In the nucleus of Saccharomyces cerevisiae, TRAMP complexes recognize and polyadenylate RNAs, which enhances RNA degradation by the exosome and may contribute to its specificity. TRAMPs contain either of two putative RNA-binding factors called Air proteins. Previous studies suggested that these proteins function interchangeably in targeting the poly(A)-polymerase activity of TRAMPs to RNAs. Experiments reported here show that the Air proteins govern separable functions. Phenotypic analysis and RNA deep-sequencing results from air mutants reveal specific requirements for each Air protein in the regulation of the levels of noncoding and coding RNAs.
Keywords: RNA surveillance, RNA exosome, TRAMP complex, noncoding RNAs, polyadenylation
Abstract
RNA surveillance systems function at critical steps during the formation and function of RNA molecules in all organisms. The RNA exosome plays a central role in RNA surveillance by processing and degrading RNA molecules in the nucleus and cytoplasm of eukaryotic cells. The exosome functions as a complex of proteins composed of a nine-member core and two ribonucleases. The identity of the molecular determinants of exosome RNA substrate specificity remains an important unsolved aspect of RNA surveillance. In the nucleus of Saccharomyces cerevisiae, TRAMP complexes recognize and polyadenylate RNAs, which enhances RNA degradation by the exosome and may contribute to its specificity. TRAMPs contain either of two putative RNA-binding factors called Air proteins. Previous studies suggested that these proteins function interchangeably in targeting the poly(A)-polymerase activity of TRAMPs to RNAs. Experiments reported here show that the Air proteins govern separable functions. Phenotypic analysis and RNA deep-sequencing results from air mutants reveal specific requirements for each Air protein in the regulation of the levels of noncoding and coding RNAs. Loss of these regulatory functions results in specific metabolic and plasmid inheritance defects. These findings reveal differential functions for Air proteins in RNA metabolism and indicate that they control the substrate specificity of the RNA exosome.
INTRODUCTION
The highly conserved RNA exosome plays a central role in RNA surveillance in the archaea and eukaryotes (Houseley et al. 2006; Doma and Parker 2007; Fasken and Corbett 2009; Lykke-Andersen et al. 2009). This process destroys RNA molecules containing certain types of mutations and those that fail in their conversion from unprocessed precursors to mature transcripts capable of forming normal ribonucleoprotein complexes. In Saccharomyces cerevisiae and humans, the exo/endo-ribonuclease Dis3/Rrp44 and the nuclear exoribonuclease Rrp6 carry out the enzymatic activities of the exosome, which functions to rapidly degrade aberrant RNAs and form the mature 3′ ends of certain RNAs, including rRNAs, snRNAs, and snoRNAs (Briggs et al. 1998; de la Cruz et al. 1998; Allmang et al. 1999; van Hoof et al. 2000). Despite the critical role the exosome plays in RNA surveillance, important questions remain about its activation and the mechanism used to distinguish between RNAs destined for processing or for degradation. The highly conserved TRAMP complex, composed of a noncanonical poly(A) polymerase (Trf4 or Trf5 in S. cerevisiae), a zinc-knuckle protein (Air1 or Air2), and an RNA helicase (Mtr4), enhances exosome degradation or processing through successive rounds of polyadenylation and subsequent recruitment of the degradation machinery (Vanacova et al. 2005; Wyers et al. 2005; Egecioglu et al. 2006; Houseley and Tollervey 2006; Houseley et al. 2007; Rougemaille et al. 2007; Ciais et al. 2008; Grzechnik and Kufel 2008; Roth et al. 2009). Thus, TRAMP plays a critical role in determining nuclear RNA fate, but the molecular details that govern its specificity remain unknown.
In S. cerevisiae at least two distinct TRAMP complexes exist: TRAMP4 and TRAMP5 (named for the presence of Trf4 or Trf5, respectively). Despite significant similarities between the complexes, recent experiments suggest differences in substrate specificities (Egecioglu et al. 2006; Ciais et al. 2008; Roth et al. 2009). For example, TRAMP5 appears to preferentially enhance the degradation of aberrant rRNA precursors (Houseley et al. 2007). Recent microarray analysis indicated that while deletions of TRF4 or TRF5 alter global gene expression, there is little overlap between the transcripts affected, suggesting a difference in substrate specificity (San Paolo et al. 2009). However, the determinants of this specificity remain elusive. Air1 and Air2, as putative RNA-binding proteins, may have unique RNA-binding specificities, accounting for the unique gene expression patterns. Accordingly, TRAMP4 preferentially contains Air2p, and TRAMP5 appears to only contain Air1p (LaCava et al. 2005; Vanacova et al. 2005; Wyers et al. 2005; Houseley and Tollervey 2006). If Air1 and Air2 do, indeed, function as RNA-binding proteins, differences in their substrate recognition specificities may explain the unique gene expression profiles seen in trf4-Δ and trf5-Δ mutants.
The genes expressing Air1 and Air2 arose from the whole genome duplication event that occurred 100 million years ago (Kellis et al. 2004). The proteins share 45% amino acid identity, with the majority of the identical amino acids concentrated in the first two-thirds of the protein. This highly conserved stretch of amino acids contains five zinc knuckle motifs, which may provide protein- or nucleic acid–binding abilities. Structural and biochemical studies indicate that the fourth and fifth zinc knuckles function in Air2 binding to Trf4, while additional biochemical analysis indicates that the zinc knuckles also function in proper binding to at least some substrates in vitro (Hamill et al. 2010; Fasken et al. 2011; Holub et al. 2012). The presumably unstructured and less conserved C-terminal region, as well as the remaining four zinc knuckles, of Air2 may contribute to RNA binding. These findings correlate well with earlier studies showing that Trf4 fails to polyadenylate substrates in vitro in the absence of Air1 and Air2 (Vanacova et al. 2005; Wyers et al. 2005). It seems possible that Air1 and Air2 function as RNA-binding proteins in context on the TRAMP complexes, and therefore TRAMP4 and TRAMP5 specificity may reflect differences in Air1 and Air2 substrate recognition. Indeed, studies using Air-GFP fusion proteins suggest slightly different subnuclear localizations for the proteins (Huh et al. 2003). However, currently available studies on relatively few RNA substrates suggest that Air1 and Air2 function interchangeably (LaCava et al. 2005; Vanacova et al. 2005; Houseley et al. 2007).
Homologs of Trf4 and Trf5 exist in eukaryotes from yeast to humans, though only the Cid14 from Schizosaccharomyces pombe, which forms a similar TRAMP complex with homologs to Air1 and Mtr4, has been shown to be functionally homologous to the Trf proteins from S. cerevisiae (Aravind and Koonin 1999; Saitoh et al. 2002; Win et al. 2006; Buhler et al. 2007; Wang et al. 2008). Humans contain two Trf homologs, PAPD5 and PAPD7, both of which exhibit 37% identity to the catalytic domain of S. cerevisiae Trf4. Although these proteins remain uncharacterized, recent experiments suggest that they may serve a similar role in RNA degradation, because short poly(A) tails exist on some mRNAs destined for degradation by the exosome (West et al. 2006). Human orthologs to Air1/2 and Mtr4 also exist; ZCCHC7 shares 35% and 34% identity to Air1 and Air2, respectively, across the five zinc knuckle–containing regions, while hMtr4/SKIV2L2 exhibits 51% total identity to Mtr4p. Indeed, recent studies showed that hMTR4 and hRRP6 precipitates contain hZCCHC7 and hPAPD5, suggesting conservation of the interaction between Air1/2 and Trf4 in humans (Lubas et al. 2011). The high degree of conservation, along with the recent identification of adenylation as a possible signal for RNA degradation in humans, strongly suggests that humans have functionally related TRAMP complexes.
