Abstract
Ammonia-oxidizing archaea (AOA) are widespread and abundant in aquatic and terrestrial habitats and appear to have a significant impact on the global nitrogen cycle. Like the ammonia-oxidizing bacteria, AOA encode a gene homologous to copper-containing nitrite reductases (nirK), which has been studied very little to date. In this study, the diversity, abundance and expression of thaumarchaeal nirK genes from coastal and marine environments were investigated using two mutually excluding primer pairs, which amplify the nirK variants designated as AnirKa and AnirKb. Only the AnirKa variant could be detected in sediment samples from San Francisco Bay and these sequences grouped with the nirK from Candidatus Nitrosopumilus maritimus and Candidatus Nitrosoarchaeum limnia. The two nirK variants had contrasting distributions in the water column in Monterey Bay and the California Current. AnirKa was more abundant in the epi- to mesopelagic Monterey Bay water column, whereas AnirKb was more abundant in the meso- to bathypelagic California Current water. The abundance and community composition of AnirKb, but not AnirKa, followed that of thaumarchaeal amoA, suggesting that either AnirKa is not exclusively associated with AOA or that commonly used amoA primers may be missing a significant fraction of AOA diversity in the epipelagic. Interestingly, thaumarchaeal nirK was expressed 10–100-fold more than amoA in Monterey Bay. Overall, this study provides valuable new insights into the distribution, diversity, abundance and expression of this alternative molecular marker for AOA in the ocean.
Keywords: ammonia-oxidizing archaea, copper-containing nitrite reductase, Monterey Bay time series, nirK, thaumarchaea
Introduction
Ammonia-oxidizing bacteria (AOB) and ammonia-oxidizing archaea (AOA) have a central role in the global nitrogen cycle by oxidizing ammonia (NH3) to nitrite (NO2−). In addition to the ammonia oxidation machinery (for example, ammonia monooxygenase), most ammonia oxidizers, interestingly, also encode a copper-containing nitrite reductase (nirK), which catalyzes the reduction of NO2− to nitric oxide (NO; Casciotti and Ward, 2001; Treusch et al., 2005; Garbeva et al., 2007; Cantera and Stein, 2007b; Bartossek et al., 2010). In AOB, NirK is involved in production of the potent greenhouse gas nitrous oxide (N2O) through ‘nitrifier denitrification' (Dundee and Hopkins, 2001; Shaw et al., 2006). AOA also produce N2O, part of which appears to be formed through a reductive pathway (for example, nitrite reduction; Santoro et al., 2011). Nitrite reduction mediated by nitrifiers contributes significantly to N2O release to the atmosphere from both terrestrial (Webster and Hopkins, 1996; Kool et al., 2011) and marine (Dore et al., 1998; Santoro et al., 2011) ecosystems.
The physiological importance of nirK in ammonia oxidizers is still unclear. Classically, ammonia oxidizers are known to use oxygen (i) for the oxidation of NH3 to hydroxylamine (NH2OH) and (ii) as terminal electron acceptor in their electron transport chain. For oxidizing NH3, dinitrogen tetroxide (N2O4), rather than O2, has been proposed to be the primary oxidant, as described in the NOx cycle hypothesis (Schmidt et al., 2001; Kampschreur et al., 2006; Schmidt, 2008) in which NO is an important intermediate: NO produced by NO2− reduction can feed into the NOx cycle and the nitrite reductase is thereby indirectly involved in ammonia oxidation. This is supported by lower growth yield and growth defects (Schmidt et al., 2004) and slower ammonia oxidation rate (Cantera and Stein, 2007a) in Nitrosomonas europaea nirK mutants. Furthermore, when NO is scavenged from the growth media, N. eutropha is unable to oxidize ammonia (Zart et al., 2000), whereas ammonia oxidation activity increases when NO is added to the media (Zart and Bock, 1998).
AOB have long been known to substitute O2 with NO2− as the electron acceptor in ‘nitrifier denitrification' under O2-limiting conditions (Abeliovich and Vonshak, 1992; Bock et al., 1995; Schmidt and Bock, 1997). This has also been suggested for AOA (Walker et al., 2010) but, unlike AOB, they only encode nirK and none of the downstream enzymes (for example, NO reductase) involved in classical denitrification (Hallam et al., 2006; Walker et al., 2010; Blainey et al., 2011). It is unknown whether nitrite reduction in AOA is linked to energy conservation. Finally, nitrite reductase has been proposed to be involved in detoxification of nitrite in AOB cultures (Beaumont et al., 2002; Cantera and Stein, 2007a). Nitrite detoxification has yet to be examined in AOA cultures, but AOA are generally more competitive in low-ammonia environments (Martens-Habbena et al., 2009; Di et al., 2010; Herrmann et al., 2011) where nitrite concentrations are unlikely to reach toxic levels.
Copper-containing nitrite reductase genes homologous to nirK genes from AOB and denitrifying bacteria have been found in thaumarchaeal soil fosmids (Treusch et al., 2005; Bartossek et al., 2010), environmental metagenomes (for example, Venter et al., 2004; Hollibaugh et al., 2011) and in sequenced genomes of two AOA cultures, Candidatus Nitrosopumilus maritimus (two gene copies; Walker et al., 2010) and Candidatus Nitrosoarchaeum limnia (Blainey et al., 2011). Curiously, the archaeal sponge symbiont ‘Cenarchaeum symbiosum' lacks the nirK homolog, but does harbor a phylogenetically related multicopper oxidase (Hallam et al., 2006; Bartossek et al., 2010). Thaumarchaeal nirK sequences derived from soil and marine environments form two highly divergent clusters that are just as distantly related to each other as to AOB nirK sequences (Bartossek et al., 2010).
Although nirK is implicated in the production of the greenhouse gas, N2O, our knowledge of the diversity of thaumarchaeal nirK in marine systems is limited to a few meta-genomic and -transcriptomic studies (for example, Venter et al., 2004; Hollibaugh et al., 2011) and a single clone library from 3480 m in the Tyrrhenian Sea (Yakimov et al., 2011). To obtain a broader picture of the occurrence of thaumarchaeal nirK in marine systems, we designed specific primers and examined the diversity, abundance and expression of the gene in samples from Monterey Bay, the California Current and San Francisco Bay. The distribution of AOA nirK was also compared with the diversity and abundance of thaumarchaeal ammonia monooxygenase subunit A (amoA) genes, and with the abundance of the Marine Group I (MGI) 16S rRNA genes.
Materials and methods
Sampling
Samples were collected from Monterey Bay aboard R/V Point Lobos along an onshore–offshore transect with three oceanographic stations; C1, M1 and M2. Monterey Bay is a >1000 m deep, non-estuarine embayment in central California that opens broadly to the coastal ocean. The mid-bay station, M1, is located in the center of the upwelling plume (Service et al., 1998), M2 is the outer-bay station and C1 is the inner-bay ‘coastal' station (see Supplementary Figure S1). Upwelling season starts in March and continues through summer. Samples were collected at discrete depths down to 200 m on May 18, June 10 and December 2 of 2010, using a 12-bottle rosette sampler with a conductivity-temperature-depth device. Samples were collected within the central California Current on cruise S310 aboard McArthur II in July 2010 down to 3000 m water depth at station 60.90 (located about 300 km offshore, position 36.608/−125.778). San Francisco Bay sediments samples were collected, and DNA extracted, as described in Mosier and Francis (2010).
