Ribosomal proteins L7 and L8 are required for some early steps in pre-rRNA processing during assembly of large subunits in yeast but are found in distinct domains within the ribosome. Here, following depletion of L7 and L8 in yeast, assembly of the large ribosomal subunit is broadly analyzed. In addition to previously described defects in rRNA processing, specific sets of proteins in the neighborhoods of L7 and L8, respectively, fail to assemble into the large subunit structure. These observations argue that localized RNP structures first assemble and then independently come together in the final ribosome structure.
Keywords: yeast ribosomal proteins L7 and L8, yeast ribosome assembly, polypeptide exit tunnel, 25S rRNA domains I and II, preribosomes
Abstract
Ribosome biogenesis is a complex multistep process that involves alternating steps of folding and processing of pre-rRNAs in concert with assembly of ribosomal proteins. Recently, there has been increased interest in the roles of ribosomal proteins in eukaryotic ribosome biogenesis in vivo, focusing primarily on their function in pre-rRNA processing. However, much less is known about participation of ribosomal proteins in the formation and rearrangement of preribosomal particles as they mature to functional subunits. We have studied ribosomal proteins L7 and L8, which are required for the same early steps in pre-rRNA processing during assembly of 60S subunits but are located in different domains within ribosomes. Depletion of either leads to defects in processing of 27SA3 to 27SB pre-rRNA and turnover of pre-rRNAs destined for large ribosomal subunits. A specific subset of proteins is diminished from these residual assembly intermediates: six assembly factors required for processing of 27SA3 pre-rRNA and four ribosomal proteins bound to domain I of 25S and 5.8S rRNAs surrounding the polypeptide exit tunnel. In addition, specific sets of ribosomal proteins are affected in each mutant: In the absence of L7, proteins bound to domain II, L6, L14, L20, and L33 are greatly diminished, while proteins L13, L15, and L36 that bind to domain I are affected in the absence of L8. Thus, L7 and L8 might establish RNP structures within assembling ribosomes necessary for the stable association and function of the A3 assembly factors and for proper assembly of the neighborhoods containing domains I and II.
INTRODUCTION
Assembly of yeast ribosomes begins in the nucleolus with transcription of the pre-35S rRNA by RNA polymerase I and pre-5S rRNA by RNA polymerase III. During and after transcription, precursor-rRNAs (pre-rRNAs) are modified and folded to form structures necessary for removal of transcribed spacer sequences and for binding of the 79 ribosomal proteins (r-proteins) (van Nues et al. 1995; Côté et al. 2002; Staley and Woolford 2009; Karbstein 2011). Initial cleavage events within transcribed spacer sequences, some of which occur cotranscriptionally (Osheim et al. 2004; Koš and Tollervey 2010), generate 43S and 66S precursors to mature 40S and 60S ribosomal subunits, respectively, which then undergo separate pathways of maturation.
In yeast, ribosome biosynthesis is facilitated by more than 200 assembly factors, as well as a large number of snoRNAs, which associate with one or more of the ribosome assembly intermediates (Decatur and Fournier 2002; Henras et al. 2008; Kressler et al. 2010). Following early steps in particle biogenesis, some assembly factors dissociate from preribosomes and others join pre-rRNPs as they transit from the nucleolus through the nucleoplasm to the cytoplasm. There, the particles undergo final stages of maturation prior to functioning in protein synthesis (Panse and Johnson 2010; Strunk et al. 2011). Most yeast assembly factors have been assigned to one or a few steps in pre-rRNA processing, nuclear export, or cytoplasmic maturation, based on mutant phenotypes. Studies are now underway to determine more precisely the mechanisms by which these factors function (Kressler et al. 2010; Panse and Johnson 2010).
The roles played by r-proteins in subunit biogenesis were first investigated by in vitro reconstitution of bacterial ribosomes from mature rRNA and r-proteins (Mizushima and Nomura 1970; Nomura 1973; Held et al. 1974; Herold and Nierhaus 1987; Nierhaus 1991; Culver 2003; Shajani et al. 2011; Woodson 2011). A subset of r-proteins binds rRNA first, recognizing prefolded RNA structures and inducing changes in them. This enables subsequent association of secondary and finally tertiary binding proteins. Thus, construction of ribosomes is hierarchical and cooperative, mediated by parallel and sequential pathways of r-protein binding, alternating with conformational changes in rRNA. The functions of r-proteins inferred from these in vitro studies are to fine-tune rRNA secondary structure, promote formation of rRNA tertiary structure, and act as RNA chaperones to prevent or correct misfolding of rRNA that could lead to kinetic traps that block assembly.
However, mechanisms of bacterial subunit assembly in vitro may not completely reflect the pathways of ribosome biogenesis in vivo in either prokaryotes or eukaryotes. Although ribosome assembly in vivo is more efficient than in vitro, it is also more complex. First, assembly occurs cotranscriptionally in vivo (Osheim et al. 2004; Koš and Tollervey 2010). Proteins bind to rRNA structures that form as the RNA is transcribed, largely in a 5′ to 3′ direction (Adilakshmi et al. 2005; Talkington et al. 2005; Shajani et al. 2011). Second, assembly is coupled with removal of the spacer sequences, which present different folding opportunities than mature rRNAs and might act as timers or intra-molecular chaperones for rRNA folding (Karbstein 2011). Third, assembly factors are present in vivo and improve the efficiency and accuracy of folding and processing of rRNA, binding of r-proteins, and intracellular transit of nascent ribosomes (Henras et al. 2008; Kressler et al. 2010).
Functional analysis of a few yeast and human r-proteins, and more recently, systematic examination, have begun to reveal roles of r-proteins in eukaryotic subunit assembly in vivo (Rotenberg et al. 1988; Moritz et al. 1990; Deshmukh et al. 1993; van Beekvelt et al. 2001; Jakovljevic et al. 2004; Léger-Silvestre et al. 2004; Ferreira-Cerca et al. 2005; Martín-Marcos et al. 2007; Rosado et al. 2007; Robledo et al. 2008; Pöll et al. 2009; Babiano and de la Cruz 2010; O'Donohue et al. 2010). Most investigations of r-protein function in vivo have focused on their participation in pre-rRNA processing. Much less is known about the roles of eukaryotic r-proteins in driving the formation and rearrangement of the pre-rRNP particles. How do r-proteins help propagate the assembly of ribosomal subunits? Which other proteins (r-proteins and assembly factors) are dependent upon r-proteins for association with preribosomes? How do the different domains of rRNA fold and assemble with r-proteins? For example, Zhang et al. (2007) showed that r-proteins L5 and L11 are required for the stable association of assembly factors Rpf2 and Rrs1, as well as 5S rRNA, with 60S preribosomes. Ferreira-Cerca et al. (2007) showed that association of r-proteins with the 3′ domain of 18S rRNA to form the head domain of 40S subunits depends on S5 and, to a lesser extent, S15.
Previously, we and others investigated a subset of assembly factors necessary for processing of 27SA3 pre-rRNA to 27SBs pre-rRNA, called A3 factors (Granneman et al. 2011; Sahasranaman et al. 2011). To complement this analysis, we have now focused on two r- proteins, L7 and L8, which are involved in early steps of ribosome biogenesis (Pöll et al. 2009). L8 is of interest because it is bound to domain I of 25S/5.8S rRNA in ribosomes, close to the proposed binding site for the A3 factor Erb1 (Ben-Shem et al. 2011; Granneman et al. 2011). Interestingly, depletion of ribosomal protein L7 has the same pre-rRNA processing defect, but L7 is bound to domain II of 25S rRNA, distal from L8 and known A3 factor binding sites. We decided to study both proteins in detail, relating defects in pre-rRNA processing to changes in preribosomal protein composition upon depletion of either one of them. These proteins assemble into preribosomes early in the pathway; they both coimmunoprecipitate 35S as well as 27SA and 27SB pre-rRNAs. In the absence of either r-protein, 27SA2 and 27SA3 pre-rRNA accumulate, and there is significant turnover of 27S pre-rRNA. Consistent with this perturbation in early steps of pre-rRNA processing, preribosomes purified from either depleted strain are enriched for early assembly intermediates, and late-assembling proteins are diminished. These abortive assembly intermediates that form when either L7 or L8 is depleted are unable to be exported to the cytoplasm. Notably, assembly factors specifically required for 27SA3 pre-rRNA processing (A3 factors) are diminished from these preribosomes. In addition, several r-proteins are absent or diminished. These include r-proteins L17, L26, L35, and L37, which bind 25S and 5.8S rRNAs at domain I and surround the polypeptide exit tunnel. Also, when L7 is depleted, r-proteins L6, L14, L20, and L33 that bind to domain II near L7 are diminished. In the absence of L8, r-proteins near L8 and bound to domain I, L13, L15, and L36, are greatly affected. We conclude that L7 and L8 are necessary for the formation of stable rRNP neighborhoods adjacent to each of them and also surrounding the polypeptide exit tunnel, thus establishing proper pre-rRNP structures required for 27SA3 pre-rRNA processing.
RESULTS
Ribosomal proteins L7 and L8 are necessary for production of 60S ribosomal subunits
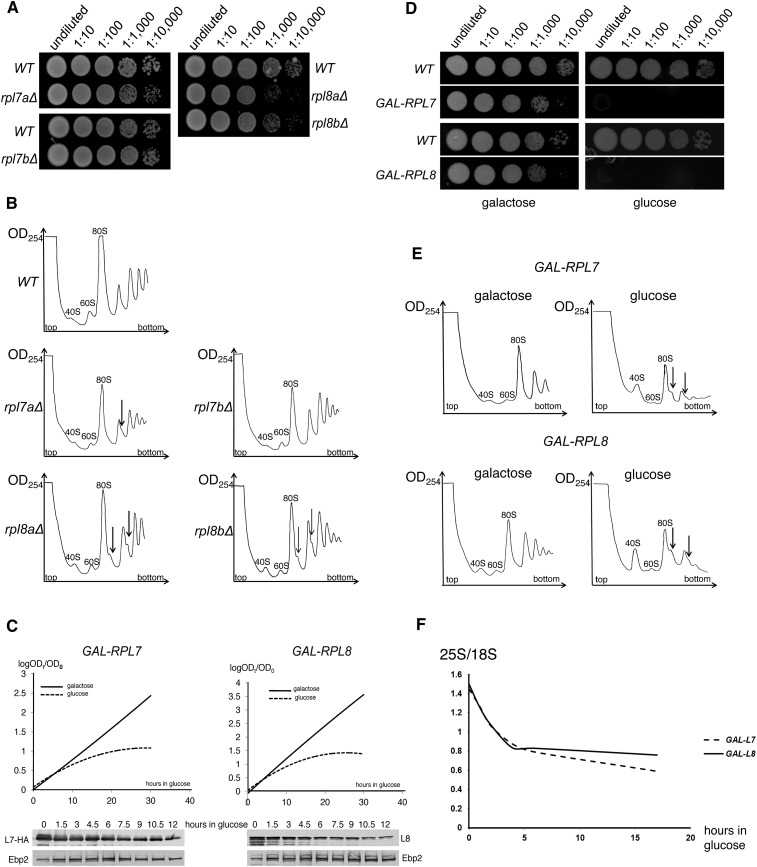
Because most r-proteins are essential, we needed to construct strains conditional for expression of L7 and L8 to examine their functions. As is the case for many r-proteins (Mager et al. 1997), both L7 and L8 are encoded by two different, unlinked genes (Arevalo and Warner 1990; Yon et al. 1991; Ohtake and Wickner 1995). Therefore, we began by deleting each copy of the RPL7 and RPL8 genes to determine which deletion of each pair had the stronger phenotype. Deletion of RPL7A had a mild defect on growth, while deletion of RPL7B had no growth defect (Fig. 1A). Deletion of either RPL8A or RPL8B had a detectable growth defect (Fig. 1A; Yon et al. 1991; Ohtake and Wickner 1995). Previously, Robledo et al. (2008) showed that knockdown of human L7 led to a decrease in 60S subunits. Consistent with these results, examination of polyribosome profiles of rpl7aΔ revealed diminished amounts of free 60S ribosomal subunits relative to free 40S subunits and accumulation of halfmer polyribosomes, while the rpl7bΔ strain had a wild-type polyribosome profile (Fig. 1B). Both the rpl8aΔ and rpl8bΔ deletion strains contained decreased levels of 60S subunits relative to 40S subunits and had halfmer polyribosomes (Fig. 1B; Ohtake and Wickner 1995).
FIGURE 1.
