The protein kinase PKR is activated by RNA to phosphorylate eIF-2α, inhibiting translation initiation. Long dsRNA activates PKR via interactions with the dsRNA-binding domain (dsRBD). Weakly structured RNA also activates PKR and does so in a 5′-triphosphate (ppp)-dependent fashion, however relatively little is known about this pathway. The authors used a mutant T7 RNA polymerase to incorporate all four triphosphate-containing nucleotides into the first position of a largely single-stranded RNA and found absence of selectivity, in that all four transcripts activate PKR. Recognition of 5′-triphosphate, but not the nucleobase at the 5′-most position, makes this RNA-mediated innate immune response sensitive to a broad array of viruses.
Keywords: PKR, dsRNA, ssRNA, triphosphate, RNA–protein interaction
Abstract
The protein kinase PKR is activated by RNA to phosphorylate eIF-2α, inhibiting translation initiation. Long dsRNA activates PKR via interactions with the dsRNA-binding domain (dsRBD). Weakly structured RNA also activates PKR and does so in a 5′-triphosphate (ppp)–dependent fashion, however relatively little is known about this pathway. We used a mutant T7 RNA polymerase to incorporate all four triphosphate-containing nucleotides into the first position of a largely single-stranded RNA and found absence of selectivity, in that all four transcripts activate PKR. Recognition of 5′-triphosphate, but not the nucleobase at the 5′-most position, makes this RNA-mediated innate immune response sensitive to a broad array of viruses. PKR was neither activated in the presence of γ-GTP nor recognized NTPs other than ATP in activation competition and ITC binding assays. This indicates that the binding site for ATP is selective, which contrasts with the site for the 5′ end of ppp-ssRNA. Activation experiments reveal that short dsRNAs compete with 5′-triphosphate RNAs and heparin for activation, and likewise gel-shift assays reveal that activating 5′-triphosphate RNAs and heparin compete with short dsRNAs for binding to PKR's dsRBD. The dsRBD thus plays a critical role in the activation of PKR by ppp-ssRNA and even heparin. At the same time, cross-linking experiments indicate that ppp-ssRNA interacts with PKR outside of the dsRBD as well. Overall, 5′-triphosphate-containing, weakly structured RNAs activate PKR via interactions with both the dsRBD and a distinct triphosphate binding site that lacks 5′-nucleobase specificity, allowing the innate immune response to provide broad-spectrum protection from pathogens.
INTRODUCTION
Host recognition of molecular patterns in RNA serves as an integral component of the innate immune response in humans. A number of RNA sensors have been identified as part of this response, including Toll-like receptors (TLRs), retinoic acid-inducible gene 1 (RIG-I), and the protein kinase PKR (Roy and Mocarski 2007). These sensors respond to specific pathogen-associated molecular patterns (PAMPs) that distinguish certain RNAs as non-self, such as long double-stranded stretches and absence of internal nucleoside modifications (Nallagatla and Bevilacqua 2008; Anderson et al. 2010). The 5′-triphosphate (ppp) has been identified as a PAMP for both RIG-I (Hornung et al. 2006; Pichlmair et al. 2006) and PKR (Nallagatla et al. 2007). The structural determinants of this activation are different for both proteins: RIG-I recognizes the 5′-triphosphate of short blunt-end dsRNA (Schlee et al. 2009), while PKR recognizes the 5′-triphosphate of primarily ssRNA with short stem–loops (Nallagatla et al. 2007). Because cellular RNA is typically capped by 7-methylguanosine at its 5′ end, the 5′-triphosphate serves as the molecular pattern to identify an RNA as potentially pathogenic.
PKR is typically activated by long stretches of dsRNA that accumulate as intermediates of viral transcription and replication, which allows PKR to phosphorylate its substrate, eIF2α, thereby disrupting GTP–GDP exchange and ultimately inhibiting translation initiation (Garcia et al. 2006). PKR activation, or autophosphorylation, is mediated by the two tandem N-terminal dsRNA-binding motifs (dsRBMs) that comprise PKR's dsRBD (Fig. 1A; Green and Mathews 1992). PKR autophosphorylation requires binding of two PKR monomers to dsRNAs of at least 33 bp, which results in dimerization of the C-terminal kinase domains and subsequent phosphorylation at Thr446 (Manche et al. 1992; Bevilacqua and Cech 1996; Lemaire et al. 2008). Given the requirement of 33 bp for activation of PKR by dsRNA, activation by ppp-ssRNA with stem–loops as short as 5 bp is surprising and suggests a distinct mechanism of activation.
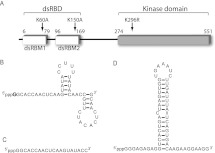
FIGURE 1.
Protein and RNA constructs used in this study. (A) Schematic of PKR primary sequence. The N-terminal dsRNA binding domain (dsRBD, also referred to as P20), composed of two tandem dsRNA-binding motifs (dsRBM 1 and 2), and the catalytic C-terminal kinase domain are indicated. The positions of point mutations used in this study are indicated. The double-mutant PKR (dmPKR) contains both K60A and K150A mutations, and the K296R mutation in the kinase domain renders PKR catalytically inactive. (B) Experimentally determined secondary structure of ssRNA-47 (Nallagatla et al. 2007). The starting nucleotide is indicated in boldface; G is shown, which is typical of transcripts made from WT T7 polymerase, but RNAs were also transcribed containing pppA, pppC, and pppU using mutant T7 polymerase. (C) Sequence of ssRNA-20. Secondary structure prediction via free energy minimization (mFold) indicates that this RNA is essentially completely unstructured. (D) Experimentally determined secondary structure of ss-dsRNA (9,11) (Zheng and Bevilacqua 2004).
We previously identified and characterized the primary RNA structural requirements for PKR activation by ppp-ssRNA in vitro and in vivo and found that all three phosphates in the triphosphate moiety are required for activation, because ssRNA with 5′-end signatures of pp and p did not activate PKR (Nallagatla et al. 2007). Moreover, we showed that dsRNA constructs have no requirement for this 5′-triphosphate for PKR activation, which is unlike RIG-I. Additionally, we demonstrated that PKR activation by ppp-ssRNA requires ∼47 nt and is optimal when the short stem–loops are placed near the middle of the RNA. The mechanistic nature of the above RNA structural requirements for ppp-ssRNA-mediated PKR activation has remained unclear, however.
One parsimonious model states that the 5′-triphosphate of ssRNA uses the known catalytic ATP-binding site. This model assumes that there is only one triphosphate-binding site within PKR, which would then serve two functions. Given the dependence of PKR activation on protein dimerization for both dsRNA and heparin activators (Lemaire et al. 2008; Anderson et al. 2011), these functions would be RNA 5′-triphosphate recognition in one protomer and ATP binding for catalysis in the other. Supporting this economical model, inspection of the crystal structure of the PKR kinase domain with the ATP analog AMP-PNP bound in the catalytic cleft reveals steric accessibility for extension of an RNA chain from the 3′-hydroxyl of AMP-PNP (Dar et al. 2005; Toroney 2010). A second model assumes that two functionally distinct triphosphate-binding sites reside within PKR.
In the present study, we seek to provide insight into the mechanism of 5′-triphosphate-dependent activation of PKR. Our study supports the second model, with distinct triphosphate-binding sites, wherein the catalytic ATP-binding site has high specificity and binding affinity, while the 5′-triphosphate-binding site has lower specificity and affinity. We see that ppp-ssRNA interacts with regions of PKR outside of the dsRBD, but that this RNA and also heparin require PKR's dsRBD for strong activation. These findings suggest that the dsRBD aids in recognition of diverse PKR activators and reinforce the importance of the dsRBD as a universal target for anti-viral therapeutics that function by up-regulating PKR.
RESULTS
In this study, the designation “ssRNA” refers to an RNA that is comprised of one strand; i.e., it does not contain a complementary strand. A ssRNA may contain some secondary structure and is numbered according to length in nucleotides (see Fig. 1B,C). Similarly, “dsRNA” refers to RNA composed of two strands and is numbered according to length in base pairs. In one specific instance, “ss-dsRNA (9,11)” refers to single-stranded RNA with a helical stem and unstructured 5′- and 3′-end tails of 9 and 11 nt, respectively (Fig. 1D).