Water samples (1 l for DNA and 1–2 l for RNA) were pressure filtered through a 0.2-μM, 25-mm diameter Supor-200 membrane filter (Pall, Ann Arbor, MI, USA) with a 10-μM polyester pre-filter (GE Water & Process Technologies, Trevose, PA, USA), using a peristaltic pump. Filters for DNA and RNA extraction were transferred to 2-ml bead-beading tubes containing about 100 μl of 0.1-mm glass beads, and immediately frozen in liquid nitrogen. The Biological Oceanography Group at Monterey Bay Aquarium Research Institute kindly provided water chemistry data.
Nucleic acid extraction and cDNA synthesis
DNA was extracted from frozen filters following the protocol in Santoro et al. (2010), with a few modifications: 700 μl Sucrose-EDTA Lysis buffer (0.75 M sucrose, 20 mM EDTA, 400 mM NaCl and 50 mM Tris) were added and the filters were agitated for 45 s at speed 5.5 in a FastPrep bead-beating machine (MP Biomedicals, Solon, OH, USA). A volume of 100 μl 10% sodium dodecyl sulfate and proteinase K (50 mg ml−1 final concentration) were added and filters were incubated at 55 °C overnight. The lysate was purified using the Qiagen Blood & Tissue DNeasy kit (Valencia, CA, USA) following the manufacturer's protocol with an additional wash step with buffer AW2. Purified DNA was quantified using a Qubit fluorometer (Invitrogen, Grand Island, NY, USA).
RNA was extracted using the mirVana miRNA isolation kit (Ambion, Grand Island, NY, USA). The initial lysis was carried out by adding 750 μl lysis/binding buffer to the filters with beads followed by vortexing at full speed for 5 min. The organic extraction and on-column RNA purification were performed according to the manufacturer's protocol. The RNA was finally eluted in 75 μl warm (95 °C) RNase-free water. Contaminating DNA was removed using the TURBO DNA-free kit (Ambion) following the manufacturer's protocol. cDNA was synthesized using the SuperScript III First-Strand Synthesis System for reverse transcription-PCR (Invitrogen) according to the manufacturer's protocol except for increasing the reverse transcription incubation step to 4 h at 50 °C. Negative reverse transcription control reactions were performed for each sample replacing the reverse transcriptase enzyme with water.
Primer design
A total of 106 thaumarchaeal nirK-like sequences were retrieved from available marine metagenomes from CAMERA (Sun et al., 2011) and NCBI using BLASTn and tBLASTx with Ca. Np. maritimus ORF 1259 (NC_010085) as query sequence. Sequences were aligned in Geneious v 5.4 (Drummond et al., 2011) by translation alignment and primers were chosen by visual inspection of the alignment. Two mutually excluding primer pairs amplifying the same ∼500 bp of the gene were necessary to target all of the currently known sequence diversity. The primer pair for AnirKa is AnirKa-58F 5′-ACB YTA TTC GGA AGY ACA TAC ACA-3′ and AnirKa-579R 5′-GYM ATT CCG TAC ATK CCG GA-3′. The primer pair for AnirKb is AnirKb-61F 5′-CTA TTC GGA RGT WCT TTY ACT GC-3′ and AnirKb-555R 5′-ACG TGT TGG TCC ATT GCT GC-3′. To determine whether a longer fragment of the gene would give higher phylogenetic resolution, an additional reverse primer (used together with AnirKa-58F) was also designed to yield a fragment of 1050 bp (AnirKa-1108R 5′-TRM TGC RTA TRC ACC TGG KTC-3′); all sequences generated with this primer are also targeted by the primer AnirKa-579R. The longer fragment did not result in higher phylogenetic resolution. The numbering of primers refers to the corresponding position in Ca. Np. maritimus ORF 1259 (NC_010085). The two primer pairs do not target any known soil-derived archaeal nirK sequences.
PCR, cloning and community analysis
The diversity of thaumarchaeal nirK was analyzed in eight water samples from Monterey Bay (stations M1 and M2 at 10, 40, 100 and 200 m water depth, May 2010), two water samples from California Current station 60.90 (30 and 3000 m) and four sediment samples from San Francisco Bay (BG20, BG30, BC11 and BA10). The diversity of thaumarchaeal amoA was analyzed in the same samples, except for Monterey Bay M2-10 m, which had too low abundance to be amplified for cloning, and M1-100 m. Diversity of thaumarchaeal nirK and amoA mRNA transcripts was analyzed in three water samples from Monterey Bay mid-bay station M1 at 10, 40 and 200 m from May 2010.
PCR amplification of AnirKa sequences was performed with the primers AnirKa-61F and AnirKa-579R, except for the DNA samples from Monterey Bay station M1 and San Francisco Bay, these were amplified with the reverse primer AnirKa-1108R instead. PCR amplification of AnirKb sequences was performed with the primers AnirKb-58F and AnirKb-555R. Both AnirKa and AnirKb sequences were PCR amplified in triplicate 25-μl reaction mixtures (12.5 μl Premix F (epicenter), 0.1 μM of each primer, 0.63 unit AmpliTaq DNA polymerase (Applied Biosystems, Carlsbad, CA, USA)) under the following thermal cycling conditions: 95 °C for 5 min, followed by 35 cycles of 94 °C for 30 s, 50 °C for 45 s, 72 °C for 1 min and finally 72 °C for 5 min. PCR amplification of AOA amoA was carried out with the primers Arch-amoAF and Arch-amoAR as described in Francis et al. (2005). Triplicate reactions were pooled, PCR purified (the QIAquick PCR purification Kit, Qiagen), cloned using the pGEM-T Easy Vector System (Promega, Madison, WI, USA) and sequenced (ABI 3100 Capillary Sequencer, ELIM Biopharm, Hayward, CA, USA).