Depletion of ribosomal protein L7 or L8 inhibits cell growth and production of 60S ribosomal subunits. (A) Growth of the rpl7aΔ, rpl7bΔ, rpl8aΔ, and rpl8bΔ deletion strains. Strains JWY9878 (rpl7aΔ), JWY7771 (rpl7bΔ), JWY8561 (rpl8aΔ), and JWY8568 (rpl8bΔ), and the wild-type control strains BY4741 and JWY6147 were grown in YEPGlu. Serial dilutions (1:10 to 1:10,000) were spotted onto YEPGlu solid medium and incubated at 30°C. (B) The rpl7aΔ, rpl8aΔ, and rpl8bΔ strains are deficient in 60S ribosomal subunit assembly. The deletion strains from A were grown in YEPGlu, extracts purified from them were loaded on 7%–47% sucrose gradients and subjected to centrifugation to resolve 40S, 60S, and 80S monosomes and polyribosomes. Halfmer polyribosomes (polysomes containing one 40S subunit bound to the AUG start site in mRNA without the partner 60S subunit) are indicated by vertical arrows. (C) Yeast cease dividing ∼6–8 h after initiating depletion of L7 or L8. Yeast strains JWY8423 (GAL-RPL7) and TY1097 (GAL-RPL8) were grown at 30°C in YEPGal liquid medium to early log phase, and cultures were split into two. One culture was maintained in YEPGal liquid medium, and the other culture was shifted to YEPGlu. Growth of cultures was continuously monitored, and cultures were diluted to ensure exponential growth. OD610 values are plotted as log (ODt/ODt0), where ODt0 is the initial reading before medium shift, and ODt is the OD at time t min after the shift (top panels). At the time points indicated, aliquots of cultures were used for Western blotting with anti-HA or anti-L8 antibodies to confirm depletion of HA-tagged L7 or L8, respectively. As a loading control, an antibody against Ebp2 was used (bottom panels). (D) Proteins L7 and L8 are essential for growth on solid medium. Strains from C were grown in YEPGal liquid medium, and serial dilutions (1:10 to 1:10,000) were spotted onto YEPGal and YEPGlu solid media and incubated at 30°C. (E) 60S ribosomal subunits decrease and halfmer polyribosomes form upon depletion of L7 or L8. Yeast strains from C were grown in YEPGal or shifted to YEPGlu for 16 h. Sucrose gradient fractionations were performed as described in B. (F) Levels of 25S rRNA decrease compared to 18S rRNA after initiating depletion of L7 or L8. Total RNA was extracted from strains used in C at the indicated times after shifting from YEPGal to YEPGlu medium and subjected to electrophoresis on a 1% agarose gel. RNA was stained with ethidium bromide, and levels of mature rRNAs were quantified using Image Quant software.
Based on these observed phenotypes, strains conditional for expression of L7 and L8 were built by replacing the promoters of RPL7A and RPL8B with the GAL promoter in strains containing deletions of RPL7B and RPL8A, respectively (see also Pöll et al. 2009). Four to six hours after shifting the GAL-RPL7 and GAL-RPL8 strains from galactose to glucose-containing medium, the rate of cell division slowed down significantly (Fig. 1C). Western blotting at time points after shifting the strains to glucose confirmed that both L7 and L8 were significantly depleted (Fig. 1C). Some L7 or L8 remained in cells even after longer shifts, due to the presence of ribosomes that assembled before shutting off expression of RPL7 or RPL8. Both strains grew on solid medium containing galactose, although less well than the corresponding wild-type strains, presumably because overexpression of L7 or L8 is slightly deleterious for cells (Fig. 1D). Alternatively, expression of one gene from the GAL promoter may not completely complement the absence of both wild-type genes. Importantly, neither strain grew on glucose-containing solid medium, as expected upon depletion of essential proteins (Fig. 1D).
Sucrose gradient fractionation of extracts from each conditional strain grown in galactose or shifted to glucose for 17 h showed that production of 60S ribosomal subunits was compromised. Amounts of free 60S subunits relative to free 40S subunits were significantly decreased, and halfmer polyribosomes were present (Fig. 1E). In both cases, these defects were greater than in strains containing a deletion of either copy of the corresponding r-protein gene. Decreased amounts of 60S subunits were evident as early as 4 h after depletion of L8 (Supplemental Fig. S1). These results were confirmed by assaying total RNA extracted from the depleted strains at different times after shifting from galactose- to glucose-containing medium. Amounts of 25S rRNA relative to 18S rRNA decreased twofold within 4 h after the shift (Fig. 1F). Taken together, these results confirm that L7 and L8 are essential for growth and are specifically required for production of 60S ribosomal subunits, as previously reported (Pöll et al. 2009).
L7 and L8 are necessary for processing of 27SA3 pre-rRNA, and in their absence aberrant assembly intermediates are rapidly turned over
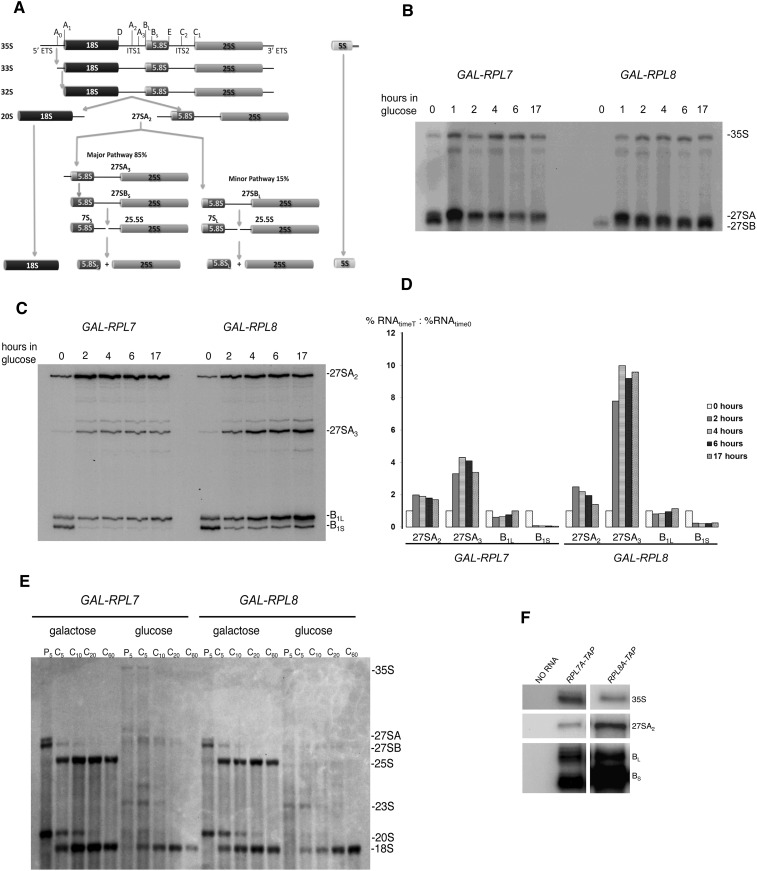
Previously, we showed that 2–4 h after turning off expression of L7 or L8, early steps in pre-rRNA processing were perturbed, resulting in accumulation of 27SA pre-rRNAs in the nucleus and in decreased amounts of downstream processing intermediates and mature 25S and 5.8S rRNAs (Pöll et al. 2009). To examine these pre-rRNA processing defects in more detail, we first assayed changes in steady-state levels of pre-rRNAs after turning off transcription of RPL7 or RPL8. Northern blot analysis of total RNA was used to detect 35S, 27SA, and 27SB processing intermediates (Fig. 2B). Significant amounts of 27SA pre-rRNA accumulated 2 h after shifting to glucose. No 27SB pre-rRNA was detected after short shifts of either strain to glucose, but some was present at later time points in the GAL-RPL8 strain. Almost no 27SB pre-rRNA was detectable in the GAL-RPL7 strain at any time point after the shift. 35S pre-rRNA, the earliest detectable rRNA precursor, also accumulated, as reported in many instances where early steps in pre-rRNA processing are affected (Venema and Tollervey 1999).
FIGURE 2.
Processing of 27SA2 and 27SA3 pre-rRNA is impaired, and pre-rRNAs are turned over upon depletion of L7 or L8. (A) The pre-rRNA processing pathway in Saccharomyces cerevisiae. (B,C) To assay the steady-state levels of pre-rRNAs upon depletion of L7 or L8, yeast strains JWY8423 (GAL-RPL7) and JWY8591 (GAL-RPL8) were grown at 30°C in YEPGal and shifted to YEPGlu. Total RNA was extracted from cultures at the indicated times after the shift, and steady-state levels of pre-rRNAs were assayed by Northern blotting (B), or primer extension and denaturing gel electrophoresis (C). The oligonucleotide probe or primer used in these experiments was complementary to sequences in ITS2 (de la Cruz et al. 1998). Pre-rRNAs and mature rRNA species are labeled. B1L and B1S represent the 5′ ends of 27SBL+7SL and 27SBS+7SS pre-rRNAs, respectively. (D) Image Gauge software was used to quantify levels of 5′ ends of pre-RNAs. The graph represents the fold change in levels of pre-rRNAs compared to initial levels, where each pre-rRNA is represented as a percentage of total RNA quantified. The sum of amounts for A2, A3, B1L, and B1S equals 100%. (E) To measure the synthesis and turnover of pre-rRNAs when L7 or L8 are depleted, GAL-RPL7 and GAL-RPL8 strains were grown in synthetic liquid media lacking methionine and containing galactose or shifted to selective media containing glucose for 17 h. Cells were pulse-labeled with [3H methyl]-methionine for 5 min and chased with an excess of unlabeled methionine. RNA extracted from cells at time 0 after the pulse-labeling and at 5, 10, 20, and 60 min after initiating the chase was subjected to denaturing gel electrophoresis, blotted to nitrocellulose, and exposed to X-ray film. Pre-rRNAs and mature rRNA species are labeled. (F) Ribosomal proteins L7 and L8 assemble early in the pathway, with 90S preribosomes containing 35S pre-rRNA. Yeast strains JWY7148 and JWY7158 expressing TAP-tagged L7A and TAP-tagged L8A, respectively, were grown in YEPGlu. Preribosomes and ribosomes were affinity-purified from whole-cell extracts. RNA was extracted from the purified particles and subjected to primer extension analysis to assay for the presence of the indicated pre-rRNAs. B1L and B1S represent the 5′ ends of 27SBL+7SL and 27SBS+7SS pre-rRNAs, respectively.
To more thoroughly assay changes in pre-rRNA intermediates, we used primer extension to measure levels of A2, A3, B1L, and B1S 5′ ends of pre-rRNAs. As shown in Figure 2C, amounts of A2 and A3 ends increased, and amounts of B1S ends decreased after shifting either strain to glucose, as expected for mutants blocked in processing of 27SA2 and 27SA3 pre-rRNAs. To pinpoint the most significant changes in levels of pre-rRNAs, primer extension ends were quantified as percent of total ends measured. The relative accumulation of the 5′ ends at each time point was calculated as %RNAt/%RNA0. Based on these results, the most significant relative accumulation over time was seen with A3 ends, and the most significant decrease was observed in amounts of B1s ends (Fig. 2D). Relative accumulation of B1L ends did not change significantly in either mutant. These results clearly indicate that depletion of L7 or L8 affects the processing of 27SA2 to 27SA3 pre-rRNAs and processing of 27SA3 to 27SBS pre-rRNAs but not the production of 27SBL pre-rRNA from 27SA2 pre-rRNAs. This pre-rRNA processing phenotype has been previously reported for a subset of assembly factors, named “A3 factors” (Dunbar et al. 2000; Harnpicharnchai et al. 2001; Pestov et al. 2001; Adams et al. 2002; Gadal et al. 2002; Oeffinger et al. 2002; Fatica et al. 2003; Oeffinger and Tollervey 2003; Miles et al. 2005; Sahasranaman et al. 2011).
The changes in amounts of 27SA2, 27SA3, and 27SBS pre-rRNAs observed upon depletion of L7 or L8 could result from (1) decreased production of 27SBS pre-rRNA from 27SA2 and 27SA3 pre-rRNA, (2) increased turnover of pre-rRNAs upon abortion of subunit maturation, or (3) a combination of both decreased production and increased turnover. To begin to distinguish among these possibilities, we assayed the synthesis and turnover of pre-rRNA by pulse-chase experiments. After shifting cells from galactose- to glucose-containing medium for 17 h, cells were pulse-labeled with [3H-methyl]-methionine for 5 min and chased with an excess of unlabeled methionine. RNA was extracted after the pulse and 5, 10, 20, and 60 min after the chase was initiated, separated by gel electrophoresis and detected by autoradiography (Fig. 2E). In cells grown in galactose, 35S pre-rRNA was rapidly converted to 20S, 27SA, and 27SB pre-rRNAs, which were efficiently processed to mature 18S and 25S rRNAs, as typically observed in wild-type cells. In contrast, in the strains depleted of L7 or L8, some 35S pre-rRNA was produced, and normal amounts of 18S rRNA accumulated, but the majority of pre-rRNAs destined for large subunits was turned over. Small amounts of 27S and 20S precursors were detected, and there was visible accumulation of the 23S rRNA precursor. This 23S rRNA species is formed when processing at sites A0, A1, and A2 is delayed, such that cleavage at the A3 site occurs before these other three cleavage steps. No mature 25S rRNA was detected in either depleted strain, as previously reported for knockdown of human r-protein L7 (Robledo et al. 2008). Mature 18S rRNA was still produced, although less was detected compared to unshifted strains.
