Micro (mi)RNAs are important regulators of gene expression in plants and animals; however, miRNAs have previously only been identified in a handful of unicellular eukaryotes. This study describes the first high-throughput sequencing of small RNAs from the unicellular social amoeba Dictyostelium discoideum, establishing the presence of miRNAs in the Amoebozoa supergroup. Identified miRNAs were further analyzed with regard to developmental regulation and biogenesis.
Keywords: microRNA, Dictyostelium discoideum, high-throughput sequencing, small RNA, development
Abstract
The RNA interference machinery has served as a guardian of eukaryotic genomes since the divergence from prokaryotes. Although the basic components have a shared origin, silencing pathways directed by small RNAs have evolved in diverse directions in different eukaryotic lineages. Micro (mi)RNAs regulate protein-coding genes and play vital roles in plants and animals, but less is known about their functions in other organisms. Here, we report, for the first time, deep sequencing of small RNAs from the social amoeba Dictyostelium discoideum. RNA from growing single-cell amoebae as well as from two multicellular developmental stages was sequenced. Computational analyses combined with experimental data reveal the expression of miRNAs, several of them exhibiting distinct expression patterns during development. To our knowledge, this is the first report of miRNAs in the Amoebozoa supergroup. We also show that overexpressed miRNA precursors generate miRNAs and, in most cases, miRNA* sequences, whose biogenesis is dependent on the Dicer-like protein DrnB, further supporting the presence of miRNAs in D. discoideum. In addition, we find miRNAs processed from hairpin structures originating from an intron as well as from a class of repetitive elements. We believe that these repetitive elements are sources for newly evolved miRNAs.
INTRODUCTION
Small silencing RNAs (20–30 nt) are exceptionally important regulators of genome defense and maintenance as well as gene expression in eukaryotic cells. These small RNAs are the key components of the evolutionarily conserved RNA interference (RNAi) system (Ghildiyal and Zamore 2009).
Although lost in some lineages, it appears that a basic RNAi machinery was present in the last common eukaryotic ancestor, responsible for generation of small interfering RNAs (siRNAs) from longer double-stranded RNAs (dsRNAs) and the subsequent inactivation of cognate sequences (Cerutti and Casas-Mollano 2006). The dsRNA source of siRNAs has many origins, such as invading viruses, retrotransposons, or other types of inverted repeats (Aravin et al. 2007).
In contrast to siRNAs, microRNAs (miRNAs) are derived from endogenous transcripts that fold into distinct hairpin structures and commonly regulate protein-coding genes. miRNA was discovered in Caenorhabditis elegans (Lee et al. 1993) and first believed to be a worm-specific phenomenon. However, miRNAs have now been found in other animals and their viruses, as well as in plants, algae, and protists (Griffiths-Jones et al. 2008). It is likely that miRNAs have evolved more than once, considering the many homologous miRNAs that are restricted to closely related species but not present in more distantly related groups of organisms, e.g., animals as compared to plants (Shabalina and Koonin 2008).
Although miRNAs can be generated through several pathways, the canonical biogenesis pathway is defined by the processing of miRNAs from longer precursors that are transcribed from independent genes (Miyoshi et al. 2010; Axtell et al. 2011). The pri-miRNAs fold into hairpin structures, which are targeted by the RNase III family proteins, Dicer (in plants) or Drosha (in animals). The resulting pre-miRNA is further processed by Dicer to an ∼21-bp miRNA/miRNA* duplex. The miRNA is subsequently incorporated into the RNA-induced silencing complex (RISC) while the miRNA* in most cases is discarded. miRNAs associate with the effector Argonaute (AGO) protein of RISC and act as guides, bringing the silencing complex to the target (mRNA). Pairing between the miRNA and the mRNA can direct cleavage of the message, which is common in plants. In metazoans, the regulation is usually carried out through mRNA destabilization, translational repression, or a combination of both (Bartel 2009; Carthew and Sontheimer 2009). In a recent study, it was shown that decreased levels of mRNAs account for most of the effect on protein production in mammals (Guo et al. 2010).
With the widespread use of high-throughput sequencing methods, a large number of microRNAs has now been discovered mainly in animals and plants (Griffiths-Jones et al. 2008). This has led to the estimate that >60% of all human protein-coding genes encode mRNAs that are direct targets for miRNAs and that most cellular processes in mammals are in some way regulated by these tiny RNAs (Friedman et al. 2009). Not much is known about the presence or the role of miRNAs in less complex eukaryotes (outside metazoans and plants). The first evidence of miRNAs in a single-cell organism came in 2007 when two groups sequenced the small RNA population in the green algae Chlamydomonas reinhardtii (Molnar et al. 2007; Zhao et al. 2007). Soon after, we reported a few miRNA candidates in Dictyostelium discoideum (Hinas et al. 2007). Recently, miRNAs were also suggested to be present in the pathogenic protists Toxoplasma gondii and Giardia lamblia (Saraiya and Wang 2008; Braun et al. 2010).
The genetically tractable amoeba D. discoideum is interesting, not least from an evolutionary perspective as it branched out before animals and fungi but after plants, and because it hovers between uni- and multicellularity (Baldauf et al. 2000; Bapteste et al. 2002). D. discoideum grows as single cells, but when challenged by starvation, up to 100,000 cells can aggregate and form a ball of spores lifted from the ground by dead stalk cells (Kessin 2001). As a model organism, D. discoideum has proven valuable in studies of features attributed to both uni- and multicellular organisms such as cytokinesis, cell motility, signal transduction, and cell differentiation (Kessin 2001). Several genes have been predicted to encode proteins involved in the RNAi machinery, i.e., two Dicer-like proteins, DrnA and DrnB, and three RNA-dependent RNA polymerases (RdRPs), RrpA, RrpB, and RrpC. There are also five genes predicted to encode PIWI-like Argonautes, AgnA-E, and a gene expressing a putative RNA helicase, HelF, a suppressor of RNAi (Martens et al. 2002; Cerutti and Casas-Mollano 2006; Popova et al. 2006).
In this study, we establish the presence of miRNAs in the Amoebozoa supergroup. We have performed the first reported high-throughput sequencing and analysis of small RNA populations from the social amoeba D. discoideum. The analysis was performed on small RNA from growing single cells and two multicellular developmental stages. Stringent search criteria were used to identify miRNAs and their cognate miRNA* sequences within the small RNA libraries. Many of the miRNAs are developmentally regulated and derived from intergenic regions, short repetitive elements, as well as, in one case, from an intron sequence. Four of the pre-miRNAs were overexpressed, and the biogenesis of the miRNAs (and three miRNA*) was shown to be strictly dependent on DrnB. Furthermore, this study indicates that miRNAs in D. discoideum have evolved independently and may originate from short repetitive elements.