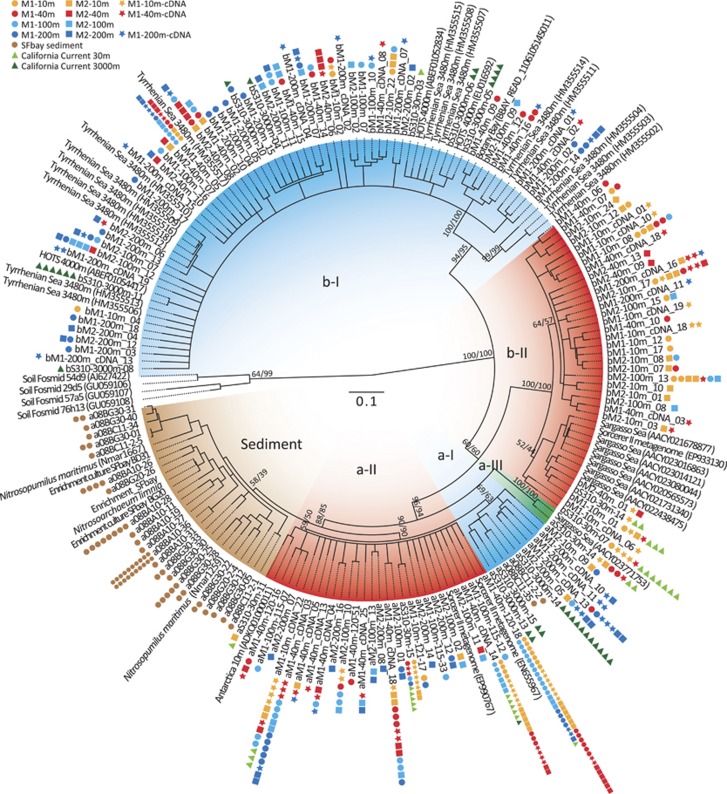
Clone library coverage was assessed by calculating rarefaction curves and coverage in the Mothur software package (Schloss et al., 2009; Table 1, Supplementary Table S1, and Supplementary Figure S2). Diversity indices (Shannon, Simpson) and richness estimators (Chao, ACE) were calculated in Mothur (Schloss et al., 2009; Table 1). Calculations were based on 477, 450 and 594 bp for AnirKa, AnirKb and AOA amoA, respectively. The archaeal nirK nucleotide sequences were aligned using translation alignment in Geneious (Drummond et al., 2011) and trimmed to the length of the shortest sequence. A consensus tree based on 453 bp was constructed from maximum likelihood (PhyML (Guindon and Gascuel, 2003) using general time-reversible substitution model) and neighbor-joining (Jukes-Cantor distance correction), both with 1000 bootstrap replicates (Figure 1). Representative sequences from an operational taxonomic unit (OTU) cutoff of 97% similarity are shown in the tree along with thaumarchaeal nirK sequences retrieved from NCBI and CAMERA (Sun et al., 2011) covering the same region of the gene. Thaumarchaeal nirK genes from soil fosmids were included as outgroups (Figure 1). AOA amoA sequences were aligned with GenBank sequences in ARB (Ludwig et al., 2004), and a nucleotide-based phylogenetic tree was constructed by neighbor-joining with Jukes-Cantor distance correction (Supplementary Figure S3).
Table 1. Richness, diversity and coverage estimators calculated for clone libraries from all environments calculated at 97/95% OTU cutoff.
| Sample | Target | n | Shannon (H) | Simpson (D) | Chao | ACE | Coverage (%) |
|---|---|---|---|---|---|---|---|
| Monterey Bay: M1 | AnirKa | 78 | 1.9/1.1 | 0.22/0.56 | 16/11 | 18/15 | 95/96 |
| AnirKb | 69 | 3.4/2.9 | 0.04/0.06 | 75/56 | 92/45 | 59/80 | |
| amoA | 35 | 2.1/1.6 | 0.16/0.30 | 16/9 | 19/13 | 83/94 | |
| Monterey Bay: M2 | AnirKa | 67 | 2.5/1.8 | 0.12/0.25 | 31/11 | 92/12 | 82/97 |
| AnirKb | 66 | 3.2/2.5 | 0.04/0.10 | 57/19 | 110/21 | 70/94 | |
| amoA | 42 | 2.2/2.0 | 0.14/0.17 | 22/14 | 36/15 | 81/90 | |
| Monterey Bay: M1 cDNA | AnirKa | 60 | 2.4/1.9 | 0.10/0.20 | 16/10 | 17/11 | 95/97 |
| AnirKb | 49 | 3.3/2.9 | 0.03/0.05 | 122/36 | 218/38 | 45/76 | |
| amoA | 60 | 2.4/1.4 | 0.10/0.36 | 14/8 | 15/9 | 97/97 | |
| California current 30 m | AnirKa | 16 | 1.4/1.0 | 0.22/0.38 | 6/5 | 6/8 | 89/89 |
| AnirKb | 13 | 0.9 | 0.40 | 3 | 4 | 92 | |
| amoA | 7 | 0.0 | 1.00 | 1 | 1 | 100 | |
| California current 3000 m | AnirKa | 16 | 1.0 | 0.48 | 4 | 5 | 94 |
| AnirKb | 15 | 1.4 | 0.22 | 5 | 6 | 93 | |
| amoA | 12 | 1.9/1.8 | 0.09/0.11 | 23/10 | 21/11 | 50/67 | |
| San Francisco Bay | AnirKa | 62 | 2.6/2.4 | 0.11/0.12 | 28/17 | 33/17 | 84/95 |
| amoA | 44 | 2.5/2.3 | 0.10/0.12 | 36/22 | 70/42 | 73/82 |
Figure 1.
Phylogeny of marine thaumarchaeal nirK sequences. Maximum likelihood (ML; PhyML, general time-reversible (GTR)+Γ) 30% cutoff consensus tree with 1000 bootstrap replicates, based on 453 bp. Major groupings were all supported by neighbor-joining (NJ, Jukes-Cantor distance correction, 1000 bootstraps). Numbers on branches are bootstrap values from ML/NJ. Only sequence representatives from a 97% similarity OTU cutoff are shown in tree. Colored symbols show total number of clones represented by a sequence (see legend). Sequence name prefix ‘a/b' refers to which primer pair the sequence was amplified with (AnirKa or AnirKb).
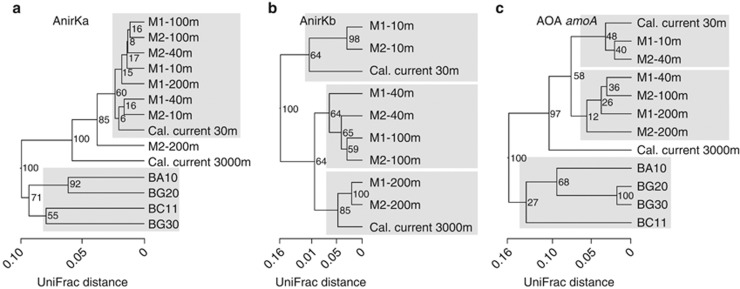
Community composition of the different samples was compared using weighted UniFrac Jackknife Environment clustering with 100 permutations (Figure 2, Supplementary Figure S5). UniFrac is a non-OTU-based metric that compares the relatedness of different environments using a phylogenetic tree as input. The trees used in the analyses were constructed separately for AnirKa, AnirKb and AOA amoA by maximum likelihood (PhyML, general time-reversible) from 480, 462 and 597 nucleotide positions, respectively, using a soil fosmid nirK sequence (AJ627422) as the outgroup.
Figure 2.
Dendrograms of hierarchical clustering of community similarities based on weighted UniFrac Jackknife Cluster Analysis with 100 permutations. (a) AnirKa sequences, (b) AnirKb sequences and (c) AOA amoA sequences. Numbers on branches are Jackknife values.
GenBank accession numbers are as follows: AnirKa, JN257263–JN257407, JN257543–JN257574, JN257603–JN257664 and JN620225–JN620284; AnirKb, JN257408–JN257542, JN257575–JN257602 and JN620285–JN620333; and AOA amoA, JN375838–JN375977 and JN624325–JN624384.
