Abstract
Common single-nucleotide polymorphisms (SNPs) in microRNAs (miRNAs) have been shown to be associated with susceptibility to several types of human cancer. We evaluated the association between three SNPs (rs11614913, rs2910164 and rs3746444) in pre-miRNAs (miR-196a2, miR-146a and miR-499) and various clinicopathological characteristics, including CpG island hypermethylation (CIHM) status and overall survival in gastric cancer (GC) patients. rs11614913 (T>C), rs2910164 (C>G) and rs3746444 (A>G) SNPs were genotyped in 127 GC patients. CIHM of p14, p16, DAP-kinase and CDH1 genes was determined by methylation-specific polymerase chain reaction in the cancer tissues. A significant marginal association was found between the rs11614913 CC genotype and polypoid or elevated type morphology in early-stage GC (OR=6.29, 95% CI 1.18–33.47, p=0.03). The rs2910164 CC and CG genotypes were associated with increased susceptibility to CIHM of DAP-kinase (CC+CG, OR=5.48, 95% CI 1.30–23.10, p=0.02; CC, OR=6.93, 95% CI 1.37–35.02, p=0.02; CG, OR=4.24, 95% CI 0.87–20.78, p=0.07). The 11614913 TT and TC genotypes were associated with a higher number of CIHM (no. of CIHM 0–1 vs. 2–4; TT+TC, OR=3.67, 95% CI 0.98–13.72, p=0.05; TC, OR=4.08, 95% CI 1.04–15.97, p=0.04). When the subjects were divided according to age group, the combined rs11614913 TT+TC genotype tended to be associated with worse overall survival than the CC genotype in patients younger than 65 years of age (p=0.05). The combined rs2910164 CG+GG genotype also tended to be associated with worse overall survival than the CC genotype in the same age group (p=0.09). It appears that rs11614913 and rs2910164 SNPs in pre-miRNAs (miR-196a2 and miR-146a) affect the clinicopathological characteristics of GC, including its morphological appearance, CIHM status and overall survival.
Keywords: pre-microRNAs, Helicobacter pylori, polymorphism, gastric cancer, rs11614913, rs2910164, miR-196a2, miR-146a, CpG island hypermethylation
Introduction
Gastric cancer (GC) is one of the most common and lethal malignancies in Japanese and East Asian populations, and the second most common cause of cancer-related death in the world (1,2). Although the incidence and mortality rate of GC located outside the cardia have been decreasing over the last few decades, a considerable percentage of patients still have advanced disease at diagnosis, and some of these patients are not candidates for curative surgery. Each GC tumor often has a different biological behavior, which leads them to have differing clinical phenotypes, prognosis and response to treatment. These differences may be partly explained by certain host genetic differences.
MicroRNAs (miRNAs) are 21- to 24-nucleotide-long small noncoding RNA gene products that regulate gene expression by base pairing with target mRNAs at the 3′-untranslated region, leading to mRNA cleavage or translational repression (3–5). It has been suggested that miRNAs are involved in various biological processes, including cell proliferation, cell death, stress resistance and fat metabolism (6). Several reports have shown that miRNAs participate in human tumorigenesis as tumor suppressors or oncogenes (7–9). For example, miR-143 and miR-145, targeting ERK4, are down-regulated in GC (10), whereas miRNA-106b, miR-93 and miR-25, clustering at MCM7 intron, were reported to be overexpressed in GC (11).
Single-nucleotide polymorphisms (SNPs), or mutations in the miRNA sequence, may alter miRNA expression and/ or maturation. Recently, Hu et al performed a screening for common SNPs in miRNA sequences and identified four SNPs (rs2910164, rs2292832, rs11614913 and rs3746444) located at the pre-miRNA regions of miR-146a, miR-149, miR-196a2 and miR-499, respectively (12). Among the above four SNPs, the rs11614913 SNP in miR-196a2 was found to be associated with shortened survival time of non-small cell lung cancer patients through the altered expression of mature miR-196a and the binding activity of target mRNA (12). Consequently, the rs11614913 SNP in miR-196a2 contributed to lung cancer susceptibility (13). In carcinomas in other organs, the rs2910164 SNP within miR-146a was associated with papillary thyroid (14) and hepatocellular carcinomas (15). Moreover, the rs11614913 SNP in miR-196a2 and the rs3746444 SNP in miR-499 were both associated with the risk of breast cancer (16). In the case of stomach cancer, a preliminary association was initially reported for rs11614913 in miRNA-146a in a Chinese population (17).
