Precursor let-7 miRNAs are degraded in undifferentiated cells and cancer cells in the presence of the small RNA-binding protein Lin28 and the Terminal Uridyl Transferase (TUTase) Zcchc11/TUTase 4. While recent efforts have focused on the role of Lin28 in the repression of let-7, little is known regarding the mechanism by which Zcchc11 mediates the uridylation of this miRNA. Here the authors perform mutational analyses in vitro to identify the domains of Zcchc11 necessary and sufficient for the uridylation of pre-let-7. The authors find that another TUTase, Zcchc6/TUTase 7, shares these critical domains and functions redundantly with Zcchc11 in vitro and in vivo to repress let-7 levels in embryonic stem cells.
Keywords: Lin-28, Lin28, let-7, microRNA (miRNA), TUTase, Zcchc11, Zcchc6, TUTase4, TUTase7, embryonic stem cells
Abstract
The pluripotency factor Lin28 recruits a 3′ terminal uridylyl transferase (TUTase) to selectively block let-7 microRNA biogenesis in undifferentiated cells. Zcchc11 (TUTase4/TUT4) was previously identified as an enzyme responsible for Lin28-mediated pre-let-7 uridylation and control of let-7 expression. Here we investigate the protein and RNA determinants for this interaction. Biochemical dissection and reconstitution assays reveal the TUTase domains necessary and sufficient for Lin28-enhanced pre-let-7 uridylation. A single C2H2-type zinc finger domain of Zcchc11 was found to be responsible for the functional interaction with Lin28. We identify Zcchc6 (TUTase7) as an alternative TUTase that functions with Lin28 in vitro, and accordingly, we find Zcchc11 and Zcchc6 redundantly control let-7 biogenesis in embryonic stem cells. Our study indicates that Lin28 uses two different TUTases to control let-7 expression and has important implications for stem cell biology as well as cancer.
INTRODUCTION
microRNAs (miRNAs) are small, 22-nucleotide (nt) noncoding RNAs that repress the expression of many target messenger RNAs (mRNAs) (Wightman et al. 1993). The canonical process of miRNA biogenesis is well understood and is characterized by successive cleavage events by RNase III enzymes (Winter et al. 2009). After transcription by RNA polymerase II, primary miRNA transcripts (pri-miRNAs) are cleaved by the Microprocessor complex consisting of the RNAse III enzyme Drosha and its essential double-stranded RNA-binding partner DGCR8, yielding a short hairpin miRNA precursor (pre-miRNA) (Denli et al. 2004; Gregory et al. 2004; Han et al. 2006). Pre-miRNAs are exported to the cytoplasm by Exportin 5 where they are processed by Dicer, another RNase III enzyme, yielding the canonical 22-nt miRNA duplex (mature miRNA) (Hutvagner et al. 2001). The mature miRNA undergoes strand selection, where one strand of the duplex (guide strand) is preferentially incorporated into the miRNA-Induced Silencing Complex (miRISC) over the other strand (passenger or miR* strand). At its center, miRISC contains an Argonaute protein, and through base-pairing between the mature miRNA and the 3′ UTR of a target mRNA mediates translational inhibition and/or mRNA decay (Bartel 2009; Fabian et al. 2010).
Proper temporal and spatial expression of miRNAs is essential for normal development and physiology, as perturbations in specific miRNAs or miRNA processing factors can lead to aberrant development and cancer (Calin and Croce 2006; Esquela-Kerscher and Slack 2006; Small and Olson 2011). In embryonic cells, the RNA-binding protein Lin28 coordinately represses the let-7 family of miRNAs by binding to the terminal loop (also known as pre-element or preE) of pre- and pri-let-7 miRNAs, thereby inhibiting let-7 biogenesis (Heo et al. 2008; Newman et al. 2008; Rybak et al. 2008; Viswanathan et al. 2008). As cells undergo differentiation, Lin28 levels decrease, leading to a corresponding increase in mature let-7, which is retained in many adult tissues (Martinez and Gregory 2010). Furthermore, Lin28 mRNA is repressed by let-7 miRNAs, leading to an inversely correlated expression pattern between let-7 and Lin28 and a double-negative feedback loop that controls cell differentiation (Wu and Belasco 2005). Lin28 is required for normal development and contributes to the pluripotent state by preventing let-7-mediated differentiation of embryonic stem cells (ESCs) (Ambros and Horvitz 1984; Moss et al. 1997; Viswanathan and Daley 2010). Lin28 overexpression or let-7 inhibition with antisense RNAs promotes reprogramming of human and mouse fibroblasts to induced pluripotent stem cells (iPSCs) (Ambros and Horvitz 1984; Yu et al. 2007; Melton et al. 2010). The Lin28/let-7 axis is also relevant to a wide variety of human cancers as well as the control of glucose homeostasis in mammals (Iliopoulos et al. 2009; Viswanathan et al. 2009; Frost and Olson 2011; Piskounova et al. 2011; Zhu et al. 2011).
In mammals there are two Lin28 paralogs, Lin28A (Lin28) and Lin28B. Lin28A recognizes pre-let-7 in the cytoplasm and recruits the terminal uridyl transferase (TUTase) Zcchc11 (TUTase4/TUT4) to add an oligouridine tail to the 3′ end of pre-let-7, blocking Dicer cleavage and leading to the degradation of the pre-miRNA (Heo et al. 2008, 2009; Hagan et al. 2009). Lin28B is predominantly localized to the nucleus and blocks pri-miRNA processing through a TUTase-independent mechanism (Piskounova et al. 2011). Most cells that express a Lin28 family member do so selectively, such that either Lin28A or Lin28B is exclusively expressed. This selective expression and differential localization of Lin28 family members allows for the repression of let-7 by distinct mechanisms in different cell and tumor types. The recently identified TUTase Zcchc11 may also regulate IL-6 levels by uridylating mature miR-26a, promote the cell-cycle-dependent degradation of a subset of histone mRNAs, and is required for the growth of Lin28A-driven cancers in vitro and in vivo (Jones et al. 2009; Piskounova et al. 2011; Schmidt et al. 2011). Given its central role in processes ranging from the inflammatory response to cell cycle regulation and Lin28-mediated repression of let-7, Zcchc11 is an important RNA-modifying enzyme that may have essential roles in diverse aspects of human biology. However, very little is known about the cis-acting elements of mammalian TUTases or how TUTases interact with their binding partners.
