Abstract
Symmetry is commonly observed in many biological systems. Here we discuss representative examples of the role of symmetry in structural molecular biology. Point group symmetries are observed in many protein oligomers whose three-dimensional atomic structures have been elucidated by x-ray crystallography. Approximate symmetry also occurs in multidomain proteins. Symmetry often confers stability on the molecular system and results in economical usage of basic components to build the macromolecular structure. Symmetry is also associated with cooperativity. Mild perturbation from perfect symmetry may be essential in some systems for dynamic functions.
Proteins are linear polymers of l-amino acids organized in a hierarchical way: amino acid sequence, helices and strands, structural motifs, globular domains, protomers, and oligomers (1, 2). At the lowest level of organization, the sequence of amino acids or primary structure is folded into α-helices (α), β-strands (β), and other secondary structures. These in turn usually form compact supersecondary structural motifs such as αα, βββ, and βαβ, most of which are dependent on higher-order interactions for their stability. Thus, at the next level of organization globular domains may comprise several such motifs, stabilized by interactions between side chains of different amino acids known as tertiary interactions. Such domains usually fold independently, probably reflecting their evolutionary origins as smaller, independent proteins in earlier organisms. The individual gene products, the protomers or subunits, may contain several such globular domains in one polypeptide chain. At the highest level of organization, oligomers, which are assemblies of such protomers, often contain several different gene products, usually organized in a symmetrical way. Because l-amino acids are enantiomers, natural proteins synthesized from them on a ribosome cannot have mirror planes or centers of inversion. However, identical or similar protein motifs, globular domains, or protomers can be related by rotational symmetries.
There are many examples of oligomers involving simple point group symmetries; Table 1 lists representative examples. Most common is 2-fold symmetry, which is found in many oligomers such as immunoglobulin, triose-phosphate isomerase, and wheat germ agglutinin. Threefold symmetry is also common; for example, it is found in bacteriochlorophyll protein and glucagon. Higher rotational symmetries are less common, although they do occur as shown in the pentraxin serum amyloid P-component (Fig. 1), which has nearly perfect 5-fold symmetry (11). Many oligomers with high rotational symmetry tend to be associated with a membrane or a surface coat of a cell or spherical virus. Alternatively, they may comprise a disc that is the basic building element of a tubular cytoskeletal protein or of a cylindrical virus; an example is the tobacco mosaic virus protein disc, which has 17-fold symmetry.
Table 1.
Representative proteins with rotational symmetry
| Protein | No. of subunits | Rotational symmetry | Reference* |
|---|---|---|---|
| HIV proteinase | 2 | 2 | 3, 4 |
| βB2 crystallin | 2 | 2 | 5 |
| Immunoglobulins | 4 | 2 | 6 |
| Purine nucleoside phosphorylase | 3 | 3 | 7 |
| Porin | 3 | 3 | 8 |
| Hemoglobin | 4 | Three orthogonal 2-fold | 9 |
| Neuraminidase | 4 | 4 | 10 |
| Pentraxins | 5 | 5 | 11, 12 |
| β-Subunit of types 1 and 2 heat-labile enterotoxins and cholera toxin | 5 | 5 | 13 |
| Insulin | 6 | 3- and 2-fold | 14, 15 |
| Limulus C-reactive protein | 6 | 6 | 16 |
| GroEL | 14 | 7 | 17 |
| Aerolysin | 7 | 7 | 18, 19 |
| 20S proteosome | 7 | 7 | 20 |
| Mandelate racemase | 8 | 8 | 21 |
| Light-harvesting complex 2 | 18 | 9 | 22 |
| GTP cyclohydrolase I | 10 | 5- and 2-fold | 23 |
| trp RNA binding attenuating protein | 11 | 11 | 24 |
| Aspartate transcarbamoylase | 12 | 3- and 2-fold | 25 |
| Phaseolin | 12 | Tetramer of trimers | 26 |
| Portal protein of bacteriophage | 13 | 13 | 27 |
| Apoferritin | 24 | 4-, 3-, and 2-fold | 28 |
| Light-harvesting complex 1 | 32 | 16 | 29 |
| Tobacco mosaic virus disc | 34 | 17 | 30 |
| Coat of tomato bushy stunt virus | 180 | 5-, 3-, and 2-fold | 31 |
Reference to crystal structure is provided if a crystal structure is available.
Figure 1.