Since SNPs in miRNAs are closely associated with GC susceptibility, it is necessary to clarify whether these SNPs are also associated with the clinical characteristics of GC. Accordingly, this study was designed to evaluate the possible association between three common SNPs (rs11614913, rs2910164 and rs3746444) in pre-miRNAs (miR-196a2, miR-146a and miR-499) that had previously been shown to contribute to human cancer susceptibility (13–17), and various clinicopathological characteristics of GC.
We also investigated the association between these SNPs and the CpG island hypermethylation (CIHM) status in tumor suppressor genes, which constitutes a distinct tumor subtype in GC (18,19). Furthermore, we investigated the association between these SNPs and overall survival in GC patients.
Materials and methods
Patients, tissue samples, DNA extraction and Helicobactor pylori infection status
The studied population comprised 127 patients with GC being treated at the Endoscopy Center of Fujita Health University Hospital. The GCs were histologically diagnosed and classified according to Lauren's classification (20). Detailed information was obtained concerning lymph node, peritoneal, liver and distant metastases. Data on venous and lymphatic invasion were also obtained in 98 resected cases. Based on this information, early-stage GC was defined as GC localized within the mucosa or submucosa, irrespective of lymph node metastasis (21), and all other cases were defined as advanced-stage GC. According to its morphological appearance in an endoscopic image or surgically resected specimen, early GC was divided into two groups: polypoid or elevated type, and depressed type. Advanced GC was also classified according to Borrmann's classification. H. pylori infection status was assessed by serologic or histological analysis, or by the urea breath test. Patients were diagnosed as infected when at least one of the diagnostic tests was positive. All patients underwent an upper endoscopy with a biopsy from both the cancer lesion and the non-pathological mucosa, and the biopsy specimens were immediately frozen and stored at −80°C. Genomic DNA was isolated from the frozen specimens using proteinase K. Patients with severe systemic diseases were not included in the study. The experimental protocol was approved by the Ethics Committee of the Fujita Health University School of Medicine, and written informed consent was obtained from all participating subjects.
Genotyping
Using genomic DNA extracted from non-pathological mucosa, the rs11614913 (C>T), rs2910164 (G>C) and rs3746444 (A>G) SNPs were determined by polymerase chain reaction (PCR)-based restriction fragment length polymorphism (RFLP) assays, as described by Hu et al (12). Genotypes were determined by evaluation of the gel images by two independent investigators, who were blinded to the names and phenotype of the patients.
Bisulfite modification and methylation-specific PCR (MSP)
To detect CIHM, four candidate promoter CpG islands were selected, whose CIHM has been reported in GC (22–27): p14, p16, DAP-kinase and CDH1. For the examination of DNA methylation, genomic DNA from the cancer lesion was treated with sodium bisulfite using the BislFast DNA Modification kit for methylated DNA detection (Toyobo Co., Ltd., Osaka, Japan). CIHM status of four candidate promoter CpG islands was examined by MSP, as described previously (28). The primer pairs and experimental conditions for MSP were the same as those in our recent studies (18,19). CIHM was defined as the presence of a positive methylation band, separated by electrophoresis in 2.5% agarose gels under UV illumination using ethidium bromide staining, showing signals approximately equivalent to or greater than that of the size marker (10 ng/μl: 100 bp DNA Ladder; Takara Bio Inc., Shiga, Japan), irrespective of the presence of unmethylated bands. Samples giving faint positive signals were analyzed a further two times, and only those samples with a consistent positive methylation band were considered to be CIHM. In addition, we measured the fluorescence intensities of methylated bands in 50 randomly selected CHIM samples using a digital densitometer (Lane Analyzer; ATTO, Tokyo, Japan) and confirmed that the fluorescence intensities of all 50 methylated bands were approximately equivalent to or greater than the size marker (data not shown).