In this study we set out to examine the mechanism by which Zcchc11 represses pre-let-7 in a Lin28-dependent manner. Mutational analyses of Zcchc11 identified domains required for activity both in the absence and presence of Lin28, and using recombinant proteins we were able to show that Lin28 and Zcchc11 proteins are sufficient for uridylation of pre-let-7 in vitro. Furthermore, we found that the single C2H2-type zinc finger at the N terminus of Zcchc11 mediates the functional interaction with Lin28. Comparing the domain architecture of Zcchc11 to other mammalian TUTases, we identified Zcchc6, another TUTase with extensive homology with Zcchc11, which also mediates Lin28-dependent uridylation of pre-let-7 in vitro. Accordingly, we found Zcchc6 depletion in embryonic cells synergized with Zcchc11 knockdown to up-regulate let-7 miRNAs, implying that these two TUTases work redundantly to repress let-7 expression. These findings provide insight into the mechanism of Lin28-mediated TUTase control of let-7 expression in development, stem cells, and cancer.
RESULTS
Domains of Zcchc11 required for Lin28-enhanced pre-miRNA uridylation
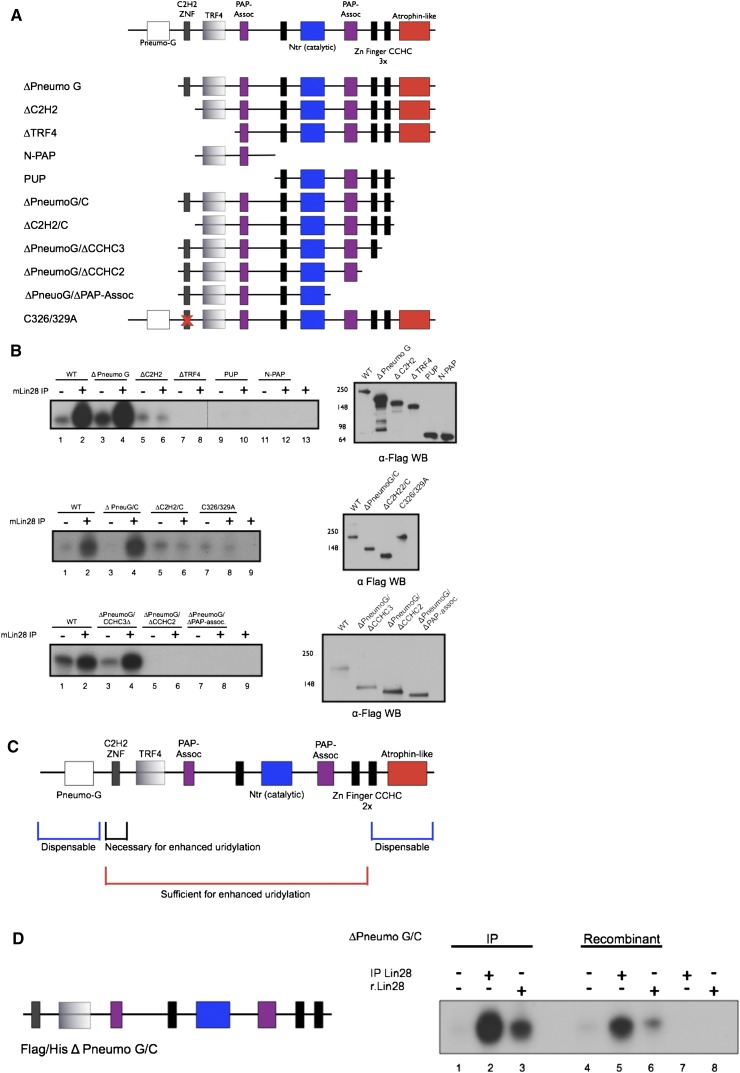
Previous work from our group and others identified Zcchc11 as a cytoplasmic Lin28-interacting TUTase in embryonic and cancer cells (Hagan et al. 2009; Heo et al. 2009; Piskounova et al. 2011). Its depletion in Lin28-expressing cells leads to the specific up-regulation of let-7 family members similar to the depletion of Lin28, and its expression is required for potent let-7 repression and rapid cell growth in Lin28A-expressing cancers (Piskounova et al. 2011). Zcchc11 encodes a 184-kDa noncanonical poly(A) polymerase that is highly conserved across vertebrates. The Zcchc11 active site is located within the Nucleotidyl Transferase (Ntr) domain, which is paired with a Poly(A)-Polymerase-Associated (PAP) domain; a common feature of noncanonical poly(A) polymerases (Saitoh et al. 2002; Kwak and Wickens 2007; Martin and Keller 2007). Catalysis requires a conserved Aspartate triad in the Ntr. and when a mutant lacking these residues is overexpressed it functions as a dominant negative (Hagan et al. 2009). Flanking the active site are three CCHC retroviral-type zinc fingers/zinc knuckles, which are implicated in nucleic acid binding. At the N terminus of the protein is a region that shares significant homology with the active site, including a proximal PAP domain; however, this region lacks one of the crucial Aspartates predicted to be necessary for catalysis. Instead, this region is most similar to the yeast TRF4 proteins, which carry out cytoplasmic poly(A) RNA polymerase activity (Saitoh et al. 2002). N-terminal to this region is a classical C2H2 zinc finger with no known function, but these motifs are known to bind DNA, RNA, or protein. Finally, at the N- and C-termini of Zcchc11 there are two domains of unknown function similar, respectively, to pneumoviridae attachment proteins and the glutamine-rich neurodegenerative disease-associated protein atrophin-1 (Fig. 1A).
FIGURE 1.
Domains of Zcchc11 required for Lin28-mediated pre-let-7 uridylation. (A) Schematic representation of Zcchc11 and truncations used for in vitro uridylation assays. (B) Uridylation assays with synthetic pre-let-7g carried out using Flag immunopurified (IP) Zcchc11 variants and IP Lin28. α-Flag Western blots show similar amounts of IP Zcchc11 within experiments. (C) Summary of Zcchc11 domain requirements from in vitro uridylation assays. (D) Reconstitution of Lin28-enhanced pre-let-7 uridylation with recombinant proteins. Zcchc11 truncation ΔPneumoG/C purified from either HEK293T (IP) or E. coli (Recombinant) was incubated with either Flag-Lin28 (IP) or 6x-His Lin28 (r.Lin28) in a uridylation assay with synthetic pre-let-7g. (Left) Schematic representation of the domains present in ΔPneumoG/C.
To understand which domains of Zcchc11 are required for uridylation activity, we generated a series of mutant cDNAs and tested the ability of the resulting Flag-immunopurified (Flag IP) proteins to uridylate synthetic pre-let-7 miRNA in vitro. Mutants were generated lacking N- and C-terminal domains or harboring point mutations in conserved residues (Fig. 1A). As described previously, Zcchc11 exhibits a low level of uridylation against pre-let-7 and this activity is strongly enhanced by the addition of recombinant or immunopurified Lin28 (Hagan et al. 2009; Heo et al. 2009). As determined by the incorporation of radiolabeled UTP, the activity of wild-type Zcchc11 was compared with each of the series of mutants (Fig. 1B). We found that after performing serial N-terminal truncations, the pneumoviridae (PneumoG) domain was dispensable for both basal level uridylation and activity enhanced by IP Flag-Lin28. Loss of the N-terminal C2H2 domain still allowed for basal activity, but this mutant could no longer support Lin28-enhanced uridylation against pre-let-7, indicating that the C2H2 zinc finger may be essential for the interaction between Lin28 and Zcchc11. Interestingly, when the TRF4 domain was deleted we could not detect either basal or Lin28-enhanced activity. This result was surprising given the prediction that the TRF4 domain was insufficient to carry out catalytic activity on its own. Indeed, when we tested a fragment of Zcchc11 containing the N-terminal TRF4 domain but lacking the NTR domain, no uridylation activity was detected (Fig. 1B, top, cf. lanes 7,8–11,12). All further mutant proteins tested lacking TRF4 failed to support any detectable uridylation activity (Fig. 1B, top, lanes 9,10; data not shown).