RESULTS
The small RNA population in D. discoideum during growth and development
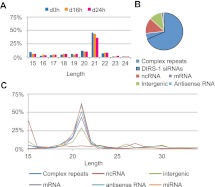
In order to investigate the population of small RNAs during different stages of the life cycle of D. discoideum, total RNA was prepared from growing single cells (0 h) and from two developmental stages: slugs (16 h after the onset of development) and mature fruiting bodies (24 h after the onset of development), where cells have aggregated and differentiated. Small RNA fractions were isolated and subjected to SOLiD System high-throughput sequencing. The three sequenced libraries are referred to as d0h, d16h, and d24h. After removing the adapter sequences and only considering reads of a length between 15 nt and 34 nt, with at least one but at the most 30 matches in the genome, the libraries contained: d0h, 1.5 × 106; d16h, 0.92 × 106; and d24h, 0.76 × 106 sequences. The great majority of the sequences in all three libraries had a length of 21 nt (Fig. 1A). Many of the sequences matched annotated loci (Fig. 1B), i.e., complex repeats, noncoding RNAs (ncRNAs) such as rRNA, tRNAs, snRNAs, SRP RNAs, and Class I RNAs (Aspegren et al. 2004; Eichinger et al. 2005; Hinas et al. 2006, 2007; Hinas and Soderbom 2007; Larsson et al. 2008; Avesson et al. 2011; Boesler et al. 2011), and protein-coding genes (mRNAs and antisense RNAs, depending on which strand they matched). Reads were also found mapping intergenic regions. Most of the small RNAs were derived from complex repeats, of which the great majority, i.e., 89%, mapped to the retrotransposon element DIRS-1 (Fig. 1B), which also constitutes centromeres in D. discoideum (Glockner and Heidel 2009). The great number of small RNAs from this element and their size distribution (Fig. 1B,C) are in accordance with our previously published data (Hinas et al. 2007). A similar size distribution, with a peak around 21 nt, was also observed for sequences from mRNAs, antisense RNAs, and intergenic regions (Fig. 1C). Reads from ncRNAs had a more even distribution, covering all different lengths, but with a peak at 15 nt that mainly comprised tRNA fragments. The overall size distribution of small RNAs did not differ significantly between growing cells and the two developmental stages, except for the 15-nt tRNA fraction which was more pronounced in the d0h library (Fig. 1A,C; data not shown).
FIGURE 1.
Size distribution and classification of small RNAs. (A) Size distribution of the small RNAs from growing cells (d0h) and from two developmental stages (d16h and d24h, respectively). (B) Classification of small RNAs (21 nt). The proportion of DIRS-1 small RNAs is marked with a black line. (C) Length distribution of different classes of small RNAs. In A and C, % depicts the relative distribution for each time point and class, respectively, of small RNA.
Taken together, these data show a strong bias for 21-nt-long RNAs in D. discoideum in growing as well as developing cells. These small RNAs are mainly derived from repetitive elements with the retrotransposon DIRS-1 as the major source.
Identification of miRNAs in D. discoideum
The main aim of this study was to identify miRNAs in D. discoideum. Hence, we focused our analysis on the 21-nt-long sequences since this is the length of the previously identified miRNA candidates in D. discoideum (Hinas et al. 2007). In addition, the great majority of small RNAs were 21 nt, and the longer and shorter sequences were mainly derived from known ncRNA, DIRS-1, or other genomic locations that do not fulfill the criteria for being sources for miRNAs (see below; data not shown). Hence, the search scheme described below is set to increase the signal to noise ratio as much as possible.
First, a number of stringent filters were applied to remove sequences from mRNAs, antisense RNAs, and from many complex repeats and ncRNAs. Most of the reads that mapped to more than nine locations belonged to repetitive elements and were, therefore, removed from further analysis (data not shown). Furthermore, we excluded sequences originating from regions where RNAs were derived from both strands, e.g., DIRS-1 (Hinas et al. 2007) and mRNA and their cognate antisense transcripts (Materials and Methods). Among the remaining sequences, we only considered those where at least five reads in one library mapped to a specific location in the genome. Although 10–20 reads are commonly used as cutoffs (Kozomara and Griffiths-Jones 2011), the relatively limited number of sequences in each library and the fact that almost 75% were derived from complex repeats, mainly from DIRS-1, caused us to use a lower cutoff. The filter scheme, the reduction of reads, and the change in distribution per filter step, is shown in Supplemental Figure S1. After filtration, the d0h library contained ∼13,000 sequences from 283 genomic locations, the d16h library contained ∼4300 sequences from 289 genomic locations, and the d24h library contained 8400 sequences from 252 genomic locations.
Next, we analyzed the potential of the small RNAs to be derived from pre-miRNA-like structures (Supplemental Fig. S2; Axtell et al. 2011). Since it was unknown whether D. discoideum pre-miRNA structures would resemble those in animals or plants, we used MiRfold for predictions (Billoud et al. 2005). For this, 200 nt upstream of and downstream from the genomic location of the 21-nt RNAs were extracted from the genome sequence (a total of 421 nt). This length includes almost all animal-like pre-miRNAs and >50% of the plant-like pre-miRNAs deposited in miRbase. Out of the total 567 genomic locations, presented in Supplemental Table S2, that matched the filtered small RNAs from the three different libraries, we found 19 pre-miRNA structures that passed the MiRfold screen, e.g., forming stable unbranched hairpin structures with few bulges.
In order to validate our search for pre-miRNA structures in D. discoideum, specificity and sensitivity were analyzed using MiRfold, applying the same free-energy properties and penalty settings as for the determined 19 pre-miRNA structures (Materials and Methods). The specificity was evaluated by generating 10,000 random 421-nt-long sequences with the same di-nucleotide frequency as for the identified 19 pre-miRNA sequences (see Materials and Methods). Only three of these generated sequences were below our set cutoff, giving a specificity of 0.9997 in our data set (Supplemental Fig. S2B). To examine the sensitivity, we investigated whether our stringent search criteria would identify known pre-miRNAs sequences present in miRBase. We analyzed 2281 sequences from six different organisms (animals, plant, and algae). The sensitivity was dependent on which species was considered and ranged between 0.3744 in C. elegans and 0.8750 in Amphimedon queenslandica. Taken together, the evaluation of the specificity and sensitivity of our search for pre-miRNA in D. discoideum suggests that there should be very few (if any) false positives, but due to the stringent cutoff settings, many miRNA precursors most likely have been disregarded.
Of the 19 pre-miRNA sequences, 18 folded into unbranched hairpin-like structures, suggesting they are more animal-like. To test this, the 19 pre-miRNA sequences were analyzed using miR-abela, a program optimized to predict human pre-miRNAs (Sewer et al. 2005). The program predicted that 17 of the pre-miRNA structures were human pre-miRNA-like (Supplemental Figs. S2A,C, S3). Twelve of these originate from intergenic sequences, one is located in an intron, and four are derived from the repetitive element thug-S (Fig. 2A). Among these, the two previously predicted miRNAs mica1190 and mica1198 (Hinas et al. 2007) named ddi-mir-1176 and ddi-mir-1177, respectively, in miRBase, were represented. Notably, none of the three false positive sequences that passed the cutoff we set using MiRfold were predicted by miR-abela to form a human pre-miRNA-like structure. This implies that the specificity of using MiRfold and, subsequently, miR-abela to identify pre-miRNA-like structure is greater than 0.9999 in our data set.
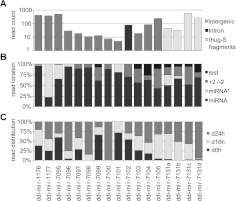
FIGURE 2.
miRNAs in Dictyostelium discoideum. (A) Genomic location and number of miRNA reads in the three libraries. (B) Relative distribution of reads for miRNAs, miRNA*s, and reads which are shifted (as compared to the defined miRNA) with, at the most, 2 nt (+2/−2), or more nt (rest). (C) Relative number of reads in growing cells (d0h) and cells developed for 16h (d16h) and 24h (d24h).
Additional criteria applied on high-throughput sequencing data can distinguish miRNA from other sequences. Multiple reads that map to the predicted hairpin structures should have a distinct size distribution with defined 5′ ends (Kozomara and Griffiths-Jones 2011). To investigate whether our miRNA candidates were in accordance with these criteria, we mapped all the reads with length 15 nt to 34 nt to the predicted pre-miRNA sequences, allowing only perfect matches (Fig. 2B; Supplemental Fig. S4). Nine miRNAs, ddi-mir-1176 and 1177, and ddi-mir-7095 to 7101, matched all the criteria referred to above. For ddi-mir-7102 and 7103, there seem to be two preferred 5′ ends for each miRNA, and for ddi-mir-7104, ddi-mir-7105, and ddi-mir-7131a to 7131d, the selection of the miRNA 5′ ends is less specific. The lowest specificity is represented by ddi-mir-7105, with 43% of the reads with the same 5′ end as the miRNA sequence and 47% with 5′ ends at positions from –2 to + 2 (Fig. 2B; Supplemental Fig. S4).