Studies implicating the Air protein subunits as the likely source of TRAMP RNA-binding specificity highlight their potential role in regulating the RNA surveillance activity of the nuclear exosome. Our understanding of the molecular details of RNA surveillance remains clouded by the idea that the Air proteins in the most advanced model system, budding yeast, function redundantly. To better define the specificities of the TRAMP complexes, we used phenotype profiling and RNA deep sequencing to identify differences in strains deficient in nuclear exosome function and lacking either Air1 or Air2. These results reveal distinct growth phenotypes and accumulation patterns of polyadenylated RNAs in these strains, indicating that Air1 and Air2 proteins each target specific transcripts for polyadenylation and degradation by the nuclear RNA exosome, resulting in profound effects on the physiology of the cell.
RESULTS
Nonredundant functions of Air1 and Air2 proteins
To investigate the functional overlap between Air1 and Air2, we constructed air1-Δ rrp6-Δ and air2-Δ rrp6-Δ strains. The strains show no apparent synthetic interaction between RRP6 and either AIR1 or AIR2, because the double-mutant strains grow comparably to rrp6-Δ and wild-type strains (Fig. 1A). We also constructed an air1-Δ air2-Δ strain but did not use it further because it grows extremely slowly and exhibits multiple colony sizes, suggesting genetic instability (Fig. 1A). Finally, we considered the possibility that the cell might balance the overall level of Air proteins by up-regulating the expression of one protein upon deletion of the other's gene. However, Western blot analysis shows no appreciable change in the amount of Air1 or Air2 upon deletion of the gene encoding the other protein (Supplemental Fig. S1). Moreover, the RNA-seq measurements described below show no significant change in AIR1 or AIR2 mRNA levels in strains deleted for the other gene.
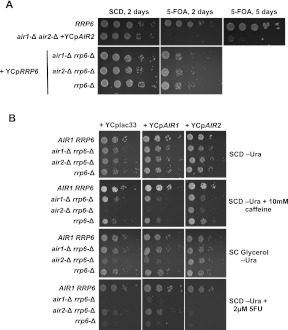
FIGURE 1.
(A) Growth phenotypes of air1-Δ rrp6-Δ, air2-Δ rrp6-Δ, and air1-Δ air2-Δ strains after loss of complementing plasmids. The indicated strains were grown overnight in YPD media, resuspended at OD600 of 0.5 and 10-fold serial dilutions were plated on synthetic complete dextrose (SCD) or 5-FOA containing media. 5-FOA selects for cells that have lost the complementing YCp plasmids. The plates were incubated at 30°C and imaged after 2 d. The plate in the top panel was imaged a second time after 5 d of growth (far right panel). (B) Differential growth phenotypes of strains lacking Air1 or Air2. Cultures of strains with the indicated genotypes were diluted to an OD600 of 0.5, serially diluted 10-fold, and spotted onto SC glycerol (2%)–URA plates or SC dextrose (2%)–URA media with no addition, or with 10 mM caffeine, or 2 μM 5-fluorouracil and incubated for 3–5 d at 30°C. (B) Plating of strains with the indicated genotypes carrying yeast CEN plasmids as in (A) onto SC dextrose (2%)–URA or SC glycerol (2%)–URA plates.
Next, we explored the potential for functional redundancy between Air1 and Air2, by screening air1-Δ rrp6-Δ and air2-Δ rrp6-Δ strains, along with all single-mutant strains, against a variety of conditions (Supplemental Table S1). Strains lacking Air2 and Rrp6 (air2-Δ rrp6-Δ) show a growth defect when grown with glycerol as the sole carbon source, while the air1-Δ rrp6-Δ and single-deletion strains grew similarly to wild-type cells (Fig. 1B). The air2-Δ rrp6-Δ strain grows slowly on ethanol, another nonfermentable carbon source, and displays a frequency (15%) of respiratory-deficient (petite) cells similar to the frequency seen for wild type, indicating that the glycerol and ethanol growth defects probably result from an overall metabolic deficiency rather than a loss of mitochondrial function (data not shown). The air2-Δ rrp6-Δ strain also displays enhanced sensitivity to media containing the purine analog caffeine compared with wild-type and rrp6-Δ or air1-Δ rrp6-Δ strains (Fig. 1B). Finally, deletion of AIR2, but not AIR1, suppresses the 5-fluorouracil (5FU) hypersensitivity previously reported for rrp6-Δ strains (Fang et al. 2004; Lum et al. 2004). Plasmid expression of AIR2, but not AIR1, complements all three of these Air2-specific growth phenotypes (Fig. 1B). Overall, the results of these phenotypic tests show that the Air1 and Air2 proteins do not function interchangeably.
RNA deep sequencing detects poly(A)+ targets of the RNA exosome
Next, we examined transcripts targeted for degradation by either Air1 or Air2 by comparing the population of poly(A)+ RNAs that accumulate in rrp6-Δ cells to the poly(A)+ RNA populations in strains also deleted for AIR1 or AIR2 (Fig. 2A). This approach allows for the identification of transcripts normally degraded by Rrp6, represented by an increase of the polyadenylated transcripts in rrp6-Δ cells compared with wild type, and reveals transcripts targeted for polyadenylation by Air1 or Air2, because the amount of such transcripts should decrease in the poly(A)+ pool in the absence of either of these proteins. We isolated poly(A)+ RNA using poly(U)-Sepharose, which binds transcripts with adenosine tails as short as 5 nt (Binder et al. 1994; Beilharz and Preiss 2007). Quantitative RT-PCR analysis of poly(A)+ coding and noncoding RNAs before and after poly(U)-Sepharose selection verified that the procedure selected the expected RNAs (Fig. 2B). We then sequenced the RNA samples from two biological replicates using the ABI SOLiD deep-sequencing platform and analyzed the results as described in Materials and Methods. Recent studies suggest that some TRAMP substrates receive as few as one to five adenosines (Jia et al. 2011; Wlotzka et al. 2011), while others indicate much longer stretches in steady-state populations (Allmang et al. 1999; van Hoof et al. 2000; Kuai et al. 2004). Technical aspects of the RNA-seq technique used here do not allow an unambiguous determination of the number of adenosines carried by transcripts. Although our detection of a wide array of known TRAMP substrates suggests that our findings are representative of TRAMP activity, we cannot exclude that the poly(U)-Sepharose selection used might have missed transcripts with very short stretches of adenosines.
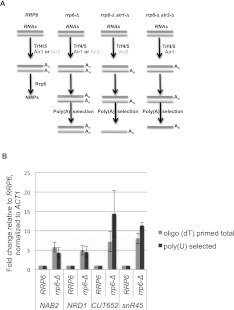
FIGURE 2.