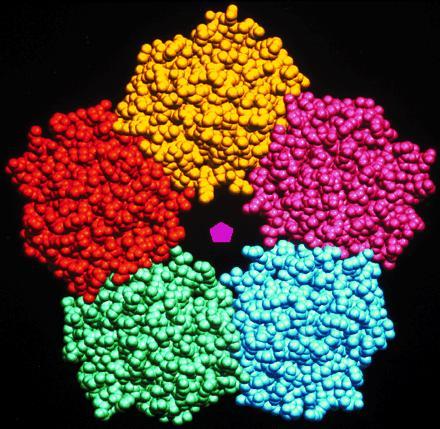
Crystal structure of pentameric human serum amyloid P-component (11) showing 5-fold symmetry.
Rotational operations are often combined together in oligomers with point group symmetry. Most common are point combinations of 2- and 3-fold symmetries, reflecting the formation of intermediate oligomers in assembly and/or evolution rotational symmetries. Thus 222 symmetry is found in concanavalin A, and 32 symmetry is found in both aspartate transcarbamoylase and the zinc insulin hexamer shown in Fig. 2, which has perfect 3-fold and approximate 2-fold symmetries (14, 15). Higher levels of organization, such as octahedral 432 symmetry found in ferritin and icosahedral 532 symmetry found in many spherical viruses, such as tomato bushy stunt virus, give rise to hollow shells that can be used to package molecules safely, in these cases iron and nucleic acid.
Figure 2.
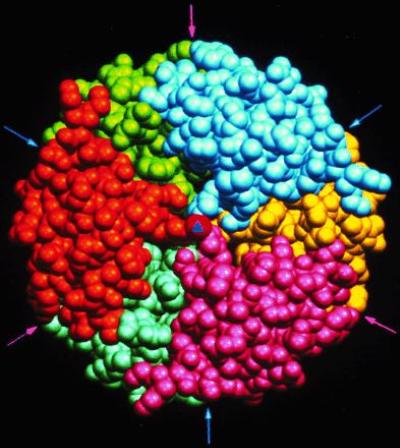
The structure of the zinc insulin hexamer as defined by Hodgkin and coworkers (14). The hexamer is viewed down the exact 3-fold axis (triangle at the center); the arrows indicate positions of approximate 2-fold axes relating pairs of protomers. Each protomer is represented in a specific color, and the zinc at the center is shown in red.
Rotational symmetries may be combined with translations to form fibrous, surface planar, or solid structures. Thus, protomers are often related by line groups in fibrous structures such as microtubules and filamentous phage, as plane groups in arrays of bacteriochlorophyll protein and other membrane proteins, and as space groups in crystalline storage granules—for example, insulin in the β cells of the endocrine pancreas. Such structures are responsible for the highly structured but dynamic organization of the cell.
In this article we focus on point group symmetries. We describe examples of exact or approximate symmetry that relate supersecondary structural motifs, domains, or whole proteins in complex multidomain proteins or oligomers.
Symmetry, Economy, and Stability
For symmetry to play a role in any branch of science, there must be multiple identical copies of certain objects. This arises in biology from the enforced economies of living systems. Building a molecular structure from several small identical proteins requires less genetic material, DNA, than constructing it from a single protein chain. This is of particular importance as the three nucleic acid bases that code for one building unit (one amino acid) of a protein involve about five times as many atoms as the amino acid itself. Thus, economy of mass may be the reason why many proteins that encircle DNA are hexamers (37) and many proteins of viral coats that completely encapsulate nucleic acids are icosahedral structures of 60 or more subunits (38, 39). Making many smaller copies is also a safer strategy, as shorter chains with single point errors can be discarded more easily. Thus, large systems such as viruses, multienzyme complexes, cell-cytoskeleta, and flagella are usually constructed from multiple copies of the same protein. In general, each of the units may be extremely complex, involving several hundred amino acids in a complicated but beautiful architecture. Nevertheless, the protein units are more or less identical, and, generally, the lowest energy state of an assembly is a symmetrical one.
“Symmetry signifies rest and binding; asymmetry motion and loosening.” These words, written of the visual arts, may equally well be used of nature. They were reflected by D’Arcy Thompson in his classic work On Growth and Form (40), which is concerned principally with symmetry and structure at the level of whole organisms and the cell. But they are also relevant to the fast-growing field of structural molecular biology.
