Abstract
The recently developed Tightly Bound Ion (TBI) model offers improved predictions for ion effect in nucleic acid systems by accounting for ion correlation and fluctuation effects. However, further application of the model to larger systems is limited by the low computational efficiency of the model. Here, we develop a new computational efficient TBI model using free energy landscape-guided sampling method. The method leads to drastic reduction in the computer time by a factor of 50 for RNAs of 50-100 nucleotides long. The improvement in the computational efficiency would be more significant for larger structures. To test the new method, we apply the model to predict the free energies and the number of bound ions for a series of RNA folding systems. The validity of this new model is supported by the nearly exact agreement with the results from the original TBI model and the agreement with the experimental data. The method may pave the way for further applications of the TBI model to treat a broad range of biologically significant systems such as tetraloop-receptor and riboswitches.
Keywords: ion-nucleic acid interaction, Tightly Bound Ion theory, RNA folding
1. Introduction
Nucleic acids (RNAs and DNAs) are negatively charged chain molecules. The negative backbone charges on nucleic acids attract cations in the solution. As a result, nucleic acid molecules are effectively dressed with ions. Ions play critical roles in stabilizing nucleic acid structures.1–18 Understanding nucleic acid folding requires quantitative predictions of ion-mediated nucleic acid energetics. Most available thermodynamic parameters for nucleic acids are based upon thermal melting data for secondary structural motifs such as helix stems and loops under a fixed standard salt condition, namely, 1M Na+.19–23 These thermodynamic parameters have led to many successful predictions for nucleic acid secondary structures and folding stabilities.19–23 However, the problem about accurate quantitative predictions for RNA folding stabilities under the different (non-standard) ionic conditions of the solution and furthermore, the ion effects in tertiary structural folding, remain unsolved. One of the key issues is about how to model the Mg2+ ion effect.
Mg2+ ions can interact with nucleic acids through diffusive nonspecific and site-specific binding.12 A variety of theories have been developed to treat Mg2+ ion binding. For example, for the nonspecific binding, the classical Debye-Huckel theory and the nonlinear Poisson-Boltzmann theory have been used to study the ion binding thermodynamics.24–30 These theories can efficiently treat the large nucleic acids system. However, these theories do not explicitly consider the potential effects of ion fluctuation and correlation and ion dehydration in ion-RNA binding.
Site-specific ion effect can also play an important role for many RNA and DNA structures. Theoretical studies based on molecular dynamics (MD) simulations have provided invaluable insights into the atomic details for the energetics and dynamic ionic contributions.18,31–38 For example, recently Chen et al used MD simulation to investigate the ion-mediated interactions in a RNA kissing loop and demonstrated the important roles of ion-specific dehydration effects in counterion binding to a kissing loops.18 In another application of the MD simulation, Auffinger et al studied the Mg2+ binding to the 5S rRNA loop E motif and illustrated two distinct binding modes for pentahydrated Mg2+ in the system.31 The above and many other studies showed the unique advantages of using MD simulations to investigate the ion-specific effects in ion-RNA interactions and the dependence on RNA structure and sequence.18,31–39 However, the accuracy and efficiency of MD simulations are restricted by the force field and the computational expense of adequately exploring the configurations of nucleic acids and the surrounding water molecules and ions.33 The problem is more pronounced for the study of the folding of large nucleic acids. A coarse-grained model, on the other hand, may provide more reliable (low-resolution) sampling for ion distribution and RNA structures. Such a model may provide a useful complement to MD simulations.
A compact tertiary structure involves significant charge build-up and strong binding of cations to the nucleic acid. The high local concentration of the counterions can potentially cause strong coupling (correlation) between the ions, i.e., ions form a network due to the strong Coulomb interactions. As a result, the electric field on a particle is a function of not only the coordinate of the particle but also the coordinates of all the other particles (ions). A correlated ion distribution can only be accounted for by discrete ion distributions (instead of the mean-field distribution). For multivalent ions, because of the strong Coulomb interactions, the correlation effect can be significant. Indeed, recent experimental findings suggested that ion correlation effect may be important for multivalent ions around nucleic acids.6,40–42
Motivated by the potential importance of ion correlation and fluctuation effects, we recently developed the tightly bound ion (called TBI) model.43,48 The key idea in the TBI model is to account for the ion correlation effect by using many-ion distribution function instead of the mean-field single-ion distribution. In the TBI model we generate an ensemble of ion distributions by enumerating all the possible discrete modes for the many-ion distribution. We then calculate the many-ion interaction energy. In this way, the model can account for the many-ion correlation effect. Comparisons with experimental data showed that, by accounting for ion correlation and fluctuations, the model can offer improved predictions for the ion-dependence of nucleic acid folding stability.44–48
In practical calculation, for a given nucleic acid structure, we first run the Poisson-Boltzmann (PB) calculation to have a crude estimate for the ion concentration. Based on the distribution of the ion concentration, we calculate the spatial distribution of the Coulomb correlation strength. The correlation strength establishes a demarcation of space into the different regions: the tightly bound (TB) and the diffusively bound (DB) regions for the the correlation strength above and below a critical value, respectively. For monovalent ion solution, the correlation effect is weak and nearly the whole solution is the DB region. For a solution containing multivalent ions, however, the TB region can become significant. The TB region usually corresponds to a thin layer surrounding the nucleic acid structure. For a divalent ion solution, the width of the TB region (layer) can vary from 0 to 3 Angstroms (depending on the multivalent ion concentration). Extensive tests have shown that the theoretical predictions are not very sensitive to the specific boundary between the TB and the DB regions.50,51,53,54 Corresponding to the two regions, we classify two types of ions: the tightly bound (TB) ions and the diffusively bound (DB) ions, for ions in the TB and the DB regions respectively. For the DB ions, the correlation is weak and the fluid model (PB theory) would be able to provide an accurate description. For the TB ions, the correlation is strong, thus, the mean-field model would fail and we need a model that can account for discrete many-ion distributions instead of a mean single-ion distribution.