(A) Experimental approach to identify Air protein targets. Strains with the genotypes indicated at the top of the figure will contain RNAs (dark or light gray) targeted for Trf4- or Trf5-catalyzed polyadenylation by Air1 (dark gray) or Air2 (light gray), respectively. Polyadenylated forms of these RNAs will accumulate in rrp6-Δ cells (left). RNAs specifically targeted for polyadenylation by Air1 or Air2 will be lost when either gene is deleted. Differential loss of the poly(A)+ RNAs is assayed by poly(U)-Sepharose selection and RNA sequencing. (B) Quantification of the levels of specific transcripts in RNA samples before and after poly(U)-Sepharose selection. Real-time qRT-PCR analysis of NRD1, NAB2 mRNA, CUT652 RNA, and snR45 RNA before [oligo(dT)-primed total] or after [poly(U)-selected) poly(U)]-Sepharose selection. All histograms represent the fold change of the transcript compared to WT (RRP6) after normalization to ACT1 mRNA. Error bars represent standard deviations calculated from two independent reactions.
The results revealed that 12% (1124 out of 9014) of all annotated transcripts increased (P-value <0.05) in the absence of RRP6 (Supplemental Table S2). This includes (1) 68% of the CUTs (cryptic unstable transcripts; 627 out of 925); (2) 27% of the SUTs (stable unannoted transcripts; 228 out of 847); (3) 31% of the noncoding RNAs (five out of 16); and (4) 60% of the snRNAs (49 out of 82), in agreement with previous results implicating Rrp6 in the normal turnover or processing of polyadenylated forms of these transcripts (Allmang et al. 1999, 2000; van Hoof et al. 2000; Phillips and Butler 2003; Xu et al. 2009). Deletion of RRP6 also results in an increase in the levels of 196 transcripts annotated as open reading frames (Supplemental Table S2). These include two transcripts: NAB2 (log2 FC = 0.93; P = 0.10) and NRD1 (log2 FC = 2.84; P = 4.3 × 10−06) regulated by Rrp6 (Fig. 2B; Roth et al. 2005; Arigo et al. 2006) and 78 other classified bona fide protein-coding genes. Gene Ontology (GO) analysis revealed no significant (P < 0.01) common biological processes for their products. Interestingly, most of the 196 elevated transcripts are annotated as dubious (36%) or putative (20%) ORFs. The 70 dubious ORFs comprise 9.5% of the 734 ORFs annotated as such, suggesting that Rrp6 may play an important role in degrading these transcripts.
Only 5% (444 out of 9014) of the transcripts decrease in rrp6-Δ cells compared with wild type. Most (412 of 444) of these are ORFs, of which 76% correspond to protein-coding genes and 18% to dubious or putative ORFs. GO analysis of the bona fide protein-coding genes revealed significant shared biological processes including generation of precursor metabolites (40 genes; P = 1.12 × 10−09), oxidation–reduction processes (54 genes; P = 8.60 × 10−07), and energy derivation by oxidation of organic compounds (30 genes; p = 4.58 × 10−07) (Supplemental Table S4). Moreover, GYP5 mRNA decreases substantially in rrp6-Δ cells, while its antisense CUT402 increases, in agreement with the findings of Camblong et al. (2009). Less than 7% of the CUTs, SUTs, snRNAs, and other noncoding transcripts decrease in the rrp6-Δ strain. Finally, the results show increases in the levels of poly(A)+ transcripts from the internal transcribed spacer (ITS1) and 5.8S portions of the rRNA loci, consistent with their degradation by Rrp6 (Kuai et al. 2004; Houseley and Tollervey 2006; Callahan and Butler 2008). These findings indicate that RNA-seq analysis of poly(U)-selected RNAs identifies coding and noncoding poly(A)+ transcripts degraded by Rrp6.
Air2 preferentially targets some snRNAs for degradation
Analysis of the distribution of fold changes for the snRNA RNA-seq reads revealed a bias for Air2 in the targeting of snRNAs for polyadenylation. Comparison of the air1-Δ rrp6-Δ and air2-Δ rrp6-Δ reads with the rrp6-Δ reads revealed that 57 of the 82 snRNAs decrease significantly (P < 0.05) in air2-Δ rrp6-Δ compared with only 27 in air1-Δ rrp6-Δ cells. Of the latter, 21 also decrease significantly in air2-Δ rrp6-Δ cells, and none decrease only in air1-Δ rrp6-Δ cells (Supplemental Table S3). Seven of the 19 snRNAs that decrease only in the air2-Δ rrp6-Δ cells are box H/ACA snoRNAs, while nine are box C/D snoRNAs, and four (LSR1, snr6, snr14, and snr19) are the spliceosomal RNAs, U2, U6, U4 and U1, respectively (Supplemental Table S3). These findings suggest that while deletion of either Air protein decreases the level of many poly(A)+ snRNAs, Air2 preferentially targets a specific subset for polyadenylation.
We used quantitative RT-PCR of poly(U)-selected RNAs to analyze changes for a subset of the snoRNAs and found that they increase in concentration in rrp6-Δ cells and decrease more in air2-Δ rrp6-Δ compared with air1-Δ rrp6-Δ cells, consistent with the RNA-seq data (Fig. 3A). Since polyadenylation of snoRNAs targets the transcripts for degradation by the exosome (Allmang et al. 1999; van Hoof et al. 2000; Phillips and Butler 2003), we used qRT-PCR and primers internal to the mature transcripts to measure the relative levels of these Air2-specific snoRNAs in total RNA samples and found that they increase more in air2-Δ rrp6-Δ cells than in air1-Δ rrp6-Δ cells (Fig. 3B). The results also suggest that deletion of AIR1 reduces the slight to moderate increase in total snRNA found in RRP6 cells. The different effects of the loss of Air1 or Air2 on the total levels of these snRNAs raise the possibility that the proteins may play opposing roles in the 3′-end formation and degradation of some snRNAs.
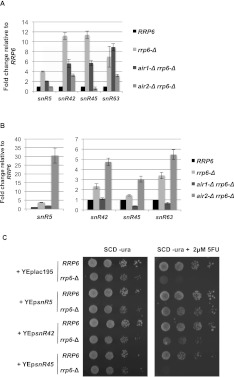
FIGURE 3.
Absence of Air2 elevates levels of some snoRNAs and suppresses the 5FU toxicity of rrp6-Δ. (A,B) Real-time qPCR validation of Air2-specific snoRNAs targets. All histograms represent the fold change of the transcript compared to WT (RRP6) after normalization to ACT1 mRNA. Error bars represent standard deviations calculated from three independent reactions. (A) Real-time qRT-PCR analysis of snoRNAs after reverse transcription of poly(U)-Sepharose selected [poly(A)+] RNA with a mix of random primers and oligo-(dT). (B) Real-time RT-PCR of total isolated RNA after reverse transcription using a mix of random primers and oligo(dT). (C) Strains with the indicated genotypes and carrying the indicated yeast expression plasmids (YEp) were diluted to an OD600 of 0.5 and 10-fold dilutions were spotted onto SCD-URA plates with or without 2 μM 5FU. Plates were imaged after a 3-d incubation at 30°C.