Ideally, high-throughput sequencing should also reveal the presence of miRNA* with a 2-nt 3′ overhang, generated from the opposite strand of the hairpin structure. We detect miRNA* for nine of the miRNAs—ddi-mir-1176, 1177, 7095, 7096, 7101, 7104, and 7131a to 7131c (Fig. 2B; Supplemental Figs. S3, S4). Allowing for reads with imperfect matches to the genome, additional miRNA* sequences were scored (see below). In summary, 17 new miRNAs were identified using stringent search criteria, thus clearly demonstrating the presence of miRNAs in D. discoideum.
Experimental validation and overexpression of miRNAs in D. discoideum
To further investigate the characteristics of miRNAs in D. discoideum, four were chosen for further analysis. In accordance with a recent validation of mammalian miRNAs (Chiang et al. 2010), the four miRNA hairpin structures were overexpressed, and generation of small RNAs investigated. Ddi-mir-1176, ddi-mir-1177, and ddi-mir-7095 were the most abundant miRNAs in our library, of which the first two had previously been reported as miRNA candidates (Hinas et al. 2007). Ddi-mir-7102 was selected since it is generated from an intron sequence, unlike all other miRNAs in this study that are derived from intergenic sequences or repetitive elements.
ddi-mir-1176 and ddi-mir-1177
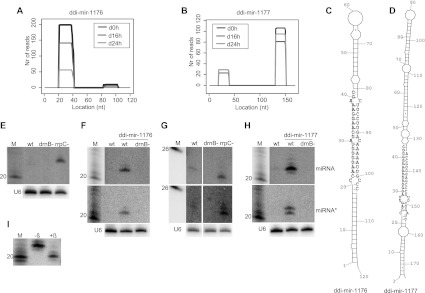
The high-throughput sequencing data showed the great majority of reads for both these miRNAs and their star sequences to have defined 5′ and 3′ ends. Only the miRNA and the miRNA* pairs were detected, i.e., no small RNAs derived from other parts of the predicted hairpin precursor could be identified (Fig. 3A,B; Supplemental Fig. S4). The miRNA* sequences display the 2-nt 3′ overhang, a signature of Dicer cleavage (Fig. 3C,D). To further examine the expression of the miRNAs, Northern blot analysis was performed. As previously reported, ddi-mir-1177 is dependent on one of the Dicer homologues, DrnB, for its maturation (Fig. 3G; Hinas et al. 2007). In addition, the miRNA was up-regulated in a strain in which one of the RdRP genes (rrpC) had been disrupted. The same pattern was observed for the miRNA* sequence (Fig. 3G). However, in the case of ddi-mir-1177, it is not possible to distinguish between miRNA and its corresponding miRNA*. The sequencing data suggests that the 21-nt RNA from the 3′ part of the stem is the miRNA sequence, based on its higher number of reads compared to those representing the one derived from the 5′ stem segment in the libraries (Fig. 3B; Supplemental Fig. S4). In contrast, detection by Northern blot suggests the opposite, since ddi-mir-1177 signals are more intense than those for its miRNA*. Differences in, e.g., labeling efficiency of the oligonucleotide probes might account for this result (the star probe has a 5′ adenosine, which tends to decrease labeling efficiency compared to other nucleotides) (van Houten et al. 1998). It is also conceivable that both small RNAs are functional miRNAs.
FIGURE 3.
Analyses of WT and overexpressed ddi-mir-1176 and ddi-mir-1177. (A,B) Histograms representing the distribution of reads mapping to the predicted pre-miRNA structures generating ddi-mir-1176 and ddi-mir1177. (C,D) Predicted hairpin structures of the sequences harboring the miRNAs. The miRNA and the miRNA* are represented by black (5′ part of the stem) and gray (3′ part of the stem) nucleotides, respectively. Note that, for ddi-mir-1177, it is uncertain which is the miRNA and which is the miRNA* (see Results). (E) Northern blot analysis showing expression of ddi-mir-1176 in wild-type, drnB− , and rrpC− strains, and (F) extrachromosomal overexpression of ddi-mir-1176 (upper panel), ddi-mir1176* (middle panel) in wild-type and drnB− strains. (G) Northern blot analysis showing expression of ddi-mir-1177 (upper panel) and ddi-mir1177* (middle panel) in wild-type, drnB−, and rrpC− strains, and (H) extrachromosomal overexpression of ddi-mir-1177 (upper panel), ddi-mir1177* (middle panel). (I) Analysis of 3′ end of ddi-mir-1177 by β-elimination assay. Each panel representing Northern blot analyses was probed and reprobed for the different RNAs. Where depicted, U6 spliceosomal RNA was probed for and used as loading control.
The other previously predicted miRNA, ddi-mir-1176, was also up-regulated in the rrpC− strain, but its expression in the wild-type strain was too low for detection in growing cells. Consequently, there is no compelling experimental evidence that ddi-mir-1176, as we would expect, is DrnB-dependent (Fig. 3E).
Since the miRNAs are expressed at low levels in normal laboratory conditions, we decided to overexpress the miRNA precursor hairpins to validate the generation of small RNAs and to approach an understanding of their biogenesis. The predicted miRNA hairpin sequences were cloned into extrachromosomal expression vectors and transformed into wild-type and drnB− D. discoideum strains. The expression in growing cells was analyzed by Northern blot. Indeed, the presumed precursor hairpins generated the expected miRNA and miRNA* sequences in wild-type strains. We estimate a 20- to 30-fold increase in miRNA level (ddi-mir-1177) as compared to untransformed wild-type cells (Fig. 3F,H). As a control, we analyzed the level of the endogenously expressed ddi-mir-1177 in strains that overexpressed miRNA ddi-mir-1176 and vice versa. This experiment showed that overexpression of the miRNAs did not significantly affect the biogenesis or levels of the endogenous miRNAs, which indicates that the factors involved in miRNA generation are not depleted under these conditions (data not shown). Furthermore, we observed that generation of ddi-mir-1176 and ddi-mir-1177 and their star sequences was strictly DrnB-dependent (Fig. 3F,H). No small RNAs above detection levels were generated from sequences flanking the miRNAs (data not shown).
ddi-mir-7095 and ddi-mir-7102
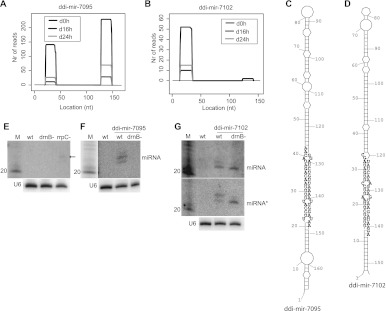
In addition, ddi-mir-7095 and ddi-mir-7102 and their star sequences have defined 5′ and 3′ ends (Fig. 4A–D; Supplemental Fig. S4). For ddi-mir-7102, its star sequence with a 2-nt 3′ overhang is found when reads with mismatches are allowed (data not shown), while it is shifted 1 or 2 nt when sequences with only perfect matches are considered. This could be due to the fact that ddi-mir-7102 is the only miRNA derived from an intron, and this may affect processing (see below). Since no ddi-mir-7102* with perfect matches was identified, we excluded ddi-mir-7102 from the list of miRNAs with star sequences (Fig. 2B). Northern blot analysis permitted detection of ddi-mir-7095 only in the rrpC− strain (although at a very low level) (Fig. 4E), whereas it failed to detect ddi-mir-7102 under all conditions (Fig. 4G; data not shown). The predicted pre-miRNA hairpins were overexpressed in wild-type and drnB− D. discoideum cells. Ddi-mir-7095 was specifically processed in a DrnB-dependent manner as demonstrated for ddi-mir-1176 and ddi-mir-1177, but its star sequence remained undetected (Fig. 4F; data not shown). Interestingly, ddi-mir-7102, which is derived from one of the introns of the DNA polymerase B gene (DDB_G0284713), showed a different processing pattern (Fig. 4G). Two small RNAs, with 1 nt difference in length, were detected by the miRNA probe, but only the longer RNA variant showed dependence on DrnB. The same processing pattern was observed for the star sequence.