Quantitative PCR
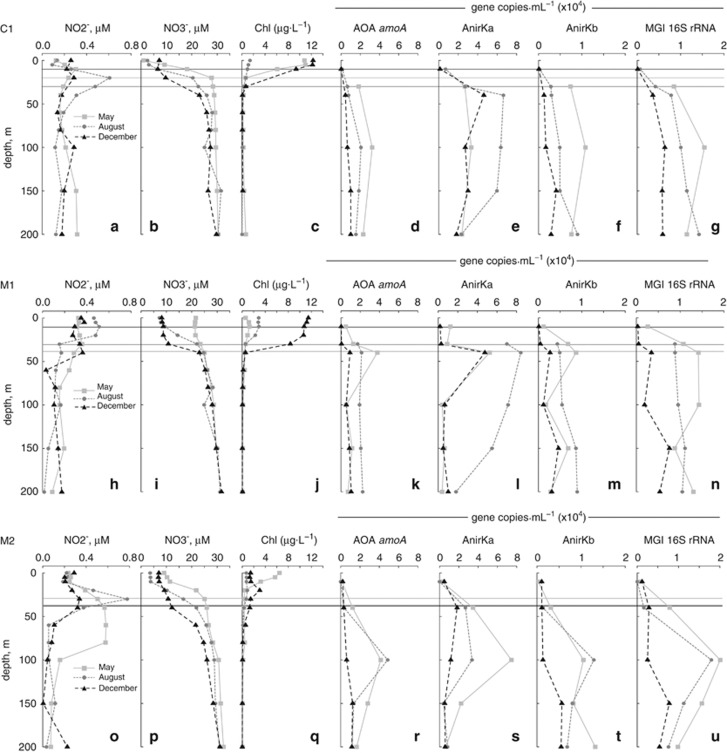
The abundance of thaumarchaeal nirK, amoA and MGI 16S rRNA genes was estimated by quantitative PCR in depth profiles from Monterey Bay station C1, M1 and M2 from May, August and December 2010 (Figure 3). The abundance of thaumarchaeal nirK and amoA was also estimated within a single depth profile from California Current station 60.90 (Supplementary Figure S6). Abundance of mRNA transcripts of thaumarchaeal nirK and amoA was estimated in samples from Monterey Bay station C1, M1 and M2 collected in May 2010 (Figure 6).
Figure 3.
Water column profiles from Monterey Bay stations C1, M1 and M2 from May, August and December 2010. Profiles show NO2−, NO3−, chlorophyll and gene copy abundances of AOA amoA, AnirKa, AnirKb and MGI 16S rRNA. Horizontal lines show the approximate depth of light penetration (1% of surface irradiance) in the different months.
The two different primer sets, AnirKa and AnirKb, were used to estimate the abundance of thaumarchaeal nirK. Both assays used the following reaction chemistry: 12.5 μl iTaq SYBR Green Supermix with ROX (Bio-Rad, Hercules, CA, USA), 400 nM of each primer and 0.3–37.3 ng template DNA or cDNA in a final volume of 25 μl. Thermal cycling conditions were as follows: 95 °C for 3 min, 35 cycles of 95 °C for 30 s, 50 °C for 45 s, 72 °C for 45 s and a detection step at 78 °C; finally, a melting curve was run from 65 to 90 °C with 0.3 °C increments.
The abundance of AOA amoA was estimated using the primers Arch-amoAF and Arch-amoAR (Francis et al., 2005) with the same reaction chemistry as used for nirK; the cycling conditions were as follows: 95 °C for 3 min, 35 cycles of 95 °C for 30 s, 56 °C for 45 s, 72 °C for 45 s and a detection step at 77 °C, finalized by a melting curve from 65 to 90 °C with 0.3 °C increments. The abundance of MGI 16S rRNA genes (thought to represent most marine AOA) was estimated using a TaqMan assay; the primers were GI_751F and GI_956R (Mincer et al., 2007) and a probe was designed for this study based on SILVA release 104 (Pruesse et al., 2007): MGI_889 (6-FAM-5′-AGT ACG TAC GCA AGT ATG AA-3′). The assay was run with the following reaction chemistry: 12.5-μl TaqMan Environmental Master Mix 2.0 (Applied Biosystems), 200 nM of each primer, 300 nM of probe and 0.5–12.4 ng of DNA template. Cycling conditions were as follows: 95 °C for 10 min, 40 cycles of 95 °C for 15 s and 56 °C for 1 min followed by detection. A clone library was generated with the primers GI_751F and GI_956R to obtain a plasmid standard.
All quantitative PCR reactions were run in triplicate using a StepOnePlus Real-Time PCR System (Applied Biosystems). Standard curves ranging from 5 to 1 × 106 copies per reaction were generated from purified, linearized plasmids obtained from the clone libraries described above. If the s.d. of a sample exceeded 10% of average value, one of the triplicates was omitted or the sample was re-run. Assay efficiencies were 73–78% for AnirKa, 85–88% for AnirKb, 79–90% for AOA amoA and 95–96% for MGI 16S rRNA genes. All assays produced consistent and reproducible results.
Statistical analysis
Spearman's rank-order correlation was used to test the correlations between gene copy numbers (Figure 4); a non-parametric method was used because the gene copy abundance data did not follow a normal distribution. Correlations between environmental parameters (NO2−, NO3−, water depth, temperature, salinity and chlorophyll) and gene copy abundances were also calculated using non-parametric Spearman's rank-order correlation (Supplementary Table S3). Change in average gene copy ratio with water depth (Figure 5a) was tested with one-way analysis of variance (ANOVA). All calculations were performed in R Commander (Fox, 2005).
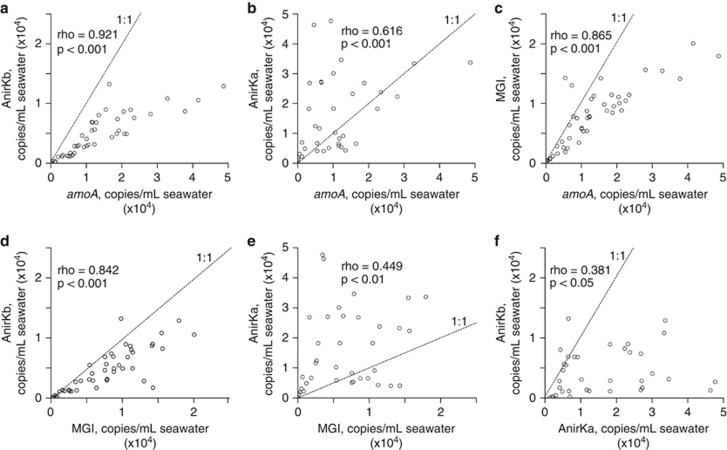
Figure 4.
Correlations of gene copy abundances of AOA amoA, AnirKa, AnirKb and MGI 16S rRNA from Monterey Bay (a–f). Spearman's rank-order correlations (rho) and significance levels are shown in each plot. Dotted lines show the 1:1 relationships.
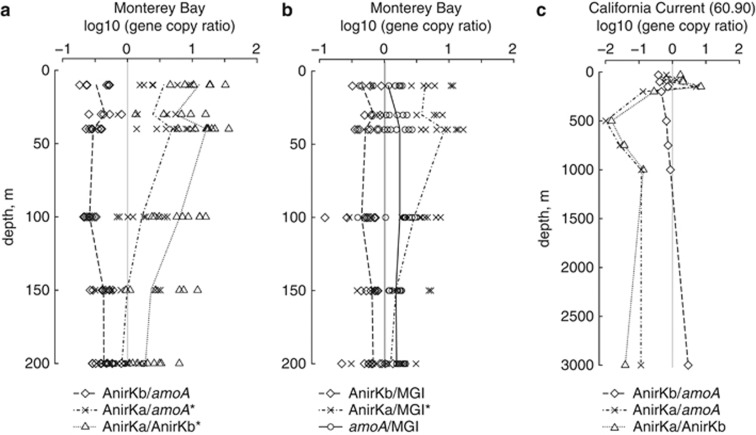
Figure 5.