To determine whether our findings on N-terminal deletions of Zcchc11 could be supported in the context of additional C-terminal truncations, we tested mutants lacking the C-terminal Atrophin-like domain in combination with ΔPneumoG and ΔC2H2 mutants. The Atrophin-like domain was dispensable in these experiments, indicating that it is not required for basal or Lin28-enhanced uridylation by Zcchc11. To confirm that the C2H2 zinc finger per se was required for Lin28-enhanced uridylation, a full-length Zcchc11 cDNA was generated bearing Cysteine to Alanine mutations in the residues predicted to be central to the C2H2 zinc finger (C326/329A). Indeed, this mutant exhibited only basal uridylation activity, as the addition of Lin28 had no impact on its catalysis in vitro. Given that this mutant phenocopied the ΔC2H2 and ΔC2H2/C mutants, we conclude that this zinc finger is required for Lin28-enhanced uridylation in vitro.
To define the minimal Zcchc11 mutant that supports Lin28-enhanced uridylation, we further examined the requirements of C-terminal domains. Compared with wild type, a mutant lacking the C-terminal-most CCHC zinc finger exhibited robust basal and Lin28-enhanced activity, whereas additionally truncating the adjacent CCHC zinc finger led to no detectable activity, implying that the three CCHC zinc fingers may be required for different aspects of RNA recognition or positioning (Fig. 1B, bottom). These studies provide insight into the basic mechanism underlying the catalytic nature of Zcchc11 (Fig. 1C).
In vitro reconstitution of Lin28-mediated pre-let-7 uridylation with recombinant proteins
The experiments described above suggest that specific domains of Zcchc11 mediate the interaction with Lin28 to uridylate pre-let-7 in vitro. To confirm that these two proteins are sufficient for activity and do not rely on contaminating or accessory factors interacting with the immunopurified proteins, we purified 6x-His Lin28 (r.Lin28) and Flag/6x-His ΔPneumo G/C Zcchc11 from Escherichia coli. Compared with immunopurified ΔPneumo G/C Zcchc11, the Flag/6x-His protein uridylated pre-let-7 at the basal level to a similar extent, indicating that the Zcchc11 expressed and purified from bacteria is catalytically active (Fig. 1D). Adding either immunopurified Flag-Lin28 or recombinant 6x-His Lin28 to the reaction similarly enhanced the uridylation of pre-let-7 by either Zcchc11 preparation. Neither of the Lin28 proteins themselves led to detectable levels of uridylated pre-let-7, indicating that labeled products originated from the enzymatic activity of Zcchc11. These experiments show that the combination of Lin28 and Zcchc11 proteins are necessary and sufficient to carry out the robust uridylation of pre-let-7 in vitro (Fig. 1D).
The let-7 preE confers Lin28-enhanced pre-miRNA uridylation by Zcchc11
To understand the role of pre-miRNA substrates in Zcchc11-mediated uridylation, we investigated which cis-acting RNA elements support uridylation enhanced by Lin28. Lin28 binding to pre-let-7 requires specific sequence and structural information in both the RNA and the protein. The cold-shock domain (CSD) of Lin28 is inserted into the terminal loop of various pre-let-7 RNAs, and the Lin28 CCHC zinc fingers dimerize to recognize a GGAG motif proximal to the Dicer cleavage site of pre-let-7 (Loughlin et al. 2011; Nam et al. 2011). However, to this point the various domains of pre-let-7 required for Zcchc11-mediated uridylation are unknown.
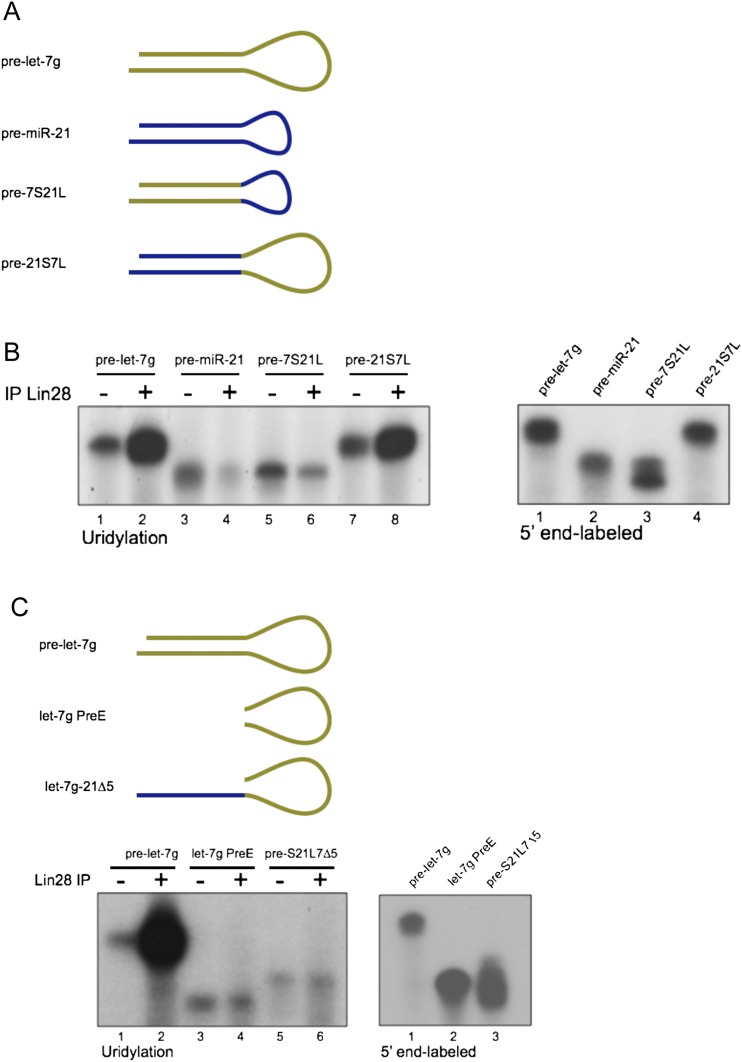
To determine the regions of pre-let-7 required for uridylation by Zcchc11, we took advantage of the understanding that the let-7 preE is bound by recombinant Lin28 as efficiently as full-length pre-let-7 (Piskounova et al. 2008). If Lin28 binding is sufficient to direct Zcchc11-mediated uridylation, then RNA substrates with divergent sequences outside of the let-7 preE should be comparable substrates to pre-let-7. To test this, we generated two synthetic pre-miRNAs; one composed of the preE of let-7g and the stem sequence of miR-21 (pre-21S7L) and another composed of the preE of miR-21 and the stem of let-7g (pre-7S21L) (Fig. 2A). We compared the uridylation activity of Zcchc11 toward these chimeric RNAs versus both pre-let-7g and pre-miR-21. As shown in Figure 2B, pre-let-7g undergoes robust uridylation with the addition of IP Flag-Lin28. miR-21 is uridylated at a basal level similar to that of pre-let-7, but the addition of IP Flag-Lin28 has no effect on uridylation levels, as described previously (Hagan et al. 2009). Zcchc11 also only exhibits basal activity toward pre-7S21L; however, when the chimeric pre-21S7L is incubated with IP Flag-Lin28, it is subjected to enhanced uridylation activity similar to that of WT pre-let-7 (Fig. 2B, cf. lanes 7 and 8 with 1 and 2). This result suggests that the effect of Lin28 binding to the preE of pre-let-7 is sufficient to allow targeting and uridylation by Zcchc11.
