K, Stone MD 2012. The C-terminal domain of Tetrahymena thermophila telomerase holoenzyme protein p65 induces multiple structural changes in telomerase RNA. RNA 18: 653–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armanios M 2009. Syndromes of telomere shortening. Annu Rev Genomics Hum Genet 10: 45–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autexier C, Greider CW 1995. Boundary elements of the Tetrahymena telomerase RNA template and alignment domains. Genes Dev 9: 2227–2239 [DOI] [PubMed] [Google Scholar]
- Autexier C, Lue NF 2006. The structure and function of telomerase reverse transcriptase. Annu Rev Biochem 75: 493–517 [DOI] [PubMed] [Google Scholar]
- Bachand F, Boisvert FM, Cote J, Richard S, Autexier C 2002. The product of the Survival of Motor Neuron (SMN) gene is a human telomerase-associated protein. Mol Biol Cell 13: 3192–3202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beernink HT, Miller K, Deshpande A, Bucher P, Cooper JP 2003. Telomere maintenance in fission yeast requires an Est1 ortholog. Curr Biol 13: 575–580 [DOI] [PubMed] [Google Scholar]
- Belfort M, Curcio MJ, Lue NF 2011. Telomerase and retrotransposons: Reverse transcriptases that shaped genomes. Proc Natl Acad Sci 108: 20304–20310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman AJ, Gooding AR, Cech TR 2010. Tetrahymena telomerase protein p65 induces conformational changes throughout telomerase RNA (TER) and rescues telomerase reverse transcriptase and TER assembly mutants. Mol Cell Biol 30: 4965–4976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn EH, Collins K 2011. Telomerase: An RNP enzyme synthesizes DNA. Cold Spring Harb Perspect Biol 3: a003558 doi: 10.1101/cshperspect.a003558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn EH, Greider CW, Szostak JW 2006. Telomeres and telomerase: The path from maize, Tetrahymena and yeast to human cancer and aging. Nat Med 12: 1133–1138 [DOI] [PubMed] [Google Scholar]
- Bosoy D, Peng Y, Mian IS, Lue NF 2003. Conserved N-terminal motifs of telomerase reverse transcriptase required for ribonucleoprotein assembly in vivo. J Biol Chem 278: 3882–3890 [DOI] [PubMed] [Google Scholar]
- Boulon S, Verheggen C, Jady BE, Girard C, Pescia C, Paul C, Ospina JK, Kiss T, Matera AG, Bordonne R, et al. 2004. PHAX and CRM1 are required sequentially to transport U3 snoRNA to nucleoli. Mol Cell 16: 777–787 [DOI] [PubMed] [Google Scholar]
- Boulon S, Marmier-Gourrier N, Pradet-Balade B, Wurth L, Verheggen C, Jady BE, Rothe B, Pescia C, Robert MC, Kiss T, et al. 2008. The Hsp90 chaperone controls the biogenesis of L7Ae RNPs through conserved machinery. J Cell Biol 180: 579–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Box JA, Bunch JT, Tang W, Baumann P 2008a. Spliceosomal cleavage generates the 3′ end of telomerase RNA. Nature 456: 910–914 [DOI] [PubMed] [Google Scholar]
- Box JA, Bunch JT, Zappulla DC, Glynn EF, Baumann P 2008b. A flexible template boundary element in the RNA subunit of fission yeast telomerase. J Biol Chem 283: 24224–24233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown Y, Abraham M, Pearl S, Kabaha MM, Elboher E, Tzfati Y 2007. A critical three-way junction is conserved in budding yeast and vertebrate telomerase RNAs. Nucleic Acids Res 35: 6280–6289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai W, Ford LP, Lenertz L, Wright WE, Shay JW 2002. Human Ku70/80 associates physically with telomerase through interaction with hTERT. J Biol Chem 277: 47242–47247 [DOI] [PubMed] [Google Scholar]
- Chang M, Arneric M, Lingner J 2007. Telomerase repeat addition processivity is increased at critically short telomeres in a Tel1-dependent manner in Saccharomyces cerevisiae. Genes Dev 21: 2485–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapon C, Cech T, Zaug A 1997. Polyadenylation of telomerase RNA in budding yeast. RNA 3: 1337–1351 [PMC free article] [PubMed] [Google Scholar]
- Chen JL, Blasco MA, Greider CW 2000. Secondary structure of vertebrate telomerase RNA. Cell 100: 503–514 [DOI] [PubMed] [Google Scholar]