Statistical analysis
Genotype frequencies were calculated by direct counting. Differences in genotype frequencies among different clinicopathological subtypes were determined by the χ2 test. The odds ratios (ORs) and 95% confidence intervals (CI) were calculated by logistic regression analysis, with adjustment for age and gender. Survival among the different genotypes was assessed using the Kaplan-Meier method and compared using the log-rank test. A probability value of <0.05 was considered statistically significant in all analyses.
Results
Characteristics of the subjects and association between SNPs in miRNAs and GC clinicopathological characteristics
The characteristics of 127 GC patients are shown in Table I, and the association between SNPs in miRNAs and clinicopathological characteristics of GC are shown in Table II. The rs11614913 (T>C), rs2910164 (C>G) and rs3746444 (A>G) SNPs were successfully genotyped in all subjects.
Table I.
Clinicopathologic characteristics of the GC patients.
| Variable (n) | |
|---|---|
| Mean age ± SD (years) | 65.2±12.1 |
| Gender (Male:Female) | 90:37 |
| Lauren's histologic subtype | |
| Intestinal type | 73 |
| Diffuse type | 54 |
| H. pylori infection status | |
| H. pylori (+) | 102 |
| H. pylori (−) | 25 |
| Stage | |
| Early cancer | 57 |
| Advanced cancer | 70 |
| Morphology (early cancer) | |
| Polypoid or elevated type | 23 |
| Depressed type | 34 |
| Morphology (advanced cancer) | |
| Borrmann type I | 4 |
| Borrmann type II | 24 |
| Borrmann type III | 34 |
| Borrmann type IV | 8 |
Table II.
Associations between rs11614913, rs2910164 and rs3746444 SNPs and clinicopathological subtypes of GC.
| Variables (n) | rs11614913 genotype
|
rs2910164 genotype
|
rs3746444 genotype
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| TT | TC | CC | CC | CG | GG | AA | AG | GG | |
| Overall GC (127) | 36 | 63 | 28 | 64 | 49 | 14 | 82 | 33 | 12 |
| Lauren's classification (127) | |||||||||
| Diffuse type (54) | 16 | 27 | 11 | 25 | 25 | 4 | 37 | 12 | 5 |
| Intestinal type (73) | 20 | 36 | 17 | 39 | 24 | 10 | 45 | 21 | 7 |
| Staging (127) | |||||||||
| Early (57) | 19 | 27 | 11 | 28 | 20 | 9 | 36 | 16 | 5 |
| Advanced (70) | 17 | 36 | 17 | 36 | 29 | 5 | 46 | 17 | 7 |
| Lymphatic invasion (98) | |||||||||
| Negative (37) | 9 | 18 | 10 | 17 | 13 | 7 | 22 | 11 | 4 |
| Positive (61) | 19 | 32 | 10 | 31 | 25 | 5 | 39 | 17 | 5 |
| Venous invasion (98) | |||||||||
| Negative (65) | 20 | 32 | 13 | 31 | 24 | 10 | 42 | 17 | 6 |
| Positive (33) | 8 | 18 | 7 | 17 | 14 | 2 | 19 | 11 | 3 |
| Lymph node metastasis (127) | |||||||||
| Negative (65) | 18 | 34 | 13 | 32 | 24 | 9 | 42 | 17 | 6 |
| Positive (62) | 18 | 29 | 15 | 32 | 25 | 5 | 40 | 16 | 6 |
| Peritoneal dissemination (127) | |||||||||
| Negative (101) | 31 | 49 | 21 | 48 | 41 | 12 | 64 | 28 | 9 |
| Positive (26) | 5 | 14 | 7 | 16 | 8 | 2 | 18 | 5 | 3 |
| Liver or distant metastasis (127) | |||||||||
| Negative (120) | 35 | 60 | 25 | 59 | 48 | 13 | 76 | 32 | 12 |
| Positive (7) | 1 | 3 | 3 | 5 | 1 | 1 | 6 | 1 | 0 |
| Morphology (early GC) (57) | |||||||||
| Polypoid or elevated type (23) | 7 | 9 | 7 | 12 | 8 | 3 | 14 | 6 | 3 |
| Depressed type (34) | 12 | 18 | 4 | 16 | 12 | 6 | 22 | 10 | 2 |
| Morphology (advanced GC) (70) | |||||||||
| Borrmann type I (4) | 1 | 2 | 1 | 2 | 2 | 0 | 3 | 0 | 1 |
| Borrmann type II (24) | 6 | 13 | 5 | 12 | 10 | 2 | 14 | 8 | 2 |
| Borrmann type III (34) | 9 | 17 | 8 | 18 | 14 | 2 | 22 | 8 | 4 |
| Borrmann type IV (8) | 1 | 4 | 3 | 4 | 3 | 1 | 7 | 1 | 0 |
rs11614913 CC vs. TT+TC: depressed type vs. polypoid or elevated type; age- and gender-adjusted OR=6.29, 95% CI 1.18–33.47, p=0.03.