Of course, a living organism is the very antithesis of “rest and binding.” Life is often defined as motion. Why then should symmetry be found in life at the molecular level? Will it not be selectively disadvantageous in evolution? The answer is, of course, that in certain respects life requires rest and binding, harmony, and stability. The zinc insulin hexamer, which has both 2- and 3-fold symmetry, as shown in Fig. 2, is a good example. In humans the hormone may “rest” for several days in the endocrine gland, the pancreas, packaged as a hexamer comprising six copies of the insulin molecule and two zinc ions. This form is thermodynamically stable and resistant to proteolytic degradation. When it is released into circulation it dissociates quickly into protomers, which have a half-life of only a few minutes; if they lasted longer the sensitivity of the regulation by the hormone would be lost, as the system would be turned on for too long. Thus, the stability of the symmetrical resting or storage form of insulin is as vital to the organism as the instability or proteolytic sensitivity of the active protomeric form. It is fascinating that this example of yin and yang in biology involves a molecule with both 2- and 3-fold symmetries, which occur in representations of this concept in Chinese and Japanese art.
Pentraxins, such as serum amyloid P-component (Fig. 1) (11) and C-reactive protein (12), have pentameric ring structures. Sequence similarity with other proteins in the family shows that this structure is likely to be conserved (36) except in the evolutionarily oldest member, from Limulus polyphemus (horseshoe crab), which is hexameric (16). This suggests that cyclic pentameric or hexameric arrangements may be important for the biological functions of pentraxins (41), which probably involve binding or linking several polysaccharide-containing cells or molecules together to form a stable multicomponent system. To achieve this, pentraxins need several equivalent binding sites, one on each subunit. Such molecules are known as lectins; there are many examples in plants as well as animals, as shown in Fig. 3. These molecules have 2- (2 and 222), 5-, and 6-fold symmetries (42), although, in the coral tree, the lectin glycosylation site interferes with the classical legume lectin dimer interface, resulting in an unusual dimer (43). Most of these are stable molecules, but this is particularly true of the pentraxins, which need to survive during the acute-phase response in the circulation. In fact, serum amyloid P-component occurs in many amyloid diseases such as amyloidosis, Alzheimer disease, and Creutzfeld–Jacob disease, where it binds tightly to the amyloid fibers, decorates them, and increases their resistance to removal by the body’s defense system (44).
Figure 3.
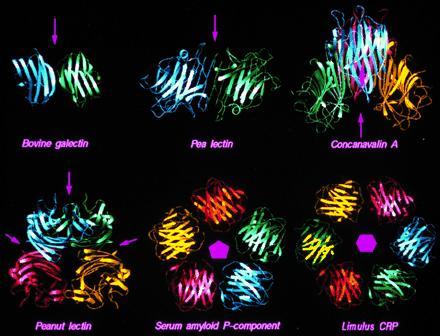
Quaternary structures of proteins with the lectin fold. The crystal structures used are pea lectin (32), bovine S-lectin (33), peanut lectin (34), jack bean concanavalin A (35), and human serum amyloid P-component (11). The hexameric Limulus C-reactive protein (Limulus CRP) structure has been generated on the basis of sequence similarity with serum amyloid P-component (36). Symmetry axes are indicated for each structure. Arrows represent 2-fold axes in the plane of the paper.
Such harmonious and symmetrical motifs occur widely in Chinese art, especially in bronze mirrors that display both rotational and mirror symmetries and reflect the search for permanent, symmetrical relationships expressed in Confucian philosophy and taken as necessary for the stability of Chinese feudal society. Christianity, once established, also led to symmetry in its visual arts and architecture. The relationship of Jesus and the twelve apostles has given rise to many circular church windows with pseudo 3- and 4-fold symmetries, which establish a mood of restfulness and permanence. A fine example of this symmetrical architecture is found in the design of the Chartres Cathedral.
Very interesting pseudosymmetries, reminiscent of the structures of oligomers, can also relate protein domains and motifs. For example, the βγ-crystallins of the vertebrate eye lens are composed of a polypeptide chain that has been internally quadruplicated in evolution so that a similar motif occurs four times in series. The three-dimensional structure (45) is arranged as two “domains” each containing two motifs related by 2-fold symmetry; the two domains are themselves related by a further approximate 2-fold symmetry as shown in Fig. 4. The crystallins are further assembled into complex multichain oligomers. Because the lens must remain transparent when it loses its nucleus, and so its ability to make new proteins, these proteins have evolved to be very stable, as they must last for the lifetime of the individual. When they become denatured, cataracts are more likely to form, an increasing problem as humans live longer than they did when they were under the selective pressure of evolution! Similar proteins (47) are secreted onto the surface of some bacterial spores to increase their survival in a hostile environment.
Figure 4.