FIGURE 4.
Analyses of ddi-mir-7095 and ddi-mir-7102. (A,B) Histograms representing the distribution of reads mapping to the predicted miRNA hairpins. (C,D) Predicted hairpin structures of sequence harboring the miRNAs. The miRNA and the miRNA* are represented by black nucleotides (3′ part of the stem for ddi-mir-7095 and 5′ part for ddi-mir-7102) and gray nucleotides (5′ part of the stem for ddi-mir-7095 and 3′ part for ddi-mir-7102), respectively. (E) Northern blot analysis showing expression of ddi-mir-7095 in wild-type, drnB−, and rrpC− strains (weak signal indicated by arrow), and (F) extrachromosomal overexpression of ddi-mir-7095 in wild-type and drnB− strains. (G) Northern blot analysis showing extrachromosomal overexpression of ddi-mir-7102 (upper panel) and ddi-mir-7102* (middle panel) in wild-type and drnB− strains. Note that the depicted ddi-mir-7102* is the sequence probed for and present in the small RNA library when allowing mismatches. U6 spliceosomal RNA was used as a loading control. Each panel representing Northern blot analyses was probed and reprobed for the different RNAs.
Taken together, these experiments confirm the high-throughput sequencing results and computational analyses, thereby strongly supporting the presence of miRNAs in D. discoideum. Dicer-dependent processing was also demonstrated for the four miRNAs analyzed and for three of their miRNA* sequences, at least when overexpressing the pre-miRNA. The overexpression experiments also suggest that DrnB-dependent processing is not depleted when subjected to an excess of pre-miRNAs.
Developmentally regulated miRNAs in D. discoideum
Next, we examined whether the identified miRNAs were differentially expressed. The high-throughput sequencing reads revealed that some of the miRNAs were expressed in all life stages analyzed, while others were only found at specific developmental stages (Fig. 2C; Supplemental Figs. S5–S7). The most striking observation was the high enrichment for many of the miRNAs during development, some of which were completely absent in growing cells. Significantly, the great majority of miRNAs derived from the thug-S complex repeats were found in the d16h and d24h libraries. These repeats belong to a class of elements that bear resemblance to miniature inverted-repeat transposable elements (MITEs) (Glockner et al. 2001). Roughly the same developmental regulation was also observed for the miRNA*, which is expected if these RNAs are generated simultaneously with the miRNA by Dicer cleavage (Supplemental Figs. S5–S7).
3′-end property of miRNAs
To further characterize miRNAs in D. discoideum, we asked whether the 3′ end of ddi-mir-1177, here used as a representative model, was modified; 3′-methylation of miRNAs and siRNAs by Hen1 proteins occurs in plants and protects from degradation (Li et al. 2005). The 3′ end of overexpressed ddi-mir-1177 was analyzed by a β-elimination assay, which removes the unprotected 3′ nucleotide from RNA (Alefelder et al. 1998). Northern blot analysis shows that ddi-mir-1177 became shortened after β-elimination, indicating that its 3′ end is not modified (Fig. 3I).
D. discoideum miRNA 5′ properties and pre-miRNA structures
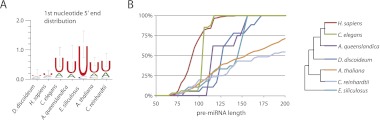
MiRNAs from a number of different organisms have been reported to display a bias toward certain nucleotides, e.g., A or U, at their 5′ ends (Seitz et al. 2011 and references within). The D. discoideum miRNAs, in contrast, show no significant preference for any particular 5′ nucleotide (Fig. 5A). As a comparison, we performed the same analysis of miRNAs deposited in miRBase from a selected number of other organisms, i.e., animals, plants, and green and brown algae. For both algae and plants, there is a clear enrichment of U at the 5′ end of the miRNAs (Fig. 5A). For miRNAs identified in human and worm, the pattern was not that clear (data not shown). We reasoned that this may be due to the fact that some of the small RNAs in miRBase are wrongly annotated as miRNAs. To address this, we reanalyzed the sequences using miRNAs in humans that are conserved in mice and miRNAs in worms that are reported in both C. elegans and Caenorhabditis briggsae (Fig. 5A). For these subsets of conserved miRNAs, there was also a preference for U at their 5′ ends, although not very pronounced in the case of human miRNAs. We also conducted a comparison of the length and size distribution of predicted pre-miRNA hairpins from D. discoideum to those from animals, plants, and green and brown algae. The results presented in Figure 5B are based on precursors generating the same set of miRNAs analyzed in Figure 5A, i.e., for human and worm miRNAs, only conserved precursors are included. Interestingly, the predicted D. discoideum miRNA hairpin precursors display a narrow size distribution, similar to animal pre-miRNA structures. In plants and algae, these structures show a higher variability in length. In particular, the pre-miRNA hairpins from D. discoideum are similar in size distribution to those from A. queenslandica, belonging to the sponges, which are considered as a branch within the oldest surviving metazoan lineage (Srivastava et al. 2010). However, if only considering the overall length of the precursors, the D. discoideum hairpins are more similar to earlier evolved organisms, e.g., plants, but also to A. queenslandica which seems to have unusually long miRNA precursors compared to other animals. For human and C. elegans miRNA precursors, similar results were obtained when all reported precursors were taken into account, although their size profiles were slightly broader, including a larger fraction of smaller precursors, indicating tight constraints of, at least, conserved miRNA precursors in these organisms (data not shown).
FIGURE 5.
Analyses of 5′ nucleotide identity and pre-miRNA length. (A,B) 5′ identity of miRNAs and relative length distribution of pre-miRNA structures (reported in miRBase), respectively, from Homo sapiens (H. sapiens), Caenorhabditis elegans (C. elegans), Amphimedon queenslandica (A. queenslandica), Dictyostelium discoideum (D. discoideum), Ectocarpus siliculosus (E. siliculosus), Chlamydomonas reinhardtii (C. reinhardtii), and Arabidopsis thaliana (A. thaliana). The miRNAs and their precursors from H. sapiens and C. elegans represent those also present in mice and Caenorhabditis briggsae, respectively. (B) The figure also includes a schematic phylogeny of the organisms.