Previous experiments showed that expression of box H/ACA, but not box C/D snoRNAs from high-copy plasmids suppresses the 5FU sensitivity of rrp6-Δ cells (Hoskins and Butler 2008). Indeed, high-copy plasmid expression of two of the Air2-specific box H/ACA snoRNAs identified here (snR5 and snR42) suppresses the rrp6-Δ 5FU sensitivity, while the box C/D snoRNA snR45 does not (Fig. 3C). These findings suggest that Air2 plays a specific role in the polyadenylation and subsequent degradation of certain snoRNAs, and loss of this pathway by deletion of AIR2 contributes to suppression of rrp6-Δ 5FU sensitivity (Fig. 1B).
Air2 and Air1 control the levels of specific sets of mRNAs
The growth defect of air2-Δ rrp6-Δ cells on some carbon sources suggests a metabolic defect specific to this genotype (Fig. 1B). Accordingly, we searched the RNA-seq results for transcripts corresponding to annotated open reading frames (ORFs) that increased or decreased (P < 0.05) in the air1-Δ rrp6-Δ or air2-Δ rrp6-Δ cells compared with rrp6-Δ cells. Each group of transcripts was then divided into bona fide genes, dubious ORFs, and putative ORFs. GO analysis queried the bona fide genes according to related cellular processes (Supplemental Table S4). Similar to the comparison of rrp6-Δ to wild-type cells, those carrying the air2-Δ rrp6-Δ deletions show a significant (P < 0.05) decrease, compared with rrp6-Δ cells, in the levels of mRNAs encoding proteins involved in generation of precursor metabolites (10 out of 40; P = 5.18 × 10−04), glycogen metabolic processes (six out of 40; P = 2.45 × 10−05), and energy derivation by oxidation of organic compounds (nine out of 40; P = 3.29 × 10−04). Indeed, quantitative RT-PCR of two of these transcripts, HXK1 and HXT4, confirms that levels of these transcripts decrease in the absence of Rrp6 and more so in the absence of both Air2 and Rrp6 (Fig. 4A). The additional effects on these classes of transcripts due to the loss of Air2 likely play a role in the poor growth of air2-Δ rrp6-Δ cells on some carbon sources (Fig. 1B; Supplemental Table S1).
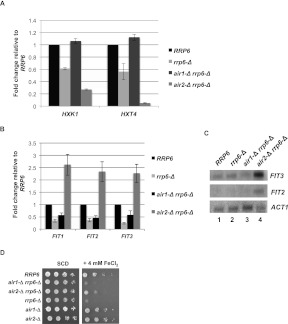
FIGURE 4.
Absence of Air2 decreases levels of mRNAs encoding proteins involved in glucose transport and metabolism while elevating levels of mRNAs involved in iron import. (A) Real-time qRT-PCR analysis of the indicated mRNAs after reverse transcription of poly(U)-Sepharose selected [poly(A)+] RNA with a mix of random primers and oligo(dT). (B) Same as in A, but with the indicated iron transport genes. (C) Northern blot analysis of the levels of FIT3, FIT2, and ACT1 mRNAs. Indicated strains were grown in YPD at 30°C, and 20 μg of total RNA was run on a 1% agarose gel and analyzed by Northern blotting for the indicated RNA species. (D) Differential growth phenotypes of 10-fold dilutions of cells with the indicated genotypes on SCD plates incubated with and without 4 mM FeCl2 for 3 d at 30°C.
The air2-Δ rrp6-Δ strain also shows a specific increase in transcripts whose products play a role in iron uptake into the cell (P = 7.44 × 10−04) (Supplemental Table S4). Quantitative RT-PCR of three of these—FIT1, FIT2, and FIT3—whose products bind iron siderophores, shows that loss of Air2 increases their levels above that found in the wild-type cells (Fig. 4B). Northern blot analysis confirms this increase (Fig. 4C). The differences in mRNA levels correlate with physiological effects, because FeCl2 inhibits growth of rrp6-Δ and air1-Δ rrp6-Δ cells but has less of an effect on air2-Δ rrp6-Δ cells, indicating that loss of Air2 dampens the sensitivity to iron caused by loss of Rrp6 (Fig. 4D).
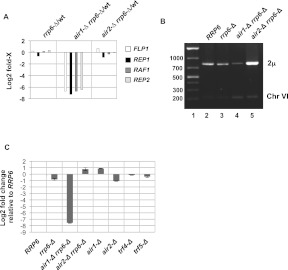
The air1-Δ rrp6-Δ strain shows a remarkable decrease (log2 FC > −6.0, P < 1 × 10−30) in the levels of four transcripts (REP2, REP3, RAF1, and FLP1) encoded by the 2μ plasmid found in most yeast strains (Fig. 5A). These transcripts encode proteins required for copy number control and segregation of the plasmid, and defects in their functions result in decreased 2μ plasmid levels in mutant strains (Jayaram et al. 1985; Murray et al. 1987; Veit and Fangman 1988). Indeed, measurement of 2μ DNA levels in our test strains shows a large decrease in the air1-Δ rrp6-Δ strain (Fig. 5B). Interestingly, the 2μ plasmid decrease requires a synergistic interaction between the air1-Δ rrp6-Δ mutations because neither deletion alone causes the same effect (Fig. 5C). Northern blot analysis of the levels of REP1, REP2, and RAF1 mRNAs confirmed the loss of the transcripts in the air1-Δ rrp6-Δ strain (Fig. 6A). Thus, loss of Air1 in rrp6-Δ cells causes a defect in the retention of 2μ plasmid in yeast cells.
FIGURE 5.
Absence of Air1 causes the loss of 2μ plasmid and transcripts. (A) Graphical comparison of the relative log2 fold change in the number of RNA-seq reads for the indicated 2μ plasmid transcripts between wild type (wt) and strains with the indicated genotypes. (B) Semiquantitative PCR analysis of 2μ plasmid DNA and chromosome VI levels for DNA from strains with the indicated genotypes. (C) Real-time qRT-PCR analysis of 2μ plasmid DNA levels from strains with the indicated genotypes. 2μ plasmid DNA levels are normalized to the amount of chromosome VI DNA determined by the same method.
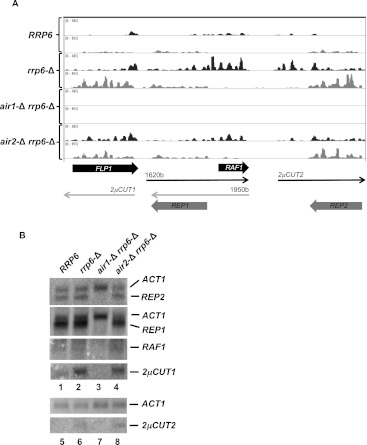
FIGURE 6.