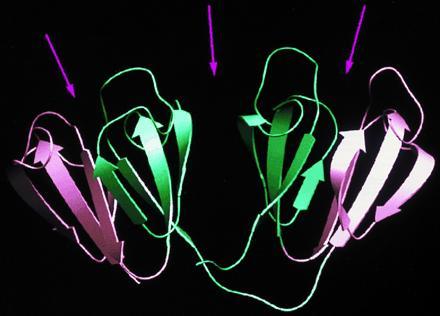
Internal symmetry in γB-crystallin (45, 46). The two symmetry-related Greek-key motifs within a domain are shown in two different colors and are related by approximate 2-fold axes (arrows). In a similar manner, the two domains are related by an approximate 2-fold axis.
Symmetry operations that relate supersecondary structural motifs within globular domains can also lead to cyclic packing of identical or similar units, resulting in highly stable “rings” (48). Thus, in triose-phosphate isomerase and many related barrel structures, eight supersecondary αβ-motifs, each comprising an α-helix folded onto a β-strand, are circularly arranged (49). An inner cylinder of parallel β-strands is surrounded by an outer cylinder of α-helices, producing an aesthetically pleasing structure. Neuraminidase (10), the β-subunit of heterotrimeric G proteins (50–52), and several other proteins are constructed as propellers with seven, eight, or more blades, made of β-strands that are related by approximate rotational symmetry. In a similar way “Rangoli” patterns, used in India as a form of worshipping Hindu gods, are almost always cyclic, thereby signifying a stable state, harmony, and peace.
Symmetry and Cooperativity
Symmetry-related subunits or domains in protein structures often communicate among themselves; they show cooperativity. This was first discovered by Haurowitz (53) for hemoglobin, where the addition of oxygen molecules increases the affinity for others. The seminal work of Perutz (54) on hemoglobin defined the molecular mechanism; it is a textbook example in which the four subunits retain approximate 222 symmetry but exist in two forms, the oxy (relaxed or R-state) and deoxy (tense or T-state) states. In the transition between the two states it is known that the spatial disposition of the subunits undergoes dramatic changes, although the mechanism is still not fully understood (55–57). By maintaining the symmetry of the tetramer, affinity is first suppressed but then sharply increased as the concentration of oxygen increases to that found in the lungs. Thus, the oxygen affinity is optimally controlled; the oxygen molecules are efficiently picked up in the lungs and deposited in the tissues. While 30% of the oxygen from free oxygen concentration of 130 μM in lungs can be transferred to tissues by an oligomeric globin, less than 20% would be transferred by a hemoglobin with only one subunit. Thus, if the cooperativity between the subunits of hemoglobin were absent, less oxygen would be transported from the lungs to the tissues (58). Symmetry thus plays a key role in the molecular regulation of important physiological processes.
Major structural rearrangements are observed in many oligomers that show cooperativity. X-ray analyses have defined the structures of R-states and T-states for four proteins in addition to hemoglobin. All show positive cooperativity in ligand binding. As described above phosphofructokinases are generally tetramers of identical polypeptide chains, related by three orthogonal 2-fold symmetry axes (222 symmetry). Each subunit is composed of two domains, which are themselves related by symmetry (59). The fructose + ATP reaction catalyzed by this enzyme appears to involve cooperativity, reflected in rearrangements within the subunits and a 7° rotation between subunits. A pronounced rearrangement of tertiary structure is also observed in the aspartate transcarbamoylase, which is involved in the formation of carbamoyl aspartate from the carbamoyl phosphate and aspartate. The enzyme consists of six copies of each of a catalytic and a regulatory polypeptide chain, resulting in a highly symmetric 32 structure. Comparison of the R-state and T-state of the enzyme reveals a remarkable 12° rotation of the catalytic trimer and 15° rotation of the regulatory dimer resulting in an 11-Å separation of the subunit surfaces (see ref. 60 for a review). Changes between symmetry-related subunits and their angle of rotation between subunits for the other three proteins are as follows: hemoglobin (15°), glycogen phosphorylase (10°), and fructose-1,6-bisphosphatase (19°) (57, 60). In all these cases symmetry is central to biological function.
Symmetry and Motion
But what of motion at the molecular level? Does nature abandon symmetry or does it distort symmetrical structures so that they reach a point of disequilibrium with resulting motion? Is there a parallel with Mao Tse Tung’s dialectical reinterpretation of Confucian relationships where the thesis and antithesis are no longer in balance (as in yin and yang), and a new synthesis takes place?
Enzymes, the biological catalysts that endow dynamic properties on living organisms, are good examples. For example, the aspartic proteinases, which include the digestive enzyme pepsin (Fig. 5a), have evolved from a protein made of two identical copies of a smaller molecule. It is now evident that such enzymes still exist in retroviruses, where the organism has to travel light; i.e., it has to package all its own nucleic acid within its capsid. Fig. 5b shows the symmetrical structure of the HIV proteinase (3, 4). Such retroviral proteinases are not efficient enzymes. Indeed, medicinal chemists have exploited their symmetry to make symmetrical drugs that bind more tightly than the substrate at the catalytic site (Fig. 5c) and so inhibit maturation of the virus (see ref. 64 for a review). They are now used as part of a cocktail of HIV antivirals that are beginning to have success in the clinic.