FIGURE 2.
The preE of let-7 is sufficient to direct both Lin28 binding and uridylation of pre-let-7. (A) Diagram of synthetic RNAs used for in vitro uridylation assays. (Pre-let-7g) Endogenous precursor let-7g miRNA sequence; (Pre-miR-21) endogenous precursor miR-21 sequence; (Pre-7S21L) synthetic RNA consisting of the miR-21 preE and let-7g stem; (Pre-21S7L) synthetic RNA consisting of the let-7g preE and miR-21 stem sequences. (B, left) Uridylation assay as in Figure 1 using WT Flag IP-mZcchc11, with or without Flag IP-mLin28, and the indicated precursor miRNAs. (Right) 5′ end-labeled RNAs showing equal amounts. (C) (Top) Diagram of synthetic RNAs used for in vitro uridylation assays. (Bottom left) Uridylation assays and (bottom right) 5′ end-labeled RNAs showing equal amounts as in B.
To further examine the RNA determinants supporting Zcchc11 and Lin28-mediated uridylation, we monitored the substrate preference of several other synthetic RNAs in in vitro uridylation assays. One explanation for the sufficiency of the let-7 preE to support uridylation is that pre-miRNA stem regions are dispensable altogether and the loop alone encodes all necessary regulatory information. To test this hypothesis we performed uridylation assays on pre-let-7g, the preE of let-7g alone, or a chimeric RNA bearing the preE of let-7g with only the 3′ stem region of miR-21 (pre-S21L7Δ5). Compared with pre-let-7g, neither let-7g preE nor pre-S21L7Δ5 underwent Lin28-enhanced uridylation, indicating the necessity of an intact pre-miRNA stem to drive this activity (Fig. 2C).
A related TUTase Zcchc6 is functionally redundant with Zcchc11 in vitro
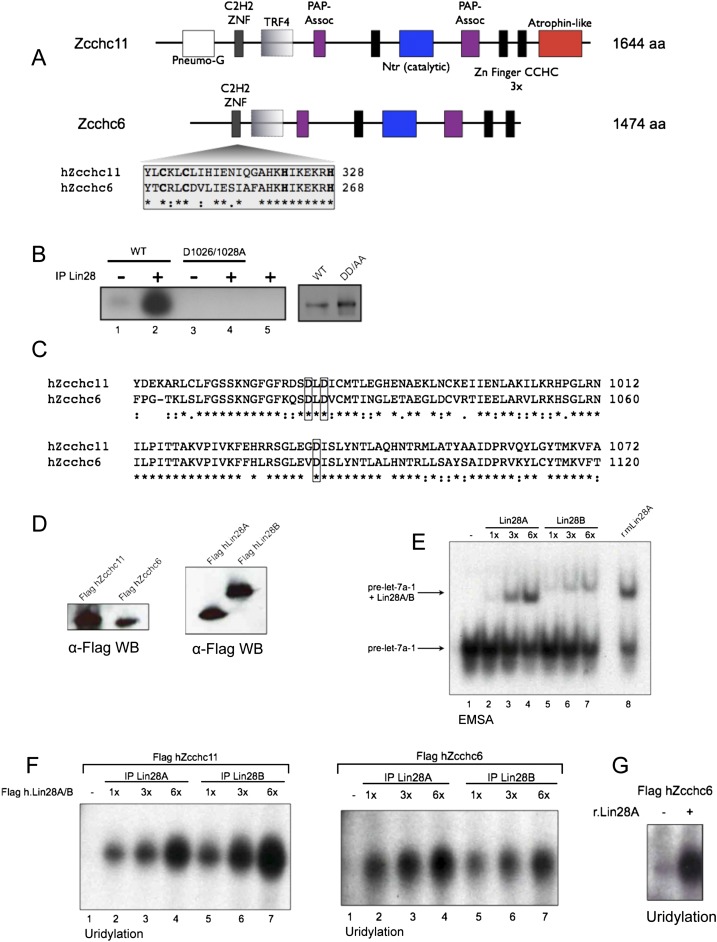
Our findings on the domains of Zcchc11 supporting Lin28-mediated uridylation in vitro led us to examine other TUTases as potential regulators of pre-miRNAs. Among the seven noncanonical poly(A) polymerases encoded in the human genome, we found that Zcchc6 (PAPD6/TUTase 7) has striking homology with Zcchc11, including the domains constituting its active site, its three CCHC zinc fingers, the N-terminal TRF4/PAP-associated domains, and C2H2 zinc finger (Fig. 3A). Importantly, there is extensive conservation between Zcchc11 and Zcchc6 at critical residues in the active site and in the C2H2 zinc finger (Fig. 3A–C). To determine whether Zcchc6 shares activity similar to Zcchc11, we tested the ability of IP Flag-hZcchc6 to uridylate pre-let-7 in vitro in the absence or presence of Lin28. Similar amounts of Flag-hZcchc11 or Flag-hZcchc6 were used in uridylation assays with Flag-hLin28A and both TUTases were stimulated to an equal extent (Fig. 3D,E). To confirm that the effects seen with Flag-hLin28A were not dependent on the paralog of Lin28 used, we also tested the stimulatory effect of Flag-hLin28B, since both Lin28 proteins act identically in vitro (Fig. 3D–F; Heo et al. 2009). In these experiments either Lin28A or Lin28B enhanced the uridylation activity of either TUTase in a dose-dependent manner. The enhancement in hZcchc6 uridylation activity was also observed using r.Lin28, indicating that this effect was not due to coimmunoprecipating proteins and that Zcchc6 and Zchc11 are functionally indistinguishable in these assays (Fig. 3G).