Recognition of 5′-triphosphate-containing RNA lacks specificity for the 5′-nucleobase
Previously, we established that PKR activation by 5′-triphosphate-dependent ssRNAs has high selectivity for the triphosphate moiety (Nallagatla et al. 2007). For example, we showed that transcripts beginning with 5′-OH and 7-methyl-guanosine abrogate PKR activation, and transcripts beginning with 5′-p and 5′-pp were also very poor activators relative to the same RNA beginning with 5′-ppp. Specificity of PKR activation for the identity of the base at position 1 of the transcript has not been explored, however, with all studies to date involving RNAs that begin with pppG. Although RNAs transcribed in vitro by T7 polymerase typically require G in the first position for transcription initiation (Milligan and Uhlenbeck 1989), bacterial and viral RNAs are initiated by G or A, and even occasionally by C (Bieger and Nierlich 1989; Luo et al. 2000; Cai et al. 2004). Base specificity of PKR activation has implications for both specificity regarding the structural nature of the triphosphate-binding site in PKR and for the functional permissiveness of PKR activation by non-self ppp-RNAs in innate immunity.
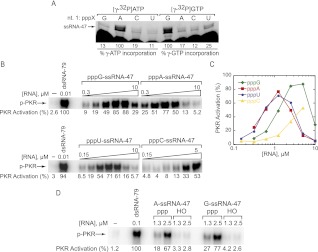
We used a mutant T7 polymerase containing a P266L change, which reduces abortive cycling during transcription owing to decreased affinity of the promoter binding site (Guillerez et al. 2005). This variant should be more tolerant to transcripts that start with nucleotides other than pppG, typically required by wild-type T7 polymerase (Milligan and Uhlenbeck 1989). We transcribed four versions of ssRNA-47 (Fig. 2A), which we have previously characterized as a strong 5′-triphosphate-dependent activator of PKR (Nallagatla et al. 2007), with pppG-, pppA-, pppC-, and pppU-starting nucleotides. We generated linearized plasmid templates for each of the four starting nucleotides and, in an effort to verify insertion of the correct starting nucleotide, transcribed each RNA in the presence of [γ-32P]ATP or [γ-32P]GTP (Fig. 2A). Because the radiolabel is located on the γ-phosphate, a band should only be visible when the labeled nucleotide is inserted at the first position. We observed preferential incorporation of [γ-32P]ATP and [γ-32P]GTP at position 1 where expected (Fig. 2A, lanes 2,5), and limited misincorporation of these nucleotides in transcripts starting with the other three nucleotides.
FIGURE 2.
All ssRNA-47 5′-triphosphate-starting nucleotides activate PKR. (A) Verification of starting nucleotide identity. RNA was transcribed from DNA templates coding for starting the transcript with pppG, pppA, pppC, and pppU, in the presence of [γ-32P]ATP or [γ-32P]GTP. The expected starting RNA nucleotide based on template (nt. 1: pppX) is indicated. Percent incorporation was calculated by normalizing the counts at the indicated mobility “ssRNA-47” to the counts from the pppA lane for [γ-32P]ATP incorporation, or the pppG lane for [γ-32P]GTP incorporation. 7 M urea gel is shown. (B) Activation of PKR by ssRNA-47 with pppG, pppA, pppU, and pppC. RNA concentrations were 0.31, 0.63, 1.3, 2.5, 5.0, and 10 μM for pppG- and pppA-ssRNA-47; 0.15, 0.31, 0.63, 1.3, 2.5, 5.0, and 10 μM for pppU-ssRNA-47; and 0.15, 0.31, 0.63, 1.3, 2.5, and 5.0 μM for pppC-ssRNA-47. Phosphorylation activities are provided under the gels and were normalized to the dsRNA-79 lane in the top gel. (C) Graphical representation of phosphorylation activities from panel B as a function of RNA concentration. (D) Activation of PKR by A-ssRNA-47 and G-ssRNA-47 are both dependent on a 5′-triphosphate. RNAs starting with 5′-triphosphate were generated by in vitro T7 transcription, while RNAs starting with 5′-OH-G and 5′-OH-A were chemically synthesized. Phosphorylation activities are provided under the gel and were normalized to the dsRNA-79 lane. For both panels B and D, 10% SDS-PAGE gels are shown, with the position of phosphorylated PKR (p-PKR) indicated.
These RNAs were then tested for activation of PKR. We observed PKR activation of similarly high potency for ssRNA-47 with all four 5′-NTPs (Figs. 2B,C). For example, pppG-ssRNA-47 activated maximally (88% the level of activation by the long 79-bp activator dsRNA-79) at 5 μM, with a bell-shaped dependence on RNA concentration, as observed previously for this RNA (Nallagatla et al. 2007). Maximal activation intensities for RNAs starting with A and U were similar as for G, but their bell-shaped activation profiles were shifted to even lower RNA concentrations (∼1.25 μM RNA), implying higher affinity (Fig. 2C). Potent activation of PKR by pppA-ssRNA-47 and pppG-ssRNA-47 is consistent with the known prevalence of viral transcripts containing 5′-G or 5′-A (see Discussion).
We also used synthetic versions of 5′-A-ssRNA-47 and 5′-G-ssRNA-47, which contain bona fide 5′-OHs, to test whether ssRNA-47 starting with a nucleotide other than G still activates PKR in a 5′-triphosphate-dependent fashion. As shown in Figure 2D, both 5′-A-ssRNA-47 and 5′-G-ssRNA-47 have similar 41- and 54-fold triphosphate dependence for activation, with the value for 5′-G-ssRNA-47 in agreement with an earlier study (Nallagatla et al. 2007). Thus, the specificity of PKR activation for 5′-triphosphate does not depend on 5′-nucleobase identity.
Activation by pppC-ssRNA-47 was within at least approximately two-thirds the maximum value of the other three RNAs, although its profile was shifted to higher RNA concentration. In summary, potent activation of PKR by ppp-ssRNA containing different bases at the 5′-position suggests that the binding site for the 5′-triphosphate of ssRNA is permissive and that PKR-mediated contacts are primarily to the triphosphate moiety of the 5′-nucleotide rather than the nucleobase.
Recognition of ATP has specificity for the nucleobase: Activation competition experiments
The parsimonious model for the 5′-triphosphate-dependent activation of PKR is that the 5′-triphosphate binds in the ATP active site. This model is supported by steric accessibility at the active site for chain extension off the 3′-hydroxyl of ATP and by the observation that the major contacts between ATP and amino acids at the active site that are necessary to position the phosphates for catalysis occur to the ribose sugar and triphosphate rather than the base (Johnson et al. 1996; Niefind et al. 1999; Dar et al. 2005; Toroney 2010). Moreover, potent activation by pppA-ssRNA-47 would seem to suggest potential binding of this RNA into the ATP-binding site.
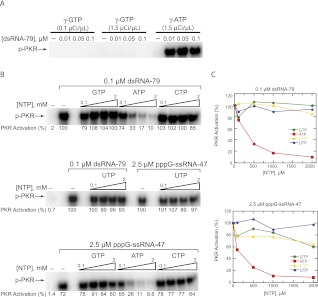
The results presented here on PKR activation by ss-RNA-47s starting with any of the four triphosphate nucleotides, and the reported 5′-triphosphate-dependent activation of PKR by several different ssRNAs with G as the starting nucleotide (Nallagatla et al. 2007) suggest that if the ATP active site also serves as the 5′-triphosphate-binding site for ssRNA, then the active site NTP may also be permissive to GTP. To test this notion, we performed a PKR activation assay in which all of the ATP (unlabeled and [γ-32P]ATP) was replaced with GTP and [γ-32P]GTP. In contrast to the parsimonious model, we observed complete abrogation of activation in the presence of GTP only (Fig. 3A, first two sets of lanes), suggesting that the ATP-binding site has high nucleotide specificity for chemistry. Furthermore, we showed that a radiolabeled γ-phosphate of a 5′-triphosphate activator is not transferred to PKR (SR Nallagatla and PC Bevilacqua, unpubl.), suggesting that the 5′-triphosphate end of ssRNA cannot fill the ATP-binding site. It thus appears that PKR has separate triphosphate-binding sites—one with base specificity and one without.
FIGURE 3.