Figure 5.


(a and b) Comparison of monomeric pepsin (a) with two symmetry-related lobes (61) and dimeric HIV proteinase (b) (4) inspired by the analyses of Blundell and coworkers (61–63). The arrows indicate the positions of approximate and perfect 2-fold axes relating domains (pepsin) and protomers (HIV proteinase), respectively. (c) The two topologically equivalent catalytic groups (the “hands” Asp-32 and Asp-215 of pepsin), which have an approximate 2-fold axis shown as an arrow.
In the meantime, evolution has accepted mutations in aspartic proteinases that have modified the details of each half while keeping the basic architecture (or tertiary structure) of the ancestral protein. The net result (compare Fig. 5 a with b) is a pseudosymmetrical (2-fold axis) molecule in which the two topologically equivalent catalytic groups (the two “hands” of the molecule) are now in distinctly different environments; one can push, while the other can pull more effectively. A much better enzyme results, which is now found in fungi, mammals, and plants. In humans the design is manifested as pepsin to digest food, as cathepsin D to process proteins within cells, and as renin to process a key hormone precursor, angiotensinogen, which controls blood pressure.
There is an analogy here with the loss of symmetry in Michelangelo’s “Creation.” God and Adam, each in human form, offer a pseudosymmetry; their hands touch in a configuration related by a mirror plane—the right hand of God reflected as the left hand of Adam. Michelangelo disturbed the symmetry gently but firmly to make it clear that the relationship is loosened and life flows strongly from God into Adam.
Saibil and coworkers (65, 66) have shown that GroEL, which is composed of two discs of 7-fold symmetric subunits (17) and binds non-native folding intermediates to assist proper folding, undergoes significant structural alterations when binding to ATP and GroES, which is also a heptamer (67, 68). This example emphasizes the significance of mobility associated with stable and highly symmetric biological systems.
Conclusions
The role of symmetry in resting or static systems is subjectively pleasing. It is, therefore, not surprising that internal symmetry has been the intuitive justification for many scientific hypotheses. The Monod symmetry model for cooperativity has been advantaged by this approach. But nature does not always choose the most restful or harmonious motif in its search for motion in life. Molecular biologists should consider Chinese bronzes, Rangoli patterns, Leonardo da Vinci’s drawings, and Escher’s prints by all means; but they should also consider Michelangelo’s “Creation” if they wish to understand life and motion at the molecular level.
Acknowledgments
We thank Prof. M. Vijayan and Dr. K. Suguna for providing the coordinate set of the high-resolution structure of peanut lectin–lactose complex (34) prior to the public release. We also thank Drs. Nagarajaram, Sowdhamini, and Dhanaraj for suggestions and comments on the manuscript. All figures have been prepared using the software setor (69). This research is supported by Wellcome Trust.
References
- 1.Schulz G E, Schirmer R H. Principles of Protein Structure. New York: Springer; 1979. [Google Scholar]
- 2.Creighton T E. Proteins: Structures and Molecular Properties. New York: Freeman; 1993. [Google Scholar]