In the comparison of genotype frequencies among different clinicopathological subtypes, we found only a significant marginal association between the rs11614913 CC genotype and polypoid or elevated type morphology in the early-stage GC (depressed type vs. polypoid or elevated type, rs11614913 CC vs. TT+TC; age- and gender-adjusted OR=6.29, 95% CI 1.18–33.47, p=0.03), while other subtypes, such as Lauren's classification, staging, lymphatic and venous invasion, lymph node, peritoneal, liver and distant metastasis, were not associated with any of the three SNPs in the miRNAs.
Association between SNPs in miRNAs and GC CIHM status
We assessed the association between the three SNPs in the miRNAs and the CIHM status of GC, which has been reported to be involved in the biological characteristics of GC (18,19). The results are shown in Table III. All 127 GC samples were available for MSP analysis. Among the subjects, CIHM for p14 was found in 59 (46.4%) patients, CIHM for p16 in 26 (20.5%), DIHM for CDH1 in 87 (68.5%) and CIHM for DAP-kinase in 114 (89.8%) patients. The rs2910164 CC and CG genotypes were associated with increased susceptibility to CIHM of DAP-kinase (DAP-kinase unmethylated vs. hypermethylated; rs2910164 CC+CG vs. GG, age- and gender-adjusted OR=5.48, 95% CI 1.30–23.10, p=0.02; rs2910164 CC vs. CG+GG, age- and gender-adjusted OR=6.93, 95% CI 1.37–35.02, p=0.02; rs2910164 CG vs. CC+GG, age- and gender-adjusted OR=4.24, 95% CI 0.87–20.78, p=0.07). The 11614913 TT and TC genotypes were also found to be associated with a higher number of CIHM (no. of CIHM 0–1 vs. 2–4; rs11614913 TT+TC vs. CC, age- and gender-adjusted OR=3.67, 95% CI 0.98–13.72, p=0.05; rs11614913 TC vs. TT+CC, age- and gender-adjusted OR=4.08, 95% CI 1.04–15.97, p=0.04).
Table III.
Associations between rs11614913, rs2910164 and rs3746444 SNPs and GC CIHM status.
| Variables (n) | rs11614913 genotype
|
rs2910164 genotype
|
rs3746444 genotype
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| TT | TC | CC | CC | CG | GG | AA | AG | GG | |
| CIHM status (127) | |||||||||
| p14 | |||||||||
| Unmethylated (68) | 19 | 35 | 14 | 37 | 22 | 9 | 47 | 17 | 4 |
| Hypermethylated (59) | 17 | 28 | 14 | 27 | 27 | 5 | 35 | 16 | 8 |
| p16 | |||||||||
| Unmethylated (101) | 30 | 48 | 23 | 50 | 39 | 12 | 64 | 27 | 10 |
| Hypermethylated (26) | 6 | 15 | 5 | 14 | 10 | 2 | 18 | 6 | 2 |
| CDH1 | |||||||||
| Unmethylated (40) | 11 | 21 | 8 | 22 | 16 | 2 | 27 | 9 | 4 |
| Hypermethylated (87) | 25 | 42 | 20 | 42 | 33 | 12 | 55 | 24 | 8 |
| DAP-kinase | |||||||||
| Unmethylated (13) | 3 | 7 | 1 | 4 | 5 | 4 | 8 | 4 | 1 |
| Hypermethylated (114) | 33 | 54 | 27 | 60 | 44 | 10 | 74 | 29 | 11 |
| No. of CIHM | |||||||||
| 0–1 (30) | 9 | 18 | 3 | 16 | 12 | 2 | 20 | 7 | 3 |
| 2–4 (97) | 27 | 45 | 25 | 48 | 37 | 12 | 62 | 26 | 9 |
rs11614913 TT+ TC vs. CC: no. of CIHM, 0–1 vs. 2–4; age- and gender-adjusted OR=3.67, 95% CI 0.98–13.72, p=0.05. rs11614913 TC vs. TT+CC: no. of CIHM, 0–1 vs. 2–4; age- and gender-adjusted OR=4.08, 95% CI 1.04–15.97, p=0.04. rs2910164 CC+CG vs. GG: DAP-kinase, unmethylated vs. hypermethylated; age- and gender-adjusted OR=5.48, 95% CI 1.30–23.10, p=0.02. rs2910164 CC vs. CG+GG: DAP-kinase, unmethylated vs. hypermethylated; age- and gender-adjusted OR=6.93, 95% CI 1.37–35.02, p=0.02. rs2910164 CG vs. CC+GG: DAP-kinase, unmethylated vs. hypermethylated; age- and gender-adjusted OR=4.24, 95% CI 0.87–20.78, p=0.07.