Taken together, these results confirm the role of L7 and L8 in the processing of 27SA2 and 27SA3 pre-rRNAs and show that both proteins are involved in the same steps of pre-rRNA processing. Furthermore, we conclude that, upon depletion of these two ribosomal proteins, there is significant turnover of pre-rRNAs. Finally, we note that the phenotypes after depletion of L7 and L8 are very similar to those in strains mutant for assembly factors involved in 27SA3 processing (Sahasranaman et al. 2011).
L7 and L8 assemble early into 90S preribosomes containing 35S pre-rRNA
Pioneering studies by Kruiswijk and colleagues in the late 1970s, using pulse-labeling of yeast spheroplasts, provided a first low-resolution view of the time course of assembly of yeast r-proteins into ribosomes (Kruiswijk et al. 1978). These experiments indicated that L8 (then named L4) assembled rather early. Results for L7 were ambiguous but suggested that L7 assembled into immature ribosomes in the nucleus. The defects in early steps in pre-rRNA processing that we observe upon depleting either of these two r-proteins suggest that both L7 and L8 associate with preribosomes early in the assembly pathway, prior to 27SA3 pre-rRNA processing.
To investigate in more detail the timing of assembly of L7 and L8 with preribosomes, we used the pre-rRNA processing intermediates as landmarks of subunit maturation and assayed with which pre-rRNAs each protein copurifies. To do so, we used TAP-tagged L7A and L8A to affinity-purify preribosomes and ribosomes containing L7 or L8 and assayed copurifying pre-rRNAs by primer extension. Both L7 and L8 coimmunoprecipitated 35S, 27SA, and 27SB pre-rRNAs (Fig. 2F). Consistent with this observation, we previously found by Western blotting that L8 is present in early ribosome assembly intermediates purified using TAP-tagged early factor Npa2 (Sahasranaman et al. 2011). Npa2 assembles into particles containing 35S pre-rRNA and exits from preribosomes early in the lifetime of particles containing 27SB pre-rRNA (Rosado et al. 2007). Thus, L7 and L8 appear to assemble early into 90S preribosomes containing 35S pre-rRNA, consistent with their subsequent role in processing of 27SA3 pre-rRNA.
L7 and L8 are necessary for association of A3 factors and a subset of ribosomal proteins with preribosomes
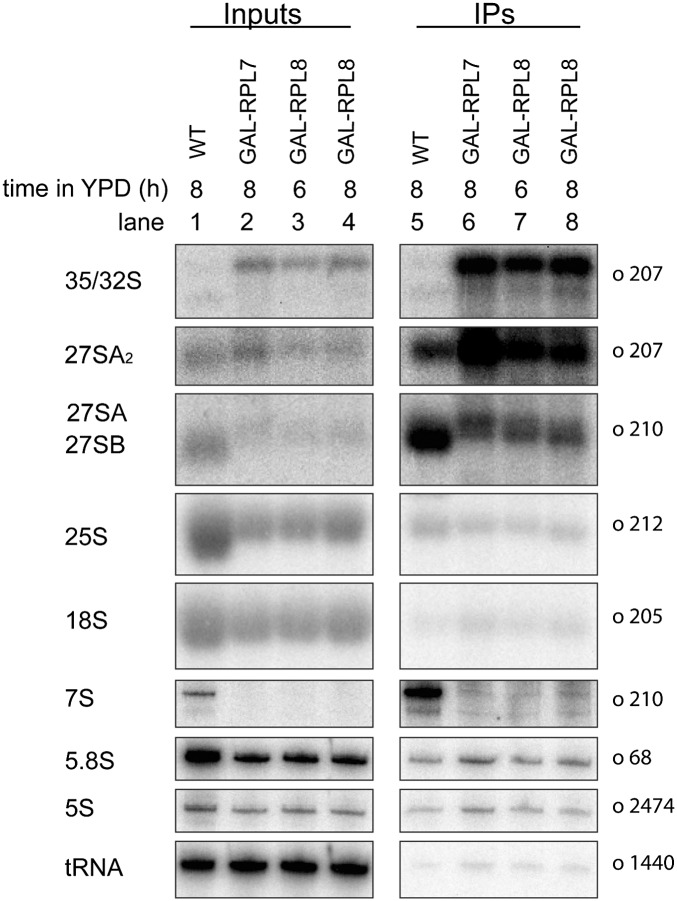
To start to understand why the absence of L7 or L8 perturbs processing of pre-rRNA, we investigated changes that occur in the protein constituents of preribosomal particles when either protein was depleted. In particular, we wanted to test whether some assembly factors and r-proteins were absent or diminished from assembly intermediates. To do so, we affinity-purified preribosomes from yeast in which L7 or L8 were either present or depleted, using TAP-tagged Rpf2. This assembly factor is present in 90S preribosomes and in each of the 66S pre-rRNP precursors to mature 60S subunits. Importantly, Rpf2 does not function in 27SA2 or 27SA3 pre-rRNA processing (Zhang et al. 2007). The effects of depleting L7 or L8 on preribosomes were qualitatively the same 4 and 17 h after shifting cells from galactose to glucose (Supplemental Fig. S2). In our purifications, we are using amounts of whole-cell extracts that saturate the IgG-coated beads; therefore, we recover similar amounts of preribosomes from undepleted and depleted strains. The preribosomes purified after depletion of either r-protein are mostly the residual particles that have not been turned over. Northern blotting of rRNAs present in these purified mutant preribosomes determined that the population of pre-rRNAs in these pre-rRNPs was identical to that seen in mutant cell extracts (Fig. 3). To globally assay changes in preribosomal protein constituents upon depletion of L7 or L8, we used iTRAQ, a semiquantitative mass spectrometric method (Fig. 4; Ross et al. 2004; Merl et al. 2010; Jomaa et al. 2011). In the case of depletion of L8, the iTRAQ data were confirmed using another tagged assembly factor, Noc2, to purify preribosomes (Supplemental Fig. S3).
FIGURE 3.
Pre-rRNAs present in total cellular extract and in preribosomes purified from L7 or L8 depleted yeast. Pre-rRNA content of total cellular extracts (Inputs, lanes 1–4) and affinity-purified fractions (IPs, lanes 5–8) were analyzed by total RNA extraction and Northern blotting (see Materials and Methods). Identical signal intensities in the Input and IP lanes reflect retention of 4% of the respective pre-rRNA loaded on the affinity matrix. Pre-RNAs and rRNAs that were detected are indicated on the left, and oligonucleotide primers used are indicated on the right.
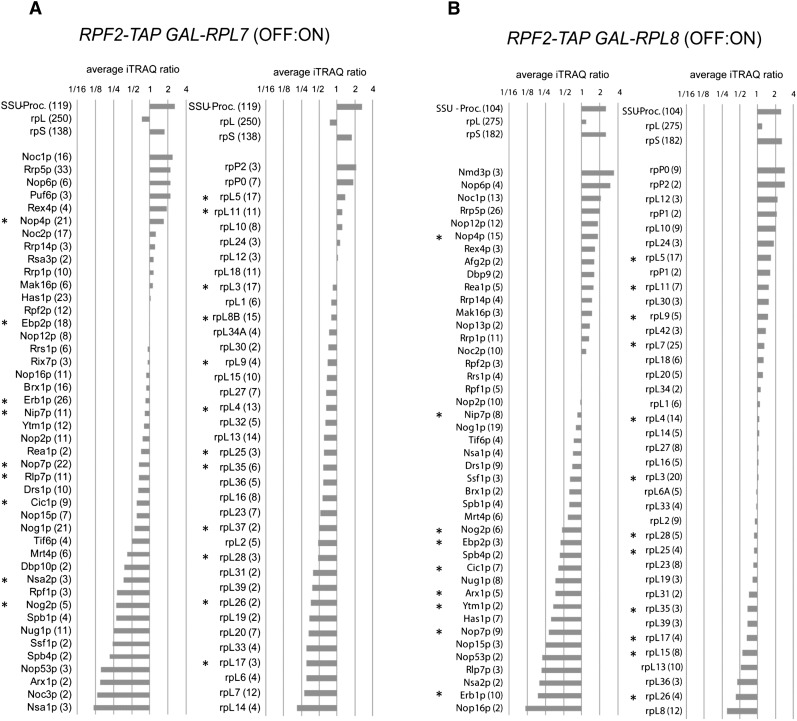
FIGURE 4.
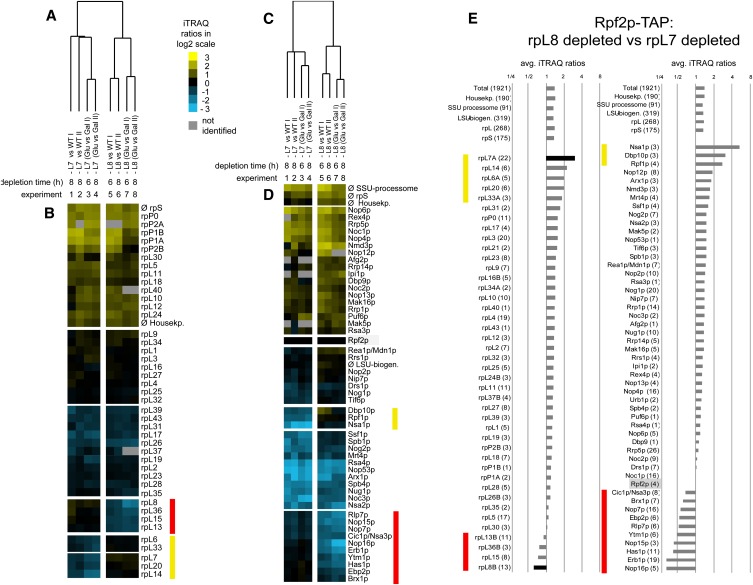
Changes in composition of preribosomal particles upon depletion of L7 or L8, assayed by iTRAQ mass spectrometry. Yeast strains JWY8309 (RPF2-TAP), JWY8492 (GAL-RPL7 RPF2-TAP), and JWY8493 (GAL-RPL8 RPF2-TAP) were cultivated overnight in galactose-containing medium (YEPGal) and were further incubated for 8 h either in YEPGal (on) or in glucose-containing medium (YEPGlu, off) to shut down expression of RPL7 or RPL8, respectively. Rpf2-TAP and associated preribosomal particles were then affinity-purified from corresponding cellular extracts and further processed for comparative protein analyses by semiquantitative mass spectrometry using iTRAQ reagents (see Materials and Methods). (A) Changes in composition of preribosomal particles when L7 is depleted. (B) Changes in composition of preribosomal particles when L8 is depleted. Average iTRAQ ratios of respective proteins identified by more than one peptide are indicated. Numbers in parentheses after protein names indicate the number of peptides by which the respective protein was identified (confidence interval >95%). The average iTRAQ ratios of all peptides identified from 90S/SSU processome components (SSU-Proc.), LSU r-proteins (rpL), and SSU r-proteins (rpS) are indicated on the left. Asterisks indicate proteins also assayed by Western blotting (see Fig. 5B).