- 3.Wlodawer A, Miller M, Jaskolski M, Sathyanarayana B K, Baldwin E, Weber I T, Selk L M, Clawson L, Schneider J, Kent S B H. Science. 1989;245:616–621. doi: 10.1126/science.2548279. [DOI] [PubMed] [Google Scholar]
- 4.Lapatto P, Blundell T, Hemmings A, Overington J, Wildersprin A, Wood S, Merson J R, Whittle P J, Danley D E, Geoghegan K F, Hawrylik S J, Lee S E, Scheld K G, Hobart P M. Nature (London) 1989;342:299–302. doi: 10.1038/342299a0. [DOI] [PubMed] [Google Scholar]
- 5.Bax B, Lapatto L, Nalini V, Driessen H, Lindley P F, Mahadevan D, Blundell T L, Slingsby C. Nature (London) 1990;347:776–780. doi: 10.1038/347776a0. [DOI] [PubMed] [Google Scholar]
- 6.Huber R, Deisenhofer J, Colman P M, Matsushima M, Palm W. Nature (London) 1976;264:415–420. doi: 10.1038/264415a0. [DOI] [PubMed] [Google Scholar]
- 7.Ealick S E, Rule S A, Carter D C, Greenhough T J, Babu Y S, Cook W J, Habash J, Helliwell J R, Stoeckler J D, Parks R D, Jr, Chen S F, Bugg C E. J Biol Chem. 1990;265:1812–1820. doi: 10.2210/pdb2pnp/pdb. [DOI] [PubMed] [Google Scholar]
- 8.Weis M S, Wacker T, Weckesser J, Welte W, Schulz G E. FEBS Lett. 1990;267:268–272. doi: 10.1016/0014-5793(90)80942-c. [DOI] [PubMed] [Google Scholar]
- 9.Frier J A, Perutz M F. J Mol Biol. 1977;112:97–112. doi: 10.1016/s0022-2836(77)80158-7. [DOI] [PubMed] [Google Scholar]
- 10.Varghese J N, Laver W G, Coleman P M. Nature (London) 1983;303:35–40. doi: 10.1038/303035a0. [DOI] [PubMed] [Google Scholar]
- 11.Emsley J, White H E, O’Hara B P, Oliva G, Srinivasan N, Tickle I J, Blundell T L, Pepys M B, Wood S P. Nature (London) 1994;367:338–345. doi: 10.1038/367338a0. [DOI] [PubMed] [Google Scholar]
- 12.Shrive A K, Cheetham G M T, Holden D, Myles D A A, Turnell W G, Volanakis J E, Pepys M B, Bloomer A C, Greenhough T J. Nat Struct Biol. 1996;3:346–354. doi: 10.1038/nsb0496-346. [DOI] [PubMed] [Google Scholar]
- 13.Vandenakker F, Sarfaty S, Twiddy E M, Connell T D, Holmes R K, Hol W G J. Structure. 1996;4:665–678. doi: 10.1016/s0969-2126(96)00073-1. [DOI] [PubMed] [Google Scholar]
- 14.Blundell T, Dodson G, Hodgkin D, Mercola D. Adv Protein Chem. 1972;26:279–402. [Google Scholar]
- 15.Baker E N, Blundell T L, Cutfield J F, Cutfield S M, Dodson E J, Dodson G G, Hodgkin D M C, Hubbard R E, Issacs N W, Reynolds C D, Sakabe K, Sakabe N, Vijayan M. Philos Trans R Soc London Ser B. 1988;319:369–456. doi: 10.1098/rstb.1988.0058. [DOI] [PubMed] [Google Scholar]
- 16.Tennent G A, Butler P J G, Hutton T, Woolfitt A R, Harvey D J, Rademacher T W, Pepys M B. Eur J Biochem. 1993;214:91–97. doi: 10.1111/j.1432-1033.1993.tb17900.x. [DOI] [PubMed] [Google Scholar]
- 17.Braig K, Otwinowski Z, Hegde R, Boisvert D C, Joachimiak A, Horwich A L, Sigler P B. Nature (London) 1994;371:578–586. doi: 10.1038/371578a0. [DOI] [PubMed] [Google Scholar]
- 18.Wilmsen H U, Leonard K R, Tichelaar W, Buckley J T, Pattus F. EMBO J. 1992;11:2457–2463. doi: 10.1002/j.1460-2075.1992.tb05310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parker M W, Buckley J T, Postma J P M, Tucker A D, Leonard K, Pattus F, Tsernoglou D. Nature (London) 1994;367:292–295. doi: 10.1038/367292a0. [DOI] [PubMed] [Google Scholar]
- 20.Lowe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 21.Neidhart D J, Kenyon G L, Gerlt J A, Petsko G A. Nature (London) 1990;347:692–694. doi: 10.1038/347692a0. [DOI] [PubMed] [Google Scholar]
- 22.McDermott G, Prince S M, Freer A A, Hawthomthwaite-Lawless A M, Papiz M Z, Cogdell R J, Issacs N W. Nature (London) 1995;374:517–521. [Google Scholar]