Identification of novel antisense CUTs from the 2μ plasmid. (A) Distribution of RNA sequence reads from the 2μ plasmid. The chart shows the density of RNA-seq reads mapped to each strand (black or gray) of the B-form of the 2μ plasmid for strains with the indicated genotypes. (Block arrows below the chart) The positions and orientations of protein-encoding transcripts and line arrows indicate noncoding transcripts. The noncoding transcripts include two previously described RNAs overlapping RAF1 (the “1620b” RNA) and REP1 (the “1950b” RNA) and two novel transcripts 2μCUT1 and 2μCUT2 antisense to FLP1 and REP2, respectively. (B) Northern blot analysis of the levels of mRNA levels from strains with the indicated genotypes. Total RNA (20 μg) from each strain was separated by electrophoresis on a 1% agarose-formaldehyde gel, transferred to GeneScreen membrane (DuPont), and probed with strand-specific 5′ 32P-labeled oligonucleotides.
Previous experiments showed that 2μ produces two long noncoding RNAs, whose effects on plasmid function remain unclear (Sutton and Broach 1985). Analysis of the distribution of the RNA-seq reads and Northern blotting confirm this and identify two novel transcripts antisense to FLP1 and REP2 (Fig. 6A,B). Interestingly, the two novel antisense RNAs increase in concentration in the absence of Rrp6, suggesting that the RNA exosome plays a role in regulating 2μ gene expression. Accordingly, we name these transcripts 2μCUT1 and 2μCUT2 (2μ cryptic unstable transcripts). Together, the analysis of poly(A)+ mRNA levels reveals that Air1 and Air2 govern the expression levels of distinct sets of mRNAs, which impacts the metabolic fitness and plasmid inheritance functions of the cell.
DISCUSSION
The results presented here provide evidence that the Air1 and Air2 components of the TRAMP complexes function in concert with the Rrp6 component of the nuclear RNA exosome to regulate the levels of specific classes of RNAs in the cell. First, phenotypic profiling revealed specific growth differences between strains lacking Rrp6 and either of the Air proteins. These include (1) defects in the ability of air2-Δ rrp6-Δ cells to grow on nonfermentable carbon sources or caffeine, (2) the ability of an air2-Δ deletion to suppress the 5FU sensitivity of an rrp6-Δ strain, and (3) suppression of rrp6-Δ-dependent iron sensitivity by air2-Δ. Second, RNA-seq analysis uncovered specific classes of polyadenylated RNAs whose levels change upon deletion of either Air1 or Air2. These include Air2-specific changes in the levels of (1) many polyadenylated snoRNAs, (2) some iron-responsive mRNAs, and (3) transcripts involved in carbohydrate transport and catabolism. Moreover, absence of Air1 causes loss of mRNAs whose products play a critical role in proper maintenance of the endogenous 2μ plasmid. While many of the transcripts analyzed in this report are known TRAMP substrates, for others we cannot discount the possibility that some of the effects of Air protein loss may reflect TRAMP-independent functions of the proteins, such as their ability to interact with the arginine methyltransferase Hmt1 (Inoue et al. 2000). Our findings clearly show a lack of redundancy in the function of Air1 and Air2 for many transcripts, yet many RNAs that accumulate as poly(A)+ species in the absence of Rrp6 are equally affected by loss of either Air1 or Air2. This agrees with previous experiments that showed that some poly(A)+ noncoding RNAs accumulate in strains lacking either protein (LaCava et al. 2005; Vanacova et al. 2005; Houseley et al. 2007). Thus, the synthetic effect of the double mutation on growth could reflect lack of Air activity on polyadenylation of some RNAs. Alternatively, activities of the Air proteins outside the context of TRAMPs, for instance, via known interactions with Hmt1, may explain the synthetic effect. Nevertheless, these results support our conclusion that Air1 and Air2 each play specific roles in the regulation of RNA levels.
Previous analysis of increases in RNA levels resulting from deletion of TRF4 or TRF5 revealed little overlap between the apparent targets of the poly(A)-polymerases (San Paolo et al. 2009). Loss of Trf4 increased the levels of noncoding RNAs, snoRNAs, and transcripts from Ty retrotransposons, while loss of Trf5 increased the levels of a diverse set of mRNAs. Our findings suggest that many of the Trf4-specific transcripts receive poly(A) tails with the guidance of either Air1 or Air2, in agreement with evidence indicating that Trf4 forms TRAMP complexes with either protein (Wyers et al. 2005). Interestingly, most of the transcripts that increased in concentration in the TRF4 deletion experiment did not increase in cells with a catalytically inactive Trf4, indicating that the observed effects occur independently of polyadenylation by Trf4 (San Paolo et al. 2009). Our experiments rely directly on differences in the polyadenylation status of RNAs, which complicates strict comparison of the two sets of data.
In light of our phenotypic evidence demonstrating that Air1 and Air2 do not function interchangeably, we determined the global specificity of Air1 and Air2 targeting of RNAs for polyadenylation. Deep sequencing of poly(A)+ RNAs revealed unique subsets of polyadenylated transcripts that decrease upon deletion of AIR1 or AIR2, suggesting that Air1 or Air2 specifically targets these transcripts for polyadenylation and subsequent degradation. Interestingly, many poly(A)+ transcripts that did not increase in concentration upon deletion of RRP6 alone, decreased upon additional deletion of AIR1 or AIR2. These transcripts may represent RNAs polyadenylated by TRAMP but degraded independently of Rrp6, by Exo10 (core exosome + Dis3/Rrp44). This view agrees with experiments that showed that Rrp6 and Exo10 can function independently in the degradation of many RNAs (Callahan and Butler 2008).
RNA-seq analysis indicated that Air2 preferentially targets the polyadenylation of many snoRNAs, because their poly(A)+ forms decrease significantly more upon loss of Air2 than Air1. The fact that many of these Air2-specific, and none of the Air1-specific snoRNAs, belong to the box H/ACA class suggests that suppression of the 5FU sensitivity of an rrp6-Δ strain by deletion of AIR2, but not AIR1, results from changes in the levels of this class of snoRNA. Indeed, previous experiments, and results reported here, show that increased expression of box H/ACA, but not box C/D snoRNAs, suppresses the 5FU sensitivity of rrp6-Δ strains (Hoskins and Butler 2008). Moreover, the decrease in the number of poly(A)+ snoRNAs correlates with an increase in the total amount of RNA for these snoRNAs. This suggests that the 5FU suppression probably requires the accumulation of poly(A)- snoRNA processing or degradation intermediates. Finally, because this effect occurs in air2-Δ rrp6-Δ, but not rrp6-Δ cells, the degradation of these snoRNAs likely requires targeting of polyadenylation by Air2 and subsequent degradation by Exo10. Thus, polyadenylation by TRAMP may play a critical role in determining snoRNA fate; functioning to direct Exo10 in 3′-end processing, or degradation of the snoRNA.