FIGURE 3.
Zcchc11 and Zcchc6 have a highly similar domain organization and can both mediate Lin28-dependent pre-let-7 uridylation in vitro. (A) Schematic showing the domain similarities between hZcchc11 and hZcchc6 with the N-terminal C2H2 zinc finger highlighted and critical zinc finger residues in bold. (B) Uridylation assay with Flag-IP WT mZcchc11 and a mutant harboring point mutations in two conserved asparates required for catalysis, with or without Flag-IP Lin28. (C) Alignment of the nucleotidyl transferase (Ntr) domains of hZcchc11 and hZcchc6. Aspartic acid residues critical for catalysis are boxed. (D) α-Flag WB showing relative amounts of Flag-hZcchc11 and Flag-hZcchc6 (left) or Flag-hLin28A and Flag-hLin28B (right). (E) EMSA showing similar amounts of functional Flag-hLin28A and Flag-hLin28B used in uridylation assays. (F) Uridylation assays using Flag-hZcchc11 or Flag-hZcchc6 with either Flag-hLin28A or Flag-hLin28B. (G) Uridylation assay with Flag-hZcchc6 and r.Lin28A.
These results in vitro suggest that both TUTases may recognize let-7 precursors in biologically relevant settings. Zcchc6 has previously been shown to have poly(U) activity in vitro (Kwak and Wickens 2007; Rissland et al. 2007), and depletion of Zcchc6 in colon cancer cells led to reduced levels of uridylated mature let-7e (Wyman et al. 2011). Zcchc6 is also a homolog of Caenorhabditis elegans CDE-1, which uridylates a subset of siRNAs bound by the Argonaute protein CSR-1, and loss of CDE-1 leads to aberrant chromosomal segregation and dysregulation of CSR-1-bound siRNAs (van Wolfswinkel et al. 2009). In spite of these data, this is the first evidence of Zcchc6 uridylating pre-miRNAs and suggests parallel activity with Zcchc11 and a role in the Lin28 pathway.
Zcchc11 and Zcchc6 redundantly control let-7 biogenesis in embryonic stem cells
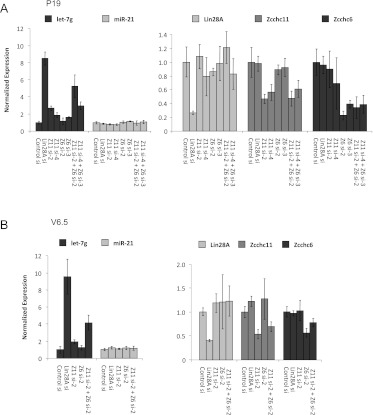
Given the findings on the activity of Zcchc6 in vitro, we investigated whether Zcchc6 functions in parallel with Zcchc11 in vivo. We have previously shown that Zcchc11 depletion in embryonic carcinoma (EC) and embryonic stem (ES) cells led to the coordinate derepression of let-7 miRNAs, while Zcchc6 depletion led to no change in mature let-7 levels (Hagan et al. 2009; Heo et al. 2009). The derepression observed upon Zcchc11 knockdown was, however, generally more modest than the depletion of Lin28 in all cell types tested (Hagan et al. 2009; Heo et al. 2009; Piskounova et al. 2011). One interpretation of this finding is that there are redundant factors working in parallel with Lin28 and Zcchc11 to repress let-7 miRNAs in undifferentiated cell types. To test whether Zcchc6 works redundantly with Zcchc11, we used siRNAs to deplete both TUTases in P19 and V6.5 cell lines (EC and ES cells, respectively). Upon Zcchc11 knockdown with two independent siRNAs there was a modest two- to threefold up-regulation of mature let-7g, as we have previously shown, whereas consistent with previous reports, depletion of Zcchc6 with two independent siRNAs led to no significant changes at the level of mature let-7g. When both TUTases were knocked down, however, we observed a consistent up-regulation in mature let-7 that was more dramatic than either individual knockdown alone (Fig. 4). This trend was specific to let-7 family members, as levels of the unrelated miRNA miR-21 were unchanged. Moreover, global profiling revealed that changes in miRNA expression were restricted to let-7 family members (data not shown). This trend was seen in both P19s and V6.5s, suggesting that Zcchc11/Zcchc6 redundancy is a general mechanism of embryonic cells. The synergistic relationship between these two related TUTases, both in vitro and in vivo, could explain the modest effects seen for depletion of either TUTase individually in Lin28-expressing cells and expands the repertoire of miRNA-modifying enzymes.
FIGURE 4.
Zcchc11 and Zcchc6 function redundantly to suppress let-7 expression in embryonic cells. (A) qRT–PCR analysis of mature let-7g and mature miR-21 levels in P19 embryonal carcinoma cells after transfection with the indicated siRNAs (left). mRNA levels of the indicated genes in P19 cells after transfection with the indicated siRNAs (right). (B) qRT–PCR as in A in V6.5 mouse embryonic stem cells. For all experiments, miRNA levels were normalized to sno-142 and mRNA levels were normalized to β-actin. Error bars represent SD of experiments in triplicate.
DISCUSSION
Recent work examining the role of miRNAs in development and cancer has revealed extensive post-transcriptional control at various levels of miRNA biogenesis (Siomi and Siomi 2010). Lin28 (Lin28A) and Lin28B have emerged as important post-transcriptional regulators of let-7 expression in stem cells, development, metabolism, and disease (Viswanathan and Daley 2010). In the case of Lin28A, this regulation involves the recruitment of a TUTase Zcchc11 to catalyze the 3′ terminal uridylation of pre-let-7 RNAs. Several studies have identified extensive nontemplated nucleotide addition to the 3′ ends of mature and precursor miRNAs (Heo et al. 2008; Jones et al. 2009; Katoh et al. 2009; Lehrbach et al. 2009; Ameres et al. 2010; Burroughs et al. 2010; Chiang et al. 2010; Berezikov et al. 2011; Newman et al. 2011). Our work here is the first extensive mechanistic analysis of one of these enzymes, Zcchc11. First, we have uncovered specific domains that are required for mediating efficient Lin28-endhanced uridylation of pre-let-7 in vitro. Of the four zinc fingers encoded in Zcchc11, the unique C2H2 zinc finger at the N terminus of the protein mediates the functional interaction with Lin28, as point mutations in conserved Cysteine residues of this zinc finger abolish Lin28-enhanced uridylation activity. The TRF4 domain at the N terminus of Zcchc11, while incapable of supporting uridylation activity on its own, is nonetheless required for activity in vitro. This essential role may explain its significant degree of conservation across taxa. Furthermore, the CCHC zinc fingers, which define a class of at least 13 mammalian proteins, are differentially required for uridylation activity in vitro. Specifically, the C-terminal-most CCHC zinc finger is dispensable for in vitro activity, while the zinc finger just C-terminal to the active site is required for any detectable activity. Finally, there are regions dispensable for Lin28-enhanced uridylation at the N- and C-termini of Zcchc11 (Fig. 1C). Both of these domains are of unknown function but remain conserved in other organisms. Since Zcchc11 has been implicated in several other biological pathways, we cannot rule out that the domains identified as dispensable are required for other processes. Indeed, a recent study has identified the N-terminal portion of Zcchc11, which lacked any catalytic regions, to be sufficient to alter the cell cycle of cultured human cancer cells (Blahna et al. 2011). Our study unveils critical domains and residues that are required for Lin28-dependent Zcchc11 activity. Though still controversial, the Lin28-mediated control of let-7 expression in C. elegans has also been reported to involve pre-let-7 uridylation (Lehrbach et al. 2009; Van Wynsberghe et al. 2011). Notably, however, the proposed Zcchc11 ortholog, PUP-2, lacks the C2H2 domain that we find mediates the functional interaction between Lin28 and Zcchc11 (Lehrbach et al. 2009). Therefore it remains unclear whether and how Lin28 in worms recruits PUP-2 to repress let-7 expression.