Water column profiles of gene copy ratios from (a, b) Monterey Bay and (c) California Current station 60.90. Vertical gray lines show the 1:1 relationships. For Monterey Bay, every data point is plotted and the mean values are connected with lines. *Average gene copy ratio changes with water depth (one-way ANOVA, P<0.001).
Results
Community composition of thaumarchaeal nirK and amoA
The phylogeny of thaumarchaeal nirK sequences is shown in Figure 1. To target the known sequence diversity in marine environments, two mutually excluding primer sets were designed and used, and the derived nirK sequence types will be referred to as AnirKa and AnirKb throughout. AnirKa sequences form four major clusters with an average nucleotide-based identity of 83%. Group a-I, a-II and a-III contain water column samples from Monterey Bay and the California Current; group a-I is primarily comprised of sequences from below 200 m depth. Sediment-derived nirK sequences from San Francisco Bay form a separate cluster along with nirK from the cultured AOA representatives Ca. Np. maritimus (two gene copies) and Ca. Na. limnia, the latter of which was originally isolated from San Francisco Bay sediments (Blainey et al., 2011). AnirKb sequences form two major clusters with an average nucleotide-based identity of 74%. Group b-I primarily contains sequences from below 100 m depth (meso- to bathypelagic zone) and group b-II primarily contains sequences from above 100 m depth (epi- to mesopelagic zone; Figure 1, Supplementary Figure S4). Most of the metagenomic sequences from the Sargasso Sea shotgun library (Venter et al., 2004) and the Sorcerer II expedition (Yooseph et al., 2007) fall in group b-II, whereas metagenomic and fosmid sequences from Hawaii Ocean Time series (Konstantinidis and DeLong, 2008) and all of the sequences from 3480 m water depth in the Tyrrhenian Sea (Yakimov et al., 2011) fall in group b-I; however, it should be noted that the primers used in the latter study do not target all sequences amplified by the AnirKa primers. The phylogeny of thaumarchaeal amoA is shown in Supplementary Figure S3; the sequences from the San Francisco Bay sediments group with other low-salinity sediment samples and with soil/sediment samples. The AOA amoA sequences from Monterey Bay fall primarily in the marine ‘shallow' group A, although the proportion of sequences associated with the ‘deep' group B increases with water depth. All AOA amoA sequences from 30 m in the California Current fall within group A and all samples from 3000 m fall within group B (Supplementary Figures S3, S4).
In Monterey Bay, the diversity (estimated by Shannon and Simpson diversity indices) of AnirKb is much greater than both AnirKa and AOA amoA (Table 1), and the OTU richness (estimated by Chao and ACE) is two to six times higher at a 95% similarity cutoff. In the California Current, the overall diversity is lower than in Monterey Bay. In San Francisco Bay sediment, the diversity and richness of AnirKa and AOA amoA are similar.
Similarities in community composition between the different samples were analyzed by weighted UniFrac cluster analysis (Figure 2). The sediment samples from San Francisco Bay have a distinct community based on AnirKa and AOA amoA (Figure 2a and c ). The water column samples from Monterey Bay and California Current show a clear clustering with water depth for AnirKb, forming three groups with samples from 10 and 30 m, 40 and 100 m and 200 and 3000 m water depths, respectively. Environment clustering with water depth is also evident for AOA amoA, with two distinct depth groupings: 10, 30 and 40 m, and 40, 100 and 200 m; in contrast, the 3000-m sample from the California Current is distinct from the remaining water column samples. For AnirKa, there is no clear environment clustering with water depth, except for the samples from California Current (3000 m) and M2-200 m, both of which are distinct from the remaining water column samples.
Clone libraries of thaumarchaeal nirK and amoA mRNA transcripts from Monterey Bay Station M1 (10, 40 and 200 m) showed similar diversity and OTU-based richness as the corresponding DNA-based clone libraries (Table 1), except for AnirKb transcript richness, which was higher. For AnirKa and AOA amoA, the gene- and transcript-based communities from all water depths are very similar, as indicated by the overall low UniFrac distance of about 0.05 (Supplementary Figure S5). For AnirKb, there is clustering of the DNA- and cDNA-based communities: the corresponding libraries from M1-200 m form one cluster and the corresponding libraries from M1-10 and 40 m form another cluster (Supplementary Figure S5).
Thaumarchaeal gene copy abundances
The abundance of thaumarchaeal nirK, amoA and MGI 16S rRNA gene sequences was estimated in depth profiles in Monterey Bay from 10 to 200 m. All four assays showed low gene abundance at 10 m and a clear increase in copy numbers below the photic zone (Figures 3d–g). At the inner- and mid-bay stations (C1 and M1), AOA abundance peaked at ∼40 m, but peaked deeper in the water column at the outer-bay station, M2. Environmental parameters (NO3−, depth, temperature, salinity and chlorophyll, all correlating with water depth) showed clear Spearman's rank correlations with AnirKb, AOA amoA and MGI 16S rRNA gene copy abundances, but not with AnirKa abundance (Supplementary Table S3).
The overall abundances of both AOA amoA and MGI 16S rRNA genes correlate strongly with AnirKb abundance (Spearman's correlations of 0.921 and 0.842, respectively), whereas the correlation with AnirKa is weak (Spearman's correlations of 0.616 and 0.449, respectively; Figures 4a). There is a strong correlation between MGI 16S rRNA and AOA amoA gene abundances (rho=0.865, Figure 4c), and a weak correlation between AnirKb and AnirKa abundances (rho=0.381, Figure 4f).
Gene copy ratios reveal different distributions of AnirKb and AnirKa in the water column (Figure 5a): AnirKb is less abundant than both AOA amoA and MGI 16S rRNA genes with average gene copy ratios of 0.4 and 0.6, respectively, and the ratios do not change with water depth (one-way ANOVA, P>0.05; Figures 5a and b). In contrast, the gene copy ratios of AnirKa to AOA amoA and MGI 16S rRNA genes vary significantly with water depth (one-way ANOVA, P<0.001), with AnirKa being five and sevenfold more abundant than AOA amoA and MGI 16S rRNA genes, respectively, in the upper 40 m of the water column and approaching ratios of 0.5 and 2 toward 150–200 m depth (Figures 5a and b). In addition, AnirKa was relatively more abundant closer to the coast, with a significantly higher ratio of AnirKa to AnirKb at the inner Monterey Bay station C1 than at station M1, M2 and California Current station 60.90 (Supplementary Figure S7).
The ratio of AnirKa to AnirKb copy numbers also varies strongly with depth (one-way ANOVA, P<0.001), with AnirKa outnumbering AnirKb 15-fold in the upper 40 m of the water column, dropping to threefold as abundant toward 200 m depth (Figure 5a). AOA amoA is on average 1.7 times more abundant than MGI 16S rRNA genes and the gene ratio shows no variation with water depth from 10 to 200 m (one-way ANOVA, P>0.05). A ratio of 1 has previously been reported from station M1 in Monterey Bay (Mincer et al., 2007), whereas a ratio >1 has been found in a number of oceanic regions, including the Gulf of California (Beman et al., 2008), the California Current (Santoro et al., 2010), and the epipelagic of the central Pacific Ocean (Church et al., 2010) and South China Sea (Hu et al., 2011).