- Chen JL, Opperman KK, Greider CW 2002. A critical stem-loop structure in the CR4-CR5 domain of mammalian telomerase RNA. Nucleic Acids Res 30: 592–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes-Rojas C, Kannan K, Tseng L, Shippen DE 2011. Two RNA subunits and POT1a are components of Arabidopsis telomerase. Proc Natl Acad Sci 108: 73–78 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Collins K 1999. Ciliate telomerase biochemistry. Annu Rev Biochem 68: 187–218 [DOI] [PubMed] [Google Scholar]
- Collins K 2008. Physiological assembly and activity of human telomerase complexes. Mech Ageing Dev 129: 91–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K. 2009. Forms and functions of telomerase RNA. In Non-protein coding RNAs, Vol. 13 (ed. NG Walter et al.), pp. 285–301. Springer-Verlag, Berlin, Germany. [Google Scholar]
- Collins K 2011. Single-stranded DNA repeat synthesis by telomerase. Curr Opin Chem Biol 15: 643–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristofari G, Adolf E, Reichenbach P, Sikora K, Terns RM, Terns MP, Lingner J 2007. Human telomerase RNA accumulation in Cajal bodies facilitates telomerase recruitment to telomeres and telomere elongation. Mol Cell 27: 882–889 [DOI] [PubMed] [Google Scholar]
- Cunningham DD, Collins K 2005. Biological and biochemical functions of RNA in the Tetrahymena telomerase holoenzyme. Mol Cell Biol 25: 4442–4454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X, Kittur N, Roy S, Shav-Tal Y, Singer RH, Meier UT 2006. Stepwise RNP assembly at the site of H/ACA RNA transcription in human cells. J Cell Biol 173: 207–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T 2010. How shelterin solves the telomere end-protection problem. Cold Spring Harb Symp Quant Biol 75: 167–177 [DOI] [PubMed] [Google Scholar]
- DeZwaan DC, Freeman BC 2010. Is there a telomere-bound ‘EST’ telomerase holoenzyme? Cell Cycle 9: 1913–1917 [DOI] [PubMed] [Google Scholar]
- Eckert B, Collins K 2012. Roles of telomerase reverse transcriptase N-terminal domain in assembly and activity of Tetrahymena telomerase holoenzyme. J Biol Chem 287: 12805–12814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan ED, Collins K 2010. Specificity and stoichiometry of subunit interactions in the human telomerase holoenzyme assembled in vivo. Mol Cell Biol 30: 2775–2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan ED, Collins K 2012. An enhanced H/ACA RNP assembly mechanism for human telomerase RNA. Mol Cell Biol 32: 2428–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Funk WD, Wang S, Weinrich SL, Avilion AA, Chiu C, Adams RR, Chang E, Allsopp RC, Yu J, et al. 1995. The RNA component of human telomerase. Science 269: 1236–1241 [DOI] [PubMed] [Google Scholar]
- Ferrezuelo F, Steiner B, Aldea M, Futcher B 2002. Biogenesis of yeast telomerase depends on the importin Mtr10. Mol Cell Biol 22: 6046–6055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher TS, Zakian VA 2005. Ku: A multifuntional protein involved in telomere maintenance. DNA Repair (Amst) 4: 1215–1226 [DOI] [PubMed] [Google Scholar]
- Fisher TS, Taggart AK, Zakian VA 2004. Cell cycle-dependent regulation of yeast telomerase by Ku. Nat Struct Mol Biol 11: 1198–1205 [DOI] [PubMed] [Google Scholar]
- Forsythe HL, Jarvis JL, Turner JW, Elmore LW, Holt SE 2001. Stable association of hsp90 and p23, but not hsp70, with active human telomerase. J Biol Chem 276: 15571–15574 [DOI] [PubMed] [Google Scholar]
- Fu D, Collins K 2003. Distinct biogenesis pathways for human telomerase RNA and H/ACA small nucleolar RNAs. Mol Cell 11: 1361–1372 [DOI] [PubMed] [Google Scholar]
- Fu D, Collins K 2006. Human telomerase and Cajal body ribonucleoproteins share a unique specificity of Sm protein association. Genes Dev 20: 531–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu D, Collins K 2007. Purification of human telomerase complexes identifies factors involved in telomerase biogenesis and telomere length regulation. Mol Cell 28: 773–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo F, Olivier C, Dandjinou AT, Wellinger RJ, Chartrand P 2008. TLC1 RNA nucleo-cytoplasmic trafficking links telomerase biogenesis to its recruitment to telomeres. EMBO J 27: 748–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo F, Laterreur N, Cusanelli E, Ouenzar F, Querido E, Wellinger RJ, Chartrand P 2011. Live cell imaging of telomerase RNA dynamics reveals cell cycle-dependent clustering of telomerase at elongating telomeres. Mol Cell 44: 819–827 [DOI] [PubMed] [Google Scholar]
- Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V 2007. RPA-like proteins mediate yeast telomere function. Nat Struct Mol Biol 14: 208–214 [DOI] [PubMed] [Google Scholar]
- Gilley D, Blackburn EH 1999. The telomerase RNA pseudoknot is critical for the stable assembly of a catalytically active ribonucleoprotein. Proc Natl Acad Sci 96: 6621–6625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis AJ, Schuller AP, Skordalakes E 2008. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature 455: 633–637 [DOI] [PubMed] [Google Scholar]
- Girard C, Verheggen C, Neel H, Cammas A, Vagner S, Soret J, Bertrand E, Bordonne R 2008. Characterization of a short isoform of human Tgs1 hypermethylase associating with small nucleolar ribonucleoprotein core proteins and produced by limited proteolytic processing. J Biol Chem 283: 2060–2069 [DOI] [PubMed] [Google Scholar]
- Gomes NM, Shay JW, Wright WE 2010. Telomere biology in metazoa. FEBS Lett 584: 3741–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandin N, Damon C, Charbonneau M 2001. Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13. EMBO J 20: 1173–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider CW 1991. Telomerase is processive. Mol Cell Biol 11: 4572–4580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH 1989. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 337: 331–337 [DOI] [PubMed] [Google Scholar]
- Grozdanov PN, Roy S, Kittur N, Meier UT 2009. SHQ1 is required prior to NAF1 for assembly of H/ACA small nucleolar and telomerase RNPs. RNA 15: 1188–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunisova S, Elboher E, Nosek J, Gorkovoy V, Brown Y, Lucier JF, Laterreur N, Wellinger RJ, Tzfati Y, Tomaska L 2009. Identification and comparative analysis of telomerase RNAs from Candida species reveal conservation of functional elements. RNA 15: 546–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinkley C, Blasco M, Funk W, Feng J, Villeponteau B, Greider C, Herr W 1998. The mouse telomerase RNA 5′-end lies just upstream of the telomerase template sequence. Nucleic Acids Res 26: 532–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoareau-Aveilla C, Bonoli M, Caizergues-Ferrer M, Henry Y 2006. hNaf1 is required for accumulation of human box H/ACA snoRNPs, scaRNPs, and telomerase. RNA 12: 832–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt SE, Aisner DL, Baur J, Tesmer VM, Dy M, Ouellette M, Trager JB, Morin GB, Toft DO, Shay JW, et al. 1999. Functional requirement of p23 and Hsp90 in telomerase complexes. Genes Dev 13: 817–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu HL, Gilley D, Blackburn EH, Chen DJ 1999. Ku is associated with the telomere in mammals. Proc Natl Acad Sci 96: 12454–12458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu M, Yu EY, Singh SM, Lue NF 2007. Mutual dependence of Candida albicans Est1p and Est3p in telomerase assembly and activation. Eukaryot Cell 6: 1330–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs SA, Podell ER, Cech TR 2006. Crystal structure of the essential N-terminal domain of telomerase reverse transcriptase. Nat Struct Mol Biol 13: 218–225 [DOI] [PubMed] [Google Scholar]
- Jády BE, Bertrand E, Kiss T 2004. Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body-specific localization signal. J Cell Biol 164: 647–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jády BE, Richard P, Bertrand E, Kiss T 2006. Cell cycle-dependent recruitment of telomerase RNA and Cajal bodies to human telomeres. Mol Biol Cell 17: 944–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain D, Cooper JP 2010. Telomeric strategies: Means to an end. Annu Rev Genet 44: 243–269 [DOI] [PubMed] [Google Scholar]
- Jamonnak N, Creamer TJ, Darby MM, Schaughency P, Wheelan SJ, Corden JL 2011. Yeast Nrd1, Nab3, and Sen1 transcriptome-wide binding maps suggest multiple roles in post-transcriptional RNA processing. RNA 17: 2011–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Park SM, Kang MR, Oh SY, Lee TH, Muller MT, Chung IK 2005. Ubiquitin ligase MKRN1 modulates telomere length homeostasis through a proteolysis of hTERT. Genes Dev 19: 776–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T, Fayet-Lebaron E, Jády BE 2010. Box H/ACA small ribonucleoproteins. Mol Cell 37: 597–606 [DOI] [PubMed] [Google Scholar]