The ATP binding site has high specificity: activation assays. (A) Only ATP supports PKR phosphorylation. Activation of PKR by dsRNA-79 using [γ-32P]GTP or [γ-32P]ATP as the phosphate source. RNA concentrations are provided. In the “γ-GTP” lanes, activation assays were performed as per standard assay conditions (see Materials and Methods) with the following exceptions: 100 μM GTP was used instead of ATP, and 0.1 or 1.5 μCi/μL [γ-32P]GTP was added instead of 1.5 μCi/μL [γ-32P]ATP. In the “γ-ATP” lanes, standard assay conditions were used. (B) NTP-competition assays reveal that only free ATP competes with ATP for activation. PKR activation by dsRNA-79 or pppG-ssRNA-47 was assayed in the presence of increasing concentrations of unlabeled ATP, GTP, CTP, and UTP. The concentrations of dsRNA-79 and pppG-ssRNA-47 were 0.1 and 2.5 μM, respectively. The concentrations of each NTP were 0.1, 0.5, 1, and 2 mM. (All of these are added concentrations and are in the background of 100 μM ATP.) The “no-RNA” and “no-competitor-NTP” lanes are included as negative and positive controls, respectively. Phosphorylation activities were normalized to the “no-competitor-NTP” lane in the middle gel. For both A and B, a 10% SDS-PAGE gel is shown, with the position of phosphorylated PKR (p-PKR) indicated. (C) Graphical representation of phosphorylation activities from panel B.
To probe these two models further, activation assays were performed in which activation by dsRNA-79, which does not require a 5′-triphosphate (Nallagatla et al. 2007), and by pppG-ssRNA-47 were challenged with each of the four NTPs. As shown in Figure 3, B and C, for each RNA activator, only ATP was able to compete effectively with PKR activation (up to ∼10-fold inhibition). (Note that these plots, which are in the background of 100 μM ATP, are consistent with the Kd value for ATP of ∼19 μM obtained by ITC; see below.) Because dsRNA-79 has no triphosphate dependence for activation, the observed inhibition is reflective of unlabeled ATP competing out radiolabeled ATP from the active site. Thus, the catalytic site is specific for ATP not only for chemistry, but also for binding. Given the apparent tolerance of PKR for different activating 5′-triphosphate groups on RNA (Fig. 2B), the lack of competition for NTPs other than ATP provides further evidence that PKR contains two separate triphosphate-binding sites.
A similar specificity of strong inhibition by ATP but not the other NTPs was seen for activation by ssRNA-47 (Fig. 3B,C). This observation suggests that the same, specific ATP-binding site is used for this activator. We do note slight (∼40%) inhibition of activation by GTP and CTP at their highest concentrations of 2 mM; this inhibition did not occur in the presence of dsRNA-79 (Fig. 3C). Weak inhibition by GTP and CTP in the presence of pppG-ssRNA-47 but not dsRNA-79 suggests that these other NTPs can compete very weakly for pppG-ssRNA-47's triphosphate-binding site and that ssRNA-47 binds rather weakly to PKR. Failure of GTP, CTP, and UTP to potently inhibit activation by pppG-ssRNA-47 suggests that 5′-triphosphate binding at this putative second triphosphate-binding site is strengthened relative to free NTPs, probably through tethering of the triphosphate to PKR via the remainder of the RNA. Interactions of the remainder of the RNA with PKR are explored below in more depth.
Recognition of ATP has specificity for the nucleobase: ITC experiments
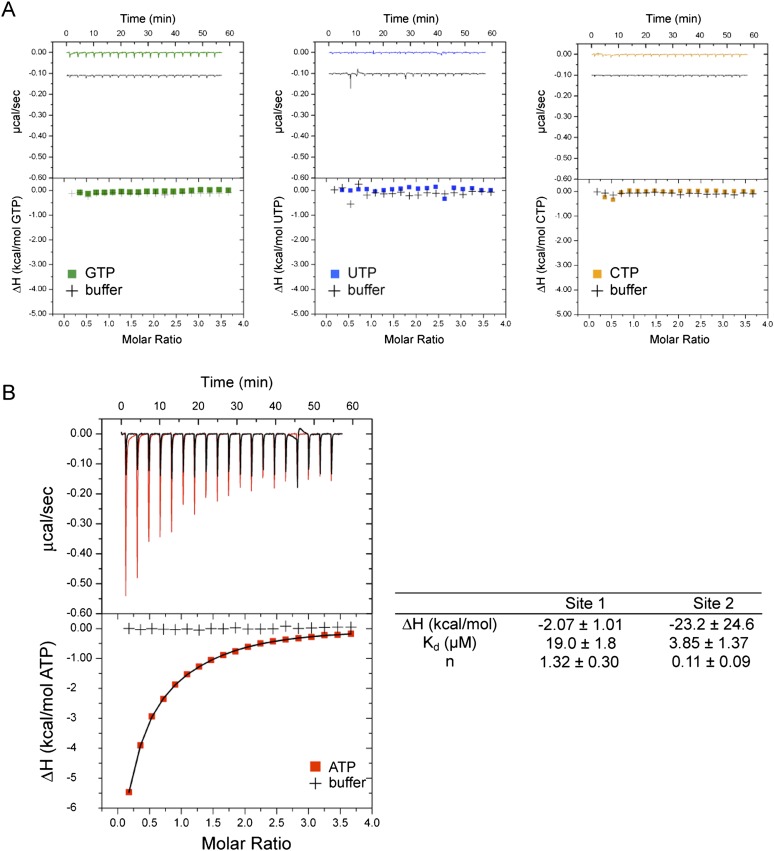
In this section, we further explore the specificity of ATP recognition by turning from activation competition experiments to isothermal titration calorimetry (ITC) experiments. To investigate further the nature of triphosphate binding, ITC was used to determine the thermodynamics of NTP binding to PKR. To prevent coupling of binding with enzymatic activity, a catalytically inactive mutant of PKR, K296R, was used (Fig. 1A). Binding was to K296R in the absence of RNA, with injections of each of the four individual NTPs in 18-fold excess over protein. As shown in Figure 4A, the titration curve for GTP revealed a very weak interaction that was barely noticeable in comparison to titration against buffer, and UTP and CTP titration curves were essentially equivalent to the buffer titration. In contrast, titration of ATP revealed a strongly exothermic interaction.
FIGURE 4.
The ATP binding site has high affinity: ITC experiments. ITC titration curves for NTP binding to K296R. (A) GTP (green trace), UTP (blue trace), and CTP (orange trace) binding to K296R. Titration of each NTP into buffer is included (black traces). NTP traces are offset from buffer traces by ∼0.1 μcal/sec in raw data (upper panels) for clarity. Legends are provided in plots. (B) ATP binding to K296R (red trace). Titration of ATP into buffer is included (black trace). ATP titration curve is fitted to a two-site binding model (see Materials and Methods), and thermodynamic parameters are provided to the right of the plot. The major contribution is with site 1 (n = 1.32 ± 0.09) and a Kd of 19 ± 1.8 μM. Note that uncertainty in ΔH is due to the lack of a good lower baseline, which is common with micromolar Kd experiments. However, this does not affect the Kd value because that is mainly based on the slope of the transition region.
The ATP titration curve presented in Figure 4B using initial concentrations of 40 μM K296R and 720 μM ATP was best fit to a two-site binding model. (Fitting these data to a one-site model [not shown] in which all of the parameters floated gave n = 0.55 and a poor fit to injections where [ATP]/[K296R] ≥ 1.5, while fitting to a one-site model with n forced to 1, gave a poor fit to all injections.) The first site had a Kd of 19 ± 1.8 μM and n = 1.32 ± 0.30, suggesting a 1:1 stoichiometry of K296R to ATP (Fig. 4B), as expected based on the crystal structure of PKR (Dar et al. 2005). This Kd is similar to that seen for ATP binding to other kinases such as PKA and MAPK (Armstrong et al. 1979; Setyawan et al. 1999), and in agreement with a value from Cole and coworkers of 20 ± 2 μM, measured for ATP binding to K296R using competition fluorescence anisotropy experiments (Lemaire et al. 2006). The second site was tighter in affinity (Kd = 3.9 ± 1.4 μM) but with a small binding stoichiometry of n = 0.11 ± 0.09 (Fig. 4B). Given the high affinity at both sites as determined by ITC and the lack of binding with the other three NTPs, it seems unlikely that one of these two sites is representative of the proposed non-specific triphosphate-binding site, which the preceding data suggest is much weaker binding than the catalytic site (Fig. 3C, bottom panel). One possible explanation for the two-site fit involves the different conformers of Mg2+-coordinated ATP, namely, the “open” conformer, in which the metal ion only coordinates the phosphate, and the “macrochelated” conformer, in which the metal ion is coordinated to the phosphate and the base (Sigel 1987). Interestingly, the macrochelated form represents ∼10% of the ATP population at physiological salt conditions and could thus represent the “site 2” observed by ITC titration (Fig. 4B; Sigel 1987). (Specificity of the ATP-binding site was also probed by a series of fluorescence competition and pulse-chase stopped-flow experiments involving the fluorophore mant-ATP and the competitors ATP, GTP, and UTP, and confirmed specificity for ATP [Toroney 2010].)