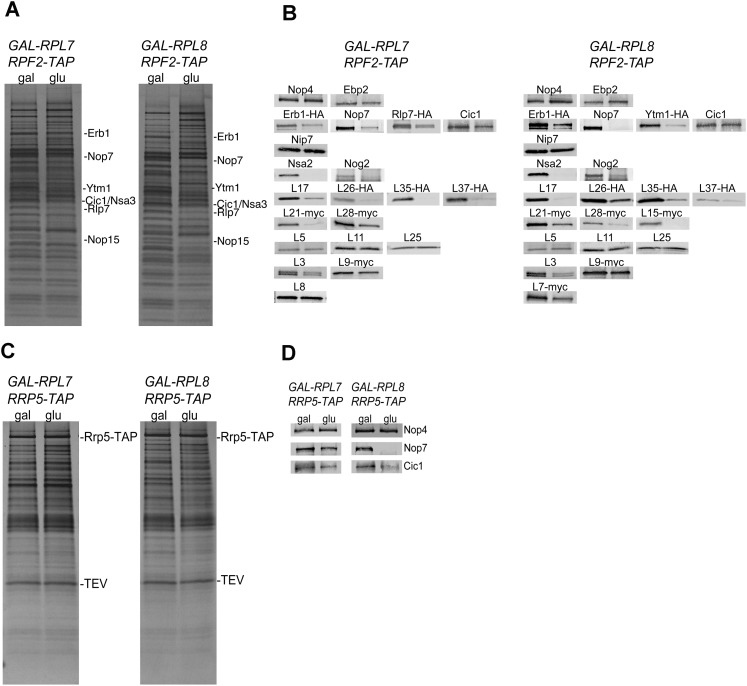
We also used SDS-PAGE to separate and display proteins present in these preribosomes (Fig. 5A). Polypeptides present in the individual gel bands were previously identified by mass spectrometry (Harnpicharnchai et al. 2001). By these means, differences in amounts of many of the ∼45 r-proteins and the ∼70 assembly factors routinely found in 66S preribosomes (for review, see Henras et al. 2008) could be surveyed. Changes in amounts of a subset of the proteins identified by either or both approaches were further tested by Western blotting (Fig. 5B).
FIGURE 5.
Changes in composition of preribosomal particles upon depletion of L7 or L8, assayed by SDS-PAGE and Western blot analysis. (A) SDS-PAGE of proteins in preribosomes from wild-type cells, or from L7- or L8-depleted yeast. Preribosomes were purified from JWY8492 (GAL-RPL7 RPF2-TAP), JWY8493 (GAL-RPL8 RPF2-TAP), and JWY8591 (GAL-RPL8 RPF2-TAP), TCA-precipitated, subjected to electrophoresis on 4%–20% Novex polyacrylamide SDS denaturing gels, and stained with silver. The six A3 factors, which decrease in both depletion strains, are labeled. Other polypeptides that either increase or decrease in pre-rRNPs from the depleted strains are included in Supplemental Table S2. (B) Western analysis of changes in preribosomal proteins upon depletion of L7 or L8. As described in A above, protein constituents of preribosomes purified from L7- or L8-depleted yeast were resolved by SDS-PAGE, transferred to nitrocellulose membranes, and detected using antibodies against specific proteins or with anti-sera against epitope tags attached to proteins. Each blot was cut into small sections based on the known mobility of each protein. For each protein shown, controls confirmed equal loading on the gel. (C) Preribosomes were purified from JWY9832 (GAL-RPL7 RRP5-TAP) and JWY9835 (GAL-RPL8 RRP5-TAP), and proteins were separated as described in A. (D) Western analysis of changes in constituents of early preribosomal particles upon depletion of L7 or L8, detected using antibodies against specific proteins or with anti-sera against epitope tags attached to proteins. Each blot was cut into small sections based on the known mobility of each protein. For each protein shown, controls confirmed equal loading on the gel.
Together, our combined approaches to survey the consequences of depleting L7 or L8 demonstrated the following specific effects on preribosomal particles: (1) Most assembly factors and r-proteins remained in preribosomes lacking either protein (Fig. 5A). (2) Assembly factors that function in subunit biogenesis before the 27SA3 pre-rRNA processing step were unchanged or accumulated in the mutant pre-rRNPs (Figs. 4, 5A,B; Supplemental Table S2). The components of early 90S particles increased, on average, more than twofold after depletion of either L7 or L8 (Fig. 4A,B). This indicates that preribosomes purified from mutant strains were enriched for early assembly intermediates, consistent with the accumulation of 35S pre-rRNA after the shift to glucose (Fig. 3, right panel). (3) The six interdependent assembly factors specifically required for processing of 27SA3 pre-rRNA, Nop7, Ytm1, Erb1, Nop15, Rlp7, and Nsa3 (Sahasranaman et al. 2011), were greatly diminished (Figs. 4, 5A,B; Supplemental Table S2), consistent with the 27SA3 pre-rRNA processing defect observed upon depletion of L7 or L8. (4) Proteins that predominantly associate with preribosomes after completion of the 27SA3 pre-rRNA processing step and are required for subsequent steps in pre-rRNA processing (Nog2, Nsa2, Nug1, and Arx1) (Baßler et al. 2001; Saveanu et al. 2001; Nissan et al. 2002; Lebreton et al. 2006) were diminished from the mutant particles (Figs. 4, 5B; Supplemental Table S2). (5) Four ribosomal proteins, L17, L26, L35, and L37, decreased in the absence of either L7 or L8 (Figs. 4, 5B). These proteins bind to domain I of 25S rRNA and surround the polypeptide exit tunnel of ribosomes (Ben-Shem et al. 2011). (6) Ribosomal proteins L6, L14, L20, and L33, adjacent to L7 and bound to domain II of 25S rRNA, were significantly decreased in the absence of L7 (Fig. 4A). (7) L13, L15, and L36, located in close proximity to each other, and L8 in domain I of rRNA in mature 60S subunits (Ben-Shem et al. 2011) decreased in pre-rRNPs in the L8 depletion strain (Figs. 4B, 5B). (8) L21 and L28 that bind to domains II and V decreased upon depletion of either L7 or L8 (Figs. 4, 5B). (9) In contrast, Western blotting confirmed that amounts of L1, L4, L5, and L11 in preribosomes were unchanged in either depleted strain (Fig. 5B; data not shown).
We also directly compared changes in protein composition of 60S assembly intermediates after depletion of L7 with those from L8-depleted yeast. Statistical analyses of eight comparative proteomic data sets revealed significant differences in the protein composition between particles isolated from strains depleted of L7 vs. L8 (Fig. 6A,C). This comparison confirmed that the strongest effect upon depletion of L7 was on r-proteins L6, L14, L20, and L33 that lie close to L7 and that proteins affected most upon depletion of L8 are L13, L15, and L36, in close proximity to L8 (Fig. 6B,E). Also, this comparison suggests that the absence of L8 affects A3 factors more than the absence of L7, consistent with the location of L8 in mature ribosomes, close to the proposed binding sites of several A3 factors (Fig. 6D,E).
FIGURE 6.
Comparison of changes in protein composition of preribosomes after in vivo depletion of L7 or L8. Preribosomes were affinity-purified from cellular extracts of yeast strains JWY8309 (RPF2-TAP), JWY8492 (GAL-RPL7 RPF2-TAP), and JWY8493 (GAL-RPL8 RPF2-TAP), and the purified fractions were further processed for semiquantitative mass spectrometric analysis as described in Materials and Methods. (A–D) Hierarchical clustering analyses of eight experimental data sets are shown. In each of the eight data sets, the composition of preribosomes affinity-purified from cells depleted of L7 (experiments 1–4, L7) or L8 (experiments 5–8, L8) was compared to preribosomes from cells expressing these proteins. Wild-type strain JWY8309 was used as a reference in experiments 1, 2, 5, and 6, and the mutant strains JWY8492 and JWY8493 grown in galactose-containing media were used as a reference in experiments 3, 4, 7, and 8. iTRAQ ratios of r-proteins or assembly factors identified in at least six out of the eight experiments were combined, and statistical clustering algorithms were applied as described in Materials and Methods. The dendograms in A and C describe the general similarities between the eight data sets. B and D are heat-map representations of relative enrichment (yellow) and depletion (blue) of the individual proteins in each of the eight experiments. (E) Direct comparison of the protein composition of Rpf2-TAP particles purified from extracts of L8-depleted cells with composition of Rpf2-TAP particles purified from extracts of L7-depleted cells. The number of peptides identified for the respective proteins is in parentheses. Yellow bars in B, D, and E mark proteins which were significantly more depleted in preribosomes derived from cells in which expression of L7 was shut down. Red bars in B, D, and E mark proteins which were significantly more depleted in preribosomes derived from cells in which expression of L8 was shut down.
SDS-PAGE and Western analysis of proteins in these preribosomes from L7- or L8-depleted cells closely resembled those previously observed upon depletion of any of the six A3 assembly factors (Sahasranaman et al. 2011), consistent with the similar pre-rRNA processing defects of these assembly factor and r-protein depleted strains. However, this “A3-like” pattern appears to be specific; it is quite different from that in mutants blocked in earlier or later steps of pre-rRNA processing. For example, in ebp2− mutants where early steps in processing are blocked, a large number of r-proteins and assembly factors are missing from preribosomes. In rpf2− mutants, in whom later steps of pre-rRNA processing are blocked, the majority of proteins are present in preribosomes (Zhang et al. 2007; Sahasranaman et al. 2011).
Preribosomes purified from wild-type cells using Rpf2 contain a mixture of early 90S preribosomes and each of the consecutive 66S pre-rRNPs. 27SB pre-rRNA is the most abundant processing intermediate in cells, indicating that 66SB pre-rRNPs containing this pre-rRNA are the longest-lived pre-rRNPs. Thus, a significant fraction of the mixture purified with Rpf2-Tap includes 27SB-containing particles, more than any other pre-rRNP. While using Rpf2-TAP purifications enabled us to survey effects on most pre-60S particles, we could not determine whether decreased amounts of certain proteins in depleted strains (Figs. 4, 5A,B) resulted from failure to assemble early in the pathway, thus preventing a downstream processing step, or else from dissociation upon a later block in assembly. In other words, is the absence of these proteins the cause or the effect of observed defects in pre-rRNA processing? This question is significant given that all A3 factors are presumed to function in 27SA3 pre-rRNA processing. Are they really absent from mutant preribosomes before the 27SA3 processing step could occur?
To address these questions, we tagged another assembly factor, Rrp5, in GAL-RPL7 and GAL-RPL8 strains. Rrp5 enters the ribosome assembly pathway early, in 90S particles containing 35S pre-rRNAs, and is released before 27SA3 pre-rRNA processing occurs (de Boer et al. 2006). Thus, we used TAP-tagged Rrp5 to purify preribosomes enriched for early particles containing mostly 35S and 27SA2 pre-rRNA. When L7 or L8 were depleted, levels of the A3 factors Nop7 and Cic1 decreased in preribosomes purified using Rrp5-Tap (Fig. 5C,D). Since the six A3 factors are interdependent for stable association with preribosomes (Sahasranaman et al. 2011), the absence of any one of them indicates that they are all absent from preribosomes. Thus, this result confirms that depletion of L7 or L8 prevents stable association of A3 factors with early preribosomes, before the pre-rRNA processing step for which they are required.
Nuclear export of 66S preribosomes is impaired upon depletion of L7 or L8
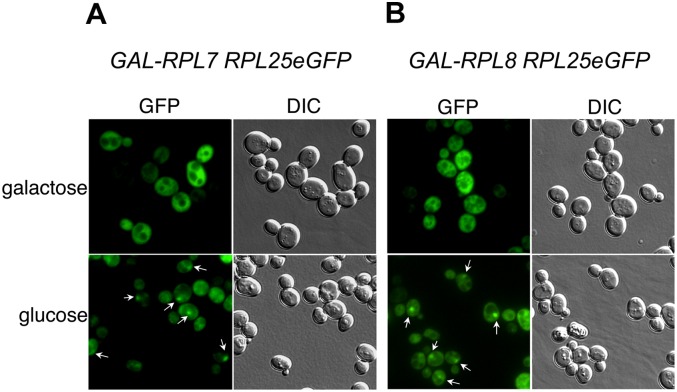
Early steps in 60S ribosomal subunit maturation, including processing of 27SA2, 27SA3, and 27SB pre-rRNA, occur in the nucleolus, followed by release of preribosomes to the nucleoplasm (Gadal et al. 2002; de la Cruz et al. 2005). Rearrangements of pre-rRNP architecture presumably must occur during and immediately after these RNA processing steps to enable subsequent association of the nuclear export machinery with nascent ribosomes (Panse and Johnson 2010). Based on the observed failure to complete early steps of subunit biogenesis in the absence of L7 or L8, we predict that the abortive assembly intermediates might not be competent for nuclear export and would accumulate in the nucleolus or nucleoplasm. To determine whether this is the case, we assayed the localization of ribosomal protein L25-eGFP, a reporter for the location of 66S preribosomes (Gadal et al. 2002), in cells depleted of L7 and L8. Importantly, L25 is stably assembled into preribosomes in the absence of L7 or L8 and thus is a valid reporter in these mutant strains (Fig. 5B). In galactose-containing medium, where L7 and L8 are expressed and ribosomes are properly assembled, eGFP-L25-tagged ribosomes were located predominantly in the cytoplasm, as expected (Fig. 7A,B, top). In contrast, when L7 or L8 were depleted by shifting GAL-RPL7 or GAL-RPL8 yeast from galactose- to glucose-containing medium for 2 h, the fluorescence signal from eGFP-L25 started to localize in the nucleus (Fig. 7A,B, bottom). We conclude that L7 and L8 are necessary for steps in 60S subunit maturation prior to attaining the capability to be exported from the nucleus, as previously reported based on nucleo-cytoplasmic fractionation of pulse-labeled cells depleted of L7 or L8 (Pöll et al. 2009).