The patterns in gene copy ratios with water depth in Monterey Bay are also found in a depth profile from California Current station 60.90 from 30 to 3000 m (Figure 5c): AnirKb is less abundant than AOA amoA throughout the water column (except at 3000 m) and the ratio varies very little with water depth. In contrast, the ratio of AnirKa to AOA amoA fluctuates greatly with water depth, with an ∼1:1 relationship in the upper 150 m of the water column and a decrease to 10–100 times less AnirKa than AOA amoA below 200 m.
Thaumarchaeal nirK and amoA transcript abundances
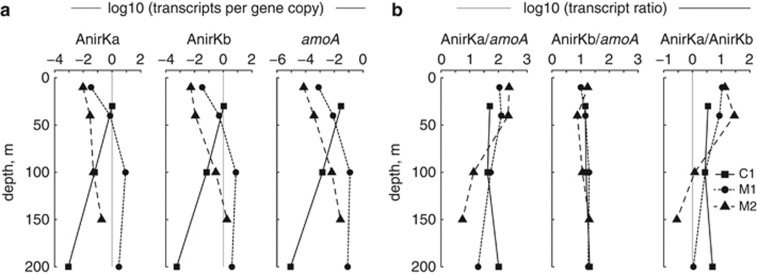
The abundance of mRNA transcripts of thaumarchaeal nirK and amoA was examined in samples from Monterey Bay Stations C1, M1 and M2 (May 2010). The mRNA abundance, in particular for AOA amoA, was low compared with the corresponding gene copy abundance (Figure 6a). AnirKb transcripts were ∼15 times more abundant than AOA amoA at all stations throughout the water column (Figure 6b). AnirKa was on average 88 times more expressed than AOA amoA, and the ratio changed with water depth: at the outer-bay station M2, and at the mid-bay station M1, AnirKa was more expressed in the upper 40 m of the water column (108–235 times) than at 100–200 m depth (6–53 times; Figure 6b). The relative abundance of AnirKa to AnirKb was also higher in the upper 40 m (3–29 times) than at 100–200 m depth (0.3–7 times) at station M1 and M2.
Figure 6.
Transcript abundances from Monterey Bay stations C1, M1 and M2 from May 2010. (a) Ratios of transcript copies (cDNA) to gene copies (DNA) for AnirKa, AnirKb and AOA amoA. (b) Ratios of transcript copies (cDNA/cDNA) of AnirKa/amoA, AnirKb/amoA and AnirKa/AnirKb. Vertical gray lines show 1:1 relationships.
Discussion
Depth distribution of thaumarchaeal nirK genes in the water column
The two thaumarchaeal nirK variants, AnirKa and AnirKb, have contrasting distributions in the marine water column, in terms of community composition, relative gene abundance, as well as gene expression. AnirKa is most abundant in the epi- to mesopelagic, and possibly coastal waters (Figures 5a and c, Supplementary Figure S7). The community composition of AnirKa shows little change with water depth (Figure 2a, Supplementary Figure S4): there is one dominant AnirKa phylotype in the epi- to mesopelagic (group a-II) and one phylotype (group a-I) comprised mostly of deep-water sequences from 3000 m in the California Current and 200 m at M2 in Monterey Bay (Figure 2a).
Both the abundance and community composition of AnirKb are highly similar to that of AOA amoA (Figures 2 and 5). The AnirKb community shifts from primarily group b-II in the upper 40 m of the water column to group b-I dominated below 40 m (Figures 1 and 2b, Supplementary Figure S4). A similar community shift is evident for AOA amoA where the proportion of sequences associated with the ‘deep' group B increases with water depth (Figure 2c, Supplementary Figures S3, S4). This follows the general distribution of thaumarchaeal amoA ecotypes in which ‘shallow' group A sequences are found throughout the water column, whereas group B sequences are more abundant in deeper water (Francis et al., 2005; Hallam et al., 2006; Beman et al., 2008; Santoro et al., 2010). The relative abundance of mRNA transcripts supports the overall picture that AnirKa is more abundant in epipelagic waters, whereas AnirKb is consistently tracking the distribution of AOA amoA genes (Figure 6b).
The divergent distributions of AnirKa and amoA suggest that the underlying mechanisms controlling the abundance of the AnirKa variant may be different from those governing the distribution of the traditional thaumarchaeal molecular marker gene, amoA. This incongruity implies that either (i) the primers targeting thaumarchaeal amoA are missing substantial diversity in the epipelagic or (ii) the AnirKa gene variant is not specific to thaumarchaeal ammonia oxidizers. The nirK gene from both Ca. Np. maritimus and Ca. Na. limnia group with the sediment-associated AnirKa sequences (Figure 1). However, none of the sequences in the three water column-associated groups are derived from cultured organisms or have genomic context, and thus cannot be conclusively linked to ammonia-oxidizing thaumarchaea at this time. Finally, differences in gene copy numbers between amoA and nirK within a given AOA genome may also contribute to the higher abundance of AnirKa in the epi- and mesopelagic: AOA are thought to only have a single amoA copy, whereas Ca. Np. maritimus has two nirK copies (Walker et al., 2010) and Ca. Na. limnia has one nirK copy (Blainey et al., 2011). However, the up to 10 times higher AnirKa abundance in surface water (Figure 5) is not likely to be due solely to differences in gene copy numbers.
Expression of thaumarchaeal nirK and amoA in Monterey Bay
Ammonia-oxidizing Thaumarchaea are thought to be a major source of N2O in marine systems, based on its natural abundance isotopic composition (Santoro et al., 2010; Santoro et al., 2011). Nitrite reductase is potentially involved in N2O production and we find thaumarchaeal nirK to be highly expressed in the marine water column (Figure 6b). A one to twofold higher expression of nirK over amoA has previously been reported in two metatranscriptomes from coastal water (Hollibaugh et al., 2011), suggesting that elevated nirK expression may be a general feature of AOA. In soil systems, where nitrifier activity can also be a significant contributor to N2O emissions (Webster and Hopkins, 1996; Kool et al., 2011), microcosm experiments did not show any correlation between thaumarchaeal nirK expression and N2O production (Bartossek et al., 2010). Similar experiments correlating thaumarchaeal nirK expression and N2O production, or natural abundance isotope composition, have yet to be carried out in marine systems.
The mRNA abundance, in particular for AOA amoA, was low compared with the corresponding gene copy abundance (Figure 6a). This has also been reported in the Black Sea (Lam et al., 2007), the Baltic Sea (Labrenz et al., 2010) and in soil (Nicol et al., 2008), whereas in the North Pacific Ocean the mRNA transcripts were 10–500-fold more abundant than gene copies in the upper 200 m of the water column (Church et al., 2010).
Thaumarchaeal nirK diversity in marine sediments
Only the AnirKa primers amplified thaumarchaeal nirK sequences from sediment samples in San Francisco Bay (Figure 1). The samples span a salinity gradient from low salinity (BG20 ∼1 and BG30 ∼0.7) to high-salinity sediments (BC11 ∼30.5 and BA10 ∼25.5; Mosier and Francis, 2010), and a distinct low-salinity AOA community has previously been identified based on amoA gene sequences (Mosier and Francis, 2008). The low-salinity ecotype of thaumarchaeal amoA was also identified in this study (Supplementary Figure S3). UniFrac cluster analysis (Figure 2c) shows a high similarity in amoA-based AOA community composition between the two low-salinity sites, BG20 and BG30; however, the pattern is not reflected in the UniFrac cluster analysis of AnirKa sequences (Figure 2a). Thus, AnirKa also has a different distribution than AOA amoA in sediments.