In sum, the results from PKR competition assays and ITC suggest that two triphosphate-binding sites exist on PKR and that they are quite different: a catalytic triphosphate binding site that binds ATP with both high specificity and high affinity, and a 5′-nucleotide triphosphate-binding site that is non-specific to 5′-nucleobase identity and low affinity.
Binding of 5′-triphosphate RNA and heparin requires the dsRBD
We previously demonstrated that the magnitude of PKR activation by ppp-ssRNA is dependent on the presence and positioning of a short ∼5-bp stem–loop, thus implicating a role for the dsRBD in PKR activation (Nallagatla et al. 2007). The optimal placement of this stem–loop was ∼20–45 nt from the 5′ end of ppp-ssRNA. Additionally, ppp-ssRNA activators displayed certain similarities to dsRNA activators, such as a requirement for length (∼47 nt of single-stranded RNA vs. ∼33 bp of double-stranded RNA) and a bell-shaped dependence on RNA concentration (Nallagatla et al. 2007). Despite these similarities, distinct differences in the 5′-end requirements of ppp-ssRNA and dsRNA exist—requiring and not requiring a 5′-triphosphate, respectively—suggesting potential mechanistic disparities between these two classes of activators. Indeed, ssRNA displays coarse structural similarity to a non-RNA activator of PKR, the polyanion heparin, which has been shown to bind basic residues near the active site in the kinase domain (Fasciano et al. 2005a). To elucidate the mechanism of activation of PKR by ppp-ssRNA, we sought to elucidate the role, if any, of the dsRBD in such activation.
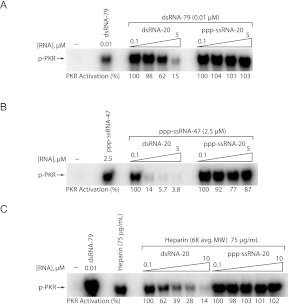
We first performed PKR competition assays, in which three activators—canonical dsRNA (dsRNA-79), pppG-ssRNA-47, and heparin—were competed by either short, 20-bp dsRNA “dsRNA-20” or short unstructured 20-nt ssRNA with a 5′-triphosphate “ppp-ssRNA-20” (Figs. 1B, 5). Both competitor RNAs have been demonstrated to bind PKR but not activate (Bevilacqua and Cech 1996; Nallagatla et al. 2007). As shown in Figure 5, A and B, dsRNA-20 effectively competed with both RNA activators (sixfold inhibition for dsRNA-79 and 26-fold for ppp-ssRNA-47), while ppp-ssRNA-20 had no effect on dsRNA-mediated activation and only a small ∼20% effect on ppp-ssRNA-47-mediated activation. For dsRNA-79, these results are consistent with the well-established model of long dsRNA interacting with the dsRBD, which drives binding of multiple PKR molecules (Fig. 5A). Short dsRNA competes with this interaction, hence leading to inhibition, while ssRNA either does not bind to the same part of PKR as dsRNA or cannot effectively compete with longer dsRNA.
FIGURE 5.
PKR activation by ppp-ssRNA and heparin is inhibited by dsRNA, but not by short ppp-ssRNA. RNA-competition assays. (A) Competition for dsRNA-79-mediated PKR activation by short dsRNA and ssRNA. PKR activation by dsRNA-79 was assayed in the presence of increasing concentrations dsRNA-20 and ppp-ssRNA-20. The concentration of dsRNA-79 was held constant at 0.1 μM, and concentrations of dsRNA-20 and ppp-ssRNA-20 were 0.1, 1, 2.5, and 5 μM. (B) Competition for ppp-ssRNA-47-mediated PKR activation by short dsRNA and ssRNA. PKR activation was assayed as in A, with the concentration of ssRNA-47 held constant at 2.5 μM. (C) Competition for heparin-mediated PKR activation by short dsRNA and ssRNA. PKR activation was assayed as in A, with the concentration of heparin (average molecular weight of 6 kDa) held constant at 75 μg/mL, and concentrations of competitor RNAs at 0.1, 1, 2.5, 5, and 10 μM. For all panels, 10% SDS-PAGE gels are shown, with the position of phosphorylated PKR (p-PKR) indicated. The “no-RNA” and “no-competitor” lanes are included as negative and positive controls, respectively. Activation in the absence of added RNA was negligible. Phosphorylation activities are provided under the gels and were normalized to the 0.1 μM competitor lane for each set of competitors; activities were normalized in this manner because in some cases the presence of competitor at low concentrations appeared to stimulate the reaction.
Competition of ppp-ssRNA-47 by dsRNA-20, on the other hand, was surprising. In fact, dsRNA-20 competes more effectively with ssRNA activator than dsRNA activator, sixfold and 26-fold, respectively. This finding suggests that ppp-ssRNA-47 makes critical contacts with the dsRBD, most likely through its short stem–loop. The fact that ppp-ssRNA-20 is at best a poor competitor for ppp-ssRNA-47, despite the fact it contains a 5′-triphosphate, may be due to absence of the short stem–loop in ssRNA-20. This notion is also consistent with the above inability of NTPs to compete for ppp-ssRNA-47.
In an effort to provide further insight into the role of the dsRBD, we considered the mechanism of PKR activation by the known activator heparin, a sulfated glycosaminoglycan (Hovanessian and Galabru 1987). Because ssRNA and heparin are both non-dsRNA polyanion activators, we thought that they might have similar mechanisms of activation. Heparin has been shown to bind clusters of basic amino acids at the entrance to the kinase active site that are non-overlapping with the dsRBD, suggesting a primarily electrostatic as opposed to structure-based mechanism of activation (Fasciano et al. 2005a). Furthermore, it has been shown that heparin can activate a truncated version of PKR that does not contain the dsRBD, although the extent of activation was strongly diminished relative to full-length PKR (Patel et al. 1994; George et al. 1996; Fasciano et al. 2005a). We thus anticipated that ppp-ssRNA activators might compete for activation by heparin while short dsRNA would not compete; indeed, such a result would suggest that the 5′-triphosphate-binding site overlaps with the cluster of basic residues that serve as the binding site for heparin.
We thus performed competition assays in which PKR activation by a constant concentration of heparin (average MW 6 kDa) was challenged with increasing concentrations of dsRNA-20 or ppp-ssRNA-20. In contrast to the above expectations, ppp-ssRNA-20 was unable to compete with heparin for activation, while dsRNA-20 was a potent inhibitor of heparin-mediated activation of PKR (Fig. 5C). Lack of inhibition by ppp-ssRNA-20 indicates that either the 5′-triphosphate does not share a binding site with heparin, or that heparin binds more tightly than this particular RNA. Even more surprising, heparin-mediated activation of PKR is strongly inhibited by dsRNA-20 (up to 7.7-fold inhibition). This observation suggests an important role for the dsRBD in activation of PKR by both heparin and ppp-ssRNA.