FIGURE 7.
Upon depletion of r-proteins L7 or L8, abortive pre-60S ribosome assembly intermediates accumulate in the nucleus. (A) JWY8592 (GAL-RPL7A pRS316-RPL25eGFP) and (B) JWY8593 (GAL-RPL8B pRS314- RPL25eGFP) strains expressing eGFP-tagged r-protein L25 were grown in selective media containing galactose or shifted to glucose for 2 h. Cells were adhered to glass cover slips coated with concanavalin A. Cell images were obtained by a Nikon Eclipse Ti inverted microscope system and analyzed using the NIS Elements image analysis software. GFP signals are shown on the left, and phase contrast images of the cells are shown on the right in each pair of images. White arrows mark the nuclear accumulation of GFP signal in cells shifted to glucose.
DISCUSSION
To begin to investigate the role of r-proteins in the assembly of eukaryotic ribosomes in vivo, we had previously surveyed effects on pre-rRNA processing and localization upon depleting each of 30 different r-proteins from the 60S subunit (Pöll et al. 2009; Zhang et al. 2007; M Gamalinda, J Jakovljevic, R Babiano, J de la Cruz, JL Woolford Jr, in prep.). The depletion phenotypes fell into four groups, affecting early, middle, or late steps in pre-rRNA processing, or having no discernible effect. The challenge before us now is to go beyond rRNA processing, to begin to understand the roles of r-proteins in the dynamics of folding of pre-rRNA, association of r-proteins and assembly factors with pre-rRNA, and how each of these facets of ribosome assembly also relate to each other. In this work, we have examined the effects of depleting the two r-proteins, L7 and L8, on protein constituents of preribosomes. Both r-proteins are required for early steps of 27S pre-rRNA processing and for the formation of stable early 66S assembly intermediates.
L7 and L8 are necessary for association of the A3 assembly factors with preribosomes
Using TAP-tagged Rpf2 to purify preribosomes enabled us to survey effects on most of the nuclear precursors to 60S ribosomal subunits when L7 or L8 were depleted and 27SA3 pre-rRNA processing was perturbed. Several assembly factors that associate with 90S preribosomes early in the subunit maturation pathway and that are thought to exit relatively early, such as Noc1 and Rrp5 (Milkereit et al. 2001; de Boer et al. 2006), are present in greater amounts in preribosomes from the depleted strains than in wild-type preribosomes (Fig. 4; Supplemental Table S2). Conversely, amounts of Nsa2, Nog2, Nug1, and Arx1, proteins that enter preribosomes after completion of 27SA3 pre-rRNA processing, are significantly decreased in preribosomes from the depleted cells (Figs. 4, 5B; Supplemental Table S2). Thus, pre-rRNP assembly intermediates that form before 27SA3 pre-rRNA processing occurs are accumulating, and the pre-rRNPs that form just before 27SB pre-rRNA processing and afterward are largely absent.
However, two classes of proteins that associate early with preribosomes are greatly diminished in pre-rRNPs in the absence of either L7 or L8: the A3 cluster of six assembly factors required for processing of 27SA3 pre-rRNA, and four r-proteins that bind primarily to domain I of 25S rRNA and surround the polypeptide exit tunnel in mature 60S subunits (Figs. 4, 5A,B, 8A).
FIGURE 8.
Ribosomal proteins bound to domain I and II are diminished upon depletion of L7 or L8. (A) The structure of the mature 60S subunit is shown with L7 and L8 indicated in dark green and dark blue, respectively. The subset of r-proteins that fail to associate with preribosomes in the absence of L7 or L8 is shown. Those that bind to domain I around the polypeptide exit tunnel and are absent when L7 and L8 are depleted are in hues of pink. R-proteins that have been shown to diminish when L7 is depleted are colored in shades of green, and those diminished when L8 is depleted are colored in shades of blue. The 5S and 5.8S rRNAs are black and dark blue, respectively. Domains I, II, III, and V of 25S rRNA are shown in light blue, green, teal, and orange, respectively. (B) The location of L7 and L8 relative to the predicted binding sites of A3 assembly factors Erb1 and Nop7. rRNAs are colored as described above. The Erb1 and Nop7 CRAC sites are represented as light blue and teal spheres, respectively; the colors correspond to the 25S rRNA domains with which they cross-link.
Several results indicate that in L8 depleted cells the decrease of the A3 factors in preribosomes might be directly triggered by the failure of Erb1 to associate with preribosomes: (1) Erb1 most likely binds immediately adjacent to L8 in preribosomes; it cross-links to helices 21 and 22 (Granneman et al. 2011), which are next to L8 in mature 60S subunits (Fig. 8B; Ben-Shem et al. 2011). L8 is located near the proximal stem formed by base-pairing between the 3′ end of 5.8S rRNA and the 5′ end of 25S rRNA (Fig. 8A,B). (2) Erb1, Ytm1, and Nop7 are adjacent to each other. These three proteins form a heterotrimeric complex (Miles et al. 2005), with Nop7 and Ytm1 bound directly to Erb1 (Tang et al. 2008). Consistent with these interactions, the RNA binding sites of Nop7 and Erb1 are near each other. Nop7 cross-links to helix 54 in domain III of 25S rRNA, which is 80–100 Å from the Erb1 binding site (Fig. 8B; Granneman et al. 2011). (3) Nop15 and Cic1 are also likely to be near L8 and the other A3 assembly factors. These two proteins bind to RNA sequences in the ITS2 pre-rRNA spacer (Granneman et al. 2011), near each other (Tarassov et al. 2008). ITS2 lies between the 3′ end of 5.8S rRNA sequences and the 5′ end of 25S rRNA sequences in pre-rRNA molecules. Thus, both ends of ITS2 lie near L8 and Erb1. Therefore, we hypothesize that binding of L8 near the proximal stem enables stable assembly of Erb1 and its interdependent A3 factors with preribosomes. Interestingly, the converse is not true; L8 is not dependent on Erb1 or other A3 factors for association with preribosomes (Sahasranaman et al. 2011).
Upon depletion of L7, the A3 factors are affected but to a lesser extent than upon depletion of L8. The effect of L7 on A3 factors might occur by a mechanism different from that when L8 is depleted. L7 binds to helices 40, 41, and 45 in domain II of 25S rRNA (Fig. 8; Ben-Shem et al. 2011) and thus is not likely to be near the known cross-linking sites of Erb1, Nop7, Nop15, or Cic1. However, the location of Ytm1 and Rlp7 within preribosomes is not known, and it is likely that we have not yet identified all of the RNA binding sites of the A3 assembly factors. Alternatively, L7 may influence binding of A3 factors to preribosomes indirectly, through long-range interactions such as those that occur in ribosome assembly in vitro (Jagannathan and Culver 2004).
Ribosomal proteins adjacent to L8 and bound to domain I of 25S rRNA fail to assemble into preribosomes in the absence of L8
Other r-proteins strongly affected by depletion of L8 are L13, L15, L21, L28, and L36 (Figs. 4B, 5B). Amounts of L7 decrease reproducibly, although to a lesser extent than the others (Fig. 5B). These seven r-proteins form a chain of protein-protein and protein-RNA interactions extending from L8 to L7, along the junction between domains I plus II and domain V of 25S rRNA (Fig. 8A). L15 and L36 contact L8 directly. L15 and L36 also bind directly to L13, which contacts L28 (Fig. 8A). L21 is adjacent to L28 and directly contacts L7. L8, L15, and L36 bind to domains I and V of 25S rRNA, L13 contacts domains I and II, and L21 and L28 bind to domains II and V (Fig. 8A). The presence of L8 (and nearby Erb1) may be critical to direct assembly of this neighborhood of r-proteins and rRNA in domains I and II and to stabilize tertiary contacts between domains I plus II and V.
Ribosomal proteins adjacent to L7 and bound to domain II of 25S rRNA are diminished from preribosomes in the absence of L7
Upon depletion of L7, amounts of r-proteins L6, L14, L20, L21, L28, and L33, bound to domain II of 25S rRNA, are diminished in preribosomes (Figs. 4A, 5B). Amounts of L8 in preribosomes are unchanged (Fig. 5B). L13, L15, and L36 are also unchanged (Figs. 4A, 6). Thus, it remains unclear whether the domino effect propagated from L8 to L7 is reciprocal. However, it is clear that L7 is important for assembly of nearby r-proteins, L6, L14, L20, and L33, in this case, to establish a stable neighborhood including domain II of 25S rRNA (Fig. 8A).
Four ribosomal proteins surrounding the polypeptide exit tunnel and bound to domain I of 25S rRNA do not assemble in the absence of L7 or L8
The four r-proteins whose association with preribosomes is strongly affected when either L7 or L8 are depleted are L17, L26, L35, and L37 (Figs. 4, 5B). These r-proteins are adjacent to each other, associated mostly with domain I of 25S/5.8S rRNAs, and surround the polypeptide exit tunnel of 60S ribosomal subunits (Fig. 8A; Ben Shem et al. 2011). L17, L26, L35, and L37 are also the r-proteins that decrease most upon depletion of the A3 assembly factors (Sahasranaman et al. 2011). Thus, it seems most likely that the failure of these four r-proteins to assemble into preribosomes when L7 or L8 are depleted is due to the inability of the A3 factors to effectively associate with pre-rRNPs. The Erb1 binding site in domain I of 25S rRNA and the Nop7 binding site in domain III are near these r-proteins (Fig. 8B; Granneman et al. 2011). However, thus far there is no evidence for direct interaction of any of these r-proteins with A3 factors; they do not interact in two-hybrid assays (J Dembowski, pers. comm.). More likely, binding of Erb1 and Nop7 significantly influences the establishment of proper conformations of rRNA domains I and III, perhaps in part by bridging these two domains (Granneman et al. 2011), to enable stable assembly of L17, L26, L35, and L37 with this neighborhood of rRNA. Alternatively, the effects of depletions of L7 or L8 could be propagated through their neighbors to these four r-proteins. For example, in the crystal structure of mature ribosomes, a long amino-terminal extension of L17 is in close proximity to L6, one of the proteins most affected in the absence of L7 (Figs. 4A, 8A). Also, a similar long extension of L35 lies very close to L13, an r-protein bound to domain I and strongly affected in the absence of L8 (Figs. 4B, 8A). Thus, the inability of L6 or L13 to properly assemble in the absence of L7 or L8, respectively, might perturb stable association of L17 or L35 with preribosomes. However, the situation is probably more complex, since L17, L26, L35, and L37 are not interdependent for their stable association with preribosomes (M Gamalinda, J Jakovljevic, R Babiano, J de la Cruz, JL Woolford Jr, in prep.).
Interestingly, not all of domain III of rRNA seems to be affected by depletion of L7 or L8. For example, L25, which binds domain III immediately adjacent to the Nop7 binding site (Ben-Shem et al. 2011) and exhibits genetic interactions with Nop7 (Oeffinger et al. 2002), is present in wild-type amounts when Nop7, L7, or L8 are depleted (Fig. 5B).
Relationships between defects in assembly and pre-rRNA processing upon depletion of L7 or L8
How might decreased amounts of A3 factors and r-proteins associated with domains I and II of rRNA affect processing and turnover of 27S pre-rRNAs when L7 or L8 are depleted? In wild-type cells, 27SA2 pre-rRNA is likely created by cleavage at the A2 site within 32S pre-rRNA by the endonuclease Rcl1 (Horn et al. 2011). Subsequent processing proceeds by two alternative pathways (Fig. 2A). Approximately 80%–90% of 27SA2 pre-rRNA molecules are cleaved by the endonuclease MRP at the A3 site in ITS1 to generate 27SA3 pre-rRNA (Schmitt and Clayton 1993; Chu et al. 1994; Lygerou et al. 1996). Subsequently, the exonucleases Rat1, Rrp17, and, under some conditions, Xrn1 remove 77 nt from the 5′ end of this RNA, stopping precisely at the B1s site (Henry et al. 1994; Oeffinger et al. 2009). About 10%–20% of 27SA2 pre-rRNAs undergo endonuclease cleavage at the B1L site, 6 nt upstream of the B1s site, to generate 27SBL pre-rRNA. Subsequently, ITS2 is removed from both 27SB pre-rRNAs.