To further enumerate the different distributions of the TB ions, we discretize the TB region. Specifically, we divide the TB region into small cells (called TB cells), each around the phosphate of a nucleotide. For an N-nt nucleic acid structure, the TB region consists of N TB cells. To enumerate the ion distributions for an ensemble of Nb TB ions, we partition the Nb ions into N TB cells. Each such distribution is called an ion distribution mode or ion binding mode (M). It is important to note that each such mode is an Nb-particle distribution instead of a single particle mean-field distribution. Mathematically, an ion distribution mode M is an Nb-ion distribution as defined by the ion occupation numbers mi (= 0 or 1) in each cell i: M = {m1; m2; · · ·; mN}. Here mi is the number of the TB ions in the ith cell. There exist totally 2N such modes for a N-nt molecule. Previous applications and tests of the TBI model suggest that the model may provide improved predictions for ion-nucleic acid interactions than other models.44–48 However, because of the rapidly increasing computational time for long chain lengths N, the application of the original TBI model has been mainly restricted to only small systems, which may have limited biological significance.
In the present study, on the basis of the recently developed all-atom TBI model,48 we develop an energy landscape-based method to effectively reduce the effective number of modes. We sample the low-energy modes (in the basins of the free energy landscape) exhaustively and calculate the exact electrostatic free energy for each of the low-energy modes. For all the other (higher energy) ion distribution modes, we use approximate free energy functions. Tests for (long-chain) RNAs of length 50-90 nts show that the new method leads to 50-fold increase in the computational efficiency over the original TBI model. The improvement is even more dramatic for longer chains. Ion electrostatic effects for RNAs of length longer than 100-nts, which cannot be treated by the original TBI model due to the exceedingly long computational time, can now be readily predicted with only 2% of the original computer time. The new method thus leads to a significant step for TBI-based prediction of the ion effects in nucleic acid folding.
2. Method
2A. The original TBI model and its low computational efficiency
Central to the free energy calculation is the partition function, which involves the sum over all the possible ion distributions. In the TBI model,43,48 the total partition function Z of the system is the sum of the partition function ZM for all the possible ion distribution modes M:
| (1) |
An ion distribution mode defines only the number (instead of the coordinates) of the TB ions in each TB cell. For a given mode, the TB ions can move around in the respective cells. We evaluate the partition function ZM for a given mode M by averaging over all the possible positions of the TB ions:
| (2) |
Here, Zid is the partition function for the uniform ion solution (without the nucleic acids), Nb is the total number of the TB ions for the given mode, and c0 is the bulk concentration of the counterion, Ri denotes the coordinate of the i_th TB ion. is the volume integral over the TB region for the TB ions. The value ΔGb and are the mean Coulombic free energy (including the phosphate groups and the TB ions) and the (Born) self-polarization energy for the charges in the TB region, ΔGd is the sum of the free energy for the DB ions (including the interactions between the DB ions and the TB ions).48 It is important to note that for monovalent ions, unless the ion concentration reaches several M's or higher, the correlation effect can be neglected. In contrast, for divalent (such as Mg2+) and higher valent ions, correlation could be important for ion concentrations at mM level. For a mixed monovalent and multivalent ion solution, because the correlation effect for the monovalent ions is negligible, we treat all the monovalent ions as DB ions which form a background for the multivalent ions.
Based on the partition function of the system (Eq. 1), we calculate the electrostatic free energy G = −kBT ln(Z/Zid). Using 1M NaCl as the reference salt solution, we can compute the excess electrostatic energy: ΔG = G − G0, where G0 is the free energy of the system in the 1M NaCl solution.
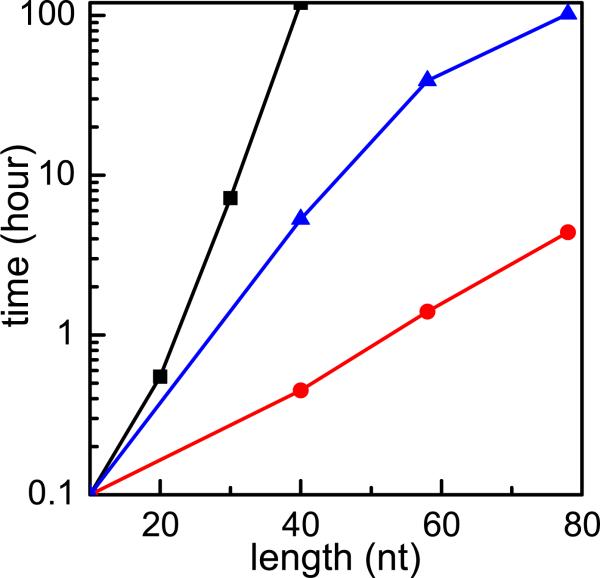
The number of ion distribution modes increases exponentially with the number of the phosphates (= the number of the nucleotides). The exceedingly long computational time prohibits the application of the TBI model to large nucleic acid molecules (see Fig. 1). In an attempt to reduce the computational time, we previously developed a hybrid method where the low-energy modes are exhaustively enumerated while the high-energy modes are randomly sampled.44 Because the method avoids exhaustive enumeration for a large portion of the modes, namely, the high energy modes, it led to notable improvement in the computational efficiency. However, the improvement of the efficiency is quite limited because a significant portion of the modes still requires exhaustive enumeration. Indeed, the method cannot treat larger structures such as those with the number of nucleotides larger than 80 (see Fig. 1). In the present paper, we report a new method that can lead to drastic improvement in the computational efficiency.