The 5′-triphosphate-binding site is located outside the dsRDB
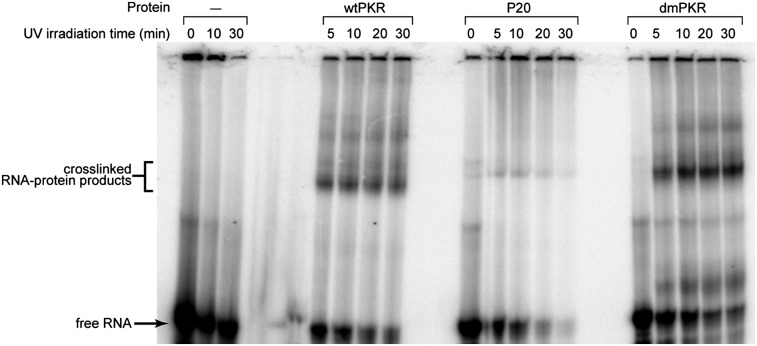
Results so far indicate that 5′-triphosphate-containing ssRNAs do not interact with the ATP-binding site, but do interact, at least in part, with the dsRBD. Our next step in defining the mechanism of 5′-triphosphate-dependent PKR activation was to investigate where the ppp-ssRNA physically associates with PKR. We transcribed ppp-ssRNA-47 (Fig. 1B) in which all uridines were replaced with 4-thiouridine, termed “ppp-4thioU-ssRNA-47.” This RNA was radiolabeled at the 5′ end and cross-linked to PKR via exposure to 365 nm light (see Materials and Methods). Several different forms of PKR were investigated: full-length (wtPKR), P20 (dsRBD only), and a mutant PKR with two point mutations in the dsRBD, K60A and K150A (dmPKR) (Fig. 1A). It has been shown previously that these two changes abrogate binding to dsRNA (Green and Mathews 1992; Green et al. 1995; Heinicke et al. 2009).
As shown in Figure 6 and Supplemental Figure 1, ppp-4thioU-ssRNA-47 cross-links to wtPKR in a protein-, 4-thioU-, and UV-dependent fashion, confirming interaction of this activator with PKR. Surprisingly, there is at most very weak cross-linking between 4thioU-ssRNA-47 and P20. To confirm that the cross-links between this RNA and PKR were outside of the dsRBD, we conducted cross-linking on dmPKR. As shown in Figure 6, we observe strong, UV-dependent interaction of ppp-4thioU-ssRNA-47 with dmPKR, which supports the notion that ppp-ssRNA-47 interacts with a region of PKR outside the dsRBD. Given the above support for interaction of ssRNA-47 with the dsRBD, poor cross-linking to the dsRBD is most likely because the quantum yield of cross-links between PKR and 4thioU-ssRNA-47 is low: There are several AU base pairs in the short stem region of ssRNA-47, which are likely broken due the thio group on the Watson–Crick face.
FIGURE 6.
Photochemical cross-linking reveals interaction of ppp-ssRNA-47 with PKR regions outside the dsRBD. 4thioU-substituted ppp-ssRNA-47 was incubated with wild-type PKR, P20, or dmPKR and exposed to 365-nm light for 0, 10, 20, or 30 min and analyzed by 7% denaturing (7 M urea) PAGE. The positions of free RNA and cross-linked products are indicated. Protein dependence of cross-linked products was confirmed by the absence of products upon irradiating 4-thioU-ppp-ssRNA-47 in buffer alone (first three lanes).
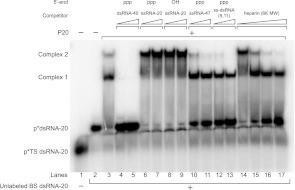
To further evaluate the role of the dsRBD in interacting with various PKR activators, we performed EMSA competitions, with P20 pre-bound to radiolabeled 20 bp dsRNA (Fig. 7). These binding-competition experiments complement the activation-competition experiments described above (Fig. 5), but also extend them because it is possible to use actual activators as competitors in the EMSAs. As expected based on previous studies characterizing interaction with short dsRNAs of various length (Bevilacqua and Cech 1996), P20 binds to trace amounts of 5′-32P-radiolabeled dsRNA-20 in two distinct complexes (Fig. 7, lane 3). A long classical dsRNA activator, dsRNA-40, competes completely for binding to both complexes, as revealed by loss of complexes 1 and 2, and by gain in free p*dsRNA-20 (Fig. 7, lanes 4,5). Moreover, in agreement with our activation assays (Fig. 5), we find that ppp- or OH-ssRNA-20 is unable to displace dsRNA-20 from P20, as revealed by absence of gain of free p*dsRNA-20. We do note, however, that ppp- or OH-ssRNA-20 appears to induce conversion of complex 1 into complex 2, indicating some type of rearrangement (Fig. 7, cf. lane 3, where complex 2 represents only ∼10% of bound product, to lanes 6–9, where complex 2 represents ∼50%–60% of products). This suggests that short, purely single-stranded RNA is capable of making contacts with the dsRBD, albeit contacts non-productive for activation. This effect is 5′-triphosphate-independent (Fig. 7, cf. lanes 6,7 to 8,9), further supporting the notion that the 5′-triphosphate binding site is not located within the dsRBD.
FIGURE 7.
Competition for P20 binding by dsRNA, ssRNA, and heparin by EMSAs. A trace amount of p*dsRNA-20 was incubated with P20 in the presence of various unlabeled competitors and analyzed by 10% native PAGE. Top strand (TS) dsRNA-20 was 5′-32P-labeled and annealed to excess unlabeled bottom strand (BS). Formation of annealed p*dsRNA-20 duplex was confirmed by the microshift of p*TS in the presence of BS (lane 2). A no-competitor mobility shift was detected upon addition of 3 μM P20, with slight formation of a second complex (lane 3). In the remaining lanes, 3 μM P20 bound to trace p*dsRNA-20 was challenged with either unlabeled RNA or heparin competitor. Competitor RNA concentrations were 5 and 10 μM, and heparin concentrations were 10, 100, 1000, and 2000 μg/mL. Mobility of p*dsRNA-20, free and bound to P20 complexes, is indicated.
Next, we find that two different 5′-triphosphate-dependent activators, ssRNA-47 and ss-dsRNA (9,11), compete with dsRNA-20 for binding to both P20 complexes. This is revealed by loss of complex 1 and especially complex 2, and by gain in free p*dsRNA-20 (Fig. 7, lanes 10–13). These observations are consistent with the dsRNA-20 competition experiments in Figure 5B, which suggested that the dsRBD plays a critical role in PKR activation by ppp-ssRNA. This binding competition, in contrast to the rearrangement of complexes induced by non-activating ssRNAs, is in agreement with a role for the small stem–loops of 5′-triphosphate-dependent RNAs in binding the dsRBD during activation. Consistent with this interpretation, ss-dsRNA (9,11), a previously characterized activator containing a 16-bp stem–loop flanked by 9-nt and 11-nt single-stranded tails (Fig. 1D; Zheng and Bevilacqua 2004; Nallagatla et al. 2007), competes more effectively for P20 than ssRNA-47, which has less double-stranded character (Fig. 1) (Fig. 7, compare gain of ∼50% free p*dsRNA-20 in lane 11 to ∼75% free p*dsRNA-20 in lane 13). It is worth noting that the effects here are not simply due to ionic strength, because ssRNA-20 offers no competition at its highest concentration, which is effectively equal to the concentration of phosphate in the lower concentration lanes for ssRNA-47 and ss-dsRNA (9,11).
Finally, we observe that heparin can also compete with dsRNA for binding (Fig. 7, lanes 14–17). This result corroborates our activation competition assays, wherein activation by heparin was effectively competed by dsRNA-20 (Fig. 5C), and further supports a key role of the dsRBD in activation of PKR by heparin. This interaction with the dsRBD appears to be dependent on the heparin concentration: At very low heparin concentrations (10 μg/mL), heparin behaves like short, non-activating ssRNAs, promoting conversion of complex 1 to complex 2 (Fig. 7, lane 14), while at higher concentrations, heparin competes out dsRNA-20 for P20 binding, resulting in free p*dsRNA-20. Overall, the EMSA competition studies provide strong support for a general role of the dsRBD in PKR activation by both classical dsRNA as well as non-dsRNA activators of ppp-ssRNA and heparin.
DISCUSSION
Surveillance of foreign RNA by PKR and other components of the innate immune response is a critical first line of defense against viral infection. While long stretches of double-stranded RNA have been recognized as a molecular signature for PKR activation, recent studies indicate that PKR can be activated by a multitude of additional RNA structural elements, including short coaxially stacked helices, RNA helices interrupted by bulges and internal loops, and internal or 5′-end modifications, such as the 5′-triphosphate (Nallagatla et al. 2011). Given this expanding landscape of RNA elements that regulate PKR, it is critical to understand the mechanisms by which PKR is specifically activated by diverse RNA structures.
We previously identified the 5′-triphosphate of weakly structured ssRNAs as a PAMP for PKR activation, which allows for discrimination between self and non-self RNA. This PAMP is also recognized by another innate immune sensor, RIG-I (Hornung et al. 2006; Pichlmair et al. 2006), suggesting that the 5′-triphosphate is a widespread signal for the innate immune system. We sought to better define the interaction of PKR and ppp-ssRNA by characterizing the 5′-triphosphate-binding pocket via a variety of functional and biophysical assays.