Association between survival curves estimated by the Kaplan-Meier method and SNPs in the miRNAs
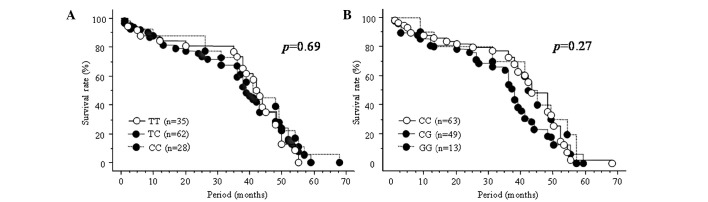
Of the 125 patients, including 21 unresectable and 104 resectable cases, overall survival, defined as the time from the date of surgery for resectable cases and the date of initial chemotherapy for unresectable cases, was characterized using the Kaplan-Meier method and was compared using the log-rank test for different genotypes of the three SNPs in miRNAs. The median follow-up period of the 125 patients was 30 months. Of the 21 unresectable patients, TS-1-based chemotherapy was performed in 18 cases and chemotherapy of other regimens was performed in the remaining three patients. No association was found between three SNPs and overall survival in any of the 125 subjects (Fig. 1A–C).
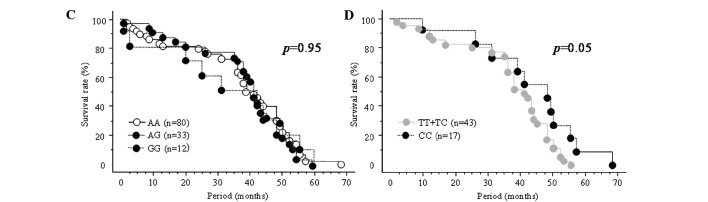
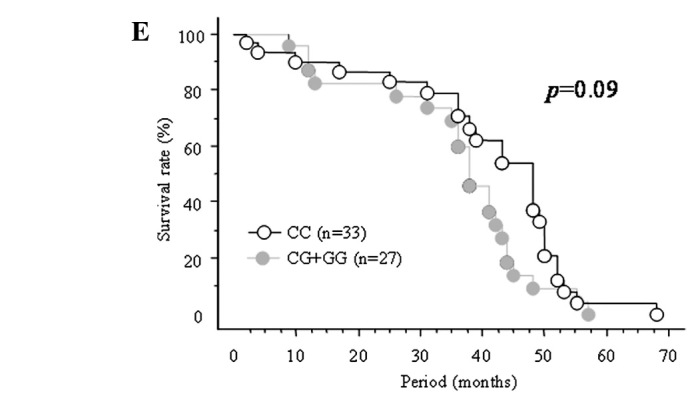
Figure 1.
Survival curves estimated by the Kaplan-Meier method for the (A) rs11614913, (B) rs2910164 and (C) rs3746444 SNPs in all the subjects (n=125), and for the (D) rs11614913 and (E) rs2910164 SNPs in subjects younger than 65 years of age. All statistical analyses were performed using the log-rank test.