Figure 1.
The computer time as a function of the sequence length for an RNA duplex in 0.02M NaCl and 0.005M MgCl2 mixed solution. The original TBI: square; the previously improved TBI: triangle; the current new TBI model: circle.
2B. A new method: energy landscape-guided sampling
The new method is based on the free energy landscape of the system, i.e., the electrostatic free energy as a function of the ion distribution. By focusing on the most important modes, namely, those in the low-energy basins on the energy landscape, we here develop a new method that can significantly enhance the sampling efficiency in the partition function calculation. In the following, we first discuss several important features of the free energy landscape.
2B-1. Free energy landscape
We consider an N-nt RNA structure immersed in a Mg2+ solution. The number of the TB ions Nb can vary from 0 to N. For each given Nb, there exist modes. We classify all the modes according to two parameters: Nb and the free energy GM = −kBT ln(ZM/Zid), the free energy of the system for the given mode M of the TB ion distribution. We then evaluate the number of modes (NOM) Ω(Nb, ΔGM) for each given (Nb, ΔGM), where ΔGM = GM −G0 is the difference between GM and the free energy of the system in the 1M NaCl solution. From the NOM Ω(Nb, ΔGM), we calculate the free energy landscape of the system as a function of the above two parameters:
Physically, ΔG(Nb, ΔGM) is the free energy for the macrostate consisting of all the modes with the same free energy ΔGM and the number of the TB ions Nb.
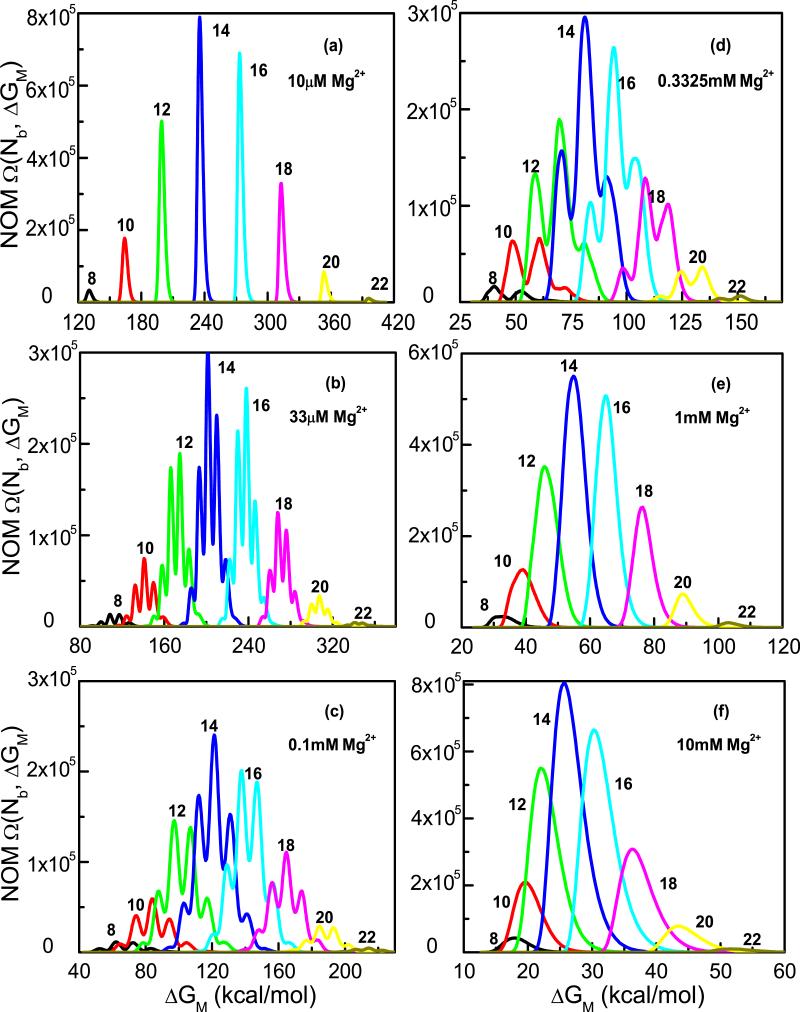
Using the BWYV system as an example, in Fig. 2 we show the NOM function of the system. The different panels in the figure correspond to the different [Mg2+] with a fixed 0.054M of Na+ background. The state of full charge neutralization (Nb = N/2) has the largest combinatorial number of partitioning the Nb ions into the N TB cells. Thus, the largest number of the modes (the peaks in Fig. 2) occurs at Nb = N/2.
Figure 2.
The distribution of the number of modes (NOM) Ω(Nb, ΔGM) for the BWYV pseudoknot in the different [Mg2+] ion solutions with a fixed 0.054 [Na+] background. The numbers (8, 10 · · ·) in the figure denote the Nb values. Not all the Nb results are shown in the figure.
Mg2+ binding causes a decrease in the translational entropy of the ion (see, e.g., the factor (c0)Nb in Eq. 2) and a resultant free energy penalty. For a low [Mg2+], Mg2+ binding results in a large free energy penalty, which, as shown in Fig. 2a, causes (a) a sharp, well separated energy distribution (NOM) and (b) higher free energies (see the x-axis values of the figures) at a low [Mg2+] such as 10μM. In contrast, for a high [Mg2+], the entropy decrease and free energy change for ion binding are much smaller. As a result, the free energy is much lower and the distribution is much broader, resulting in a significant overlapping in the NOM profile for the different Nb values (see Fig. 2f). For both the low and high Mg2+ concentrations, the different modes for a given Nb are distributed in a Gaussian fashion. However, for [Mg2+] in the intermediate regime, the energy distribution shows a non-Gaussian profile (see Figs. 2b-e). Such a complex energy distribution is due to the competition between the Na+ and the Mg2+ binding and the energy variation for the different modes.