The simple “one-triphosphate site” model is one in which the catalytic binding cleft in the kinase domain serves as a common site for binding both ATP for catalysis and the 5′-triphosphate of ssRNA for activation. This model is suggested by the fact that PKR and RIG-I, which also contains an ATP-binding cleft, are both able to bind a common RNA ligand despite lacking sequence and structural homology. An alternative is the “two-triphosphate site” model, in which the 5′-triphosphate-binding site is located either elsewhere in the kinase domain or in the dsRBD and is separate from the catalytic ATP site. The data provided herein support the two-triphosphate site model. There is low specificity for the 5′-nucleobase in 5′-ppp-ssRNA-mediated activation of RNA (Fig. 2), but high specificity for ATP, because neither GTP, CTP, nor UTP can compete efficiently for ATP in activation (Fig. 3). Moreover, ATP is the only NTP that gives detectable heat of binding to PKR (Fig. 4). An early study observed ATP and GTP competition for photo-affinity labeling of PKR at a site proposed to represent either the catalytic site or a secondary allosteric regulatory site (Bischoff and Samuel 1985). The ATP specificity at the catalytic site presented herein supports the possibility that these investigators may have probed the secondary triphosphate site. Indeed, selectivity at the PKR catalytic site for ATP is consistent with the behavior of most kinases (Shugar 1996; Smith et al. 2012). This binding specificity likely serves as a selective pressure to avoid catalysis by GTP, which could have detrimental consequences for PKR regulation in the cell. Conversely, lack of nucleobase selectivity for the proposed 5′-triphosphate site allows for activation of PKR by 5′-triphosphate RNA activators that start with any of the four nucleotides. Indeed, bacterial and viral RNAs can begin with A, C, or G (Bieger and Nierlich 1989; Luo et al. 2000; Cai et al. 2004), suggesting that lack of specificity at this nucleobase position allows a broad immune response. In particular potent activation of PKR by pppA-ssRNA-47 supports recognition of the highly conserved pppA present in influenza A, B, and C viruses (Moss et al. 1978; Desselberger et al. 1980), consistent with the 5′-triphosphate dependence of PKR activation by influenza B virus (Dauber et al. 2009).
We have previously shown that 5′-triphosphate-mediated activation of PKR is dependent on the characteristics of the downstream RNA structure, such as length and presence and positioning of short stem–loops (Nallagatla et al. 2007). Activating 5′-triphosphate RNAs interact with PKR outside the dsRBD (Fig. 6), raising the question of whether the dsRBD still plays a role in recognition of 5′-triphosphate activators. Surprisingly, we find that the dsRBD plays important roles in binding and activation of PKR by both ppp-ssRNA and heparin (Figs. 5, 7). Despite extensive structural differences between canonical dsRNA, 5′-ppp-ssRNA, and heparin, all three require interactions with the dsRBD for optimal activation. Given that PKR activation has a bell-shaped dependence on concentration of ppp-ssRNA (Fig. 2) and heparin (Anderson et al. 2011; data not shown), the dsRBD may drive dimerization of PKR on ppp-ssRNA and heparin as well, although these non-dsRNA activators also take advantage of additional contacts outside the dsRBD. Indeed, a clear mechanistic basis for the bell-shaped dependence of PKR activation on heparin concentration has been elusive (Anderson et al. 2011). We are continuing work to define the specific PKR residues involved in these non-dsRBD-mediated contacts.
It has been demonstrated that PKR recognizes the 5′-triphosphate of influenza B viral RNA in human cells (Dauber et al. 2009), and other studies have shown that PKR mediates the anti-proliferative action of heparin treatment for atherosclerosis (Fasciano et al. 2005b). As such, defining regions of PKR that contact disparate activators, such as key regions of PKR's dsRBD, may facilitate construction of therapeutic RNAs capable of activating PKR during viral infection or may provide insight into the mechanism of heparin therapeutics.
MATERIALS AND METHODS
Sequences of RNA and protein
ssRNA
ssRNA-47: Different versions of ssRNA-47 were prepared, starting with various 5′-end nucleotides, which are indicated in parentheses.
5′-(pppG, pppA, pppC, pppU, pppA, OH-G, OH-A)GGCACCAACUCAAGUAUACCUUUUAUACAACCGUUCUACACUCAACG
ssRNA-20:
5′-GGCACCAACUCAAGUAUACC
dsRNA
Top strands (TS) are provided below; the bottom strand is the Watson–Crick complement to the top strand. Double-stranded RNAs were prepared by annealing equimolar concentrations of the two strands.
dsRNA-79TS (Zheng and Bevilacqua 2004):
5′-GGGUUUUCCCAGUCACGACGUUGUAAAACGACGGCCAGUGAAUUCGAGCUCGGUACCCGGGGAUCCUCUAGAGUCGACC
dsRNA-20 (Bevilacqua and Cech 1996):
5′-GGGUUCCCUGGUUUCGGUCU
dsRNA-40 (Heinicke et al. 2010):
5′-GGACCUGUGCGUGAUCCCUGGAGCAUCCUCUGUUACGUCC
ss-dsRNA
ss-dsRNA (9,11) (Zheng and Bevilacqua 2004):
5′-GGGAGAGAGGUCACUGACUAAGUUGGUGAAAUCUUGAUUUAUCAGUGACAAGAAGGAAGG
(Single-stranded tails of 9 and 11 nt are underlined.)
Protein
To generate K60A and K150A point mutations in PKR, which are known to abrogate dsRNA binding (Green and Mathews 1992; Green et al. 1995; Heinicke et al. 2009), site-directed mutagensis (QuikChange, Agilent Tech) was performed on pET28a-wtPKR using the following primers: K60A, 5′-CCAGAAGGTGAAGGTAGATCAGCGAAGGAAGC-3′ and K150A, 5′-CAGGTTCTACTGCACAGGAAGCAAAACAATTGG-3′, where the mutated codon is in bold. To generate the double mutant (dmPKR), point mutations were made sequentially, starting with K60A.
RNA preparation
As described previously, dsRNA-79 was prepared by transcribing the two strands of pUC19 separately and later annealing (Zheng and Bevilacqua 2004). Wild-type (pppG-) ssRNA-47 (first nucleotide is G) was prepared in vitro by a standard T7 run-off transcription from a linearized pUC19 plasmid (BstU1 digested) containing a T7 promoter, as previously described (Nallagatla et al. 2007). However, because WT T7 polymerase very strongly favors RNAs that start with a G (Milligan and Uhlenbeck 1989), a different strategy had to be used to get transcripts to begin with the other 5′-triphosphate nucleotides. The QuikChange site-directed mutagenesis kit was used to generate plasmids with mutations at the first templating position. In an effort to obtain pppA-, pppC, and pppU-ssRNA-47, plasmid sequences were verified by dideoxy sequencing following maxipreps and were digested as per the pppG-ssRNA-47 template. For each RNA, 0.2 μg/μL linearized plasmid was combined with 40 mM Tris (pH 8), 25 mM MgCl2, 2 mM DTT, 1 mM spermidine, 3 mM each NTP, and 1 μM T7 polymerase containing a P266L mutation (Milligan and Uhlenbeck 1989; Guillerez et al. 2005).
Transcription reactions were incubated for 4 h at 37°C and quenched by addition of one volume of 95% (v/v) formamide loading buffer (FLB). RNA was purified by fractionating on a polyacrylamide denature gel (7 M urea, 1× TBE). The transcript was identified by UV shadowing, excised from the gel, and eluted overnight at 4°C in 1× TEN250. The RNA was ethanol-precipitated, resuspended in 1× TE buffer (pH 7.5), and stored at −20°C. RNA concentration was determined spectrophotometrically.
To verify the identity of the starting nucleotide as pppG-, pppA-, pppC-, or pppU- in ssRNA-47, these RNAs were also transcribed in the presence of trace amounts of [γ-32P]ATP or [γ-32P]GTP. Because the radiolabel is on the γ-phosphate, a band should only be observed when the radioactive nucleotide is incorporated in the first position of the transcript.
RNAs starting with a bona fide 5′-OH (A-ssRNA-47 and G-ssRNA-47) were chemically synthesized by Dharmacon.