- Kuehner JN, Pearson EL, Moore C 2011. Unravelling the means to an end: RNA polymerase II transcription termination. Nat Rev Mol Cell Biol 12: 283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CK, Mitchell JR, Collins K 2001. RNA binding domain of telomerase reverse transcriptase. Mol Cell Biol 21: 990–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CK, Miller MC, Collins K 2002. Template boundary definition in Tetrahymena telomerase. Genes Dev 16: 415–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latrick CM, Cech TR 2010. POT1-TPP1 enhances telomerase processivity by slowing primer dissociation and aiding translocation. EMBO J 29: 924–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lattmann S, Stadler MB, Vaughn JP, Akman SA, Nagamine Y 2011. The DEAH-box RNA helicase RHAU binds an intramolecular RNA G-quadruplex in TERC and associates with telomerase holoenzyme. Nucleic Acids Res 39: 9390–9404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GE, Yu EY, Cho CH, Lee J, Muller MT, Chung IK 2004. DNA-protein kinase catalytic subunit-interacting protein KIP binds telomerase by interacting with human telomerase reverse transcriptase. J Biol Chem 279: 34750–34755 [DOI] [PubMed] [Google Scholar]
- Lee J, Mandell EK, Rao T, Wuttke DS, Lundblad V 2010. Investigating the role of the Est3 protein in yeast telomere replication. Nucleic Acids Res 38: 2279–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardi J, Box JA, Bunch JT, Baumann P 2008. TER1, the RNA subunit of fission yeast telomerase. Nat Struct Mol Biol 15: 26–33 [DOI] [PubMed] [Google Scholar]
- Linger BR, Morin GB, Price CM 2011. The Pot1a-associated proteins Tpt1 and Pat1 coordinate telomere protection and length regulation in Tetrahymena. Mol Biol Cell 22: 4161–4170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Cech TR, Hughes TR, Lundblad V 1997a. Three ever shorter telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proc Natl Acad Sci 94: 11190–11195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR 1997b. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 276: 561–567 [DOI] [PubMed] [Google Scholar]
- Livengood AJ, Zaug AJ, Cech TR 2002. Essential regions of Saccharomyces cerevisiae telomerase RNA: Separate elements for Est1p and Est2p interaction. Mol Cell Biol 22: 2366–2374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludérus ME, van Steensel B, Chong L, Sibon OC, Cremers FF, de Lange T 1996. Structure, subnuclear distribution, and nuclear matrix association of the mammalian telomeric complex. J Cell Biol 135: 867–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lue NF 2010. Plasticity of telomere maintenance mechanisms in yeast. Trends Biochem Sci 35: 8–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihalusova M, Wu JY, Zhuang X 2011. Functional importance of telomerase pseudoknot revealed by single-molecule analysis. Proc Natl Acad Sci 108: 20339–20344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MC, Collins K 2002. Telomerase recognizes its template by using an adjacent RNA motif. Proc Natl Acad Sci 99: 6585–6590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min B, Collins K 2009. An RPA-related sequence-specific DNA-binding subunit of telomerase holoenzyme is required for elongation processivity and telomere maintenance. Mol Cell 36: 609–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min B, Collins K 2010. Multiple mechanisms for elongation processivity within the reconstituted Tetrahymena telomerase holoenzyme. J Biol Chem 285: 16434–16443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JR, Collins K 2000. Human telomerase activation requires two independent interactions between telomerase RNA and telomerase reverse transcriptase in vivo and in vitro. Mol Cell 6: 361–371 [DOI] [PubMed] [Google Scholar]
- Mitchell JR, Cheng J, Collins K 1999. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol Cell Biol 19: 567–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell M, Gillis A, Futahashi M, Fujiwara H, Skordalakes E 2010. Structural basis for telomerase catalytic subunit TERT binding to RNA template and telomeric DNA. Nat Struct Mol Biol 17: 513–518 [DOI] [PubMed] [Google Scholar]