- 23.Meining W, Bacher A, Bachmann L, Schmid C, Weinkauf S, Huber R, Nar H. J Mol Biol. 1995;253:208–218. doi: 10.1006/jmbi.1995.0545. [DOI] [PubMed] [Google Scholar]
- 24.Antson A A, Otridge J, Brzozowski A M, Dodson E J, Dodson G G, Wilson K S, Smith T M, Yang M, Kurecki T, Gollnick P. Nature (London) 1995;374:693–700. doi: 10.1038/374693a0. [DOI] [PubMed] [Google Scholar]
- 25.Warren S G, Edwards B F P, Evans D R, Wiley D C, Lipscomb W N. Proc Natl Acad Sci USA. 1973;70:1117–1121. doi: 10.1073/pnas.70.4.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lawrence M C, Suzuki E, Varghese J N, Davies P C, Van Donkelaar A, Tulloch P A, Coleman P M. EMBO J. 1990;9:9–15. doi: 10.1002/j.1460-2075.1990.tb08074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dube P, Taveres P, Lurz R, Van Heel M. EMBO J. 1993;12:1303–1309. doi: 10.1002/j.1460-2075.1993.tb05775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoare R J, Harrison P M, Hoy T G. Nature (London) 1975;255:653–654. doi: 10.1038/255653a0. [DOI] [PubMed] [Google Scholar]
- 29.Karrasch S, Bullough P A, Ghosh R. EMBO J. 1995;14:631–638. doi: 10.1002/j.1460-2075.1995.tb07041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bloomer A C, Champness J N, Bricogne G, Staden R, Klug A. Nature (London) 1978;276:362–368. doi: 10.1038/276362a0. [DOI] [PubMed] [Google Scholar]
- 31.Winkler F K, Schutt C E, Harrison S C, Bricogne G. Nature (London) 1977;265:509–513. doi: 10.1038/265509a0. [DOI] [PubMed] [Google Scholar]
- 32.Einspahr H, Parks E H, Suguna K, Subramanian E, Suddath F L. J Biol Chem. 1986;261:6518–6527. [PubMed] [Google Scholar]
- 33.Liao D-I, Kapadia G, Ahmed H, Vasta G R, Herzberg O. Proc Natl Acad Sci USA. 1994;91:1428–1432. doi: 10.1073/pnas.91.4.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Banerjee R, Das K, Ravishankar R, Suguna K, Surolia A, Vijayan M. J Mol Biol. 1996;259:281–296. doi: 10.1006/jmbi.1996.0319. [DOI] [PubMed] [Google Scholar]
- 35.Naismith J H, Emmerich C, Habash J, Harrop S J, Helliwell J R, Hunter W N, Raftery J, Kalb A J, Yariv J. Acta Crystallogr D. 1994;50:847–858. doi: 10.1107/S0907444994005287. [DOI] [PubMed] [Google Scholar]
- 36.Srinivasan N, White H E, Emsley J, Wood S P, Pepys M B, Blundell T L. Structure. 1994;2:1017–1027. doi: 10.1016/S0969-2126(94)00105-7. [DOI] [PubMed] [Google Scholar]
- 37.Kelman Z, Finkelstein J, Odonnell M. Curr Biol. 1995;5:1239–1242. doi: 10.1016/s0960-9822(95)00247-8. [DOI] [PubMed] [Google Scholar]
- 38.Crick F H C, Watson J D. Nature (London) 1956;177:473–475. doi: 10.1038/177473a0. [DOI] [PubMed] [Google Scholar]
- 39.Casper D L D, Klug A. Cold Spring Harbor Symp Quant Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 40.Thompson D. On Growth and Form. Cambridge, U.K.: Cambridge Univ. Press; 1952. [Google Scholar]
- 41.Wood S P, Oliva G, O’Hara B P, White H E, Blundell T L, Perkins S J, Sardharwalla I, Pepys M B. J Mol Biol. 1988;202:169–173. doi: 10.1016/0022-2836(88)90529-3. [DOI] [PubMed] [Google Scholar]
- 42.Srinivasan N, Rufino S D, Pepys M B, Wood S P, Blundell T L. Chemtracts Biochem Mol Biol. 1996;6:149–164. [Google Scholar]
- 43.Shaanan B, Lis H, Sharon N. Science. 1991;254:862–866. doi: 10.1126/science.1948067. [DOI] [PubMed] [Google Scholar]
- 44.Pepys M B. In: Samter’s Immunological Diseases. 5th Ed. Frank M M, Austen K F, Claman H N, Unanue E R, editors. Boston: Little Brown; 1994. pp. 637–655. [Google Scholar]
- 45.Blundell T, Lindley P, Miller L, Moss D, Slingsby C, Tickle I, Turnell B, Wistow G. Nature (London) 1981;289:771–777. doi: 10.1038/289771a0. [DOI] [PubMed] [Google Scholar]