DISCUSSION
In this study, we report the identification of developmentally regulated miRNAs in the unicellular social amoeba D. discoideum, establishing the presence of miRNAs in the supergroup Amoebozoa. In a previous study, we described the presence of two miRNA candidates in D. discoideum (Hinas et al. 2007). Although both of these, at the time, fulfilled the criteria for being annotated as miRNAs (Ambros et al. 2003), their relatively low abundance (single reads in a cDNA library), and the fact that D. discoideum is evolutionarily very distant from all other organisms in which miRNAs had been identified, prompted us to search for further evidence. Here, we used high-throughput sequencing of small RNAs from growing cells as well as from cells during different stages of multicellular development to verify the presence of miRNAs in D. discoideum. The sequenced small RNA libraries were screened for miRNAs using a set of stringent search criteria such as (1) disregarding small RNAs that matched the genome at more than nine locations, (2) only considering RNAs with multiple reads in each library, (3) the small RNAs should only be expressed from one DNA strand, (4) the RNAs should be derived from hairpin-structures as predicted by programs trained on plant and animal miRNA precursors, (5) no additional reads from nearby sequences (500 nt) of the hairpin, and (6) relatively uniform length and defined 5′ ends. After these stringent filters, 17 small RNAs remained and were defined as miRNAs with high confidence. Furthermore, for nine of the miRNAs, we were also able to identify the corresponding miRNA* sequences. Hence, the new miRNAs are well in agreement with the presently accepted miRNA annotation criteria (Ambros et al. 2003; Meyers et al. 2008; Kozomara and Griffiths-Jones 2011).
As a proof of principle, we verified the specific processing of four miRNAs in an overexpression system from which the predicted pre-miRNA hairpin was expressed. Overexpression of the miRNAs was confirmed in all cases. In addition, we could detect the miRNA* in three cases and also show that biogenesis of the miRNA and the miRNA* was strictly dependent on DrnB. The rationale for this approach was a recent report on deep sequencing of mammalian miRNAs. This study showed that overexpression of precursor hairpins of previously annotated miRNA, which did not fulfill all the criteria that define miRNAs, failed to yield small RNAs, whereas miRNAs defined by more stringent criteria were generated in this experimental set-up (Chiang et al. 2010).
One of the overexpressed pre-miRNAs is derived from an intron of the DNA polymerase B gene. It appears not to be a canonical mirtron since the splice sites do not coincide with the base of the predicted hairpin (Ruby et al. 2007). In addition to the DrnB-dependent miRNA and miRNA* sequences, the hairpin also generated a slightly shorter RNA which was independent of DrnB (Fig. 4G). We hypothesize that the biogenesis of this small RNA is dependent on DrnA, the other Dicer homolog in D. discoideum (see below). Similar examples have been reported in other organisms. For example, in Arabidopsis, the same precursor can be the source of different small RNA species where canonical miRNAs are generated by the Dicer-like (DCL) proteins typically required for miRNA biogenesis, while longer siRNAs (or longer miRNAs) are dependent on another DCL involved in siRNA production. Furthermore, in plants, it has also been shown that recently evolved miRNA genes consistently give rise to longer si/miRNA species, which are processed by all four DCL proteins (Vazquez et al. 2008; Chellappan et al. 2010). Hence, it is possible that the ddi-mir-7102 precursor in D. discoideum is an evolutionary intermediate recognized by both Dicer proteins.
Another surprising feature of miRNAs from D. discoideum is that they are up-regulated in strains in which the gene encoding RrpC, one of the three RdRPs, has been knocked out. This was demonstrated by Northern blot analyses of miRNAs in an rrpC− background (Figs. 3, 4). The same phenomenon was previously observed for small RNAs generated from a predicted hairpin structure from the Skipper retrotransposon in D. discoideum (Hinas et al. 2007). However, the effect of RrpC on small RNAs is further complicated since different siRNAs derived from the retrotransposon DIRS-1 are either not affected or down-regulated in the absence of RrpC, depending on from which part they are derived (Hinas et al. 2007; data not shown). RdRP-dependent amplifying effects have previously been reported and can be obtained by using a complementary RNA as template for siRNA generation (Nishikura 2001; Pak and Fire 2007; Sijen et al. 2007). However, the increased levels of miRNAs in the absence of RdRP are harder to explain. One explanation might be that DrnB and RrpC are part of the same complex that acts on different targets. If so, then depletion of RrpC might permit more DrnB recruitment for miRNA production. This hypothesis is consistent with the observation that the two Dicer proteins in D. discoideum lack the helicase domain that is present in the Dicers of most other organisms. Helicase domains are, instead, found in the RdRPs in D. discoideum, suggesting domain swapping between the two classes of proteins (Martens et al. 2002). Hence, the helicase could be present in the same complex as Dicers, which may be important for some Dicer-dependent processes. Dicers and RdRPs have been shown to be part of the same complex and to be functionally coupled in RNAi-related processes in Tetrahymena thermophila and fission yeast (Colmenares et al. 2007; Lee and Collins 2007).
The discovery of miRNAs in D. discoideum, hence establishing the presence of miRNAs in the supergroup of Amoebozoa, is interesting in an evolutionary context. This group of organisms branched out after plants but before animals (Eichinger et al. 2005). This raises the intriguing question of whether the miRNA machinery has evolved independently in Amoebozoa. We were unable to identify any D. discoideum miRNA homologs in miRbase and not even in the genome of the related amoebozoan, Discoideum purpureum. However, although D. discoideum and D. purpureum belong to the same group of Dictyostelia, they shared their last common ancestor some 400 million years ago (Parikh et al. 2010).
How can miRNAs have evolved independently in Amoebozoa, e.g., D. discoideum? The miRNAs derived from repetitive elements may shed some light on this question. MITEs (see above) have been suggested to play a role in the birth of new miRNAs and as a link between siRNAs and miRNAs (Piriyapongsa and Jordan 2007). The thug-S element in the D. discoideum genome resembles MITEs and could be the source for new miRNAs. Four miRNAs identified in this study originate from thug-S terminal repeats, which are predicted to form pre-miRNA hairpin structures. In addition, one of these miRNAs can be derived from eight different genomic locations (thug-S elements), where the predicted pre-miRNA structures are all different.
The protein components of the RNAi machinery in D. discoideum neither conform to a canonical plant nor animal composition (Martens et al. 2002; Cerutti and Casas-Mollano 2006; data not shown). It is also unresolved what role the majority of these different proteins play in D. discoideum. We have shown that the Dicer-like protein DrnB is strictly required for miRNA biogenesis, since all miRNAs analyzed here were absent in the drnB− strain. Furthermore, DrnB was recently demonstrated to be located mainly in the nucleus (Dubin and Nellen 2010). Since we fail to observe any accumulation of longer transcripts corresponding to pre-miRNAs in drnB− cells, this may indicate that DrnB processes the miRNA hairpins in two steps, as in plants, where only one Dicer is responsible for generating mature miRNAs (Voinnet 2009). The abundant siRNA population originating from repetitive elements has not been reported to be affected in drnB− cells, suggesting that the other Dicer, DrnA, is required for their biogenesis. In previous studies, DrnA was shown not to be essential for miRNA and siRNA generation. However, it is now clear that the drnA gene is located in a duplicated region of the genome, and in spite of several attempts by different laboratories including ours, deletion of both copies has been unsuccessful (Kuhlmann et al. 2005; Hinas et al. 2007).