Linking thaumarchaeal nirK and amoA sequence types
A direct link between nirK and amoA sequence types can be obtained through the available genomes of the sediment-derived AOA; Ca. Np. maritimus (Walker et al., 2010) and Ca. Na. limnia (Blainey et al., 2011). The amoA sequences from these two organisms share 89% nucleotide identity and belong to two different sequence clusters; Ca. Np. maritimus amoA groups with environmental sequences from marine/estuarine sediments and aquarium biofilters, whereas Ca. Na. limnia amoA groups with sequences from low-salinity sediments (Supplementary Figure S3). Ca. Np. maritimus has two nirK copies and their nucleotide identities with Ca. Na. limnia nirK are 82 and 85%. All three nirK gene sequences cluster with the sediment group (Figure 1) where they fall into two different subgroups; however, a more extensive study of thaumarchaeal nirK from diverse sediment types is necessary to identify putative nirK ecotypes within sediments analogous to those typically found for amoA genes (Mosier and Francis, 2008).
In the marine water columns examined in this study, the similar distribution of the nirK group b-I and amoA group B sequence types suggests that these genes could be linked within the same organisms. AOA amoA group A sequence types are present throughout the water column, but predominating in the upper 10–100 m (Supplementary Figures S3 and S4). This distribution is consistent with a mixture of two nirK sequence types: group b-II, present in the upper water column, and a-II, present throughout the water column. Although a conclusive link between thaumarchaeal nirK and amoA sequences from the water column cannot be made with the present data alone, this could indeed be achieved via single-cell-based molecular techniques and/or metagenomic sequencing of samples (or cultures) highly enriched in marine planktonic AOA.
Acknowledgments
We thank Francisco Chavez and Tim Pennington from the Biological Oceanography Group at Monterey Bay Aquarium Research Institute (MBARI, Moss Landing, CA, USA) and the crew of R/V Point Lobos for allowing our participation in their cruises on Monterey Bay and for supplying us with ancillary physical/chemical data from the water column. The work was supported in part by the National Science foundation grants OCE-0825363 and OCE-0847266 to CAF.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Abeliovich A, Vonshak A. Anaerobic metabolism of Nitrosomonas europaea. Arch Microbiol. 1992;158:267–270. [Google Scholar]
- Bartossek R, Nicol GW, Lanzen A, Klenk H-P, Schleper C. Homologues of nitrite reductases in ammonia-oxidizing archaea: diversity and genomic context. Environ Microbiol. 2010;12:1075–1088. doi: 10.1111/j.1462-2920.2010.02153.x. [DOI] [PubMed] [Google Scholar]
- Beaumont H, Hommes N, Sayavedra-Soto L, Arp D, Arciero D, Hooper A, et al. Nitrite reductase of Nitrosomonas europaea is not essential for production of gaseous nitrogen oxides and confers tolerance to nitrite. J Bacteriol. 2002;184:2557–2560. doi: 10.1128/JB.184.9.2557-2560.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beman JM, Popp BN, Francis CA. Molecular and biogeochemical evidence for ammonia oxidation by marine Crenarchaeota in the Gulf of California. ISME J. 2008;2:429–441. doi: 10.1038/ismej.2007.118. [DOI] [PubMed] [Google Scholar]
- Blainey PC, Mosier AC, Potanina A, Francis CA, Quake SR. Genome of a low-salinity ammonia-oxidizing Archaeon determined by single-cell and metagenomic analysis. PLoS One. 2011;6:e16626. doi: 10.1371/journal.pone.0016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock E, Schmidt I, Stüven R, Zart D.1995Nitrogen loss caused by denitrifying Nitrosomonas cells using ammonium or hydrogen as electron donors and nitrite as electron acceptor Arch Microbiol 16316–20.7646315 [Google Scholar]
- Cantera J, Stein LY. Role of nitrite reductase in the ammonia-oxidizing pathway of Nitrosomonas europaea. Arch Microbiol. 2007a;188:349–354. doi: 10.1007/s00203-007-0255-4. [DOI] [PubMed] [Google Scholar]
- Cantera J, Stein LY. Molecular diversity of nitrite reductase genes (nirK) in nitrifying bacteria. Environ Microbiol. 2007b;9:765–776. doi: 10.1111/j.1462-2920.2006.01198.x. [DOI] [PubMed] [Google Scholar]
- Casciotti K, Ward B. Dissimilatory nitrite reductase genes from autotrophic ammonia-oxidizing bacteria. Appl Environ Microbiol. 2001;67:2213–2221. doi: 10.1128/AEM.67.5.2213-2221.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church MJ, Wai B, Karl DM, DeLong EF. Abundances of crenarchaeal amoA genes and transcripts in the Pacific Ocean. Environ Microbiol. 2010;12:679–688. doi: 10.1111/j.1462-2920.2009.02108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di HJ, Cameron KC, Shen J-P, Winefield CS, O'callaghan M, Bowatte S, et al. Ammonia-oxidizing bacteria and archaea grow under contrasting soil nitrogen conditions. FEMS Microbiol Ecol. 2010;72:386–394. doi: 10.1111/j.1574-6941.2010.00861.x. [DOI] [PubMed] [Google Scholar]
- Dore J, Popp B, Karl D, Sansone F. A large source of atmospheric nitrous oxide from subtropical North Pacific surface waters. Nature. 1998;396:63–66. [Google Scholar]
- Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Duran C, et al. 2011 ). http://www.geneious.com/ .