We also examined the requirements of pre-let-7 that mediate Lin28-enhanced activity, and we found the preE of let-7g in the context of an intact pre-miRNA to be sufficient to direct this activity. Although Zcchc11 recognizes and uridylates the 3′ end of pre-let-7 family members and other miRNAs, this occurs through a mechanism that is independent of sequence information proximal to the site of uridylation. Instead, Lin28 bound to an intact preE sequence is sufficient to direct robust uridylation of the pre-miRNA. Although the preE in our studies contains the Lin28-binding motif of GGAG, this sequence was previously shown to be insufficient in directing uridylation activity toward pre-let-7, as gain-of-function experiments indicated that the positioning of the motif relative to the Dicer cleavage site was also essential (Heo et al. 2009). The chimeric pre-21S7L, however, has the GGAG motif positioned not in the preferred position ending 4 nt before the Dicer cleavage site, but only 2 nt away from this point, suggesting that there are other sequence or structural determinants directing Zcchc11-mediated uridylation against pre-let-7 miRNAs. Furthermore, although the preE of let-7 is sufficient for Lin28-enhanced uridylation activity, an intact pre-miRNA structure is required, since a viable substrate lacking one arm of the pre-miRNA stem was no longer targeted for robust uridylation. This requirement likely comes from RNA–protein interactions between Zcchc11 and duplex-form RNA, because recent structural studies have shown the conserved domains of Lin28 interacting exclusively with the PreE of several let-7 family members. The role of the protein domains of Zcchc11 mediating this dsRNA interaction warrant further research. We rule out the role of other protein factors giving further specificity to pre-let-7 uridylation since the reaction could be reconstituted from recombinant proteins produced in bacteria, but what defines this level of specificity remains unknown. Recent structural studies have uncovered the degree to which the let-7 preE is altered by bound Lin28, revealing a partial unwinding of the duplex region near the site of Dicer cleavage (Nam et al. 2011). While it is unknown how far this melting proceeds into the stem of pre-let-7, this structural change could alter the RNA so that it is a preferred substrate of Zcchc11. Indeed, a recent structural study showed that the CCHC zinc fingers of Lin28 preferentially bind the single-stranded heptad sequence of AGGAGAU in the stem of pre-let-7, providing evidence of sequence-specific RNA binding by zinc finger-containing proteins (Loughlin et al. 2011). Alternatively, pre-let-7-bound Lin28 may undergo a conformational change, and this may provide a suitable protein–protein interaction surface between the Lin28–let-7 complex and Zcchc11 (Nam et al. 2011). More detailed RNA mutagenesis and/or structural studies examining the interplay between pre-let-7 and these two RNA-binding proteins should provide additional insight into precisely how Lin28 functionally enhances Zcchc11 recognition of the let-7 preE. The uridylation and adenylation of mature miRNAs by Zcchc11 has also been reported (Jones et al. 2009; Wyman et al. 2011). Since Zcchc11 exhibits similar basal activity toward unrelated pre-miRNAs (Fig. 2) there may be other sequence-specific recognition factors that guide Zcchc11 activity toward other RNA substrates including mature miRNAs.
The findings in our mutational analysis led us to investigate other putative TUTases and to the identification of Zcchc6 as a regulator of let-7 expression. One study investigating the potential redundancy between Zcchc11 and Zcchc6 found that only Zcchc11 was capable of binding stem–loop containing histone mRNAs, while Zcchc6 appeared to lack this capacity (Schmidt et al. 2011). In the case of Lin28 and let-7, however, we found Zcchc6 functioned identically to Zcchc11 in vitro, as its enzymatic activity against a synthetic let-7 precursor was enhanced by either Lin28A or Lin28B, as was previously shown for Zcchc11 (Heo et al. 2009). We furthermore found Zcchc6 to be crucial in efficiently repressing mature let-7 miRNAs in embryonic cells. Although the double knockdown of Zcchc11 and Zcchc6 led to more dramatic let-7 derepression than the loss of either TUTase alone, it still did not reach the levels observed upon Lin28A knockdown. This could be explained by incomplete knockdown of both TUTases or the activity of other as-yet unidentified let-7 repressive factors. The identification of a second TUTase regulating let-7 turnover may provide valuable insight into the control of let-7 expression in cancer and embryonic stem cell biology. The expression pattern and localization of Zcchc6 are unknown, but we anticipate that the relative expression levels of these two redundant TUTases will determine the relative contribution of Zcchc6 and Zcchc11 in the Lin28A-mediated control of let-7 expression. In this regard, we have recently shown that Zcchc11 inhibition in Lin28A-driven cancers can block tumor growth in vitro and in vivo (Piskounova et al. 2011). It will be important to explore the relevance of Zcchc6 in this context. To our knowledge there have been no studies examining expression patterns of these two TUTases in human disease, and it will be important to determine whether either or both of these enzymes is correlated with malignancies characterized by repressed let-7 levels and/or increased Lin28A expression. Whereas Lin28 proteins are likely difficult chemotherapeutic targets due to their nonenzymatic activity, Zcchc11 (and potentially Zcchc6) poses an intriguing possibility as a drug target because of its defined active site and the available structural data regarding noncanonical poly(A) polymerases (Stagno et al. 2010). Moreover, our ability to reconstitute this regulatory pathway with recombinant proteins provides an opportunity to perform in vitro screening to identify small molecule inhibitors of TUTase activity as potential new chemotherapeutic agents. These possibilities pose future areas of study and expand upon the novel centrality of uridylation in stem cell maintenance and tumor development.