The RNA-seq results showed that the levels of some mRNAs that encode proteins involved in iron import and assimilation increase significantly only in air2-Δ rrp6-Δ cells. TRAMP4 and Rrp6 function in 3′-end processing of TIS11/CTH2 mRNA, whose transcription is activated by iron deprivation and whose gene product binds to and activates the degradation of transcripts encoding proteins involved in iron-dependent processes under conditions of iron deficiency (Puig et al. 2005; Ciais et al. 2008). TIS11/CTH2 mRNA levels actually increase 1.8-fold in air2-Δ rrp6-Δ compared with rrp6-Δ cells, yet some transcripts subject to Cth2-targeted degradation increase in the air2-Δ rrp6-Δ strain, while most do not change significantly in the rrp6-Δ strain or the double mutant. Thus, the increase in iron-responsive mRNA levels in the air2-Δ rrp6-Δ strain most likely occurs independently of the mRNA-destabilizing activity of Cth2. We propose that the iron response results from the Air2-specific defect in carbohydrate metabolism caused by a decrease in many transcripts encoding proteins required for sugar uptake and catabolism, which reflects itself in the growth defect of air2-Δ rrp6-Δ cells on some carbon sources. In this view, the iron response results from an increased requirement for iron import and assimilation needed to support the production of iron-containing proteins involved in respiration. Recent findings point to a regulated balance between the expression of genes involved in carbohydrate metabolism and iron assimilation, and evidence exists that Rrp6 plays a Cth2-independent role in regulating the expression of some iron-responsive genes (Lee et al. 2005; Puig et al. 2008).
Analysis of mRNA levels also revealed a specific requirement for Air1 in the control of 2μ plasmid levels. Stable segregation and copy number control of this plasmid require the activity of four plasmid-encoded proteins: Rep1, Rep2, Raf1, and Flp1. Rep1 and Rep2 act together to repress expression of their own genes and that of Flp1. Raf1 acts as an antirepressor of Rep1/Rep2, leading to balanced expression of Flp1, Rep1, and Rep2. Flp1 functions to enhance plasmid replication, thereby maintaining constant copy number (Jayaram et al. 2004). The plasmid also produces transcripts overlapping and antisense to these genes, but the contribution of these RNAs to the regulation of 2μ plasmid levels remains obscure. The RNA-seq data revealed that the air1-Δ rrp6-Δ double mutation causes a remarkable decrease (log2 FC > −6.0, P < 1 × 10−30) in the levels of the four transcripts (Figs. 5, 6). Indeed, measurement of 2μ DNA levels in our test strains shows a significant decrease in plasmid levels in only the air1-Δ rrp6-Δ strain (Fig. 5). Northern blot analysis of the levels of REP1, REP2, and RAF1 mRNAs confirmed loss of the transcripts in the air1-Δ rrp6-Δ strain and identified novel antisense RNAs—2μCUT1 and 2μCUT2—degraded by Rrp6 (Fig. 6). Thus, loss of Air1 in rrp6-Δ cells causes the loss of 2μ plasmid. Perhaps the novel 2μ CUTs function to regulate the expression of their respective antisense mRNAs in a manner similar to that suggested for GYP5 and its antisense RNA (Camblong et al. 2009). Humans contain a single apparent Air protein homolog, ZCCHC7, and segregation of Epstein-Barr virus episomes appears to follow similar rules to that of the yeast 2μ plasmid (Kapoor et al. 2001; Kapoor and Frappier 2003). Thus it is tempting to speculate that a similar mechanism may govern the inheritance of yeast plasmids and viral genomes in metazoans. Additional research will determine if the loss of the 2μ plasmid results from deregulation of the plasmid-encoded RNAs or if plasmid and RNA loss results from changes in the level of some unlinked factor. Nevertheless, these findings reveal that Air1 and Air2 govern the expression levels of distinct sets of mRNAs, which impacts metabolic and plasmid inheritance functions of the cell.
In summary, the experimental results reported here show that the Air protein components of the TRAMP complex play distinct roles in some aspects of RNA surveillance. The identification of specific sets of transcripts whose polyadenylation and degradation results from the action of Air1 or Air2 provides substrates and phenotypes that set the stage for dissection of the structural features of the RNAs and proteins that guide the specificity of nuclear RNA surveillance.
MATERIALS AND METHODS
Strains and plasmid construction
Manipulations of S. cerevisiae used standard procedures (Sherman 1991). Cells were grown in YPD media (1% yeast extract, 2% peptone, and 2% dextrose) or in SC medium (synthetic complete: 0.67% Bacto-yeast nitrogen base without amino acids, 2% dextrose [SCD], ethanol [SCE], or glycerol [SCG] and amino acid supplements as required). Strains used in this study are listed in Supplemental Table S5. The air1-Δ rrp6-Δ and air2-Δ rrp6-Δ strains were made from parental BY4742 by transforming with rrp60∷LEU2, transforming in a URA3 CEN RRP6 plasmid, followed by transformation with an air10∷bleR or air20∷bleR fragment made by PCR amplification from the pUG66 bleR cassette (Gueldener et al. 2002). The resulting strains were then plated on 5-fluoroorotic acid to select for cells that lost the URA3 CEN RRP6 plasmid. All disruptions were confirmed by PCR.
For serial dilution plating, strains were grown in YPD or appropriate selective media, and 10-fold serial dilutions, starting with 104 cells, were spotted onto YPD or SCD-ura plates supplemented with compounds tested (Supplemental Table S1). Plates were incubated for 3–5 d at 30°C (unless otherwise noted). To test for petite frequency, strains were grown overnight in YPD media, and cells were harvested and resuspended in water, and 1 × 103 cells were plated onto YP with 2% glycerol and 0.1% dextrose.
Poly(A)+ RNA selection and deep sequencing
Strains were grown in YPD to an A600 of 2.0–3.1, and total RNA was isolated as described previously (Patel and Butler 1992). Polyadenylated RNAs were selected on poly(U)-Sepharose according to published protocol, with modification (Beilharz and Preiss 2007). A final concentration of 50 mM Tris-HCl was used in both the elution buffer and the high salt binding buffer. Total cellular RNA was isolated as previously described (Burkard and Butler 2000), and 150 μg of RNA was added to 150 μL of poly(U)-Sepharose equilibrated in high salt binding buffer. Poly(A)+ RNA was eluted by resuspending in a 600-μL elution buffer for 5 min at 45°C. After collection of the resin by centrifugation at 3300g for 30 sec, the supernatant was removed with a syringe and pelleted at 3300g for an additional 30 sec, and was gradually concentrated and desalted into sterile ddH2O using an Amicon Ultra-4 Centrifugal Filter Device (Millipore). After determination of poly(A)+ RNA concentration by spectrophotometry, the RNA was submitted to the University of Rochester Functional Genomics Core for SOLiD Sequencing (Applied Biosystems). Preparation of fragment library and bead populations and subsequent sequencing were performed according to the manufacturer's instructions.