- Moser BA, Nakamura TM 2009. Protection and replication of telomeres in fission yeast. Biochem Cell Biol 87: 747–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser BA, Chang YT, Kosti J, Nakamura TM 2011. Tel1ATM and Rad3ATR kinases promote Ccq1-Est1 interaction to maintain telomeres in fission yeast. Nat Struct Mol Biol 18: 1408–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouaikel J, Verheggen C, Bertrand E, Tazi J, Bordonne R 2002. Hypermethylation of the cap structure of both yeast snRNAs and snoRNAs requires a conserved methyltransferase that is localized to the nucleolus. Mol Cell 9: 891–901 [DOI] [PubMed] [Google Scholar]
- Mozdy AD, Podell ER, Cech TR 2008. Multiple yeast genes, including Paf1 complex genes, affect telomere length via telomerase RNA abundance. Mol Cell Biol 28: 4152–4161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noël JF, Larose S, Abou Elela S, Wellinger RJ 2012. Budding yeast telomerase RNA transcription termination is dictated by the Nrd1/Nab3 non-coding RNA termination pathway. Nucleic Acids Res 40: 5625–5636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor CM, Collins K 2006. A novel RNA binding domain in Tetrahymena telomerase p65 initiates hierarchical assembly of telomerase holoenzyme. Mol Cell Biol 26: 2029–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor CM, Lai CK, Collins K 2005. Two purified domains of telomerase reverse transcriptase reconstitute sequence-specific interactions with RNA. J Biol Chem 280: 17533–17539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan RJ, Karlseder J 2010. Telomeres: Protecting chromosomes against genome instability. Nat Rev Mol Cell Biol 11: 171–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterhage JL, Friedman KL 2009. Chromosome end maintenance by telomerase. J Biol Chem 284: 16061–16065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterhage JL, Talley JM, Friedman KL 2006. Proteasome-dependent degradation of Est1p regulates the cell cycle-restricted assembly of telomerase in Saccharomyces cerevisiae. Nat Struct Mol Biol 13: 720–728 [DOI] [PubMed] [Google Scholar]
- Pardue ML, DeBaryshe PG 2011. Retrotransposons that maintain chromosome ends. Proc Natl Acad Sci 108: 20317–20324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennock E, Buckley K, Lundblad V 2001. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104: 387–396 [DOI] [PubMed] [Google Scholar]
- Peterson SE, Stellwagen AE, Diede SJ, Singer MS, Haimberger ZW, Johnson CO, Tzoneva M, Gottschling DE 2001. The function of a stem-loop in telomerase RNA is linked to the DNA repair protein Ku. Nat Genet 27: 64–67 [DOI] [PubMed] [Google Scholar]
- Pfingsten JS, Goodrich KJ, Taabazuing C, Ouenzar F, Chartrand P, Cech TR 2012. Mutually exclusive binding of telomerase RNA and DNA by Ku alters telomerase recruitment model. Cell 148: 922–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podlevsky JD, Chen JJ 2012. It all comes together at the ends: Telomerase structure, function, and biogenesis. Mutat Res 730: 3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prathapam R, Witkin KL, O'Connor CM, Collins K 2005. A telomerase holoenzyme protein enhances telomerase RNA assembly with telomerase reverse transcriptase. Nat Struct Mol Biol 12: 252–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H, Zakian VA 2000. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase α and the telomerase-associated Est1 protein. Genes Dev 14: 1777–1788 [PMC free article] [PubMed] [Google Scholar]
- Qi X, Xie M, Brown AF, Bley CJ, Podlevsky JD, Chen JJ 2012. RNA/DNA hybrid binding affinity determines telomerase template-translocation efficiency. EMBO J 31: 150–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao F, Cech TR 2008. Triple-helix structure in telomerase RNA contributes to catalysis. Nat Struct Mol Biol 15: 634–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon S, Reichenbach P, Lingner J 2007. Protein-RNA and protein-protein interactions mediate association of human EST1A/SMG6 with telomerase. Nucleic Acids Res 35: 7011–7022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichenbach P, Hoss M, Azzalin CM, Nabholz M, Bucher P, Lingner J 2003. A human homolog of yeast Est1 associates with telomerase and uncaps chromosome ends when overexpressed. Curr Biol 13: 568–574 [DOI] [PubMed] [Google Scholar]