MATERIALS AND METHODS
Cloning
All mZcchc11 mammalian-expression mutants were cloned into the XhoI and SalI sites of pBK_2x Flag EF1 vector. C326/329A mutant Zcchc11 was generated by site-directed mutagenesis using the QuickChange kit (Stratagene). hZcchc6 was amplified from HEK293 cDNA and cloned into the HindIII and BamHI sites of pFLAG-CMV2 (Sigma). For recombinant protein expression ΔPneumoG/C mZcchc11 was cloned into the SalI and NotI sites of pETDUET-1. Expression constructs for Flag-m.Lin28A, Flag-Lin28A, Flag-Lin28B, recombinant His-Lin28A, Flag-hZcchc11 wild type, and D1026/1028A mutant were described previously (Viswanathan et al. 2008; Piskounova et al. 2008, 2011; Hagan et al. 2009). Cloning primers are listed in Supplemental Table 1.
Immunoprecipitation and recombinant protein production
Expression plasmids for Flag-Zcchc11, Flag-Zccch6, Flag-Lin28A, or Flag-Lin28B were transfected into HEK293T cells using Lipofectamine 2000 (Invitrogen). Cells were harvested in lysis buffer (20 mM Tris-HCl at pH 8.0, 137 mM NaCl, 1 mM EDTA, 1% Triton x100, 10% Glycerol, 1.5 mM MgCl2, 5 mM DTT, 0.2 mM PMSF). Protein was purified using anti-Flag M2 beads (Sigma), eluted using Flag peptide (Sigma) and confirmed by Western Blot analysis with a mouse anti-Flag antibody (Sigma). For recombinant protein production: Transformed BL21-CodonPlus Competent bacteria (Stratagene) were grown to an OD600nm of 0.4–0.6. Recombinant protein expression (r.Lin28A, and r.Zcchc11) was induced with 100 μM IPTG for 2–3 h. Cell pellets were resuspended in cold lysis buffer (20 mM imidazole at pH 8.0 in PBS, 0.1% Phenylmethyl sulfonyl fluoride [PMSF]) and sonicated. Cleared lysates were incubated with Ni-NTA beads, and after a 90-min incubation at 4°C the beads were washed with 80 bead volumes wash buffer (10 mM Tris at pH 7.8, 50 mM imidazole at pH 8.0, 500 mM NaCl, 0.1% PMSF, 1 mM DTT). Bound His-tagged proteins were eluted from the column with 1 volume elution buffer (10 mM Tris at pH 7.8, 500 mM imidazole at pH 8.0, 500 mM NaCl, 1 mM DTT, 0.1% fresh PMSF) and dialyzed overnight against BC100 (20 mM Tris-HCl at pH 7.8, 100 mM KCl, 0.2 mM EDTA, 10% glycerol). Proteins were further purified by size exclusion chromatography using a Superose 6 gel-filtration column (20 mM Tris-HCl at pH 7.8, 500 mM KCl, 0.2 mM EDTA, 0.2% NP40, 10% glycerol), and peak fractions were dialyzed overnight against BC100 and stored at 4°C.
In vitro uridylation assay
Purified proteins were incubated with 4 pmol of unlabeled synthetic RNA (Dharmacon) for 1 h at 37°C in a 30-μL reaction mixture containing 100 mM KCl, 20 mM Tris-HCl (pH 7.6), 10% Glycerol, 125 nM [α-32P]UTP, 3.2 mM MgCl2, 40 units of RNasin ribonuclease inhibitor (Promega). Products were resolved on 15% denaturing polyacrylamide gels and bands were detected by autoradiography.
Electrophoretic mobility shift analysis
EMSA with purified His-Lin28A was performed with end-labeled synthetic pre-let-7 as described but without competitor yeast tRNA (Piskounova et al. 2008). Briefly, reactions were set up in binding buffer (50 mM Tris at pH7.5, 100 mM NaCl, 10 mM βMe, 20 units of RNasin [Promega]) with 0.5 or 5 nM end-labeled pre-let-7g and incubated for 60 min at room temperature. Bound complexes were resolved on native 5% polyacrylamide gels and visualized by autoradiography.
In vivo knockdowns and quantitative RT–PCR
The indicated siRNAs (see Supplemental Table 2) were reverse transfected in either P19 or feeder-free V6.5 mouse embryonic stem cells using Lipofectamine2000 in 6-well plates according to the manufacturer's protocol (Invitrogen). Total RNA was isolated 60 h post-transfection using TriZol reagent (Invitrogen). To analyze relative mRNA levels, 2 μg of total RNA was reverse transcribed using random hexamers and SuperScriptIII (Invitrogen). MiRNAs were reverse transcribed from 10 ng of total RNA using gene-specific stem–loop RT primers (Applied Biosystems). Relative levels of miRNAs were determined by TaqMan-based real-time PCR, snoRNA-142 for normalization. For quantitative analysis of mRNA levels real-time RT–PCR was performed with either SYBR green or Taqman assays. Actin was used as control. For global microRNA profiling the TaqMan Rodent MicroRNA A Array v2.0 was used with 350 ng of total RNA as starting material for the multiplex RT with preamplification according to manufacturer's directions (Applied Biosystems). The resulting data were normalized to the U6 snRNA.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Robert LaPiere and John Hagan for technical assistance. J.E.T. is supported by a pre-doctoral fellowship from the National Science Foundation. R.I.G. was supported by grants from the US National Institute of General Medical Sciences (NIGMS) (R01GM086386) and The American Cancer Society (121635-RSG-11-175-01-RMC). R.I.G. is a Pew Research Scholar.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.034538.112.