RNA-seq mapping and statistical analysis
The color space FASTA files and quality score files from the RNA-seq runs were used as input to Bowtie (version 0.12.7) for mapping with the running options “-all -f -tryhard -sam -e 70 -color -best -quals” (Langmead et al. 2009). Then SAMtools (version 0.1.17) was used to convert SAM files to sorted and indexed BAM files and sort and index them (Li et al. 2009). Reads unambiguously mapped inside a single gene were counted and used as weights to divide each multimapped read to all of the genes to which it mapped. Reference genome and gene annotation files were downloaded from the Saccharomyces Genome Database (July 2011 version) for mapping. After the read counts were summarized by gene, Spearman correlations between any two of the eight samples were calculated. The Bioconductor package edgeR (version 2.2.5) was used for TMM (trimmed mean of M values) normalization followed by statistical testing to find genes differentially expressed (Supplemental Table S6; Robinson et al. 2010). The correlation coefficients for the replicate sample reads were (1) wt.1 versus wt.2 = 0.999, (2) rrp6-Δ.1 versus rrp6-Δ.2 = 0.996, (3) air1-Δ rrp6-Δ.1 versus air1-Δ rrp6-Δ.2 = 0.996, and (4) air1-Δ rrp6-Δ.1 versus air1-Δ rrp6-Δ.2 = 0.996. Analysis of sequence reads from the wild-type and rrp6-Δ cells revealed an anomalously high read count from the rRNA loci in one of the wild-type biological replicates. Accordingly, further analysis of rrp6-Δ versus wild-type strains used only one set of wild-type data. The validity of this decision is supported by the agreement between this analysis and the qRT-PCR data reported in Results. Comparisons between the other strains used in the study were unaffected by disqualification of the one wild-type data set.
Real-time PCR
cDNA was synthesized using a qScript cDNA Supermix or Flex cDNA kit using oligo(dT) primers (Quanta Biosciences). Quantitative PCR (qPCR) was performed using SYBR GreenER master mix (Invitrogen) with a model 7500 fast real-time PCR system and software (Applied Biosystems). RNAs were quantified using primers whose sequences are available upon request.
Northern blot analysis
The listed strains were grown in YPD to an A600 of 2.0–3.5, total RNA was isolated as described previously (Patel and Butler 1992), and 20 μg was separated by electrophoresis on 1% agarose or 8% polyacrylamide 8 M urea gels. After transfer to a GeneScreen membrane (PerkinElmer Life Sciences), the blots were probed with 5′ 32P-labeled oligonucleotides specific for each transcript (sequences available upon request) as previously described (Briggs et al. 1998). Levels of specific RNAs were normalized to ACT1 RNA levels after quantification by storage PhosphorImager analysis (Molecular Dynamics).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this paper.
ACKNOWLEDGMENTS
We are grateful to Dr. Steven Welle of the University of Rochester Functional Genomics Core Facility for help with the RNA-seq experiments; Mark Dumont, Elizabeth Grayhack, Eric Phizicky, and members of our laboratory for helpful discussions; and Milo Fasken and Anita Corbett for providing plasmids. This work was supported by grants from the National Science Foundation (MCB-0817324 to J.S.B.) and the National Institutes of Health (R01-HG004002 to D.H.M. and NIH T-32 GM068411-05 to K.S.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.033431.112.
REFERENCES
- Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D 1999. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J 18: 5399–5410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang C, Mitchell P, Petfalski E, Tollervey D 2000. Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res 28: 1684–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Koonin EV 1999. DNA polymerase beta-like nucleotidyltransferase superfamily: Identification of three new families, classification and evolutionary history. Nucleic Acids Res 27: 1609–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arigo JT, Carroll KL, Ames JM, Corden JL 2006. Regulation of yeast NRD1 expression by premature transcription termination. Mol Cell 21: 641–651 [DOI] [PubMed] [Google Scholar]
- Beilharz TH, Preiss T 2007. Widespread use of poly(A) tail length control to accentuate expression of the yeast transcriptome. RNA 13: 982–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder R, Horowitz JA, Basilion JP, Koeller DM, Klausner RD, Harford JB 1994. Evidence that the pathway of transferrin receptor mRNA degradation involves an endonucleolytic cleavage within the 3′ UTR and does not involve poly(A) tail shortening. EMBO J 13: 1969–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs MW, Burkard KT, Butler JS 1998. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8S rRNA 3′ end formation. J Biol Chem 273: 13255–13263 [DOI] [PubMed] [Google Scholar]
- Buhler M, Haas W, Gygi SP, Moazed D 2007. RNAi-dependent and -independent RNA turnover mechanisms contribute to heterochromatic gene silencing. Cell 129: 707–721 [DOI] [PubMed] [Google Scholar]
- Burkard KT, Butler JS 2000. A nuclear 3′–5′ exonuclease involved in mRNA degradation interacts with Poly(A) polymerase and the hnRNA protein Npl3p. Mol Cell Biol 20: 604–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan KP, Butler JS 2008. Evidence for core exosome independent function of the nuclear exoribonuclease Rrp6p. Nucleic Acids Res 36: 6645–6655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camblong J, Beyrouthy N, Guffanti E, Schlaepfer G, Steinmetz LM, Stutz F 2009. Trans-acting antisense RNAs mediate transcriptional gene cosuppression in S. cerevisiae. Genes Dev 23: 1534–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciais D, Bohnsack MT, Tollervey D 2008. The mRNA encoding the yeast ARE-binding protein Cth2 is generated by a novel 3′ processing pathway. Nucleic Acids Res 36: 3075–3084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J, Kressler D, Tollervey D, Linder P 1998. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J 17: 1128–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma MK, Parker R 2007. RNA quality control in eukaryotes. Cell 131: 660–668 [DOI] [PubMed] [Google Scholar]
- Egecioglu DE, Henras AK, Chanfreau GF 2006. Contributions of Trf4p- and Trf5p-dependent polyadenylation to the processing and degradative functions of the yeast nuclear exosome. RNA 12: 26–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang F, Hoskins J, Butler JS 2004. 5-Fluorouracil enhances exosome-dependent accumulation of polyadenylated rRNAs. Mol Cell Biol 24: 10766–10776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasken MB, Corbett AH 2009. Mechanisms of nuclear mRNA quality control. RNA Biol 6: 237–241 [DOI] [PubMed] [Google Scholar]
- Fasken MB, Leung SW, Banerjee A, Kodani MO, Chavez R, Bowman EA, Purohit MK, Rubinson ME, Rubinson EH, Corbett AH 2011. Air1 zinc knuckles 4 and 5 and a conserved IWRxY motif are critical for the function and integrity of the TRAMP RNA quality control complex. J Biol Chem 286: 37429–37445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzechnik P, Kufel J 2008. Polyadenylation linked to transcription termination directs the processing of snoRNA precursors in yeast. Mol Cell 32: 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueldener U, Heinisch J, Koehler GJ, Voss D, Hegemann JH 2002. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res 30: e23 doi: 10.1093/nar/30.6.e23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamill S, Wolin SL, Reinisch KM 2010. Structure and function of the polymerase core of TRAMP, a RNA surveillance complex. Proc Natl Acad Sci 107: 15045–15050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holub P, Lalakova J, Cerna H, Pasulka J, Sarazova M, Hrazdilova K, Arce MS, Hobor F, Stefl R, Vanacova S 2012. Air2p is critical for the assembly and RNA-binding of the TRAMP complex and the KOW domain of Mtr4p is crucial for exosome activation. Nucleic Acids Res 40: 5679–5693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins J, Butler JS 2008. RNA-based 5-fluorouracil toxicity requires the pseudouridylation activity of Cbf5p. Genetics 179: 323–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, Tollervey D 2006. Yeast Trf5p is a nuclear poly(A) polymerase. EMBO Rep 7: 205–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, LaCava J, Tollervey D 2006. RNA-quality control by the exosome. Nat Rev Mol Cell Biol 7: 529–539 [DOI] [PubMed] [Google Scholar]