- Richard P, Darzacq X, Bertrand E, Jády BE, Verheggen C, Kiss T 2003. A common sequence motif determines the Cajal body-specific localization of box H/ACA scaRNAs. EMBO J 22: 4283–4293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard P, Kiss AM, Darzacq X, Kiss T 2006. Cotranscriptional recognition of human intronic box H/ACA snoRNAs occurs in a splicing-independent manner. Mol Cell Biol 26: 2540–2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards RJ, Wu H, Trantirek L, O'Connor CM, Collins K, Feigon J 2006. Structural study of elements of Tetrahymena telomerase RNA stem–loop IV domain important for function. RNA 12: 1475–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robart AR, Collins K 2010. Investigation of human telomerase holoenzyme assembly, activity, and processivity using disease-linked subunit variants. J Biol Chem 285: 4375–4386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robart AR, Collins K 2011. Human telomerase domain interactions capture DNA for TEN domain-dependent processive elongation. Mol Cell 42: 308–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robart AR, O'Connor CM, Collins K 2010. Ciliate telomerase RNA loop IV nucleotides promote hierarchical RNP assembly and holoenzyme stability. RNA 16: 563–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage SA, Bertuch AA 2010. The genetics and clinical manifestations of telomere biology disorders. Genet Med 12: 753–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeftner S, Blasco MA 2009. A ‘higher order’ of telomere regulation: telomere heterochromatin and telomeric RNAs. EMBO J 28: 2323–2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto AG, Zaug AJ, Sobel SG, Wolin SL, Cech TR 1999. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature 401: 177–180 [DOI] [PubMed] [Google Scholar]
- Seto AG, Umansky K, Tzfati Y, Zaug AJ, Blackburn EH, Cech TR 2003. A template-proximal RNA paired element contributes to Saccharomyces cerevisiae telomerase activity. RNA 9: 1323–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton AN, Collins K 2011. The 5′ guanosine tracts of human telomerase RNA are recognized by the G-quadruplex binding domain of the RNA helicase DHX36 and function to increase RNA accumulation. Mol Cell Biol 31: 736–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M, Wang Z, Koo BK, Patel A, Cascio D, Collins K, Feigon J 2012. Structural basis for telomerase RNA recognition and RNP assembly by the holoenzyme La family protein p65. Mol Cell 47: 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow BE, Erdmann N, Cruickshank J, Goldman H, Gill RM, Robinson MO, Harrington L 2003. Functional conservation of the telomerase protein Est1p in humans. Curr Biol 13: 698–704 [DOI] [PubMed] [Google Scholar]
- Song K, Jung D, Jung Y, Lee SG, Lee I 2000. Interaction of human Ku70 with TRF2. FEBS Lett 481: 81–85 [DOI] [PubMed] [Google Scholar]
- Stellwagen AE, Haimberger ZW, Veatch JR, Gottschling DE 2003. Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes Dev 17: 2384–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern JL, Bryan TM 2008. Telomerase recruitment to telomeres. Cytogenet Genome Res 122: 243–254 [DOI] [PubMed] [Google Scholar]
- Stern JL, Zyner KG, Pickett HA, Cohen SB, Bryan TM 2012. Telomerase recruitment requires both TCAB1 and Cajal bodies independently. Mol Cell Biol 32: 2384–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JA, Chaiken MF, Wang F, Price CM 2012. Maintaining the end: Roles of telomere proteins in end-protection, telomere replication and length regulation. Mutat Res 730: 12–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone MS, Mihalusova M, O'Connor CM, Prathapam R, Collins K, Zhuang X 2007. Stepwise protein-mediated RNA folding directs assembly of telomerase ribonucleoprotein. Nature 446: 458–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taggart AK, Teng SC, Zakian VA 2002. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science 297: 1023–1026 [DOI] [PubMed] [Google Scholar]