When subjects were divided according to age group, the combined rs11614913 TT+TC genotype tended to be associated with worse overall survival than the CC genotype among patients younger than 65 years of age (p=0.05, Fig. 1D).
Also, the combined rs2910164 CG+GG genotype tended to be associated with worse overall survival than the CC genotype in the same age group (p=0.09, Fig. 1E). On the other hand, no such association was found for the rs3746444 genotype (data not shown).
Discussion
Although a number of studies have evaluated the association between SNPs in protein-coding genes and susceptibility to GC and its prognosis or response to treatment, association studies regarding SNPs in miRNA genes are relatively rare. However, it is increasingly recognized that SNPs in miRNAs may have important phenotypic consequences in human diseases.
Here, we evaluated the effect of three selected SNPs (rs11614913, rs2910164 and rs3746444) in pre-miRNAs (miR-196a2, miR-146a and miR-499), which were shown to contribute to human cancer susceptibility in recent association studies (12–17) on various subtypes of GC, including CIHM status and overall survival. In various clinicopathological subtypes, a significant association between the rs11614913 CC genotype and polypoid or elevated type morphology in early-stage GC was found. The morphological appearance of both early and advanced GC has been reported to be closely related to its histological subtypes, prognosis and survival, and these differences have also been characterized by several genetic and epigenetic alterations (19). Although no association was found between the rs11614913 CC genotype and other clinicopathological subtypes, which provide more distinct biological characteristics of GC such as lymphatic and venous invasion or metastasis, our data provide initial evidence that the rs11614913 CC genotype in miR-196a2 may contribute to the characterization of certain phenotypes of GC. Polypoid or elevated type morphology in early-stage GC usually shows well-differentiated histopathology (29) and is unlikely to present lymph node metastasis in small size, compared to depressed type (30). In this context, the rs11614913 CC genotype may be associated with a rather mild phenotype of GC.
We also investigated whether the three SNPs (rs11614913, rs2910164 and rs3746444) are associated with the CIHM status of GC. We found that the rs2910164 CC and CG genotypes were associated with increased susceptibility to CIHM of DAP-kinase. In addition, the 11614913 TT and TC genotypes were associated with a higher number of CIHM.
CIHM is now accepted as an important mechanism in gene silencing, and CIHM of tumor suppressor genes is also highly involved in gastric carcinogenesis; CIHM of p14, p16, CDH1 and DAP-kinase genes assessed in this study frequently occur in GC tissue, as well as in pre-malignant lesions (3–8). Therefore, they are thought to be susceptible candidate genes for CIHM in GC. Furthermore, it has been suggested that this epigenetic change may also constitute a certain distinct biological behavior of GC.
CIHM of DAP-kinase and a higher number of CIHM have been associated with worse survival of GC patients (19). These epigenetic alterations have also been correlated with the morphological appearance of GC (18).
Moreover, a higher number of CIHM in neoplastic gastric mucosa has been closely associated with the severity of H. pylori-related gastritis (31) and GC occurrence (32), suggesting that rs2910164 and 11614913 SNPs may be an important influencing factor for CIHM-related gastric carcinogenesis.
We also demonstrated that combined rs11614913 TT+TC genotypes were weakly associated with worse overall survival than the CC genotype in patients younger than 65 years of age. A similar trend was also found for the combined rs2910164 CG+GG genotype for patients in the same age group, suggesting that these genotypes may be predictors of worse prognosis of GC, particularly for younger patients. Due to its variable biological behavior, GC often presents various clinical phenotypes, prognosis and response to treatment. Therefore, in order to focus on disease heterogeneity, it is necessary for the physician to more appropriately conduct a clinical evaluation for each patient. Our preliminary results showed that more longitudinal studies are required to investigate the clinical usefulness of SNPs in miRNAs as a molecular biomarker for the prediction of prognosis in GC patients.
In conclusion, we demonstrated that the rs11614913 and rs2910164 SNPs in pre-miRNAs (miR-196a2 and miR-146a) may have an effect on the clinicopathological characteristics of GC, including its morphological appearance, CIHM status and overall survival. However, it should be noted that our associations were found in a subgroup stratification analysis of a small sample; thus, the statistical power was not sufficient. In addition, the association of CIHM with subtypes of GC was found to vary in other studies, according to the different CpG islands assessed, even in the same genes (33,34). Moreover, our study is best viewed as hypothesis-generating rather than hypothesis-testing, as both the underlying in vitro and in vivo mechanisms of the three miRNAs in carcinogenesis of the stomach are largely unknown. Further characterization of miRNA SNPs, miRNAs, target mRNAs and their compensational or redundancy role in GC are required to confirm our results.