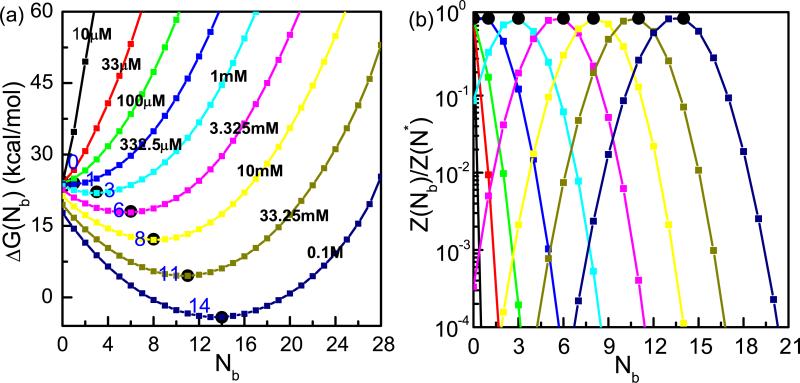
To conveniently visualize the free energy landscape, we plot a one-dimensional free energy profile: the free energy as a function of Nb, the number of the TB ions. For a given Nb (= 0, 1, · · ·, N for an N-nt RNA), we define the partition function Z(Nb) by summing over all the possible modes that have Nb TB ions. The electrostatic free energy for a given Nb can be calculated from Z(Nb) as G(Nb) = −kBT ln Z(Nb) and ΔG(Nb) = G(Nb)−G0. Fig. 3 shows our predicted ΔG(Nb) with varying [Mg2+] in a fixed background of [Na+] = 0.054M at 25°C. The energy landscape shows several important features.
As shown in Fig. 3a, the electrostatic free energy ΔG(Nb) is a smooth function of Nb. This is because a given Nb corresponds to an ensemble of modes and the ensemble average over all the modes may smooth out the energy change between the different modes. A smooth free energy landscape suggests the possibility of fitting the free energy using an analytical function.
For a given [Mg2+], the free energy landscape has a single minimum, corresponding to the (single) most stable ion distribution.
The state of the free energy minimum is sensitive to the ionic condition. As [Mg2+] is increased, the electro-static free energy of the system is lowered and , the number of the TB ions for the most stable ion distribution (at the free energy bottom), increases from zero at very low [Mg2+] (no Mg2+-induced charge neutralization) to about N/2 at higher [Mg2+] (full charge neutralization from Mg2+). The larger number of the TB ions at a higher [Mg2+] shows that the TB ions play a more important role for higher Mg2+ concentrations. Furthermore, the (nearly) full charge neutralization at high [Mg2+] results in a less sensitive dependence of the electrostatic energy on . On the free energy landscape, this is manifested as the wider and shallower basin around the minima.
The partition function is overwhelmingly dominated by the ion distribution modes around (the most stable state). Fig. 3b shows the ratio between Z(Nb) for an arbitrary Nb and for the lowest free energy mode. It is clear that Z(Nb) falls drastically as Nb deviates away from . As Nb deviates away from by 5-6 ions, the partition function Z(Nb) falls below 10−4 of .
Figure 3.
(a) The reduced (1D) electrostatic energy landscape ΔG(Nb) for the BWYV pseudoknot in the different ion solutions with the fixed [Na+]=0.054M and varying [Mg2+] (in M) (labeled on the curves). (b)The partition functions as a function of Nb for the different solutions. The filled squares and the labels denote the locations of the free energy minima and the corresponding .
2B-2. Energy landscape-guided mode reduction
The above features of the energy landscape suggest that in the partition function calculation, we can safely leave out the modes whose Nb's are far away from . How to determine the effective range of Nb for the partition function calculation? The effective range of Nb is determined by the width of the free energy basin at the global minimum, which, due to the aforementioned correlation between the size of the TB region and the width of the free energy basin, can be loosely related to the thickness t of the TB region.
In order to derive a general rule for the selection of the modes, we have performed calculations for a series of structures: DNA and RNA duplexes, RNA hairpin, BWYV pseudoknot(PDB : 437D49), rRNA fragments(PDB code: 1HC850), tRNA(PDB code: 1TRA51) and tetraloop-receptor RNA (PDB code: 2I7Z and 2ADT52) in the various Mg2+/Na+ solution with [Mg2+] ranging from 10μM to 0.1M. For each structure in a given ionic condition, we compute the partition function for the different ionic conditions using (a) the exhaustive enumeration of all the possible modes and (b) local sampling around the free energy minimum. Through systematic calibration, we find the following relationship between the range of the effective for the partition function and the average thickness (t) of the TB region:
| (3) |
The parameters A, B, C and D are tunable in accordance with the specific nucleic acids structures. In the tested DNAs and RNAs, we find A=C=0.23, B=0.32 and D=0.08. For a given structure in a specific ionic solution, if the (or ) value given by Eq. 3 is too close to or too far away from the most stable state , from the partition function Z(Nb) curve (see Fig. 3b), we can determine the value by setting the partition function at to be equal to 10−4 (see Fig. 3(b)). In practice, since the partition functions for Nb near the boundaries and are very small, the total partition function for all the different modes is not very sensitive to the specific values of the A, B, C and D coefficients in the above equation.