Protein preparation
Full-length PKR, K296R (catalytically inactive mutant), and P20 (dsRBD of PKR) containing N-terminal His6 tags were purified from Escherichia coli BL21(DE3) Rosetta cells (Novagen) as described (Bevilacqua and Cech 1996; Matsui et al. 2001; Zheng and Bevilacqua 2004). Briefly, cells were lysed by sonication, and protein was purified by FPLC with a Ni2+-NTA column (Invitrogen) followed by dialysis in a protein storage buffer (PSB) consisting of 10 mM Tris (pH 7.6), 50 mM KCl, 2 mM MgCl2, 10% glycerol, and 7 mM β-mercaptoethanol.
A dsRNA-binding deficient double mutant of PKR (dmPKR) was also purified in a similar fashion, with the following modifications to ensure removal of nucleic acids from the prep. The mutations in dmPKR are K60A in dsRBM1 and K150A in dsRBM2, which have been shown previously to interfere with binding of dsRNA even as single mutations (Green and Mathews 1992; Green et al. 1995; Heinicke et al. 2009). After sonication of cells containing overexpressed dmPKR, 5% polyethyleneimine (PEI) was added dropwise to the cleared cell lysate for a final concentration of 0.025% PEI. The lysate was then spun down at 20,000 rpm (Beckman) for 30 min, and the supernatant was collected. The protein was ammonium sulfate precipitated by slowly adding solid ammonium sulfate to the supernatant to a final percentage of 60% (w/v) followed by stirring for 30 min at 4°C. After centrifuging at 11,500 rpm for 30 min, the supernatant was decanted, and the pellet was redissolved in 50 mM sodium phosphate (pH 8.0), 700 mM NaCl, 5 mM imidizole, 7 mM BME, and 0.1 mM PMSF. Purification of dmPKR via FPLC and Ni2+-NTA then proceeded as described above.
The P266L mutant T7 polymerase was also prepared from E. coli ENS0134T BL21 cells (provided by Marc Dreyfus of Laboratoire de Genetique Moleculair) and purified on a Ni-NTA (Invitrogen) column, as described previously (He et al. 1997; Guillerez et al. 2005). Protein concentrations were determined spectrophotometrically.
PKR activation assays
The ssRNA-47 RNAs containing each of the four starting nucleotide triphosphates were tested for their ability to activate PKR autophosphorylation. Activation assays were identical to those conducted with canonical dsRNA activators. PKR was first dephosphorylated by treatment with λ-PPase (NEB) for 1 h at 30°C, followed by addition of freshly made phosphatase inhibitor, sodium orthovanadate (Matsui et al. 2001). Next, 15 μCi of [γ-32P]ATP (Perkin-Elmer), 0.8 μM dephosphorylated PKR, and RNA at various concentrations were incubated in 20 mM HEPES (pH 7.5), 4 mM MgCl2, 50 mM KCl, and 100 μM ATP (Ambion) for 10 min at 30°C. To determine specificity for the catalytic ATP-binding site, 15 μCi of [γ-32P]GTP and 100 μM GTP were added instead of [γ-32P]ATP and ATP. All reactions were quenched by addition of SDS loading buffer. Samples were heated for 5 min at 95°C and loaded on 10% SDS-PAGE gels (Pierce). After electrophoresis, gels were exposed to a storage PhosphorImager screen, and labeled band intensities were quantified on a PhosphorImager (Molecular Dynamics). A background value averaged from different portions of the gel was subtracted from each band before normalization. The 5′-triphosphate dependence for activation was calculated by first subtracting the amount of phosphorylation in the absence of added RNA from the bands in the presence of a 5′-triphosphate-containing RNA and the presence of a non-5′-triphosphate-containing RNA, and then dividing the resultant values (Nallagatla et al. 2007).
PKR activation competition assays
Two PKR activation competition assays were conducted: NTP-competition assays and RNA-competition assays. In the NTP-competition assays, each of the four NTPs was tested for its ability to compete for triphosphate-binding sites during PKR activation by dsRNA or ppp-ssRNA. Activation assays were conducted as described above, with the following exceptions: Various concentrations of ATP, CTP, GTP, and UTP (all complexed with equimolar Mg2+) were incubated along with other reaction components (see above). In the RNA-competition assays, the PKR activator, dsRNA, ssRNA, or heparin was competed by short dsRNA or ssRNA. In the RNA-competition assays, the activating RNA or heparin was held constant while competitors were added in a range of concentrations. All gels were run and analyzed as above.
Isothermal titration calorimetry
ITC was used to obtain thermodynamic parameters for binding of NTPs to K296R. Data were collected using a MicroCal Auto-iTC200 (GE Healthcare). The sample cell contained 40 μM K296R in PSB, while the syringe contained 720 μM ATP, GTP, CTP, or UTP dissolved in PSB as 100 mM stocks and diluted with further PSB. Titrations were performed at 30°C and involved 19 2-μL injections into the 200-μL sample cell (Sokoloski and Bevilacqua 2012). An ATP titration curve was fitted to a model for two binding sites using MicroCal Origin software (Version 7.0) with the average value of the last three points of the saturated portion of the curve used to baseline-correct the integrated data.
UV cross-linking
To induce cross-links between RNA and protein, the photoreactive nucleotide analog 4-thio-UTP was fully incorporated into ppp-ssRNA-47 at all uridine positions. This transcript was then 5′-32P-end-labeled, and 1 nM RNA was incubated with 4 μM wtPKR, P20, or dmPKR in 10 mM Tris (pH 7.6), 50 mM KCl, and 2 mM Mg(OAc)2•4H2O in a 20-μL reaction volume. Cross-linking reactions were immediately placed directly on the surface of a handheld UV lamp (UVP model UVGL-25), which had been turned upside-down and covered with Saran Wrap. Reaction samples were irradiated with 365-nm light (Mineralight lamp multiband UV-254/365). At various time points, 3-μL aliquots were quenched with 2× FLB. Samples were then fractionated on a 7%, 1× TBE, 7 M urea denaturing gel for 45 min, and the gel was dried and exposed to a PhosphorImager screen overnight.
Electrophoretic mobility shift assays (EMSAs)
To analyze binding to P20, excess protein (3 μM) and trace amounts of 5′-32P-end-labeled dsRNA-20 (∼2 nM) were incubated along with various unlabeled competitor RNAs or heparin and 1 mg/mL herring sperm DNA, 10 mM NaCl, 25 mM HEPES (pH 7.5), 5 mM DTT, 0.1 mM EDTA, 5% glycerol, 0.1 mg/mL bovine serum albumin, and 0.01% NP-40 for 30 min at 22°C (Bevilacqua and Cech 1996). Samples were then fractionated on 10% 0.5× TBE native gels (29:1 cross-link) for 2.5 h at 16°C. Gels were dried and exposed to storage PhosphorImager screens overnight.
SUPPLEMENTAL MATERIAL
Supplemental material containing controls for cross-linking experiments is available for this article.
ACKNOWLEDGMENTS
We thank C.K. Kwok for help with processing ITC data and for helpful discussions. We also thank O. Uhlenbeck for suggesting use of the P266L mutant T7 polymerase, as well as Marc Dreyfus at Laboratoire de Genetique Moleculair for the generous gift of ENS0134T BL21 cells containing plasmid encoding T7 RNAP with the P266L mutation. We thank S.R. Nallagatla for sharing unpublished data. This work is supported by National Institutes of Health Grant R01-58709.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.034520.112.