Perhaps the most intriguing question is how the D. discoideum miRNAs regulate their targets and what these targets are. The miRNAs are clearly not essential under laboratory conditions since drnB− cells grow and develop normally. It is conceivable that miRNAs in D. discoideum only have a role in fine-tuning of gene expression and/or that they function under conditions so far not examined. Notably, several of the miRNAs in D. discoideum exhibit a strict developmental-specific expression, suggesting a function during these particular developmental stages. We used different methods to find targets for the identified miRNAs. In order to predict plant-like targets, we used a computational approach similar to the one used to successfully predict targets in C. reinhardtii (Zhao et al. 2007). We identified a few nearly perfect plant-like target sites within the coding regions of several mRNAs. However, in contrast to the target search in C. reinhardtii, we detected approximately the same number of miRNA targets in a generated sequence library containing the same number of sequences with the same length and nucleotide composition as the coding regions. This suggests that miRNAs in D. discoideum have very few, if any, plant-like targets. Some of the predicted targets were analyzed by rapid amplification of 5′ cDNA ends (5′ RACE) in order to identify specific miRNA induced cleavage sites, as might be expected from the action of miRNAs in a situation reminiscent of that seen for plant miRNAs (Rhoades et al. 2002). However, we have so far failed to find evidence for cleavage (data not shown). If target recognition is similar to that in animals, where short target sites, often consisting of ∼6 to 7 contiguous nucleotides, are present in untranslated regions (mainly 3′ UTRs) of the mRNA, successful prediction of targets becomes much more difficult (Lewis et al. 2003). We performed different computational searches for animal-like targets for the miRNAs presented in this study (data not shown). As expected, a great number of putative mRNA targets were predicted for each miRNAs. We are in the process of performing transcriptome analyses to pinpoint true target mRNAs.
In conclusion, the discovery of miRNAs in D. discoideum adds the supergroup Amoebozoa to the list of organisms harboring a miRNA machinery. We expect the number of miRNAs in D. discoideum to be larger than presented in this study due to our stringent search criteria, the relatively low numbers of reads in the RNA libraries, and the large number of siRNAs from DIRS-1 obscuring other small RNAs. Ongoing sequencing projects of close relatives of D. discoideum will help us to further understand the evolution and function of the miRNA machinery in this group of organisms. Furthermore, this study emphasizes the unique possibility to answer questions concerning the function of small RNAs during the transition from uni- to multicellularity.
MATERIALS AND METHODS
Oligonucleotides
DNA oligonucleotides (Invitrogen) are listed in Supplemental Table S1.
Growth and development
Axenically growing D. discoideum cells; AX4, AX2, and derivatives, were cultivated in HL-5 medium supplemented with 10 μL/mL penicillin-streptomycin (Gibco). Cells transformed with expression plasmids were grown in media supplemented with 10 μg/mL G418 (Gibco). Cells were developed in PDF buffer on nitrocellulose membranes at 22°C (Sussman 1987).
Strains
The sequenced D. discoideum strain AX4 was used to extract RNA for the small RNA library construction. The rrpC deletion strain (AX2 background) was a kind gift from S. Wiegand and C. Hammann. The drnB deletion and miRNA overexpression strains (AX2 background) are described below. AX2 lacks the large duplication which is present on chromosome 2 in AX4.
RNA preparation
Total RNA from axenically growing cells and from the different developmental stages was prepared from 5 × 107 cells using TRIzol, according to the manufacturer's recommendation (Invitrogen).
High-throughput sequencing
The SOLiD (Applied Biosystems) sequencing was performed at Rudbeck Laboratory (Uppsala). Briefly, total RNA (∼20 μg) from growing cells (0 h), slugs (16 h), and fully developed fruiting bodies (24 h) was subjected to FlashPAGE fractionation (Ambion) to extract RNAs smaller than 150 nt. Subsequently, the SOLiD Small RNA Expression system (Applied Biosystems) was used for preparation of cDNA and multiplexing. Since the construction of the cDNA is dependent on ligation to nucleotides with 5′ monophosphates, the library is enriched for processed transcripts such as Dicer products.
Mapping of reads to the genome
The gff files (generated 2009-04-29) containing the annotation and genome sequence of D. discoideum (Eichinger et al. 2005) were downloaded from dictyBase (Chisholm et al. 2006; Fey et al. 2009) (www.dictyBase.org). Extra annotation for complex repeats was carried out as described in Larsson et al. (2008). To determine size distribution, the 3′ adapter sequences, provided by the Rudbeck Laboratory sequencing facility, were mapped to the SOLiD-reads using SHRiMP 1.3.1 (Rumble et al. 2009) and trimmed away. The size-separated sequences were mapped to the genome. For reads above 18 nt, one mismatch in the alignment was allowed and up to 3 nt mismatches in the ends of the alignment allowing for sequences that had been edited or nucleotides that had been added post-transcriptionally to the ends, e.g., the CCA tail on tRNAs. For sequences between 15 nt and 18 nt, only perfect matches were considered. If one read mapped to multiple locations, only the locations with the highest score were kept for further analysis. If one read matched multiple locations with the same score, all locations were kept for further analysis. To ensure that each small RNA was equally represented, each read with multiple matches in the genome was normalized with the total number of genome matches for those, i.e., a sequence that matched 20 locations in the genome was counted as 1/20 read for each genomic location.
miRNA analysis
To identify previously annotated miRNAs, all reads were mapped, allowing only perfect matches, to all miRNAs present in miRbase release 18 (www.mirbase.org) (Griffiths-Jones 2006) using SHRiMP 1.3.1. In order to remove siRNAs from the analysis, sequences originating from regions where RNAs were derived from both strands, e.g., DIRS-1 (Hinas et al. 2007), and mRNA and their cognate antisense transcripts were excluded by adding the constraint that the number of reads that mapped to a specific location on one strand should be at least 20 times higher than all the reads that mapped to a region between 500 nt upstream and downstream on the opposite strand. To identify pre-miRNA structures, sequences from 200 nt upstream of to 200 nt downstream from a mapped location were extracted and analyzed using MiRfold (Billoud et al. 2005; Boccara et al. 2007) with standard settings (default temperature 22°C, the growth temperature for D. discoideum). Pre-miRNA structures identified by MiRfold with a penalty <5 and a ΔG-value per nt <−0.4 kcal/mol were considered as pre-miRNA candidates and were subsequently reanalyzed with miR-abela (Sewer et al. 2005) (http://www.mirz.unibas.ch/cgi/pred_miRNA_genes.cgi). Further analyses are described in Results.
To determine the false positive rate of MiRfold, 10,000 sequences of 421 nt in length with the same di-nucleotide frequency as the pre-miRNA candidates were generated and analyzed by MiRfold. To determine the false negative rate of MiRfold, 1513 pre-miRNAs from Homo sapiens, 446 from C. elegans, eight from A. queenslandica, 26 from Ectocarpus siliculosus, 32 from C. reinhardtii, and 256 from Arabidopsis thaliana were downloaded from mirBase and analyzed by MiRfold. If the reported pre-miRNA sequence was shorter than 421 nt in miRbase, we added nucleotides with the same nucleotide composition as the pre-miRNA sequence, upstream and downstream, to generate a final RNA of 421 nt, with the reported miRNA sequence centered. Pre-miRNA sequences longer than 421 nt were shortened to 421 nt, keeping the reported pre-miRNA sequence in the middle. All pre-miRNA and miRNA sequences registered in mirBase were downloaded (2012-02-10) from miRBase release 18. To analyze and visualize nucleotide preferences for the miRNAs, weblogo 3.0 was used (Crooks et al. 2004).
drnB deletion
The drnB deletion construct was assembled by using pLPBLP as the host vector (Faix et al. 2004). Parts of the drnB gene adjacent to one of the RNase III domains were amplified from genomic DNA (AX2), prepared as described elsewhere (Charette and Cosson 2004), by PCR using primers 600 and 601 for the 5′ region and 602 and 603 for the 3′ region. The construct is thus designed so that one of the RNase III domains of drnB is removed during the recombination event. The 3′ fragment was first cloned using the BamHI and PstI sites, and the 5′ fragment was cloned using the ClaI and SmaI sites on each side of the Blasticidin S resistance cassette of pLPBLP. Thirty μg of the deletion vector were digested with KpnI, NotI, and ScaI and purified on a PCR purification column (Fermentas) before transformation into AX2 D. discoideum cells by electroporation. Single clone transformants were isolated on Klebsiella aerogenes and analyzed by PCR using primers 605 and 169, 457 and 458, 630 and 631. The deletion was also confirmed by Southern blot. The Bsr cassette was subsequently removed as described elsewhere (Faix et al. 2004).
miRNA overexpression strains
Four predicted miRNA precursors, pre-ddi-mir-1176, pre-ddi-mir-1177, pre-ddi-mir-7095, and pre-ddi-mir-7102, were PCR-amplified from genomic DNA (AX2) by PCR using primers 641 and 642, 643 and 644, 645 and 646, 639 and 640, respectively.