The use of only the modes with Nb between and leads to a drastic reduction in the effective number of modes from 2N to a much smaller number . The reduction is more striking for low ion concentrations in which case the average thickness t of the TB region is small and hence the free energy basin is narrow. For example, for 58-nt rRNA fragment (PDB code: 1HC850) in a Mg2+/K+ mixed solution with [K+] fixed at 0.02M, thickness t is below 1 Angstrom for [Mg2+] below 0.15mM. Correspondingly, the mode-reduction and the computer time-reduction are more significant. For the rRNA fragment in the Mg2+/K+ mixed solution, the computational time is reduced from about 43.0 hours for the original TBI model to 24 minutes for the current new TBI model.
For higher (multivalent) ion concentration, the average thickness t of the TB region is larger. For example, for the 58-nt rRNA fragment in Mg2+/K+ solution, t can exceed 1.5 Angstrom for [Mg2+] above 0.35mM. The larger t value leads to the larger number of the selected Nb's. For example, for t ~1.5 Angstrom, the range of the Nb's to be used in the calculation can reach 0.4N. For a long sequence (large N), the reduced number of the modes can still be large and the computational time can still be demanding. In such a case, we use further approximations for the free energy landscape to enhance the computational efficiency. The smooth shape of the landscape ΔG(Nb) (Fig. 3) suggests a reliable fitting of the electrostatic free energy ΔG(Nb) as a polynomial of Nb. We randomly select 24 Nb's in the range between and and compute the electrostatic energy ΔG(Nb) for each selected Nb. From the 24 data points, we then fit ΔG(Nb) with an 8th-degree polynomial of Nb. Our tests for 96-nt DNA duplex, tRNA, tetraloop-receptor RNA, show that the 24-point fitting causes relative error less than 1% and hence is reliable.
In the practical calculation, from the computed TB region, we estimate the mean thickness t and calculate and from Eq. 3. If there exist less than 24 Nb values between and , we calculate the partition function Z(Nb) and the electrostatic energy ΔG(Nb) for every Nb located in the range. If the Nb range exceeds 24, we randomly select 24 different Nb's in the range and calculate the Z(Nb) and ΔG(Nb) for each selected Nb. From the 24 data points, we then fit the ΔG(Nb) curve with an 8th-degree polynomial of Nb. From the ΔG(Nb) curve, we predict the binding fraction and electrostatic free energy of the system.
3. Application and Discussion
The purpose here is to test the new method through comparisons with the experimental data. In addition, through comparisons with the previous TBI method, we show the remarkable improvement in the computational efficiency (see Fig. 1). In order to make direct comparison with the experiments, we calculate the fractional number of bound ions. The fractional number of the bound ions fMg2+ per nucleotide consists of the diffusively bound (DB) ions and the tightly bound (TB) ions:46
| (4) |
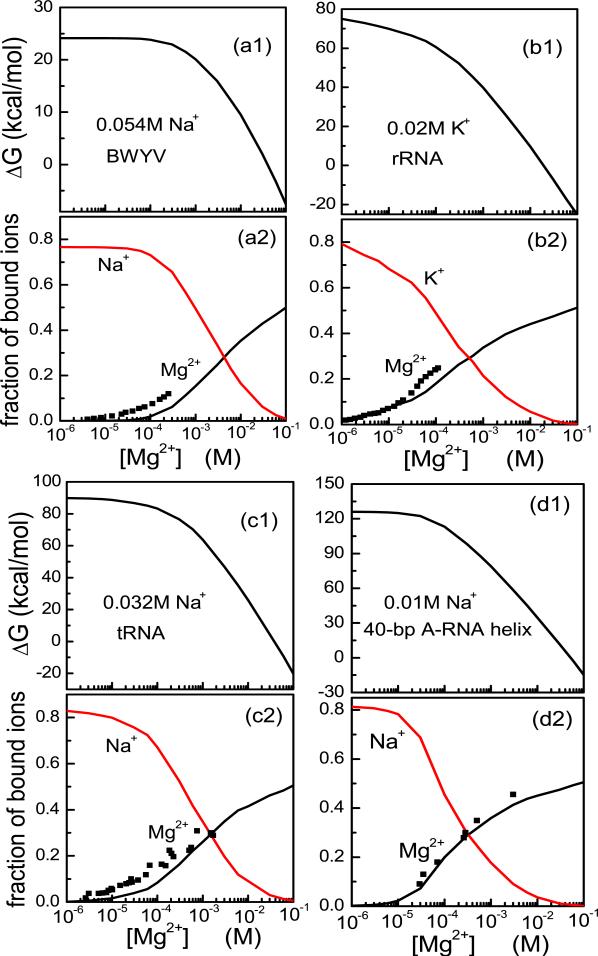
Here, the term involving is the number of the TB ions averaged over all the possible ion binding modes, the integral terms give the numbers of bound ions from the excess ion concentration,43 and cMg2+ and cNa+ are the local concentrations of the diffusively bound Mg2+ and Na+ ions, respectively. In Fig. 4a, we show the predicted electrostatic free energy for BWYV pseudoknot. For comparison, the binding fractions determined in the experimental measurements are also shown by black symbols in the figure. The theoretical predictions from the current mode-reduction method show agreements with the experimental data. Moreover, the current new method gives nearly exactly the same results as the original TBI predictions which are based on the brute force mode enumeration.48
Figure 4.
The electrostatic free energy ΔG (upper panels) and the Mg2+ and Na+ (or K+) binding fractions per nucleotide (lower panel) for various RNA tertiary structures: (a) BWYV pseudoknot fragment, (b) 58-nt rRNA fragment, (c) Yeast tRNAPhe. (d) 40-bp RNA duplex. The corresponding experimental data are from Refs.13,53–55 respectively. In the lower panels, the black symbols are the experimental data and solid lines are the predicted results.