- Talley JM, DeZwaan DC, Maness LD, Freeman BC, Friedman KL 2011. Stimulation of yeast telomerase activity by the ever shorter telomere 3 (Est3) subunit is dependent on direct interaction with the catalytic protein Est2. J Biol Chem 286: 26431–26439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Kannan R, Blanchette M, Baumann P 2012. Telomerase RNA biogenesis involves sequential binding by Sm and Lsm complexes. Nature 484: 260–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira MT, Gilson E 2005. Telomere maintenance, function and evolution: The yeast paradigm. Chromosome Res 13: 535–548 [DOI] [PubMed] [Google Scholar]
- Teixeira MT, Forstemann K, Gasser SM, Lingner J 2002. Intracellular trafficking of yeast telomerase components. EMBO Rep 3: 652–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tejera AM, Stagno d'Alcontres M, Thanasoula M, Marion RM, Martinez P, Liao C, Flores JM, Tarsounas M, Blasco MA 2010. TPP1 is required for TERT recruitment, telomere elongation during nuclear reprogramming, and normal skin development in mice. Dev Cell 18: 775–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theimer CA, Feigon J 2006. Structure and function of telomerase RNA. Curr Opin Struct Biol 16: 307–318 [DOI] [PubMed] [Google Scholar]
- Theimer CA, Jády BE, Chim N, Richard P, Breece KE, Kiss T, Feigon J 2007. Structural and functional characterization of human telomerase RNA processing and Cajal body localization signals. Mol Cell 27: 869–881 [DOI] [PubMed] [Google Scholar]
- Ting NS, Yu Y, Pohorelic B, Lees-Miller SP, Beattie TL 2005. Human Ku70/80 interacts directly with hTR, the RNA component of human telomerase. Nucleic Acids Res 33: 2090–2098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K, Cooper JP 2008. Fission yeast Ccq1 is telomerase recruiter and local checkpoint controller. Genes Dev 22: 3461–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson RL, Ziegler TD, Supakorndej T, Terns RM, Terns MP 2006. Cell cycle-regulated trafficking of human telomerase to telomeres. Mol Biol Cell 17: 955–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson RL, Abreu EB, Ziegler T, Ly H, Counter CM, Terns RM, Terns MP 2008. Telomerase reverse transcriptase is required for the localization of telomerase RNA to Cajal bodies and telomeres in human cancer cells. Mol Biol Cell 19: 3793–3800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuzon CT, Wu Y, Chan A, Zakian VA 2011. The Saccharomyces cerevisiae telomerase subunit Est3 binds telomeres in a cell cycle- and Est1-dependent manner and interacts directly with Est1 in vitro. PLoS Genet 7: e1002060 doi: 10.1371/journal.pgen.1002060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski KT, Shu MD, Kukoyi A, Steitz JA 2009. A conserved WD40 protein binds the Cajal body localization signal of scaRNP particles. Mol Cell 34: 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfati Y, Fulton TB, Roy J, Blackburn EH 2000. Template boundary in a yeast telomerase specified by RNA structure. Science 288: 863–867 [DOI] [PubMed] [Google Scholar]
- Tzfati Y, Knight Z, Roy J, Blackburn EH 2003. A novel pseudoknot element is essential for the action of a yeast telomerase. Genes Dev 17: 1779–1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venteicher AS, Meng Z, Mason PJ, Veenstra TD, Artandi SE 2008. Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell 132: 945–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venteicher AS, Abreu EB, Meng Z, McCann KE, Terns RM, Veenstra TD, Terns MP, Artandi SE 2009. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science 323: 644–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walbott H, Machado-Pinilla R, Liger D, Blaud M, Rety S, Grozdanov PN, Godin K, van Tilbeurgh H, Varani G, Meier UT, et al. 2011. The H/ACA RNP assembly factor SHQ1 functions as an RNA mimic. Genes Dev 25: 2398–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Meier UT 2004. Architecture and assembly of mammalian H/ACA small nucleolar and telomerase ribonucleoproteins. EMBO J 23: 1857–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M 2007. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature 445: 506–510 [DOI] [PubMed] [Google Scholar]