The primers were designed to amplify the hairpin precursor. However, due to the high AT content, suitable annealing sites were difficult to find. Hence, some of the precursors carry up to 20 additional upstream and downstream nucleotides. In the case of ddi-mir-1177, the 5′ primer was located ∼90 nt upstream of the predicted precursor. Primers included BglII and SpeI restriction sites for cloning into the Dictyostelium extrachromosomal expression vector pDM304 downstream from an act15 promoter (Veltman et al. 2009). The correct insert was verified by sequencing (Macrogen). 107 AX2 cells were transformed with 2–10 μg of plasmid DNA by electroporation. Ten μg/mL G418 (Invitrogen) was added for selection 18–24 h after transformation. Colonies were normally visible after 3 d. Cells were collected after 7 d and transferred to HL5 shaking cultures.
Northern hybridization
Twenty to thirty μg total RNA from D. discoideum wt (AX2) and miRNA overexpressing strains or RNA from β-elimination assay were analyzed by northern hybridization as described previously (Aspegren et al. 2004).
β-elimination assay
Thirty μg total RNA from AX2 cells overexpressing ddi-mir-1177 were incubated in borax/boric acid buffer pH 8.6 (30 mM borax, 30 mM boric acid) and 16 mM NaIO4 for 10 min at room temperature in the dark. The reaction was stopped by the addition of 0.3 μg/μL glycerol. The RNA was dried under vacuum, dissolved in 50 μL borax/boric acid pH 9.5, and incubated at 45°C for 90 min. The RNA was subsequently precipitated with EtOH and analyzed by northern hybridization using oligo 312 to detect ddi-mir-1177.
DATA DEPOSITION
Small RNAs referred to in this study have been deposited in the Gene Expression Omnibus database under accession number GSE3836.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Illona Urbarova, Naresh Thatikonda, and Anna Åsman for technical assistance; and Christian Hammann and Stephan Wiegand for the RrpC deletion strain. This work was supported by The Swedish Research Council (to F.S. and to Uppsala RNA Research Center) and The Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (FORMAS).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.033175.112.
REFERENCES
- Alefelder S, Patel BK, Eckstein F 1998. Incorporation of terminal phosphorothioates into oligonucleotides. Nucleic Acids Res 26: 4983–4988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, et al. 2003. A uniform system for microRNA annotation. RNA 9: 277–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Hannon GJ, Brennecke J 2007. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 318: 761–764 [DOI] [PubMed] [Google Scholar]
- Aspegren A, Hinas A, Larsson P, Larsson A, Soderbom F 2004. Novel non-coding RNAs in Dictyostelium discoideum and their expression during development. Nucleic Acids Res 32: 4646–4656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avesson L, Schumacher HT, Fechter P, Romby P, Hellman U, Söderbom F 2011. Abundant class of non-coding RNA regulates development in the social amoeba Dictyostelium discoideum. RNA Biol 8: 1094–1104 [DOI] [PubMed] [Google Scholar]
- Axtell M, Westholm J, Lai E 2011. Vive la différence: Biogenesis and evolution of microRNAs in plants and animals. Genome Biol 12: 221 doi: 10.1186/gb-2011-12-4-221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldauf SL, Roger AJ, Wenk-Siefert I, Doolittle WF 2000. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science 290: 972–977 [DOI] [PubMed] [Google Scholar]
- Bapteste E, Brinkmann H, Lee JA, Moore DV, Sensen CW, Gordon P, Duruflé L, Gaasterland T, Lopez P, Müller M, et al. 2002. The analysis of 100 genes supports the grouping of three highly divergent amoebae: Dictyostelium, Entamoeba, and Mastigamoeba. Proc Natl Acad Sci 99: 1414–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP 2009. MicroRNAs: Target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billoud B, De Paepe R, Baulcombe D, Boccara M 2005. Identification of new small non-coding RNAs from tobacco and Arabidopsis. Biochimie 87: 905–910 [DOI] [PubMed] [Google Scholar]
- Boccara M, Sarazin A, Billoud B, Jolly V, Martienssen R, Baulcombe D, Colot V 2007. New approaches for the analysis of Arabidopsis thaliana small RNAs. Biochimie 89: 1252–1256 [DOI] [PubMed] [Google Scholar]
- Boesler C, Kruse J, Soderbom F, Hammann C 2011. Sequence and generation of mature ribosomal RNA transcripts in Dictyostelium discoideum. J Biol Chem 286: 17693–17703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun L, Cannella D, Ortet P, Barakat M, Sautel CF, Kieffer S, Garin J, Bastien O, Voinnet O, Hakimi MA 2010. A complex small RNA repertoire is generated by a plant/fungal-like machinery and effected by a metazoan-like Argonaute in the single-cell human parasite Toxoplasma gondii. PLoS Pathog 6: e1000920 doi: 10.1371/journal.ppat.1000920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ 2009. Origins and mechanisms of miRNAs and siRNAs. Cell 136: 642–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti H, Casas-Mollano J 2006. On the origin and functions of RNA-mediated silencing: From protists to man. Curr Genet 50: 81–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charette SJ, Cosson P 2004. Preparation of genomic DNA from Dictyostelium discoideum for PCR analysis. Biotechniques 36: 574–575 [DOI] [PubMed] [Google Scholar]
- Chellappan P, Xia J, Zhou X, Gao S, Zhang X, Coutino G, Vazquez F, Zhang W, Jin H 2010. siRNAs from miRNA sites mediate DNA methylation of target genes. Nucleic Acids Res 38: 6883–6894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, et al. 2010. Mammalian microRNAs: Experimental evaluation of novel and previously annotated genes. Genes Dev 24: 992–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm RL, Gaudet P, Just EM, Pilcher KE, Fey P, Merchant SN, Kibbe WA 2006. dictyBase, the model organism database for Dictyostelium discoideum. Nucleic Acids Res 34: D423–D427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmenares SU, Buker SM, Buhler M, Dlakić M, Moazed D 2007. Coupling of double-stranded RNA synthesis and siRNA generation in fission yeast RNAi. Mol Cell 27: 449–461 [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE 2004. WebLogo: A sequence logo generator. Genome Res 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubin M, Nellen W 2010. A versatile set of tagged expression vectors to monitor protein localisation and function in Dictyostelium. Gene 465: 1–8 [DOI] [PubMed] [Google Scholar]
- Eichinger L, Pachebat JA, Glockner G, Rajandream MA, Sucgang R, Berriman M, Song J, Olsen R, Szafranski K, Xu Q, et al. 2005. The genome of the social amoeba Dictyostelium discoideum. Nature 435: 43–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faix J, Kreppel L, Shaulsky G, Schleicher M, Kimmel AR 2004. A rapid and efficient method to generate multiple gene disruptions in Dictyostelium discoideum using a single selectable marker and the Cre-loxP system. Nucleic Acids Res 32: e143 doi: 10.1093/nar/gnh136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fey P, Gaudet P, Curk T, Zupan B, Just EM, Basu S, Merchant SN, Bushmanova YA, Shaulsky G, Kibbe WA, et al. 2009. dictyBase—a Dictyostelium bioinformatics resource update. Nucleic Acids Res 37: D515–D519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19: 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD 2009. Small silencing RNAs: An expanding universe. Nat Rev Genet 10: 94–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glockner G, Heidel AJ 2009. Centromere sequence and dynamics in Dictyostelium discoideum. Nucleic Acids Res 37: 1809–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glockner G, Szafranski K, Winckler T, Dingermann T, Quail MA, Cox E, Eichinger L, Noegel AA, Rosenthal A 2001. The complex repeats of Dictyostelium discoideum. Genome Res 11: 585–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S 2006. miRBase: The microRNA sequence database. Methods Mol Biol 342: 129–138 [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ 2008. miRBase: Tools for microRNA genomics. Nucleic Acids Res 36: D154–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP 2010. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466: 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinas A, Soderbom F 2007. Treasure hunt in an amoeba: Non-coding RNAs in Dictyostelium discoideum. Curr Genet 51: 141–159 [DOI] [PubMed] [Google Scholar]