To further test the improvement in the computational efficiency for the mode-reduction method developed here, we calculate the ion electrostatics for RNAs having long sequences (longer than 50-nts). For example, for the 58-nt fragment of ribosomal RNA (PDB code: 1HC8;50 see Fig.4b) immersed in a KCl/MgCl2 solution with a fixed [KCl] = 0.02M, the mode reduction method leads to a dramatic reduction in the computational time from about 43 hours (average time for the different Mg2+ concentrations) for the previous TBI model to 48 minutes for the current new model. For larger RNAs in higher Mg2+ concentration, we employ the aforementioned 24-point fitting method for the free energy profile ΔG(Nb). For example, in Fig. 4c, we predict the thermodynamics of the 76-nt yeast tRNAPhe (PDB code: 1TRA;51) by using of the mode-reduction method in combination with the 24-point fitting method. The average time for the different Mg2+ concentration is about 2 hours, which is about 50 times shorter than the 93 hours required by the previous TBI model. In Fig. 4d, we calculate the thermodynamic properties for an 80-nt RNA duplex using the current energy landscape-guided sampling method. The average time for the different Mg2+ concentration is about 3 hours, which is about 52 times shorter than the 156 hours required by the previous TBI model based on the enumeration of the ion binding modes.
We note that the theoretical predictions (Fig. 4a) are not perfect. One of the possible sources of the error may come from the monovalent ions as explained below. The charge density of some discrete local regions of a a compact tertiary RNA structure may become so high that even monovalent ions in these regions can become correlated. In the current form of the theory, all the monovalent ions are assumed to be (weakly correlated) diffusive ions. This approximation may require further examination for compact RNA tertiary structures.
4. Conclusion
By accounting for ion correlation and fluctuation effects, the tightly bound ion (TBI) model can provide reliable predictions for the ion effects in a variety of nucleic acid systems. However, applications of the model to large nucleic structures have been limited by the low computational efficiency of the model. In the present study, we develop a new method that shows remarkable improvement in the computational efficiency for the TBI model. The new method is based on the free energy landscape: a single-valley, smooth energy landscape as a function of the the number of the tightly bound ions. The steep single valley in the landscape allows us to reduce the complete ensemble of the ion distribution modes down to a set of low-lying modes around the energy minimum. In addition, the smooth landscape allows us to fit the landscape through random sampling of a finite number of ion distribution modes. Tests for a series of RNA systems indicate that the new method can indeed lead to remarkable increase in the computational efficiency by a factor of 50. The enhancement in the computational efficiency would be more drastic for larger structures. This new method may open up a new door for the TBI-based predictions for ion effects in biologically significant systems such as the Mg2+-induced riboswitch,56 tetraloop-receptor structure,57 and the different types of ribozymes.58
Future development of the model should address several key issues. Besides the problem of the possible correlation effects for monovalent ions in RNA tertiary structures, the accuracy of the model is also limited by the inability to treat site-specific ion effect such as the sequence-specific binding of the structurally bound and partially dehydratedions. For many nucleic acid structures and sequences, divalent ion binding can be affected by non-electrostatic effects and site-specific ion binding can be critical31–33 for nucleic acid stability. For such systems, MD simulations would be particularly useful.31–33
Supplementary Material
Acknowledgments
This research was supported by NSF grants MCB0920067 and MCB0920411 and NIH grant GM063732. Most of the numerical calculations involved in this research were performed on the HPC resources at the University of Missouri Bioinformatics Consortium (UMBC).
Footnotes
Supporting Information
The free energy landscape ΔG(Nb, ΔGM) for the BWYV pseudoknot in the different ionic solutions. This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Brion P, Westhof E. Hierarchy and dynamics of RNA folding. Annu. Rev. Biophys. Biomol. Struct. 1997;26:113–137. doi: 10.1146/annurev.biophys.26.1.113. [DOI] [PubMed] [Google Scholar]
- 2.Tinoco I, Jr, Bustamante C. How RNA folds. J. Mol. Biol. 1999;293:271–281. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- 3.Sosnick TR, Pan T. RNA folding: models and perspectives. Curr. Opin. Struct. Biol. 2003;13:309–316. doi: 10.1016/s0959-440x(03)00066-6. [DOI] [PubMed] [Google Scholar]