- 46.White H E, Driessen H P C, Slingsby C, Moss D S, Lindley P F. J Mol Biol. 1989;207:217–235. doi: 10.1016/0022-2836(89)90452-x. [DOI] [PubMed] [Google Scholar]
- 47.Wistow G, Summers L, Blundell T. Nature (London) 1985;315:771–773. doi: 10.1038/315771a0. [DOI] [PubMed] [Google Scholar]
- 48.Antson A A, Dodson E J, Dodson G G. Curr Opin Struct Biol. 1996;6:142–150. doi: 10.1016/s0959-440x(96)80067-4. [DOI] [PubMed] [Google Scholar]
- 49.Banner D W, Bloomer A C, Petsko G A, Phillips D C, Pogson C I, Wilson I A, Corran P H, Furth A J, Milman J D, Offord R E, Priddle J D, Waley S G. Nature (London) 1975;255:609–614. doi: 10.1038/255609a0. [DOI] [PubMed] [Google Scholar]
- 50.Wall M A, Coleman D E, Lee E, Iniguezlluhi J A, Posner B A, Gilman A G, Sprang S R. Cell. 1995;83:1047–1058. doi: 10.1016/0092-8674(95)90220-1. [DOI] [PubMed] [Google Scholar]
- 51.Sondek J, Bohm A, Lambright D G, Hamm H E, Sigler P B. Nature (London) 1996;379:369–374. doi: 10.1038/379369a0. [DOI] [PubMed] [Google Scholar]
- 52.Lambright D G, Sondek J, Bohm A, Skiba N P, Hamm H E, Sigler P B. Nature (London) 1996;379:311–319. doi: 10.1038/379311a0. [DOI] [PubMed] [Google Scholar]
- 53.Haurowitz F. Hoppe-Seyler’s Z Physiol Chem. 1938;254:266–274. [Google Scholar]
- 54.Perutz M F. Proteins and Nucleic Acids. Amsterdam: Elsevier; 1962. [Google Scholar]
- 55.Franzen S, Lambrey J C, Bohn B, Poyart C, Martin J L. Nat Struct Biol. 1994;1:230–233. doi: 10.1038/nsb0494-230. [DOI] [PubMed] [Google Scholar]
- 56.Neet K E. Methods Enzymol. 1995;249:519–567. doi: 10.1016/0076-6879(95)49048-5. [DOI] [PubMed] [Google Scholar]
- 57.Jardetzky, O. (1996) Prog. Biophys. Mol. Biol., in press.
- 58.Stryer L. Biochemistry. San Francisco: Freeman; 1975. [Google Scholar]
- 59.Evans P R, Farrants G W, Hudson P J. Philos Trans R Soc London, Ser B. 1981;293:52–63. doi: 10.1098/rstb.1981.0059. [DOI] [PubMed] [Google Scholar]
- 60.Lipscomb W N. Chemtracts Biochem Mol Biol. 1991;2:1–15. [Google Scholar]
- 61.Cooper J B, Khan G, Taylor G, Tickle I J, Blundell T L. J Mol Biol. 1990;214:199–222. doi: 10.1016/0022-2836(90)90156-G. [DOI] [PubMed] [Google Scholar]
- 62.Tang J, James M N G, Hau I-N, Jenkins J A, Blundell T L. Nature (London) 1978;271:618–621. doi: 10.1038/271618a0. [DOI] [PubMed] [Google Scholar]
- 63.Pearl L H, Taylor W R. Nature (London) 1987;329:351–354. doi: 10.1038/329351a0. [DOI] [PubMed] [Google Scholar]
- 64.Whittle P J, Blundell T L. Annu Rev Biophys Biomol Struct. 1994;23:349–375. doi: 10.1146/annurev.bb.23.060194.002025. [DOI] [PubMed] [Google Scholar]
- 65.Saibil H R, Zheng D, Roseman A M, Hunter A S, Watson G M F, Chen S, Dermaur A A, O’Hara B P, Wood S P, Mann N H, Barnett L K, Ellis R J. Curr Biol. 1993;3:265–273. doi: 10.1016/0960-9822(93)90176-o. [DOI] [PubMed] [Google Scholar]
- 66.Chen S, Roseman A M, Hunter A S, Wood S P, Burston S G, Ranson N A, Clarke A R, Saibil H R. Nature (London) 1994;371:261–264. doi: 10.1038/371261a0. [DOI] [PubMed] [Google Scholar]
- 67.Hunt J F, Weaver A J, Landry S J, Gierasch L, Deisenhofer J. Nature (London) 1996;379:39–45. doi: 10.1038/379037a0. [DOI] [PubMed] [Google Scholar]
- 68.Mande S C, Mehra V, Bloom B R, Hol W G J. Science. 1996;271:203–207. doi: 10.1126/science.271.5246.203. [DOI] [PubMed] [Google Scholar]
- 69.Evans S V. J Mol Graphics. 1993;11:134–138. doi: 10.1016/0263-7855(93)87009-t. [DOI] [PubMed] [Google Scholar]