- Hinas A, Larsson P, Avesson L, Kirsebom LA, Virtanen A, Soderbom F 2006. Identification of the major spliceosomal RNAs in Dictyostelium discoideum reveals developmentally regulated U2 variants and polyadenylated snRNAs. Eukaryot Cell 5: 924–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinas A, Reimegard J, Wagner EG, Nellen W, Ambros VR, Soderbom F 2007. The small RNA repertoire of Dictyostelium discoideum and its regulation by components of the RNAi pathway. Nucleic Acids Res 35: 6714–6726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessin R. 2001. Dictyostelium—evolution, cell biology, and the development of multicellularity. Cambridge University Press, Cambridge, UK. [Google Scholar]
- Kozomara A, Griffiths-Jones S 2011. miRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 39: D152–D157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlmann M, Borisova BE, Kaller M, Larsson P, Stach D, Na J, Eichinger L, Lyko F, Ambros V, Soderbom F, et al. 2005. Silencing of retrotransposons in Dictyostelium by DNA methylation and RNAi. Nucleic Acids Res 33: 6405–6417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson P, Hinas A, Ardell DH, Kirsebom LA, Virtanen A, Soderbom F 2008. De novo search for non-coding RNA genes in the AT-rich genome of Dictyostelium discoideum: Performance of Markov-dependent genome feature scoring. Genome Res 18: 888–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SR, Collins K 2007. Physical and functional coupling of RNA-dependent RNA polymerase and Dicer in the biogenesis of endogenous siRNAs. Nat Struct Mol Biol 14: 604–610 [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854 [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB 2003. Prediction of mammalian microRNA targets. Cell 115: 787–798 [DOI] [PubMed] [Google Scholar]
- Li J, Yang Z, Yu B, Liu J, Chen X 2005. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr Biol 15: 1501–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens H, Novotny J, Oberstrass J, Steck TL, Postlethwait P, Nellen W 2002. RNAi in Dictyostelium: The role of RNA-directed RNA polymerases and double-stranded RNase. Mol Biol Cell 13: 445–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Axtell MJ, Bartel B, Bartel DP, Baulcombe D, Bowman JL, Cao X, Carrington JC, Chen X, Green PJ, et al. 2008. Criteria for annotation of plant MicroRNAs. Plant Cell 20: 3186–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K, Miyoshi T, Siomi H 2010. Many ways to generate microRNA-like small RNAs: Non-canonical pathways for microRNA production. Mol Genet Genomics 284: 95–103 [DOI] [PubMed] [Google Scholar]
- Molnar A, Schwach F, Studholme DJ, Thuenemann EC, Baulcombe DC 2007. miRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii. Nature 447: 1126–1129 [DOI] [PubMed] [Google Scholar]
- Nishikura K 2001. A short primer on RNAi: RNA-directed RNA polymerase acts as a key catalyst. Cell 107: 415–418 [DOI] [PubMed] [Google Scholar]
- Pak J, Fire A 2007. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science 315: 241–244 [DOI] [PubMed] [Google Scholar]
- Parikh A, Miranda E, Katoh-Kurasawa M, Fuller D, Rot G, Zagar L, Curk T, Sucgang R, Chen R, Zupan B, et al. 2010. Conserved developmental transcriptomes in evolutionarily divergent species. Genome Biol 11: R35 doi: 10.1186/gb-2010-11-3-r35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piriyapongsa J, Jordan IK 2007. A family of human microRNA genes from miniature inverted-repeat transposable elements. PLoS ONE 2: e203 doi: 10.1371/journal.pone.0000203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova B, Kuhlmann M, Hinas A, Soderbom F, Nellen W 2006. HelF, a putative RNA helicase acts as a nuclear suppressor of RNAi but not antisense mediated gene silencing. Nucleic Acids Res 34: 773–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP 2002. Prediction of plant microRNA targets. Cell 110: 513–520 [DOI] [PubMed] [Google Scholar]
- Ruby JG, Jan CH, Bartel DP 2007. Intronic microRNA precursors that bypass Drosha processing. Nature 448: 83–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumble SM, Lacroute P, Dalca AV, Fiume M, Sidow A, Brudno M 2009. SHRiMP: Accurate mapping of short color-space reads. PLoS Comput Biol 5: e1000386 doi: 10.1371/journal.pcbi.1000386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraiya AA, Wang CC 2008. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog 4: e1000224 doi: 10.1371/journal.ppat.1000224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz H, Tushir JS, Zamore PD 2011. A 5′-uridine amplifies miRNA/miRNA* asymmetry in Drosophila by promoting RNA-induced silencing complex formation. Silence 2: 4 doi: 10.1186/1758-907X-2-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewer A, Paul N, Landgraf P, Aravin A, Pfeffer S, Brownstein MJ, Tuschl T, van Nimwegen E, Zavolan M 2005. Identification of clustered microRNAs using an ab initio prediction method. BMC Bioinformatics 6: 267 doi: 10.1186/1471-2105-6-267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalina SA, Koonin EV 2008. Origins and evolution of eukaryotic RNA interference. Trends Ecol Evol 23: 578–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Steiner FA, Thijssen KL, Plasterk RHA 2007. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science 315: 244–247 [DOI] [PubMed] [Google Scholar]
- Srivastava M, Simakov O, Chapman J, Fahey B, Gauthier ME, Mitros T, Richards GS, Conaco C, Dacre M, Hellsten U, et al. 2010. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466: 720–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman M 1987. Cultivation and synchronous morphogenesis of Dictyostelium under controlled experimental conditions. Methods Cell Biol 28: 9–29 [DOI] [PubMed] [Google Scholar]
- van Houten V, Denkers F, van Dijk M, van den Brekel M, Brakenhoff R 1998. Labeling efficiency of oligonucleotides by T4 polynucleotide kinase depends on 5′-nucleotide. Anal Biochem 265: 386–389 [DOI] [PubMed] [Google Scholar]
- Vazquez F, Blevins T, Ailhas J, Boller T, Meins F Jr 2008. Evolution of Arabidopsis MIR genes generates novel microRNA classes. Nucleic Acids Res 36: 6429–6438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltman DM, Akar G, Bosgraaf L, Van Haastert PJ 2009. A new set of small, extrachromosomal expression vectors for Dictyostelium discoideum. Plasmid 61: 110–118 [DOI] [PubMed] [Google Scholar]
- Voinnet O 2009. Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687 [DOI] [PubMed] [Google Scholar]
- Zhao T, Li G, Mi S, Li S, Hannon GJ, Wang X-J, Qi Y 2007. A complex system of small RNAs in the unicellular green alga Chlamydomonas reinhardtii. Genes Dev 21: 1190–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]