REFERENCES
- Ambros V, Horvitz HR 1984. Heterochronic mutants of the nematode Caenorhabditis elegans. Science 226: 409–416 [DOI] [PubMed] [Google Scholar]
- Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, Zamore PD 2010. Target RNA-directed trimming and tailing of small silencing RNAs. Science 328: 1534–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP 2009. MicroRNAs: Target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E, Robine N, Samsonova A, Westholm JO, Naqvi A, Hung JH, Okamura K, Dai Q, Bortolamiol-Becet D, Martin R, et al. 2011. Deep annotation of Drosophila melanogaster microRNAs yields insights into their processing, modification, and emergence. Genome Res 21: 203–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blahna MT, Jones MR, Quinton LJ, Matsuura KY, Mizgerd JP 2011. Terminal uridyltransferase enzyme Zcchc11 promotes cell proliferation independent of its uridyltransferase activity. J Biol Chem 286: 42381–42389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs AM, Ando Y, de Hoon MJ, Tomaru Y, Nishibu T, Ukekawa R, Funakoshi T, Kurokawa T, Suzuki H, Hayashizaki Y, et al. 2010. A comprehensive survey of 3′ animal miRNA modification events and a possible role for 3′ adenylation in modulating miRNA targeting effectiveness. Genome Res 20: 1398–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Croce CM 2006. MicroRNA signatures in human cancers. Nat Rev Cancer 6: 857–866 [DOI] [PubMed] [Google Scholar]
- Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, et al. 2010. Mammalian microRNAs: Experimental evaluation of novel and previously annotated genes. Genes Dev 24: 992–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ 2004. Processing of primary microRNAs by the Microprocessor complex. Nature 432: 231–235 [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ 2006. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer 6: 259–269 [DOI] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N, Filipowicz W 2010. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem 79: 351–379 [DOI] [PubMed] [Google Scholar]
- Frost RJ, Olson EN 2011. Control of glu–21080.cose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proc Natl Acad Sci 108: 21075–21080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R 2004. The Microprocessor complex mediates the genesis of microRNAs. Nature 432: 235–240 [DOI] [PubMed] [Google Scholar]
- Hagan JP, Piskounova E, Gregory RI 2009. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol 16: 1021–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN 2006. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 125: 887–901 [DOI] [PubMed] [Google Scholar]
- Heo I, Joo C, Cho J, Ha M, Han J, Kim VN 2008. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell 32: 276–284 [DOI] [PubMed] [Google Scholar]
- Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN 2009. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 138: 696–708 [DOI] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293: 834–838 [DOI] [PubMed] [Google Scholar]
- Iliopoulos D, Hirsch HA, Struhl K 2009. An epigenetic switch involving NF-κB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell 139: 693–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MR, Quinton LJ, Blahna MT, Neilson JR, Fu S, Ivanov AR, Wolf DA, Mizgerd JP 2009. Zcchc11-dependent uridylation of microRNA directs cytokine expression. Nat Cell Biol 11: 1157–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh T, Sakaguchi Y, Miyauchi K, Suzuki T, Kashiwabara S, Baba T 2009. Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev 23: 433–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JE, Wickens M 2007. A family of poly(U) polymerases. RNA 13: 860–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrbach NJ, Armisen J, Lightfoot HL, Murfitt KJ, Bugaut A, Balasubramanian S, Miska EA 2009. LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans. Nat Struct Mol Biol 16: 1016–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughlin FE, Gebert LF, Towbin H, Brunschweiger A, Hall J, Allain FH 2011. Structural basis of pre-let-7 miRNA recognition by the zinc knuckles of pluripotency factor Lin28. Nat Struct Mol Biol 19: 84–89 [DOI] [PubMed] [Google Scholar]
- Martin G, Keller W 2007. RNA-specific ribonucleotidyl transferases. RNA 13: 1834–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez NJ, Gregory RI 2010. MicroRNA gene regulatory pathways in the establishment and maintenance of ESC identity. Cell Stem Cell 7: 31–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton C, Judson RL, Blelloch R 2010. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature 463: 621–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss EG, Lee RC, Ambros V 1997. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell 88: 637–646 [DOI] [PubMed] [Google Scholar]
- Nam Y, Chen C, Gregory RI, Chou JJ, Sliz P 2011. Molecular basis for interaction of let-7 microRNAs with Lin28. Cell 147: 1080–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, Thomson JM, Hammond SM 2008. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA 14: 1539–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, Mani V, Hammond SM 2011. Deep sequencing of microRNA precursors reveals extensive 3′ end modification. RNA 17: 1795–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskounova E, Viswanathan SR, Janas M, LaPierre RJ, Daley GQ, Sliz P, Gregory RI 2008. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J Biol Chem 283: 21310–21314 [DOI] [PubMed] [Google Scholar]
- Piskounova E, Polytarchou C, Thornton JE, Lapierre RJ, Pothoulakis C, Hagan JP, Iliopoulos D, Gregory RI 2011. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell 147: 1066–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissland OS, Mikulasova A, Norbury CJ 2007. Efficient RNA polyuridylation by noncanonical poly(A) polymerases. Mol Cell Biol 27: 3612–3624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG 2008. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol 10: 987–993 [DOI] [PubMed] [Google Scholar]
- Saitoh S, Chabes A, McDonald WH, Thelander L, Yates JR, Russell P 2002. Cid13 is a cytoplasmic poly(A) polymerase that regulates ribonucleotide reductase mRNA. Cell 109: 563–573 [DOI] [PubMed] [Google Scholar]
- Schmidt MJ, West S, Norbury CJ 2011. The human cytoplasmic RNA terminal U-transferase ZCCHC11 targets histone mRNAs for degradation. RNA 17: 39–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H, Siomi MC 2010. Posttranscriptional regulation of microRNA biogenesis in animals. Mol Cell 38: 323–332 [DOI] [PubMed] [Google Scholar]
- Small EM, Olson EN 2011. Pervasive roles of microRNAs in cardiovascular biology. Nature 469: 336–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagno J, Aphasizheva I, Bruystens J, Luecke H, Aphasizhev R 2010. Structure of the mitochondrial editosome-like complex associated TUTase 1 reveals divergent mechanisms of UTP selection and domain organization. J Mol Biol 399: 464–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wolfswinkel JC, Claycomb JM, Batista PJ, Mello CC, Berezikov E, Ketting RF 2009. CDE-1 affects chromosome segregation through uridylation of CSR-1-bound siRNAs. Cell 139: 135–148 [DOI] [PubMed] [Google Scholar]
- Van Wynsberghe PM, Kai ZS, Massirer KB, Burton VH, Yeo GW, Pasquinelli AE 2011. LIN-28 co-transcriptionally binds primary let-7 to regulate miRNA maturation in Caenorhabditis elegans. Nat Struct Mol Biol 18: 302–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ 2010. Lin28: A microRNA regulator with a macro role. Cell 140: 445–449 [DOI] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI 2008. Selective blockade of microRNA processing by Lin28. Science 320: 97–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng TL, Toffanin S, O'Sullivan M, Lu J, Phillips LA, Lockhart VL, et al. 2009. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet 41: 843–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G 1993. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75: 855–862 [DOI] [PubMed] [Google Scholar]
- Winter J, Jung S, Keller S, Gregory RI, Diederichs S 2009. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol 11: 228–234 [DOI] [PubMed] [Google Scholar]
- Wu L, Belasco JG 2005. Micro-RNA regulation of the mammalian lin-28 gene during neuronal differentiation of embryonal carcinoma cells. Mol Cell Biol 25: 9198–9208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman SK, Knouf EC, Parkin RK, Fritz BR, Lin DW, Dennis LM, Krouse MA, Webster PJ, Tewari M 2011. Post-transcriptional generation of miRNA variants by multiple nucleotidyl transferases contributes to miRNA transcriptome complexity. Genome Res 21: 1450–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. 2007. Induced pluripotent stem cell lines derived from human somatic cells. Science 318: 1917–1920 [DOI] [PubMed] [Google Scholar]
- Zhu H, Shyh-Chang N, Segre AV, Shinoda G, Shah SP, Einhorn WS, Takeuchi A, Engreitz JM, Hagan JP, Kharas MG, et al. 2011. The Lin28/let-7 axis regulates glucose metabolism. Cell 147: 81–94 [DOI] [PMC free article] [PubMed] [Google Scholar]