Abbreviations:
- miRNA
microRNA
- GC
gastric cancer
- H. pylori
Helicobacter pylori
- PCR
polymerase chain reaction
- RFLP
restricted fragment length polymorphism
- CIHM
CpG island hypermethylation
- MSP
methylation-specifc PCR
References
- 1.Parkin DM, Whelan SL, Ferlay J, Raymond L, Young J. Volume VII. IARC Press; Lyon: 1997. Cancer Incidence in Five Continents. [Google Scholar]
- 2.Parkin DM, Pisani P, Ferlay J. Estimates of the worldwide incidence of 25 major cancers in 1990. Int J Cancer. 1999;80:827–841. doi: 10.1002/(sici)1097-0215(19990315)80:6<827::aid-ijc6>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 3.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 4.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 5.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 6.Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673–676. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 7.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 8.Esquela-Kerscher A, Slack FJ. Oncomirs – microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 9.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 10.Petrocca F, Visone R, Onelli MR, et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272–286. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 11.Takagi T, Iio A, Nakagawa Y, Naoe T, Tanigawa N, Akao Y. Decreased expression of microRNA-143 and -145 in human gastric cancers. Oncology. 2009;77:12–21. doi: 10.1159/000218166. [DOI] [PubMed] [Google Scholar]
- 12.Hu Z, Chen J, Tian T, et al. Genetic variants of miRNA sequences and non small cell lung cancer survival. J Clin Invest. 2008;118:2600–2608. doi: 10.1172/JCI34934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tian T, Shu Y, Chen J, et al. A functional genetic variant in microRNA-196a2 is associated with increased susceptibility of lung cancer in Chinese. Cancer Epidemiol Biomarkers Prev. 2009;18:1183–1187. doi: 10.1158/1055-9965.EPI-08-0814. [DOI] [PubMed] [Google Scholar]
- 14.Jazdzewski K, Murray EL, Franssila K, Jarzab B, Schoenberg DR, de la Chapelle A. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc Natl Acad Sci USA. 2008;105:7269–7274. doi: 10.1073/pnas.0802682105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu T, Zhu Y, Wei QK, et al. A functional polymorphism in the miR-146a gene is associated with the risk for hepatocellular carcinoma. Carcinogenesis. 2008;29:2126–2131. doi: 10.1093/carcin/bgn195. [DOI] [PubMed] [Google Scholar]
- 16.Hu Z, Liang J, Wang Z, et al. Common genetic variants in premicroRNAs were associated with increased risk of breast cancer in Chinese women. Hum Mutat. 2009;30:79–84. doi: 10.1002/humu.20837. [DOI] [PubMed] [Google Scholar]
- 17.Peng S, Kuang Z, Sheng C, et al. Association of microRNA-196a-2 gene polymorphism with gastric cancer risk in a Chinese population. Dig Dis Sci. 2010;55:2288–2293. doi: 10.1007/s10620-009-1007-x. [DOI] [PubMed] [Google Scholar]
- 18.Tahara T, Shibata T, Arisawa T, et al. CpG island promoter methylation (CIHM) status of tumor suppressor genes correlates with morphological appearances of gastric cancer. Anticancer Res. 2010;30:239–244. [PubMed] [Google Scholar]
- 19.Tahara T, Shibata T, Nakamura M, et al. Association between cyclin D1 polymorphism with CpG island promoter methylation status of tumor suppressor genes in gastric cancer. Dig Dis Sci. 2010 Apr 16; doi: 10.1007/s10620-010-1206-5. (E-pub ahead of print). [DOI] [PubMed] [Google Scholar]
- 20.Lauren P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. An attempt at a histo-clinical classification. Acta Pathol Microbiol Scand. 1965;64:31–49. doi: 10.1111/apm.1965.64.1.31. [DOI] [PubMed] [Google Scholar]