- 4.Woodson SA. Metal ions and RNA folding: a highly charged topic with a dynamic future. Curr. Opin. Chem. Biol. 2005;9:104–109. doi: 10.1016/j.cbpa.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 5.Chu VB, Herschlag D. Unwinding RNAs secrets: advances in the biology, physics, and modeling of complex RNAs. Curr. Opin. Struct. Biol. 2008;18:305–314. doi: 10.1016/j.sbi.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Draper DE. RNA folding: thermodynamic and molecular descriptions of the roles of ions. Biophys. J. 2008;95:5489–5495. doi: 10.1529/biophysj.108.131813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen SJ. RNA folding: conformational statistics, folding kinetics, and ion electrostatics. Annu. Rev. Biophys. 2008;37:197–214. doi: 10.1146/annurev.biophys.37.032807.125957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rook MS, Treiber DK, Williamson JR. An optimal Mg2+ concentration for kinetic folding of the Tetrahymena ribozyme. Proc. Natl. Acad. Sci. USA. 1999;96:12471–12476. doi: 10.1073/pnas.96.22.12471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takamoto K, He Q, Brenowitz M. Monovalent cations mediate formation of native tertiary structure of the Tetrahymena thermophila ribozyme. Nat. Struct. Biol. 2002;9:928–933. doi: 10.1038/nsb871. [DOI] [PubMed] [Google Scholar]
- 10.Thirumalai D, Hyeon C. RNA and Protein Folding: Common Themes and Variations. Biochemistry. 2005;44:49574970. doi: 10.1021/bi047314+. [DOI] [PubMed] [Google Scholar]
- 11.Koculi E, Hyeon C, Woodson SA. Charge density of divalent metal cations determines RNA stability. J. Am. Chem. Soc. 2007;129:2676–2682. doi: 10.1021/ja068027r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soto AM, Misra V, Draper DE. Tertiary structure of an RNA pseudoknot is stabilized by diffuse Mg2+ ions. Biochemistry. 2007;46:2973–2983. doi: 10.1021/bi0616753. [DOI] [PubMed] [Google Scholar]
- 13.Stellwagen E, Dong Q, Stellwagen NC. Quantitative analysis of monovalent counterion binding to random-sequence, double-stranded DNA using the replacement ion method. Biochemistry. 2007;46:2050–2058. doi: 10.1021/bi062132w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiu X, Andresen K, Pollack L. Abrupt transition from a free, repulsive to a condensed, attractive DNA phase, induced by multivalent polyamine cations. Phys. Rev. Lett. 2008;101:228101. doi: 10.1103/PhysRevLett.101.228101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schlatterer JC, Kwok LW, Pollack L. Hinge stiffness is a barrier to RNA folding. J. Mol. Biol. 2008;379:859–870. doi: 10.1016/j.jmb.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lipfert J, Sim AY, Doniach S. Dissecting electrostatic screening, specific ion binding, and ligand binding in an energetic model for glycine riboswitch folding. RNA. 2010;16:708–719. doi: 10.1261/rna.1985110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen AA, Pappu RV. Quantitative characterization of ion pairing and cluster formation in strong 1:1 electrolytes. J. Phys. Chem. B. 2007;111:64696478. doi: 10.1021/jp0708547. [DOI] [PubMed] [Google Scholar]
- 18.Chen AA, Draper DE, Pappu RV. Molecular simulation studies of monovalent counterion-mediated interactions in a model RNA kissing loop. J. Mol. Biol. 2009;390:805819. doi: 10.1016/j.jmb.2009.05.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Serra MJ, Turner DH. Predicting thermodynamic properties of RNA. Methods Enzymol. 1995;259:242–261. doi: 10.1016/0076-6879(95)59047-1. [DOI] [PubMed] [Google Scholar]
- 20.SantaLucia John., Jr A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl. Acad. Sci. USA. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mathews DH, Sabina J, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 22.Chen SJ, Dill KA. RNA folding energy landscapes. Proc. Natl. Acad. Sci. USA. 2000;97:646–651. doi: 10.1073/pnas.97.2.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zuker M. MFold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Debye P, Huckel E. Zur Theorie der Elektrolyte. I. Gefrierpunktserniedrigung undverwandte Erscheinungen. Physikalische Zeitschrift. 1923;24:185–206. [Google Scholar]
- 25.Misra V, Draper DE. The interpretation of Mg2+ binding isotherms for nucleic acids using Poisson-Boltzmann theory. J. Mol. Biol. 1999;17:1135–1147. doi: 10.1006/jmbi.1999.3334. [DOI] [PubMed] [Google Scholar]
- 26.Baker NA. Improving implicit solvent simulations: A Poisson-centric view. Curr. Opin. Struct. Biol. 2005;15:137–143. doi: 10.1016/j.sbi.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 27.Baker NA, Sept D, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou H-X. Macromolecular electrostatic energy within the nonlinear Poisson-Boltzmann equation. J. Chem. Phys. 1994;100:3152–3162. [Google Scholar]
- 29.Tjong H, Zhou H-X. The dependence of electrostatic solvation energy on dielectric constants in Poisson-Boltzmann calculations. J. Chem. Phys. 2006;125:206101. doi: 10.1063/1.2393243. [DOI] [PubMed] [Google Scholar]
- 30.Tjong H, Zhou H-X. GBr6NL: a generalized Born method for accurately reproducing solvation energy of the nonlinear Poisson-Boltzmann equation. J. Chem. Phys. 2007;126:195102. doi: 10.1063/1.2735322. [DOI] [PubMed] [Google Scholar]
- 31.Auffinger P, Bielecki L, Westhof E. The Mg2+ Binding Sites of the 5S rRNA Loop E Motif as Investigated by Molecular Dynamics Simulations. Chem. Biol. 2003;10:551–561. doi: 10.1016/s1074-5521(03)00121-2. [DOI] [PubMed] [Google Scholar]
- 32.Krasovska MV, Sefcikova J, Sponer J. Cations and Hydration in Catalytic RNA: Molecular Dynamics of the Hepatitis Delta Virus Ribozyme. Biophys. J. 2006;91:626–638. doi: 10.1529/biophysj.105.079368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dong F, Olsen B, Baker NA. Computational Methods for Biomolecular Electrostatics. Methods in Cell Biology. 2008;84:843–870. doi: 10.1016/S0091-679X(07)84026-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Joung I, Cheatham TE. Molecular dynamics simulations of the dynamic and energetic properties of alkali and halide ions using water-model specific ion parameters. J. Phys. Chem. B. 2009;113:13279–13290. doi: 10.1021/jp902584c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuczera K, Jas G, Elber R. The kinetics of helix unfolding: Molecular dynamics simulations with Milestoning. J. Phys. Chem. A. 2009;113:7431–7473. doi: 10.1021/jp900407w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen A, Marucho M, Baker NA, Pappu R. Simulations of RNA Interactions with Monovalent Ions. Methods in Enzymology. 2009;469:411–432. doi: 10.1016/S0076-6879(09)69020-0. [DOI] [PubMed] [Google Scholar]