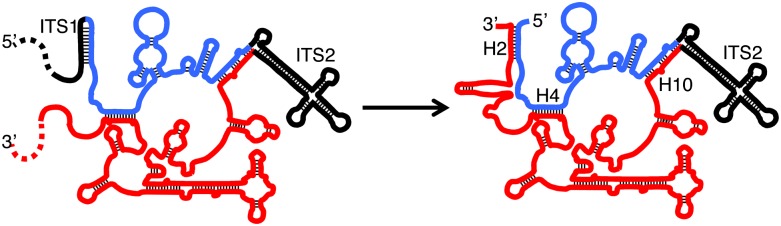
When L7 or L8 are depleted, the relative amounts of 27SA2 and 27SA3 pre-rRNA are increased, and amounts of 27SBS pre-rRNA are decreased. We speculate that the increased steady-state amounts of 27SA3 might result from decreased efficiency of processing of 27SA3 pre-rRNA to 27SBS pre-rRNA. Successful processing may require a conformational switch in sequences near the 5′ end of the 27SA3 pre-rRNA. The ITS1 sequences removed from the 5′ end of 27SA3 pre-rRNA are predicted to base-pair with nucleotides of what becomes the 5′ end of 5.8S rRNA (Fig. 9, left; Yeh et al. 1990; van Nues et al. 1994). In mature 60S subunits, these same sequences at the 5′ end of 5.8S rRNA base-pair with nucleotides in 25S rRNA to form helix 2, which is bound to r-protein L17 (Fig. 9, right; Ben-Shem et al. 2011). This conformational switch to form helix 2, as well as proper folding of the rest of domain I, including base-pairing between 5.8S and 25S rRNAs to form helices 4 and 10 (Fig. 9), may require the presence of Erb1, other A3 factors, and the r-proteins that bind to domain I of 25S rRNA. In the absence of these proteins, ITS1 sequences may not be displaced from pairing with the 5′ end of 5.8S rRNA and thus might not be presented as an efficient substrate for processing.
FIGURE 9.
Model for creation of helix 2 by removing ITS1. The ITS1 sequences removed from the 5′ end of 27SA3 pre-rRNA are predicted to base-pair with nucleotides of what becomes the 5′ end of 5.8S rRNA (left). In mature 60S subunits, these same sequences at the 5′ end of 5.8S rRNA base-pair with nucleotides in 25S rRNA to form helix 2 (right). This conformational switch to form helix 2, as well as proper folding of the rest of domain I, may require the presence of A3 factors and the r-proteins that bind to domains I and II of 25S rRNA. 5.8S rRNA is colored in blue, and 25S rRNA is colored in red. H2, H4, and H10 are helix 2, helix 4, and helix 10, respectively. Base-pairing is represented by black lines. Dashed lines indicate that the sequences continue further upstream or downstream.
Greater amounts of A3 ends relative to A2 and B1S ends observed upon depletion of L7 or L8 might also result from a switch from cotranscriptional to post-transcriptional processing of pre-rRNA (Osheim et al. 2004; Koš and Tollervey 2010). The higher levels of 23S pre-rRNA observed in pulse-chase experiments with either depleted strain indicate that the relative rate of production of 27SA3 increases. 23S rRNA results from cleavage at the A3 site before processing at the A0, A1, and A2 sites. The absence of the A3 factors and L7 and L8 from the early assembly intermediates could influence the relative timing of pre-rRNA processing events at the A0, A1, A2, and A3 sites to increase the fraction of 23S and 27SA3 pre-rRNA produced relative to other precursors. This timing may be dictated by the relative frequency of cotranscriptional processing at the A2 site prior to cleavage at A3 versus post-transcriptional cleavage of 35S pre-rRNA at the A3 site before the cleavage at A0, A1, and A2 sites. One possibility is that, in wild-type cells, the A3 factors normally might act as negative regulators of cleavage at the A3 site to delay cleavage until after proper processing at the A0, A1, and A2 sites. Alternatively, abortive intermediates lacking the A3 factors and r-proteins normally bound to domain I might sequester factors required for efficient processing at the A0, A1, and A2 sites. We note that amounts of 23S pre-rRNA also increase upon depletion or inactivation of Nop7, Ytm1, Erb1, Nop15, Nsa3/Cic1, L17, L35, and L37 (Dunbar et al. 2000; Harnpicharnchai et al. 2001; Pestov et al. 2001; Adams et al. 2002; Gadal et al. 2002; Oeffinger et al. 2002; Fatica et al. 2003; Oeffinger and Tollervey 2003; Miles et al. 2005; Sahasranaman et al. 2011; M Gamalinda, J Jakovljevic, R Babiano, J de la Cruz, JL Woolford Jr, in prep.).
The substantial decrease in B1S 5′ ends upon depletion of L7 or L8 most likely results from a combination of slightly less efficient processing and substantial turnover of 27SA3 or 27SB pre-rRNAs. Our results cannot distinguish which of these pre-rRNAs is turned over. However, it seems more likely that 27SA3 pre-rRNA is degraded, since little 27SB was detected, but some 27SA pre-rRNA was evident by pulse fractionation described in Pöll et al. (2009) and in our pulse-chase experiments (Fig. 2E). Turnover may be triggered by misfolding of rRNA domains I and II in the absence of Erb1, other A3 factors, and r-proteins normally positioned there. The Rat1 nuclease may participate in degradation of this pre-rRNA. Rat1 is present in preribosomes even when A3 factors are absent (Sahasranaman et al. 2011), so it is likely to be present when L7 or L8 are depleted. Normally, Rat1-mediated 5′ to 3′ exonucleolytic processing of 27SA3 pre-rRNA halts precisely at the B1S site, perhaps due to the presence of rRNA secondary structure, including helix 2, and r-proteins bound to and stabilizing these RNA structures. In the absence of these proteins and secondary structures, Rat1 might switch from a processing enzyme to a turnover machine, proceeding past the B1S site to degrade the aberrant pre-rRNA substrate (Pöll et al. 2009; Sahasranaman et al. 2011).
The absence of assembly factors and r-proteins normally bound to ITS2 and domains I or II might expose these RNA sequences to endonucleases to create target sites for turnover of 27SA pre-rRNAs. In addition, turnover might also occur via 3′ exonucleases. The 3′ end of 25S rRNA (which is the same as the 3′ end of 27S pre-rRNA) lies near the neighborhood surrounding L7 and L17.
CONCLUSIONS
We believe that the RNA processing and turnover phenotypes observed upon depletion of L7 or L8 result from small effects on the timing and efficiency of early processing steps and from much more significant effects on the stability of early assembly intermediates containing 27SA3 pre-rRNA. The primary functions of L7 and L8 may be to establish and stabilize rRNA structures required for the stable association of nearby r-proteins and the A3 assembly factors. In turn, these other proteins may stabilize the secondary and tertiary structure of domains I and II of 25S rRNA and bridge interactions with adjacent domains III and IV. Four of the six A3 factors are bound to domains I and III, and to ITS2, and most likely function in a similar way to establish stable functional structures of these rRNAs and to form bridges between domains of rRNA during subunit assembly.
Upon depletion of L7 or L8, the six A3 factors and r-proteins that normally bind domains I and II fail to stably associate with early precursor particles. Nevertheless, pre-rRNAs in these pre-rRNPs can still undergo cleavage at the A3 site, as well as eventual processing at the A0, A1, and A2 sites to produce 18S rRNA and 40S ribosomal subunits. However, the failure of domains I or II to assemble with r-proteins signals turnover of pre-60S assembly intermediates, presumably by a quality control surveillance machinery.
MATERIALS AND METHODS
Construction and growth of yeast strains
Yeast strains used in this study were derived from JWY6147 (Mata ura3-52 trp1-Δ101 lys2-801 his3-Δ200 leu2-Δ1) or BY4741 (MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0) and are listed in Supplemental Table S1. Unless otherwise noted, yeast were grown at 30°C in YEPGlu (2% dextrose, 2% peptone, and 1% yeast extract), or in YEPGal medium (2% galactose, 2% peptone, and 1% yeast extract). Cells were harvested in mid-logarithmic phase growth, at 3–5 × 107 cells/mL, except where otherwise indicated.
Ribosomal protein genes RPL7A (YGL076c), RPL7B (YPL198w), RPL8A (YHL033c), and RPL8B (YLL045c) were deleted by precisely replacing their ORFs with KANMX6 expressing resistance to geneticin (G418). The KANMX6 gene was amplified by PCR as described in Longtine et al. (1998). The resulting PCR products were transformed into yeast strain JWY6147, and G418-resistant transformants were identified. Correct gene disruptions were verified by genomic PCR. Yeast containing a deletion of RPL7A was purchased from Euroscarf.
Yeast strains conditional for expression of L7 or L8 were constructed either as described in Pöll et al. (2009), or as described in Longtine et al. (1998). Briefly, sequences containing the selectable marker TRP1 (for RPL7) or HIS3 (for RPL8), plus the GAL1 promoter sequence followed by an ATG and codons encoding 3HA were amplified by PCR. The PCR products were transformed into yeast strains JWY7771 (rpl7bΔ) or JWY8563 (rpl8aΔ RPF2-TAP), and the strains were grown in selective media containing galactose. Trp+ or His+ transformants were screened for correct integration of the GAL1 promoter and the 3HA tag upstream and in-frame with the r-protein genes by Western blotting with anti-HA antisera.
Yeast strains expressing C-terminal TAP-tagged Rpf2 or Rrp5, C-terminal triple hemagglutinin (3HA)-tagged proteins, or C-terminal 13Myc-tagged proteins were created by PCR of the tag sequence and a selectable marker (URA3 for the TAP tag, and HIS3 or TRP1 for other tags), transformation, and selection as described in Rigaut et al. (1999) and Longtine et al. (1998). Transformants were screened by Western blotting for those expressing the tagged proteins. Strains containing tagged proteins were monitored for any growth defect that would arise because the tagged protein is not functional; no defects were observed.
Depletion of ribosomal proteins L7 or L8
Strains JWY8423 (GAL1-RPL7), TY1097 (GAL-RPL8), and JWY8591 (GAL-RPL8) were grown at 30°C in YEPGal liquid medium, pelleted, suspended in YEPGlu, and shifted for indicated times to deplete ribosomal proteins L7 or L8 in vivo. As a control, cells were also harvested after growth in YEPGal medium. Cell growth was monitored using a spectrophotometer Spectronic Genesys 5. For the initial screen for growth in liquid medium to confirm that L7 and L8 are essential, cultures were continuously diluted to ensure exponential growth. Samples were collected at indicated times, proteins were extracted, and Western blots were performed to detect HA-tagged L7 or L8. Antibody against assembly factor Ebp2, not affected by depletion of L7 or L8, was used as a loading control.
Sucrose gradient assays of ribosomes and polyribosomes
Preribosomes, ribosomes, and polyribosomes were fractionated from 40 OD254 units of whole-cell extracts on 7%–47% (w/v) sucrose gradients as described in Deshmukh et al. (1993), with the following modifications. Cycloheximide (5 mg) was added to cultures 20 min before harvesting cells. A Teledyne ISCO Foxy R1 density gradient fractionator was used to fractionate and analyze gradients with continuous monitoring at OD254.
Assaying pre-rRNA processing and turnover
Steady-state levels of pre-rRNA and rRNA were assayed by Northern blotting and primer extension, and synthesis and turnover of pre-rRNA and rRNA were measured by pulse-chase assays, as described in Horsey et al. (2004), with the following modifications. A Nano Drop 2000C spectrophotometer (Thermo Scientific) was used to quantify amounts of total RNA in samples. Five μg of RNA per sample were used for each reaction. The oligonucleotide used for primer extension and as a probe for 35S and 27S pre-rRNA by Northern hybridization is complementary to sequences within ITS2 (Oligo 7, Fig. 1A in de la Cruz et al. 1998). The oligonucletide used for primer extension to detect the 5′ end of 35S pre-rRNA is described in Oakes et al. (1999). The annealing step during primer extension reactions was decreased from 90 min to 15 min.
RNA extractions and Northern analyses of a part of the cellular extracts (1%) and bead fractions shown in Figure 4 were done as described in Pöll et al. (2009). The sequence identity of oligos used for detection of different pre-rRNAs is indicated in Figure 4.