REFERENCES
- Anderson BR, Muramatsu H, Jha BK, Silverman RH, Weissman D, Kariko K 2010. Nucleoside modifications in RNA limit activation of 2′-5′-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res 39: 9329–9338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson E, Pierre-Louis WS, Wong CJ, Lary JW, Cole JL 2011. Heparin activates PKR by inducing dimerization. J Mol Biol 413: 973–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong RN, Kondo H, Kaiser ET 1979. Cyclic AMP-dependent ATPase activity of bovine heart protein kinase. Proc Natl Acad Sci 76: 722–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevilacqua PC, Cech TR 1996. Minor-groove recognition of double-stranded RNA by the double-stranded RNA-binding domain from the RNA-activated protein kinase PKR. Biochemistry 35: 9983–9994 [DOI] [PubMed] [Google Scholar]
- Bieger CD, Nierlich DP 1989. Distribution of 5′-triphosphate termini on the mRNA of Escherichia coli. J Bacteriol 171: 141–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff JR, Samuel CE 1985. Mechanism of interferon action. The interferon-induced phosphoprotein P1 possesses a double-stranded RNA-dependent ATP-binding site. J Biol Chem 260: 8237–8239 [PubMed] [Google Scholar]
- Cai Z, Liang TJ, Luo G 2004. Effects of mutations of the initiation nucleotides on hepatitis C virus RNA replication in the cell. J Virol 78: 3633–3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dar AC, Dever TE, Sicheri F 2005. Higher-order substrate recognition of eIF2α by the RNA-dependent protein kinase PKR. Cell 122: 887–900 [DOI] [PubMed] [Google Scholar]
- Dauber B, Martinez-Sobrido L, Schneider J, Hai R, Waibler Z, Kalinke U, Garcia-Sastre A, Wolff T 2009. Influenza B virus ribonucleoprotein is a potent activator of the antiviral kinase PKR. PLoS Pathog 5: e1000473 doi: 10.1371/journal.ppat.1000473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desselberger U, Racaniello VR, Zazra JJ, Palese P 1980. The 3′ and 5′-terminal sequences of influenza A, B and C virus RNA segments are highly conserved and show partial inverted complementarity. Gene 8: 315–328 [DOI] [PubMed] [Google Scholar]
- Fasciano S, Hutchins B, Handy I, Patel RC 2005a. Identification of the heparin-binding domains of the interferon-induced protein kinase, PKR. FEBS J 272: 1425–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasciano S, Patel RC, Handy I, Patel CV 2005b. Regulation of vascular smooth muscle proliferation by heparin: Inhibition of cyclin-dependent kinase 2 activity by p27kip1. J Biol Chem 280: 15682–15689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia MA, Gil J, Ventoso I, Guerra S, Domingo E, Rivas C, Esteban M 2006. Impact of protein kinase PKR in cell biology: From antiviral to antiproliferative action. Microbiol Mol Biol Rev 70: 1032–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George CX, Thomis DC, McCormack SJ, Svahn CM, Samuel CE 1996. Characterization of the heparin-mediated activation of PKR, the interferon-inducible RNA-dependent protein kinase. Virology 221: 180–188 [DOI] [PubMed] [Google Scholar]
- Green SR, Mathews MB 1992. Two RNA-binding motifs in the double-stranded RNA-activated protein kinase, DAI. Genes Dev 6: 2478–2490 [DOI] [PubMed] [Google Scholar]
- Green SR, Manche L, Mathews MB 1995. Two functionally distinct RNA-binding motifs in the regulatory domain of the protein kinase DAI. Mol Cell Biol 15: 358–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillerez J, Lopez PJ, Proux F, Launay H, Dreyfus M 2005. A mutation in T7 RNA polymerase that facilitates promoter clearance. Proc Natl Acad Sci 102: 5958–5963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Rong M, Lyakhov D, Gartenstein H, Diaz G, Castagna R, McAllister WT, Durbin RK 1997. Rapid mutagenesis and purification of phage RNA polymerases. Protein Expr Purif 9: 142–151 [DOI] [PubMed] [Google Scholar]
- Heinicke LA, Wong CJ, Lary J, Nallagatla SR, Diegelman-Parente A, Zheng X, Cole JL, Bevilacqua PC 2009. RNA dimerization promotes PKR dimerization and activation. J Mol Biol 390: 319–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinicke LA, Nallagatla SR, Hull CM, Bevilacqua PC 2010. RNA helical imperfections regulate activation of the protein kinase PKR: Effects of bulge position, size, and geometry. RNA 17: 957–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, et al. 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science 314: 994–997 [DOI] [PubMed] [Google Scholar]
- Hovanessian AG, Galabru J 1987. The double-stranded RNA-dependent protein kinase is also activated by heparin. Eur J Biochem 167: 467–473 [DOI] [PubMed] [Google Scholar]
- Johnson LN, Noble ME, Owen DJ 1996. Active and inactive protein kinases: Structural basis for regulation. Cell 85: 149–158 [DOI] [PubMed] [Google Scholar]
- Lemaire PA, Tessmer I, Craig R, Erie DA, Cole JL 2006. Unactivated PKR exists in an open conformation capable of binding nucleotides. Biochemistry 45: 9074–9084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire PA, Anderson E, Lary J, Cole JL 2008. Mechanism of PKR activation by dsRNA. J Mol Biol 381: 351–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G, Hamatake RK, Mathis DM, Racela J, Rigat KL, Lemm J, Colonno RJ 2000. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J Virol 74: 851–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manche L, Green SR, Schmedt C, Mathews MB 1992. Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol Cell Biol 12: 5238–5248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Tanihara K, Date T 2001. Expression of unphosphorylated form of human double-stranded RNA-activated protein kinase in Escherichia coli. Biochem Biophys Res Commun 284: 798–807 [DOI] [PubMed] [Google Scholar]
- Milligan JF, Uhlenbeck OC 1989. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol 180: 51–62 [DOI] [PubMed] [Google Scholar]
- Moss B, Keith JM, Gershowitz A, Ritchey MB, Palese P 1978. Common sequence at the 5′ ends of the segmented RNA genomes of influenza A and B viruses. J Virol 25: 312–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallagatla SR, Bevilacqua PC 2008. Nucleoside modifications modulate activation of the protein kinase PKR in an RNA structure-specific manner. RNA 14: 1201–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallagatla SR, Hwang J, Toroney R, Zheng X, Cameron CE, Bevilacqua PC 2007. 5′-Triphosphate-dependent activation of PKR by RNAs with short stem–loops. Science 318: 1455–1458 [DOI] [PubMed] [Google Scholar]
- Nallagatla SR, Toroney R, Bevilacqua PC 2011. Regulation of innate immunity through RNA structure and the protein kinase PKR. Curr Opin Struct Biol 21: 119–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niefind K, Putter M, Guerra B, Issinger OG, Schomburg D 1999. GTP plus water mimic ATP in the active site of protein kinase CK2. Nat Struct Biol 6: 1100–1103 [DOI] [PubMed] [Google Scholar]
- Patel RC, Stanton P, Sen GC 1994. Role of the amino-terminal residues of the interferon-induced protein kinase in its activation by double-stranded RNA and heparin. J Biol Chem 269: 18593–18598 [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314: 997–1001 [DOI] [PubMed] [Google Scholar]
- Roy CR, Mocarski ES 2007. Pathogen subversion of cell-intrinsic innate immunity. Nat Immunol 8: 1179–1187 [DOI] [PubMed] [Google Scholar]
- Schlee M, Roth A, Hornung V, Hagmann CA, Wimmenauer V, Barchet W, Coch C, Janke M, Mihailovic A, Wardle G, et al. 2009. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity 31: 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setyawan J, Koide K, Diller TC, Bunnage ME, Taylor SS, Nicolaou KC, Brunton LL 1999. Inhibition of protein kinases by balanol: Specificity within the serine/threonine protein kinase subfamily. Mol Pharmacol 56: 370–376 [DOI] [PubMed] [Google Scholar]
- Shugar D 1996. The NTP phosphate donor in kinase reactions: Is ATP a monopolist? Acta Biochim Pol 43: 9–23 [PubMed] [Google Scholar]
- Sigel H 1987. Isomeric equilibria in complexes of adenosine 5′-triphosphate with divalent metal ions. Solution structures of M(ATP)2- complexes. Eur J Biochem 165: 65–72 [DOI] [PubMed] [Google Scholar]
- Smith CA, Toth M, Frase H, Byrnes LJ, Vakulenko SB 2012. Aminoglycoside 2′-phosphotransferase IIIa (APH(2′)-IIIa) prefers GTP over ATP: Structural templates for nucleotide recognition in the bacterial aminoglycoside-2′ kinases. J Biol Chem 287: 12893–12903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokoloski JE, Bevilacqua PC 2012. Analysis of RNA folding and ligand binding by conventional and high-throughput calorimetry. Methods Mol Biol 905: 145–174 [DOI] [PubMed] [Google Scholar]
- Toroney R. 2010 “Investigating roles of RNA primary, secondary, and tertiary motifs in regulation of the protein kinase PKR.” PhD thesis, Pennsylvania State University, University Park. [Google Scholar]
- Zheng X, Bevilacqua PC 2004. Activation of the protein kinase PKR by short double-stranded RNAs with single-stranded tails. RNA 10: 1934–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]