- 37.Sklenovsky P, Florova P, Banas P, Reblova K, Lankas F, Otyepka M, Sponer J. Understanding RNA Flexibility Using Explicit Solvent Simulations: The Ribosomal and Group I Intron Reverse Kink-Turn Motifs. J. Chem. Theory Comput. 2011;7:2963. doi: 10.1021/ct200204t. [DOI] [PubMed] [Google Scholar]
- 38.Do TN, Ippoliti E, Parrinello M. Counterion Redistribution upon Binding of a Tat-Protein Mimic to HIV-1 TAR RNA. J. Chem. Theory Comput. 2012 doi: 10.1021/ct2005769. Article ASAP. [DOI] [PubMed] [Google Scholar]
- 39.Kirmizialtin S, Pabit SA, Meisburger SP, Pollack L, Elber R. RNA and Its Ionic Cloud: Solution Scattering Experiments and Atomically Detailed Simulations. Biophys. J. 2012;102:819–828. doi: 10.1016/j.bpj.2012.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bai Y, Greenfeld M, Herschlag D. Quantitative and comprehensive decomposition of the ion atmosphere around nucleic acids. J. Am. Chem. Soc. 2007;129:14981–14988. doi: 10.1021/ja075020g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grochowski P, Trylska J. Continuum molecular electrostatics, salt effects, and counterion binding–a review of the Poisson-Boltzmann theory and its modifications. Biopolymers. 2008;89:93113. doi: 10.1002/bip.20877. [DOI] [PubMed] [Google Scholar]
- 42.Wang K, Yu YX, Gao GH. Density functional study on the structural and thermodynamic properties of aqueous DNA-electrolyte solution in the framework of cell model. J. Chem. Phys. 2008;128:185101. doi: 10.1063/1.2918342. [DOI] [PubMed] [Google Scholar]
- 43.Tan ZJ, Chen SJ. Electrostatic correlations and fluctuations for ion binding to a finite length polyelectrolyte. J. Chem. Phys. 2005;122:44903. doi: 10.1063/1.1842059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan ZJ, Chen SJ. Predicting ion binding properties for RNA tertiary structures. Biophys. J. 2006;91:518–536. doi: 10.1016/j.bpj.2010.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tan ZJ, Chen SJ. Electrostatic free energy landscape for nucleic acid helix assembly. Nucleic Acids Res. 2006;34:6629–6639. doi: 10.1093/nar/gkl810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tan ZJ, Chen SJ. Minor groove RNA triplex in the crystal structure of a ribosomal frameshifting viral pseudoknot. Biophys. J. 2007;92:3615–3632. doi: 10.1038/6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan ZJ, Chen SJ. Salt dependence of nucleic acid hairpin stabilit. Biophys. J. 2008;95:738–752. doi: 10.1529/biophysj.108.131524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan ZJ, Chen SJ. Predicting ion binding properties for RNA tertiary structures. Biophys. J. 2010;99:1–12. doi: 10.1016/j.bpj.2010.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Su L, Chen L, Rich A. Minor groove RNA triplex in the crystal structure of a ribosomal frameshifting viral pseudoknot. Nat. Struct. Biol. 1999;6:285–292. doi: 10.1038/6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Conn GL, Gittis AG, Draper DE. A compact RNA tertiary structure contains a buried backbone-K+ complex. J. Mol. Biol. 2002;318:963–973. doi: 10.1016/S0022-2836(02)00147-X. [DOI] [PubMed] [Google Scholar]
- 51.Westhof E, Sundaralingam M. Restrained refinement of the monoclinic form of yeast phenylalanine transfer RNA. Temperature factors and dynamics, coordinated waters, and base-pair propeller twist angles. Biochemistry. 1986;25:4868–4878. doi: 10.1021/bi00365a022. [DOI] [PubMed] [Google Scholar]
- 52.Davis JH, Foster TR, Tonelli M, Butcher SM. Transposition of two amino acids changes a promiscuous RNA binding protein into a sequence-specific RNA binding protein. RNA. 2007;13:1–11. doi: 10.1261/rna.633808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grilley D, Misra V, Draper DE. Importance of partially unfolded conformations for Mg2+-induced folding of RNA tertiary structure: structural models and free energies of Mg2+ interactions. Biochemistry. 2007;46:10266–10278. doi: 10.1021/bi062284r. [DOI] [PubMed] [Google Scholar]
- 54.Romer R, Hach R. tRNA conformation and magnesium binding. A study of a yeast phenylalanine-specific tRNA by a fluorescent indicator and differential melting curves. Eur. J. Biochem. 1975;55:271–284. doi: 10.1111/j.1432-1033.1975.tb02160.x. [DOI] [PubMed] [Google Scholar]
- 55.Krakauer H. The binding of Mg2+ ions to polyadenylate, polyuridylate, and their complexes. Biopolymers. 1971;10:2459–2490. doi: 10.1002/bip.360101209. [DOI] [PubMed] [Google Scholar]
- 56.Cromie M, Shi Y, Croisman EA. An RNA sensor for intracellular Mg2+. Cell. 2006;125:71–84. doi: 10.1016/j.cell.2006.01.043. [DOI] [PubMed] [Google Scholar]
- 57.Hodak JH, Fiore J, Pardi A. Docking kinetics and equilibrium of a GAAA tetraloop-receptor motif probed by single-molecule FRET. Proc. Natl. Acad. Sci. 2005;102:10505–10510. doi: 10.1073/pnas.0408645102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martha S,R, Daniel K,T, James R,W. An optimal Mg2+ concentration for kinetic folding of the Tetrahymena ribozyme. Proc. Natl. Acad. Sci. 1999;96:12471–12476. doi: 10.1073/pnas.96.22.12471. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.