32P-labeled DNA oligonucleotide probes used are the following:
O 68 TTTCGCTGCGTTCTTCAT
O 205 CATGGCTTAATCTTTGAGAC
O 207 TGTTACCTCTGGGCCC
O 210 GGCCAGCAATTTCAAGTTA
O 212 CTCCGCTTATTGATATGC
O 1440 CGGTCTCCACGGTGAAAGC
O 2474 TTAACTACAGTTGATCGG
Affinity purification of preribosomes
Ribosome assembly intermediates were affinity-purified from whole-cell extracts with magnetic Dynabeads (Invitrogen), using TAP-tagged assembly factors Rpf2 or Rrp5, as described in Sahasranaman et al. (2011).
Protein extraction, SDS-PAGE, and Western blot analysis
Proteins in whole-cell extracts were prepared for gel electrophoresis by previously established methods, as described in Ausubel et al. (1994). Proteins were recovered from affinity-purified preribosomes by precipitation with 10% TCA and were subsequently suspended in SDS sample buffer. Proteins were resolved by SDS-PAGE on 4%–20%Tris-Glycine Novex precast gels (Invitrogen), and stained with silver by standard methods. To assay Nog2 protein by Western blotting, NuPage 4%–12% Bis-Tris gels (Invitrogen) were used, since Nog2 comigrates with IgG on 4%–20% Novex gels. Proteins from whole-cell extracts or from purified preribosomes were assayed by Western blot analysis (Ausubel et al. 1994), with the following modification. To enable detection of multiple different proteins on one blot and to conserve anti-serum by using a lower volume of blotting buffer, after electroblotting, nitrocellulose membranes were cut into smaller sections based on the previously established mobility of the different proteins. TAP-tagged proteins were detected using alkaline phosphatase conjugated to IgG (Pierce). HA-tagged proteins were identified with mouse monoclonal antibody 12CA5 and myc-tagged proteins with 9e10 antibody. Otherwise, antibodies specific for r-proteins or ribosomal assembly factors were used. AP-conjugated anti-mouse or anti-rabbit secondary antibodies (Promega) were used, and colorimetric detection was performed using NBT and BCIP (Promega).
Affinity-purification of preribosomes and subsequent semiquantitative mass spectrometric analyses
Rpf2-TAP and associated preribosomal particles were purified from total cellular extracts in one step using rabbit IgG coupled to magnetic beads as described (Oeffinger et al. 2007). The cell pellet corresponding to 1 L yeast culture with OD600 = 0.8–1.0 was resuspended in 1.5 mL of cold MB buffer (20 mM Tris HCl pH 8, 200 mM KCl, 5 mM MgOAc, 2 mM Benzamidine, 1 mM PMSF, and 0.04 U/μL RNasin) per g of cell pellet; 0.8 mL of this cell suspension was added to 1.4 mL glass beads (Ø 0.75–1 mm) and divided into 2-mL reaction tubes. A cell lysate was prepared by vigorous shaking of the cell suspension in an IKA-Vibrax VXR shaker at 4°C for 15 min, followed by 2 min on ice. This procedure was repeated twice. The cell lysate was cleared from cell debris by two centrifugation steps, 1 × 5 min at 14,000 rpm and 1 × 10 min at 14,000 rpm. The protein concentration of the cleared lysate was determined using the Bradford assay. Triton X-100 (0.5%) and Tween 20 (0.1%) was added to the cell lysate. The whole amount of cell lysate (typically 1 mL with 30–50 mg of total protein) was incubated for 1 h at 4°C with 200 μL of an IgG (rabbit serum, I5006-100MG, Sigma)-coupled magnetic beads slurry (1 μm BcMag, FC-102, Bioclone) equilibrated in MB buffer containing 0.5% Triton X-100 and 0.1% Tween. The beads were washed four times with 1 mL cold MB buffer with 0.5% Triton X-100 and 0.1% Tween 20. Twenty percent of the suspension was taken out for RNA analyses by Northern blotting, shown in Figure 3. The other part of the suspension was washed two times with AC buffer (100 mM NH4OAc pH 7.4, 0.1 mM MgCl2) to remove remaining salt from the sample. Bound proteins were eluted two times with 500 μL of freshly prepared 500 mM NH4OH solution for 20 min at RT. Both eluate fractions were pooled, lyophilized overnight, and further processed for semiquantitative mass spectrometric protein analyses as described (Jakob et al. 2012). Hierarchical clustering analysis of semiquantitative mass spectrometry data sets derived from several experiments was done as in Jakob et al. (2012), using cluster 3.0 software (Eisen et al. 1998). Cluster analysis and display of genome-wide expression patterns was performed according to Eisen et al. (1998). Data were normalized before cluster analyses by setting the respective Rpf2p-TAP iTRAQ ratios to one and using the “log2 transform data” settings in cluster 3.0. The “median center array” option was used for processing of data shown in Figure 4, A and B. City block distance and centroid linkage settings were used for array clustering shown in Figure 4, A and C. Cluster visualization was done with Java Treeview (see http://www.eisenlab.org/eisen/?page_id=42).
Fluorescence microscopy
The reporter system described in Gadal et al. (2002) was used to screen for nucleolar release and nuclear export of preribosomes upon depletion of L7 or L8. GAL-RPL7 (JWY8423) and GAL-RPL8 (JWY8591) strains were transformed with plasmids pRS316-RPL25eGFP and pRS314-RPL25eGFP, respectively. These plasmids bear an e-GFP fusion of r-protein L25 and either the URA3 gene (pRS316) or the TRP1 gene (pRS314). Transformants positive for the selectable marker were grown in appropriate minimal media containing galactose and shifted to minimal media containing glucose for 2 h. Cells were imaged using a Nikon Eclipse Ti inverted microscope system and analyzed using NIS Elements image analysis software.
Coimmunoprecipitation of pre-rRNA and rRNA
Yeast strains expressing TAP-tagged r-protein L7A or L8A (JWY7148 or JWY7158, respectively) were grown in rich medium containing galactose, harvested, and subjected to affinity-purification, as described in Sahasranaman et al. (2011). RNA enriched from these purified preribosomes and ribosomes was extracted, as described in Sahasranaman et al. (2011), and was assayed by primer extension or Northern hybridization, as described in Horsey et al. (2004).
PyMOL
PyMOL images of the structure of yeast mature 60S ribosomal subunits were generated using PDB file 3U5D and 3U5E (Ben-Shem et al. 2011).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
This work is dedicated in memory of Dr. Maria Rodrígues-Mateos, our friend and fellow scientist. We thank the following people for their generous gifts of antibodies: Jesús de la Cruz and Patrick Linder (Has1), David Goldfarb (Nip7), Janine Maddock (Nog1), Michael McAlear (Ebp2), Cosmin Saveanu and Micheline Fromont-Racine (Rlp24, Nsa2, Tif6, and Nog2), Juan Pedro Ballesta (rpL12), Arlen Johnson (rpL8), Francois Lacroute (rpL1), Lasse Lindahl (rpL4), Sabine Rospert (rpL17), K. Siegers (rpL25), Maurice Swanson (rpL39), Elizabeth Tosta (Cic1/Nsa3), Francois Lacroute (rpL1), Jeremy Thorner (Fpr3), and Jonathan Warner (rpL3). We also thank Kara Bernstein for use of her microscope and Jesús de la Cruz and members of our laboratory for critical discussions and reading of the manuscript. This work was supported in part by the Deutsche Forschungsgemeinschaft in the research unit FOR1068 (http://www.dfg.de), by NSF grant MCB0818534 to J.L.W, and by The Richard King Mellon Foundation Presidential Graduate Fellowship in the Life Sciences and the Semon H. Stupakoff Scholarship to J.T.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.032540.112.
REFERENCES
- Adams CC, Jakovljevic J, Roman J, Harnpicharnchai P, Woolford JL Jr 2002. Saccharomyces cerevisiae nucleolar protein Nop7p is necessary for biogenesis of 60S ribosomal subunits. RNA 8: 150–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adilakshmi T, Ramaswamy P, Woodson SA 2005. Protein-independent folding pathway of the 16S rRNA 5′ domain. J Mol Biol 351: 508–519 [DOI] [PubMed] [Google Scholar]
- Arevalo SG, Warner JR 1990. Ribosomal protein L4 of Saccharomyces cerevisiae: The gene and its protein. Nucleic Acids Res 18: 1447–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K 1994. Current protocols in molecular biology. Wiley, New York [Google Scholar]
- Babiano R, de la Cruz J 2010. Ribosomal protein L35 is required for 27SB pre-rRNA processing in Saccharomyces cerevisiae. Nucleic Acids Res 38: 5177–5192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baßler J, Grandi P, Gadal O, Lessmann T, Petfalski E, Tollervey D, Lechner J, Hurt E 2001. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol Cell 8: 517–529 [DOI] [PubMed] [Google Scholar]
- Ben-Shem A, de Loubresse NG, Melnikov S, Jenner L, Yusupov M 2011. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science 334: 1524–1529 [DOI] [PubMed] [Google Scholar]
- Chu S, Archer RH, Zengel JM, Lindahl L 1994. The RNA of RNase MRP is required for normal processing of ribosomal RNA. Proc Natl Acad Sci 91: 659–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côté CA, Greer CL, Peculis BA 2002. Dynamic conformational model for the role of ITS2 in pre-rRNA processing in yeast. RNA 8: 786–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culver GM 2003. Assembly of the 30S ribosomal subunit. Biopolymers 68: 234–249 [DOI] [PubMed] [Google Scholar]
- de Boer P, Vos HR, Faber AW, Voc JC, Raué HA 2006. Rrp5p, a trans-acting factor in yeast ribosome biogenesis, is an RNA-binding proteins with a pronounced preference for U-rich sequences. RNA 12: 263–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J, Kressler D, Rojo M, Tollervey D, Linder P 1998. Spb4p, an essential putative RNA helicase, is required for a late step in the assembly of 60S ribosomal subunits in Saccharomyces cerevisiae. RNA 4: 1268–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J, Sanz-Martínez E, Remacha M 2005. The essential WD-repeat protein Rsa4p is required for rRNA processing and intra-nuclear transport of 60S ribosomal subunits. Nucleic Acids Res 33: 5728–5739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decatur WA, Fournier MJ 2002. rRNA modifications and ribosome function. Trends Biochem Sci 7: 344–351 [DOI] [PubMed] [Google Scholar]
- Deshmukh M, Tsay YF, Paulovich AG, Woolford JL Jr 1993. Yeast ribosomal protein L1 is required for the stability of newly synthesized 5S rRNA and the assembly of 60S ribosomal subunits. Mol Cell Biol 13: 2835–2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbar DA, Dragon F, Lee SJ, Baserga SJ 2000. A nucleolar protein related to ribosomal protein L7 is required for an early step in large ribosomal subunit biogenesis. Proc Natl Acad Sci 97: 13027–13032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D 1998. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A, Oeffinger M, Tollervey D, Bozzoni I 2003. Cic1p/Nsa3p is required for synthesis and nuclear export of 60S ribosomal subunits. RNA 9: 1431–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira-Cerca S, Pöll G, Gleizes PE, Tschochner H, Milkereit P 2005. Roles of eukaryotic ribosomal proteins in maturation and transport of pre-18S rRNA and ribosome function. Mol Cell 20: 263–275 [DOI] [PubMed] [Google Scholar]
- Ferreira-Cerca S, Pöll G, Kühn H, Neueder A, Jakob S, Tschochner H, Milkereit P 2007. Analysis of the in vivo assembly pathway of eukaryotic 40S ribosomal proteins. Mol Cell 28: 446–457 [DOI] [PubMed] [Google Scholar]
- Gadal O, Strauss D, Petfalski E, Gleizes PE, Gas N, Tollervey D, Hurt E 2002. Rlp7p is associated with 60S pre-ribosomes, restricted to the granular component of the nucleolus, and required for pre-rRNA processing. J Cell Biol 157: 941–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman S, Petfalski E, Tollervey D 2011. A cluster of ribosome synthesis factors regulate pre-rRNA folding and 5.8S rRNA maturation by the Rat1 exonuclease. EMBO J 30: 4006–4019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harnpicharnchai P, Jakovljevic J, Horsey E, Miles T, Roman J, Rout M, Meagher D, Imai B, Guo Y, Brame C, et al. 2001. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol Cell 8: 505–515 [DOI] [PubMed] [Google Scholar]
- Held WA, Ballou B, Mizushima S, Nomura M 1974. Assembly mapping of 30S ribosomal proteins from Escherichia coli. J Biol Chem 249: 3103–3110 [PubMed] [Google Scholar]