- 21.Gotoda T, Yanagisawa A, Sasako M, et al. Incidence of lymph node metastasis from early gastric cancer: estimation with a large number of cases at two large centers. Gastric Cancer. 2000;3:219–225. doi: 10.1007/pl00011720. [DOI] [PubMed] [Google Scholar]
- 22.Toyota M, Ahuja N, Suzuki H, et al. Aberrant methylation in gastric cancer associated with the CpG island methylator phenotype. Cancer Res. 1999;59:5438–5442. [PubMed] [Google Scholar]
- 23.Kang GH, Shim YH, Jung HY, Kim WH, Ro JY, Rhyu MG. CpG island methylation in premalignant stages of gastric carcinoma. Cancer Res. 2001;61:2847–2851. [PubMed] [Google Scholar]
- 24.Chan AO, Lam SK, Wong BC, et al. Promoter methylation of E-cadherin gene in gastric mucosa associated with Helicobacter pylori infection and in gastric cancer. Gut. 2003;52:502–506. doi: 10.1136/gut.52.4.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raveh T, Kimchi A. DAP kinase – a proapoptotic gene that functions as a tumor suppressor. Exp Cell Res. 2001;264:185–192. doi: 10.1006/excr.2000.5134. [DOI] [PubMed] [Google Scholar]
- 26.Schildhaus HU, Krockel I, Lippert H, Malfertheiner P, Roessner A, Schneider-Stock R. Promoter hypermethylation of p16INK4a, E-cadherin, O6-MGMT, DAPK and FHIT in adenocarcinomas of the esophagus, esophagogastric junction and proximal stomach. Int J Oncol. 2005;26:1493–1500. [PubMed] [Google Scholar]
- 27.Waki T, Tamura G, Sato M, Terashima M, Nishizuka S, Motoyama T. Promoter methylation status of DAP-kinase and RUNX3 genes in neoplastic and non-neoplastic gastric epithelia. Cancer Sci. 2003;94:360–364. doi: 10.1111/j.1349-7006.2003.tb01447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang GH, Lee HJ, Hwang KS, Lee S, Kim JH, Kim JS. Aberrant CpG island hypermethylation of chronic gastritis, in relation to aging, gender, intestinal metaplasia, and chronic inflammation. Am J Pathol. 2003;163:1551–1556. doi: 10.1016/S0002-9440(10)63511-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song SY, Kim S, Kim DS, Son HJ, Rhee JC, Kim YI. Abnormal expression of E-cadherin in early gastric carcinoma: its relationship with macroscopic growth patterns and catenin alpha and beta. J Clin Gastroenterol. 2004;38:252–259. doi: 10.1097/00004836-200403000-00011. [DOI] [PubMed] [Google Scholar]
- 30.Namieno T, Koito K, Higashi T, Takahashi M, Yamashita K, Kondo Y. Assessing the suitability of gastric carcinoma for limited resection: endoscopic prediction of lymph node metastases. World J Surg. 1998;22:859–864. doi: 10.1007/s002689900482. [DOI] [PubMed] [Google Scholar]
- 31.Tahara T, Arisawa T, Shibata T, et al. Increased number of methylated CpG islands correlates with Helicobacter pylori infection, histological and serological severity of chronic gastritis. Eur J Gastroenterol Hepatol. 2009;21:613–619. doi: 10.1097/MEG.0b013e32830e28b2. [DOI] [PubMed] [Google Scholar]
- 32.Tahara T, Shibata T, Nakamura M, et al. Increased number of CpG island hypermethylation in tumor suppressor genes of nonneoplastic gastric mucosa correlates with higher risk of gastric cancer. Digestion. 2010;82:27–36. doi: 10.1159/000252766. [DOI] [PubMed] [Google Scholar]
- 33.Zhang KL, Sun Y, Li Y, et al. Increased frequency of CpG island methylator phenotype and CDH1 methylation in a gastric cancer high-risk region of China. Transl Oncol. 2008;1:28–35. doi: 10.1593/tlo.07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo D, Zhang B, Lv L, et al. Methylation of CpG islands of p16 associated with progression of primary gastric carcinomas. Lab Invest. 2006;86:591–598. doi: 10.1038/labinvest.3700415. [DOI] [PubMed] [Google Scholar]