- Wang Y, Ghosh G, Hendrickson EA 2009. Ku86 represses lethal telomere deletion events in human somatic cells. Proc Natl Acad Sci 106: 12430–12435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JM, Riha K 2010. Comparative biology of telomeres: Where plants stand. FEBS Lett 584: 3752–3759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb CJ, Zakian VA 2012. Schizosaccharomyces pombe Ccq1 and TER1 bind the 14-3-3-like domain of Est1, which promotes and stabilizes telomerase-telomere association. Genes Dev 26: 82–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinrich SL, Pruzan R, Ma L, Ouellette M, Tesmer VM, Holt SE, Bodnar AG, Lichsteiner S, Kim NW, Trager JB, et al. 1997. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat Genet 17: 498–502 [DOI] [PubMed] [Google Scholar]
- Witkin KL, Collins K 2004. Holoenzyme proteins required for the physiological assembly and activity of telomerase. Genes Dev 18: 1107–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkin KL, Prathapam R, Collins K 2007. Positive and negative regulation of Tetrahymena telomerase holoenzyme. Mol Cell Biol 27: 2074–2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Zakian VA 2011. The telomeric Cdc13 protein interacts directly with the telomerase subunit Est1 to bring it to telomeric DNA ends in vitro. Proc Natl Acad Sci 108: 20362–20369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt HD, West SC, Beattie TL 2010. InTERTpreting telomerase structure and function. Nucleic Acids Res 38: 5609–5622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Podlevsky JD, Qi X, Bley CJ, Chen JJ 2010. A novel motif in telomerase reverse transcriptase regulates telomere repeat addition rate and processivity. Nucleic Acids Res 38: 1982–1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin H, Liu D, Wan M, Safari A, Kim H, Sun W, O'Connor MS, Songyang Z 2007. TPP1 is a homologue of ciliate TEBP-β and interacts with POT1 to recruit telomerase. Nature 445: 559–562 [DOI] [PubMed] [Google Scholar]
- Yamazaki H, Tarumoto Y, Ishikawa F 2012. Tel1ATM and Rad3ATR phosphorylate the telomere protein Ccq1 to recruit telomerase and elongate telomeres in fission yeast. Genes Dev 26: 241–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Isaac C, Wang C, Dragon F, Pogacic V, Meier UT 2000. Conserved composition of mammalian box H/ACA and box C/D small nucleolar ribonucleoprotein particles and their interaction with the common factor Nopp140. Mol Biol Cell 11: 567–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen WF, Chico L, Lei M, Lue NF 2011. Telomerase regulatory subunit Est3 in two Candida species physically interacts with the TEN domain of TERT and telomeric DNA. Proc Natl Acad Sci 108: 20370–20375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X, Tesmer VM, Savre-Train I, Shay JW, Wright WE 1999. Both transcriptional and posttranscriptional mechanisms regulate human telomerase template RNA levels. Mol Cell Biol 19: 3989–3997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu EY 2012. Telomeres and telomerase in Candida albicans. Mycoses 55: e48–e59 [DOI] [PubMed] [Google Scholar]
- Zappulla DC, Cech TR 2006. RNA as a flexible scaffold for proteins: Yeast telomerase and beyond. Cold Spring Harb Symp Quant Biol 71: 217–224 [DOI] [PubMed] [Google Scholar]
- Zappulla DC, Goodrich K, Cech TR 2005. A miniature yeast telomerase RNA functions in vivo and reconstitutes activity in vitro. Nat Struct Mol Biol 12: 1072–1077 [DOI] [PubMed] [Google Scholar]
- Zaug AJ, Podell ER, Nandakumar J, Cech TR 2010. Functional interaction between telomere protein TPP1 and telomerase. Genes Dev 24: 613–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z, Min B, Huang J, Hong K, Yang Y, Collins K, Lei M 2011. Structural basis for Tetrahymena telomerase processivity factor Teb1 binding to single-stranded telomeric-repeat DNA. Proc Natl Acad Sci 108: 20357–20361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Kim NK, Feigon J 2011. Architecture of human telomerase RNA. Proc Natl Acad Sci 108: 20325–20332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Abreu E, Kim J, Stadler G, Eskiocak U, Terns MP, Terns RM, Shay JW, Wright WE 2011. Processive and distributive extension of human telomeres by telomerase under homeostatic and nonequilibrium conditions. Mol Cell 42: 297–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong F, Savage SA, Shkreli M, Giri N, Jessop L, Myers T, Chen R, Alter BP, Artandi SE 2011. Disruption of telomerase trafficking by TCAB1 mutation causes dyskeratosis congenita. Genes Dev 25: 11–16 [DOI] [PMC free article] [PubMed] [Google Scholar]