- Henras AK, Soudet J, Gérus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y 2008. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci 65: 2334–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry Y, Wood H, Morissey JP, Petfalski E, Kearsey S, Tollervey D 1994. The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J 13: 2452–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold M, Nierhaus KH 1987. Incorporation of six additional proteins to complete the assembly map of the 50S subunit from Escherichia coli ribosomes. J Biol Chem 263: 8826–8833 [PubMed] [Google Scholar]
- Horn DM, Mason SL, Karbstein K 2011. Rcl1 protein, a novel nuclease for 18 S ribosomal RNA production. J Biol Chem 286: 34082–34087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsey EW, Jakovljevic J, Miles TD, Harnpicharnchai P, Woolford JL Jr 2004. Role of the yeast Rrp1 protein in the dynamics of pre-ribosome maturation. RNA 5: 813–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannathan I, Culver G 2004. Ribosomal protein-dependent orientation of the 16S rRNA environment of S15. J Mol Biol 335: 1173–1185 [DOI] [PubMed] [Google Scholar]
- Jakob S, Ohmayer U, Neueder A, Hierlmeier T, Perez-Fernandez J, Hochmuth E, Deutzmann R, Griesenbeck J, Tschochner H, Milkereit P 2012. Interrelationships between yeast ribosomal protein assembly events and transient ribosome biogenesis factors interactions in early pre-ribosomes. PLoS ONE 7: e32552 doi: 10.1371/journal.pone.0032552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakovljevic J, de Mayolo PA, Miles TD, Nguyen TM, Léger-Silvestre I, Gas N, Woolford JL Jr 2004. The carboxy-terminal extension of yeast ribosomal protein S14 is necessary for maturation of 43S pre-ribosomes. Mol Cell 14: 331–342 [DOI] [PubMed] [Google Scholar]
- Jomaa A, Stewart G, Mears JA, Kireeva I, Brown ED, Ortega J 2011. Cryo-electron microscopy structure of the 30S subunit in complex with the YjeQ biogenesis factor. RNA 17: 2026–2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbstein K 2011. Inside the 40S ribosome assembly machinery. Curr Opin Chem Biol 15: 657–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koš M, Tollervey D 2010. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol Cell 37: 809–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler D, Hurt E, Bassler J 2010. Driving ribosome assembly. Biochim Biophys Acta 1803: 673–683 [DOI] [PubMed] [Google Scholar]
- Kruiswijk T, Planta RJ, Krop JM 1978. The course of the assembly of ribosomal subunits in yeast. Biochim Biophys Acta 517: 378–389 [DOI] [PubMed] [Google Scholar]
- Lebreton A, Saveanu C, Decourty L, Jacquier A, Fromont-Racine M 2006. Nsa2 is an unstable, conserved factor required for the maturation of 27SB pre-rRNAs. J Biol Chem 281: 27099–27108 [DOI] [PubMed] [Google Scholar]
- Léger-Silvestre I, Milkereit P, Ferreira-Cerca S, Saveanu C, Rousselle JC, Choesmel V, Guinefoleau C, Gas N, Gleizes PE 2004. The ribosomal protein Rps15p is required for nuclear exit of the 40S subunit precursors in yeast. EMBO J 23: 2336–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Arndt B, Philippsen P, Pringle J 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Lygerou Z, Allmang C, Tollervey D, Seraphin B 1996. Accurate processing of a eukaryotic precursor ribosomal RNA by ribonuclease MRP in vitro. Science 272: 268–270 [DOI] [PubMed] [Google Scholar]
- Mager WH, Planta RJ, Ballesta JG, Lee JC, Mizuta K, Suzuki K, Warner JR, Woolford J 1997. A new nomenclature for the cytoplasmic ribosomal proteins of Saccharomyces cerevisiae. Nucleic Acids Res 25: 4872–4875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Marcos P, Hinnebusch AG, Tamame M 2007. Ribosomal protein L33 is required for ribosome biogenesis, subunit joining, and repression of GCN4 translation. Mol Cell Biol 27: 5968–5985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merl J, Jakob S, Ridinger K, Hierlmeier T, Deutzmann R, Milkereit P, Tschochner H 2010. Analysis of ribosome biogenesis factor-modules in yeast cells depleted from pre-ribosomes. Nucleic Acids Res 38: 3068–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles TD, Jakovljevic J, Horsey EW, Harnpicharnchai P, Tang L, Woolford JL Jr 2005. Ytm1, Nop7, and Erb1 form a complex necessary for maturation of yeast 66S pre-ribosomes. Mol Cell Biol 25: 10419–10432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milkereit P, Gadal O, Podtelejnikov A, Trumtel S, Gas N, Petfalski E, Tollervey D, Mann M, Hurt E, Tschochner H 2001. Maturation and intranuclear transport of pre-ribosomes requires Noc proteins. Cell 105: 499–509 [DOI] [PubMed] [Google Scholar]
- Mizushima S, Nomura M 1970. Assembly mapping of 30S ribosomal proteins in E. coli. Nature 226: 1214–1218 [DOI] [PubMed] [Google Scholar]
- Moritz M, Paulovich AG, Tsay YF, Woolford JL Jr 1990. Depletion of yeast ribosomal proteins L16 and rp59 disrupts ribosome assembly. J Cell Biol 111: 2261–2274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierhaus KH 1991. The assembly of prokaryotic ribosomes. Biochimie 73: 739–755 [DOI] [PubMed] [Google Scholar]
- Nissan TA, Bassler J, Petfalski E, Tollervey D, Hurt E 2002. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J 21: 5539–5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura M 1973. In vitro reconstitution systems facilitate study of ribosome structure, function, and assembly. Science 179: 864–8734569247 [Google Scholar]
- Oakes M, Siddiqi I, Vu L, Aris J, Nomura M 1999. Transcription factor UAF, expansion and contraction of ribosomal DNA (rDNA) repeats, and RNA polymerase switch in transcription of yeast rDNA. Mol Cell Biol 19: 8559–8569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donohue MF, Choesmel V, Faubladier M, Fichant G, Gleizes PE 2010. Functional dichotomy of ribosomal proteins during the synthesis of mammalian 40S ribosomal subunits. J Cell Biol 190: 853–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeffinger M, Tollervey D 2003. Yeast Nop15p is an RNA-binding protein required for pre-rRNA processing and cytokinesis. EMBO J 22: 6573–6583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeffinger M, Leung A, Lamond A, Tollervey D 2002. Yeast pescadillo is required for multiple activities during 60S ribosomal subunit synthesis. RNA 8: 626–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeffinger M, Wei KE, Rogers R, DeGrasse JA, Chait BT, Aitchison JD, Rout MP 2007. Comprehensive analysis of diverse ribonucleoprotein complexes. Nat Methods 4: 951–956 [DOI] [PubMed] [Google Scholar]
- Oeffinger M, Zenklusen D, Ferguson A, Wei KE, El Hage A, Tollervey D, Chait BT, Singer RH, Rout MP 2009. Rrp17 is a eukaryotic exonuclease required for 5′ end processing of pre-60S ribosomal RNA. Mol Cell 36: 768–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtake Y, Wickner RB 1995. Yeast virus propagation depends critically on free 60S ribosomal subunit concentration. Mol Cell Biol 15: 2772–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osheim YN, French SL, Keck KM, Champion EA, Spasov K, Dragon F, Baserga SJ, Beyer AL 2004. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol Cell 16: 943–954 [DOI] [PubMed] [Google Scholar]
- Panse VG, Johnson AW 2010. Maturation of eukaryotic ribosomes: Acquisition of functionality. Trends Biochem Sci 35: 260–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestov DG, Stockelman MG, Strezoska Z, Lau LF 2001. ERB1, the yeast homolog of mammalian Bop1, is an essential gene required for maturation of the 25S and 5.8S ribosomal RNAs. Nucleic Acids Res 29: 3621–3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöll G, Braun T, Jakovljevic J, Neueder A, Jakob S, Woolford JL Jr, Tschochner H, Milkereit P 2009. rRNA maturation in yeast cells depleted of large ribosomal subunit proteins. PLoS ONE 4: e8249 doi: 10.1371/journal.pone.0008249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol 17: 1030–1032 [DOI] [PubMed] [Google Scholar]
- Robledo S, Idol RA, Crimmins DL, Ladenson JH, Mason PJ, Bessler M 2008. The role of human ribosomal proteins in the maturation of rRNA and ribosome production. RNA 14: 1918–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado IV, Kressler D, de la Cruz J 2007. Functional analysis of Saccharomyces cerevisiae ribosomal protein Rpl3p in ribosome synthesis. Nucleic Acids Res 35: 4203–4213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, et al. 2004. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics 3: 1154–1169 [DOI] [PubMed] [Google Scholar]
- Rotenberg M, Moritz M, Woolford JL Jr 1988. Depletion of Saccharomyces cerevisae ribosomal protein L16 causes a decrease in 60S ribosomal subunits and formation of halfmer polyribosomes. Genes Dev 2: 160–172 [DOI] [PubMed] [Google Scholar]
- Sahasranaman A, Dembowski J, Strahler J, Andrews P, Maddock J, Woolford JL Jr 2011. Assembly of Saccharomyces cerevisiae 60S ribosomal subunits: Role of factors required for 27S pre-rRNA processing. EMBO J 30: 4020–4032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saveanu C, Bienvenu D, Namane A, Gleizes PE, Gas N, Jacquier A, Fromont-Racine M 2001. Nog2p, a putative GTPase associated with pre-60S subunits and required for late 60S maturation steps. EMBO J 20: 6475–6484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt ME, Clayton ME 1993. Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae. Mol Cell Biol 13: 7935–7941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shajani Z, Sykes MT, Williamson JR 2011. Assembly of bacterial ribosomes. Annu Rev Biochem 80: 1–28 [DOI] [PubMed] [Google Scholar]
- Staley JP, Woolford JL Jr 2009. Assembly of ribosomes and spliceosomes: Complex ribonucleoprotein machines. Curr Opin Cell Biol 21: 109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunk BS, Loucks CR, Su M, Vashisth H, Cheng S, Schilling J, Brooks CL 3rd, Karbstein K, Skiniotis G 2011. Ribosome assembly factors prevent premature translation initiation by 40S assembly intermediates. Science 333: 1449–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkington MWT, Siuzdak G, Williamson JR 2005. An assembly landscape for the 30S ribosomal subunit. Nature 438: 628–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L, Sahasranaman A, Jakovljevic J, Schleifman E, Woolford JL Jr 2008. Interactions among Ytm1, Erb1 and Nop7 required for assembly of the Nop7-subcomplex in yeast pre-ribosomes. Mol Biol Cell 19: 2844–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarassov K, Messier V, Landry CR, Radinovic S, Molina MMS, Shames I, Malitskaya Y, Vogel J, Bussey H, Michnick SW 2008. An in vivo map of the yeast protein interactome. Science 320: 1465–1470 [DOI] [PubMed] [Google Scholar]
- van Beekvelt CA, de Graaff-Vincent M, Faber AW, van't Riet J, Venema J, Raué HA 2001. All three functional domains of the large ribosomal subunit protein L25 are required for both early and late pre-rRNA processing steps in Saccharomyces cerevisiae. Nucleic Acids Res 29: 5001–5008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nues RW, Rientjes JM, van der Sande CA, Zerp SF, Sluiter C, Venema J, Planta RJ, Raué HA 1994. Separate structural elements within internal transcribed spacer 1 of Saccharomyces cerevisiae precursor ribosomal RNA direct the formation of 17S and 26S rRNA. Nucleic Acids Res 22: 912–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nues RW, Venema J, Rientjes JM, Dirks-Mulder A, Raué HA 1995. Processing of eukaryotic pre-rRNA: The role of the transcribed spacers. Biochem Cell Biol 73: 789–801 [DOI] [PubMed] [Google Scholar]
- Venema J, Tollervey D 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu Rev Genet 33: 261–311 [DOI] [PubMed] [Google Scholar]
- Woodson SA 2011. RNA folding pathways and the self-assembly of ribosomes. Acc Chem Res 44: 1312–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh LC, Thweatt R, Lee JC 1990. Internal transcribed spacer 1 of the yeast precursor ribosomal RNA. Higher order structure and common structural motifs. Biochemistry 29: 5911–5918 [DOI] [PubMed] [Google Scholar]
- Yon J, Giallongo A, Fried M 1991. The organization and expression of the Saccharomyces cerevisiae L4 ribosomal protein genes and their identification as the homologues of the mammalian ribosomal protein gene L7a. Mol Gen Genet 227: 72–80 [DOI] [PubMed] [Google Scholar]
- Zhang J, Harnpicharnchai P, Jakovljevic J, Tang L, Guo Y, Oeffinger M, Rout MP, Hiley SL, Hughes T, Woolford JL Jr 2007. Assembly factors Rpf2 and Rrs1 recruit 5S rRNA and ribosomal proteins rpL5 and rpL11 into nascent ribosomes. Genes Dev 21: 2580–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]