- Dundee L, Hopkins D. Different sensitivities to oxygen of nitrous oxide production by Nitrosomonas europaea and Nitrosolobus multiformis. Soil Biol Biochem. 2001;33:1563–1565. [Google Scholar]
- Fox J. The R commander: a basic-statistics graphical user interface to R. J Stat Softw. 2005;14:1–42. [Google Scholar]
- Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci USA. 2005;102:14683–14688. doi: 10.1073/pnas.0506625102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbeva P, Baggs EM, Prosser JI. Phylogeny of nitrite reductase (nirK) and nitric oxide reductase (norB) genes from Nitrosospira species isolated from soil. FEMS Microbiol Lett. 2007;266:83–89. doi: 10.1111/j.1574-6968.2006.00517.x. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hallam SJ, Konstantinidis KT, Putnam N, Schleper C, Watanabe Y-I, Sugahara J, et al. Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. Proc Natl Acad Sci USA. 2006;103:18296–18301. doi: 10.1073/pnas.0608549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann M, Scheibe A, Avrahami S, Küsel K. Ammonium availability affects the ratio of ammonia-oxidizing bacteria to ammonia-oxidizing archaea in simulated creek ecosystems. Appl Environ Microbiol. 2011;77:1896–1899. doi: 10.1128/AEM.02879-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollibaugh JT, Gifford S, Sharma S, Bano N, Moran MA. Metatranscriptomic analysis of ammonia-oxidizing organisms in an estuarine bacterioplankton assemblage. ISME J. 2011;5:866–878. doi: 10.1038/ismej.2010.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu A, Jiao N, Zhang CL. Community structure and function of planktonic Crenarchaeota: changes with depth in the South China Sea. Microb Ecol. 2011;62:549–563. doi: 10.1007/s00248-011-9866-z. [DOI] [PubMed] [Google Scholar]
- Kampschreur MJ, Tan NCG, Picioreanu C, Jetten MSM, Schmidt I, van Loosdrecht MCM. Role of nitrogen oxides in the metabolism of ammonia-oxidizing bacteria. Biochem Soc T. 2006;34:179–181. doi: 10.1042/BST0340179. [DOI] [PubMed] [Google Scholar]
- Konstantinidis KT, DeLong EF. Genomic patterns of recombination, clonal divergence and environment in marine microbial populations. ISME J. 2008;2:1052–1065. doi: 10.1038/ismej.2008.62. [DOI] [PubMed] [Google Scholar]
- Kool DM, Dolfing J, Wrage N, Van Groenigen JW. Nitrifier denitrification as a distinct and significant source of nitrous oxide from soil. Soil Biol Biochem. 2011;43:174–178. [Google Scholar]
- Labrenz M, Sintes E, Toetzke F, Zumsteg A, Herndl GJ, Seidler M, et al. Relevance of a crenarchaeotal subcluster related to Candidatus Nitrosopumilus maritimus to ammonia oxidation in the suboxic zone of the central Baltic Sea. ISME J. 2010;4:1496–1508. doi: 10.1038/ismej.2010.78. [DOI] [PubMed] [Google Scholar]
- Lam P, Jensen MM, Lavik G, McGinnis DF, Müller B, Schubert CJ, et al. Linking crenarchaeal and bacterial nitrification to anammox in the Black Sea. Proc Natl Acad Sci USA. 2007;104:7104–7109. doi: 10.1073/pnas.0611081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens-Habbena W, Berube PM, Urakawa H, La Torre De JR, Stahl DA. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature. 2009;461:976–979. doi: 10.1038/nature08465. [DOI] [PubMed] [Google Scholar]
- Mincer T, Church M, Taylor L, Preston C, Karl D, DeLong E. Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific Subtropical Gyre. Environ Microbiol. 2007;9:1162–1175. doi: 10.1111/j.1462-2920.2007.01239.x. [DOI] [PubMed] [Google Scholar]
- Mosier A, Francis C. Relative abundance and diversity of ammonia-oxidizing archaea and bacteria in the San Francisco Bay estuary. Environ Microbiol. 2008;10:3002–3016. doi: 10.1111/j.1462-2920.2008.01764.x. [DOI] [PubMed] [Google Scholar]
- Mosier AC, Francis CA. Denitrifier abundance and activity across the San Francisco Bay estuary. Environmental Microbiology Reports. 2010;2:667–676. doi: 10.1111/j.1758-2229.2010.00156.x. [DOI] [PubMed] [Google Scholar]
- Nicol G, Leininger S, Schleper C, Prosser J. The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ Microbiol. 2008;10:2966–2978. doi: 10.1111/j.1462-2920.2008.01701.x. [DOI] [PubMed] [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs B. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro AE, Buchwald C, McIlvin MR, Casciotti KL. Isotopic signature of N2O produced by marine ammonia-oxidizing Archaea. Science. 2011;333:1282–1285. doi: 10.1126/science.1208239. [DOI] [PubMed] [Google Scholar]
- Santoro AE, Casciotti KL, Francis CA. Activity, abundance and diversity of nitrifying archaea and bacteria in the central California Current. Environ Microbiol. 2010;12:1989–2006. doi: 10.1111/j.1462-2920.2010.02205.x. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt I. Nitric oxide: interaction with the ammonia monooxygenase and regulation of metabolic activities in ammonia oxidizers. Method Enzymol. 2008;7:121–135. doi: 10.1016/S0076-6879(07)00807-5. [DOI] [PubMed] [Google Scholar]
- Schmidt I, Bock E. Anaerobic ammonia oxidation with nitrogen dioxide by Nitrosomonas eutropha. Arch Microbiol. 1997;167:106–111. [PubMed] [Google Scholar]
- Schmidt I, Bock E, Jetten MS. Ammonia oxidation by Nitrosomonas eutropha with NO2 as oxidant is not inhibited by acetylene. Microbiology. 2001;147:2247–2253. doi: 10.1099/00221287-147-8-2247. [DOI] [PubMed] [Google Scholar]
- Schmidt I, van Spanning RJM, Jetten MSM. Denitrification and ammonia oxidation by Nitrosomonas europaea wild-type, and NirK- and NorB-deficient mutants. Microbiology. 2004;150:4107–4114. doi: 10.1099/mic.0.27382-0. [DOI] [PubMed] [Google Scholar]
- Service S, Rice J, Chavez F. Relationship between physical and biological variables during the upwelling period in Monterey Bay, CA. Deep-Sea Res II. 1998;45:1669–1685. [Google Scholar]
- Shaw LJ, Nicol GW, Smith Z, Fear J, Prosser JI, Baggs EM. Nitrosospira spp. can produce nitrous oxide via a nitrifier denitrification pathway. Environ Microbiol. 2006;8:214–222. doi: 10.1111/j.1462-2920.2005.00882.x. [DOI] [PubMed] [Google Scholar]
- Sun S, Chen J, Li W, Altintas I, Lin A, Peltier S, et al. Community cyberinfrastructure for advanced microbial ecology research and analysis: the CAMERA resource. Nucleic Acids Res. 2011;39:D546–D551. doi: 10.1093/nar/gkq1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treusch AH, Leininger S, Kletzin A, Schuster SC, Klenk H-P, Schleper C. Novel genes for nitrite reductase and Amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling. Environ Microbiol. 2005;7:1985–1995. doi: 10.1111/j.1462-2920.2005.00906.x. [DOI] [PubMed] [Google Scholar]
- Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- Walker CB, la Torre de JR, Klotz MG, Urakawa H, Pinel N, Arp DJ, et al. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc Natl Acad Sci USA. 2010;107:8818–8823. doi: 10.1073/pnas.0913533107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster E, Hopkins D. Nitrogen and oxygen isotope ratios of nitrous oxide emitted from soil and produced by nitrifying and denitrifying bacteria. Biol Fert Soils. 1996;22:326–330. [Google Scholar]
- Yakimov MM, Cono VL, Smedile F, DeLuca TH, Juárez S, Ciordia S, et al. Contribution of crenarchaeal autotrophic ammonia oxidizers to the dark primary production in Tyrrhenian deep waters (Central Mediterranean Sea) ISME J. 2011;5:945–961. doi: 10.1038/ismej.2010.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yooseph S, Sutton G, Rusch DB, Halpern AL, Williamson SJ, Remington K, et al. The Sorcerer II Global Ocean Sampling expedition: expanding the universe of protein families. PLoS Biol. 2007;5:e16. doi: 10.1371/journal.pbio.0050016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zart D, Schmidt I, Bock E. Significance of gaseous NO for ammonia oxidation by Nitrosomonas Eutropha. Anton Leeuw. 2000;77:49–55. doi: 10.1023/a:1002077726100. [DOI] [PubMed] [Google Scholar]
- Zart D, Bock E. High rate of aerobic nitrification and denitrification by Nitrosomonas eutropha grown in a fermentor with complete biomass retention in the presence of gaseous NO2 or NO. Arch Microbiol. 1998;169:282–286. doi: 10.1007/s002030050573. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.