- Houseley J, Kotovic K, El Hage A, Tollervey D 2007. Trf4 targets ncRNAs from telomeric and rDNA spacer regions and functions in rDNA copy number control. EMBO J 26: 4996–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK 2003. Global analysis of protein localization in budding yeast. Nature 425: 686–691 [DOI] [PubMed] [Google Scholar]
- Inoue K, Mizuno T, Wada K, Hagiwara M 2000. Novel RING finger proteins, Air1p and Air2p, interact with Hmt1p and inhibit the arginine methylation of Npl3p. J Biol Chem 275: 32793–32799 [DOI] [PubMed] [Google Scholar]
- Jayaram M, Sutton A, Broach JR 1985. Properties of REP3: A cis-acting locus required for stable propagation of the Saccharomyces cerevisiae plasmid 2μm circle. Mol Cell Biol 5: 2466–2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaram M, Mehta S, Uzri D, Voziyanov Y, Velmurugan S 2004. Site-specific recombination and partitioning systems in the stable high copy propagation of the 2μm yeast plasmid. Prog Nucleic Acid Res Mol Biol 77: 127–172 [DOI] [PubMed] [Google Scholar]
- Jia H, Wang X, Liu F, Guenther UP, Srinivasan S, Anderson JT, Jankowsky E 2011. The RNA helicase Mtr4p modulates polyadenylation in the TRAMP complex. Cell 145: 890–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor P, Frappier L 2003. EBNA1 partitions Epstein-Barr virus plasmids in yeast cells by attaching to human EBNA1-binding protein 2 on mitotic chromosomes. J Virol 77: 6946–6956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor P, Shire K, Frappier L 2001. Reconstitution of Epstein-Barr virus-based plasmid partitioning in budding yeast. EMBO J 20: 222–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M, Birren BW, Lander ES 2004. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428: 617–624 [DOI] [PubMed] [Google Scholar]
- Kuai L, Fang F, Butler JS, Sherman F 2004. Polyadenylation of rRNA in Saccharomyces cerevisiae. Proc Natl Acad Sci 101: 8581–8586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D 2005. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121: 713–724 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A, Henras AK, Chanfreau G 2005. Multiple RNA surveillance pathways limit aberrant expression of iron uptake mRNAs and prevent iron toxicity in S. cerevisiae. Mol Cell 19: 39–51 [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubas M, Christensen MS, Kristiansen MS, Domanski M, Falkenby LG, Lykke-Andersen S, Andersen JS, Dziembowski A, Jensen TH 2011. Interaction profiling identifies the human nuclear exosome targeting complex. Mol Cell 43: 624–637 [DOI] [PubMed] [Google Scholar]
- Lum PY, Armour CD, Stepaniants SB, Cavet G, Wolf MK, Butler JS, Hinshaw JC, Garnier P, Prestwich GD, Leonardson A, et al. 2004. Discovering modes of action for therapeutic compounds using a genome-wide screen of yeast heterozygotes. Cell 116: 121–137 [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen S, Brodersen DE, Jensen TH 2009. Origins and activities of the eukaryotic exosome. J Cell Sci 122: 1487–1494 [DOI] [PubMed] [Google Scholar]
- Murray JA, Scarpa M, Rossi N, Cesareni G 1987. Antagonistic controls regulate copy number of the yeast 2μ plasmid. EMBO J 6: 4205–4212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D, Butler JS 1992. Conditional defect in mRNA 3′ end processing caused by a mutation in the gene for poly(A) polymerase. Mol Cell Biol 12: 3297–3304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips S, Butler JS 2003. Contribution of domain structure to the RNA 3′ end processing and degradation functions of the nuclear exosome subunit Rrp6p. RNA 9: 1098–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig S, Askeland E, Thiele DJ 2005. Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell 120: 99–110 [DOI] [PubMed] [Google Scholar]
- Puig S, Vergara SV, Thiele DJ 2008. Cooperation of two mRNA-binding proteins drives metabolic adaptation to iron deficiency. Cell Metab 7: 555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK 2010. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth KM, Wolf MK, Rossi M, Butler JS 2005. The nuclear exosome contributes to autogenous control of NAB2 mRNA levels. Mol Cell Biol 25: 1577–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth KM, Byam J, Fang F, Butler JS 2009. Regulation of NAB2 mRNA 3′-end formation requires the core exosome and the Trf4p component of the TRAMP complex. RNA 15: 1045–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougemaille M, Gudipati RK, Olesen JR, Thomsen R, Seraphin B, Libri D, Jensen TH 2007. Dissecting mechanisms of nuclear mRNA surveillance in THO/sub2 complex mutants. EMBO J 26: 2317–2326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh S, Chabes A, McDonald WH, Thelander L, Yates JR, Russell P 2002. Cid13 is a cytoplasmic poly(A) polymerase that regulates ribonucleotide reductase mRNA. Cell 109: 563–573 [DOI] [PubMed] [Google Scholar]
- San Paolo S, Vanacova S, Schenk L, Scherrer T, Blank D, Keller W, Gerber AP 2009. Distinct roles of non-canonical poly(A) polymerases in RNA metabolism. PLoS Genet 5: e1000555 doi: 10.1371/journal.pgen.1000555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F 1991. Getting started with yeast. Methods Enzymol 194: 3–21 [DOI] [PubMed] [Google Scholar]
- Sutton A, Broach JR 1985. Signals for transcription initiation and termination in the Saccharomyces cerevisiae plasmid 2μm circle. Mol Cell Biol 5: 2770–2780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W 2005. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol 3: e189 doi: 10.1371/journal.pbio.0030189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A, Lennertz P, Parker R 2000. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol Cell Biol 20: 441–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veit BE, Fangman WL 1988. Copy number and partition of the Saccharomyces cerevisiae 2μm plasmid controlled by transcription regulators. Mol Cell Biol 8: 4949–4957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SW, Stevenson AL, Kearsey SE, Watt S, Bahler J 2008. Global role for polyadenylation-assisted nuclear RNA degradation in posttranscriptional gene silencing. Mol Cell Biol 28: 656–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S, Gromak N, Norbury CJ, Proudfoot NJ 2006. Adenylation and exosome-mediated degradation of cotranscriptionally cleaved pre-messenger RNA in human cells. Mol Cell 21: 437–443 [DOI] [PubMed] [Google Scholar]
- Win TZ, Draper S, Read RL, Pearce J, Norbury CJ, Wang SW 2006. Requirement of fission yeast Cid14 in polyadenylation of rRNAs. Mol Cell Biol 26: 1710–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlotzka W, Kudla G, Granneman S, Tollervey D 2011. The nuclear RNA polymerase II surveillance system targets polymerase III transcripts. EMBO J 30: 1790–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, et al. 2005. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121: 725–737 [DOI] [PubMed] [Google Scholar]
- Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Munster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM 2009. Bidirectional promoters generate pervasive transcription in yeast. Nature 457: 1033–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]