Abstract
Disruption of the gut microbiota by high-fat diet (HFD) has been implicated in the development of obesity. It remains to be elucidated whether the HFD-induced shifts occur at the phylum level or whether they can be attributed to specific phylotypes; additionally, it is unclear to what extent the changes are reversible under normal chow (NC) feeding. One group (diet-induced obesity, DIO) of adult C57BL/6J mice was fed a HFD for 12 weeks until significant obesity and insulin resistance were observed, and then these mice were switched to NC feeding for 10 weeks. Upon switching to NC feeding, the metabolic deteriorations observed during HFD consumption were significantly alleviated. The second group (control, CHO) remained healthy under continuous NC feeding. UniFrac analysis of bar-coded pyrosequencing data showed continued structural segregation of DIO from CHO on HFD. At 4 weeks after switching back to NC, the gut microbiota in the DIO group had already moved back to the CHO space, and continued to progress along the same age trajectory and completely converged with CHO after 10 weeks. Redundancy analysis identified 77 key phylotypes responding to the dietary perturbations. HFD-induced shifts of these phylotypes all reverted to CHO levels over time. Some of these phylotypes exhibited robust age-related changes despite the dramatic abundance variations in response to dietary alternations. These findings suggest that HFD-induced structural changes of the gut microbiota can be attributed to reversible elevation or diminution of specific phylotypes, indicating the significant structural resilience of the gut microbiota of adult mice to dietary perturbations.
Keywords: dietary perturbations, gut microbiota, obesity, resilience
Introduction
Obesity and related metabolic diseases, such as type 2 diabetes, angiocardiopathy and nonalcoholic fatty liver, have become a devastating epidemic worldwide (Popkin, 2007). A number of recent studies have demonstrated that high-fat diet (HFD)-induced alterations of the gut microbiota structure have a pivotal role in the development of obesity-related diseases, possibly via two different but complementary pathways.
First, members of the gut microbiota that were enriched in response to HFD feeding may allow the host to harvest more energy from food (Ley et al., 2005; Ley et al., 2006b; Turnbaugh et al., 2006, 2008). A division-wide change in the ratio of Firmicutes:Bacteroidetes may be associated with obesity and body weight loss upon dietary intervention (Ley et al., 2006b; Turnbaugh et al., 2006). The provision of an obesogenic ‘Western' diet to wild-type mice resulted in an overall decrease in the diversity of the gut microbiota, particularly a decrease in the prevalence of Bacteroidetes species and an increase in the prevalence of a single class of Firmicutes (Mollicutes). The body weight and adiposity of the mice were reduced after they were switched from a Western-type diet to a low-fat or low-carbohydrate diet, and a significant reduction in the relative abundance of Mollicutes species was also observed (Turnbaugh et al., 2008). But other studies suggested that the changes in the ratio of Firmicutes:Bacteroidetes in obese vs lean subjects is not clear cut, for example, despite weight loss there was no change in the ratio of Firmicutes:Bacteroidetes (Duncan et al., 2008; Schwiertz et al., 2010). The reason for these conflicting reports may be that the variation of the gut microbiota associated with diet did not occur at the division level but at the specific phylotype levels (Turnbaugh et al., 2008; De Filippo et al., 2010; Murphy et al., 2010; Zhang et al., 2010). Thus, whether the HFD-induced changes of gut microbiota relevant to obesity development occurs at the division level or is attributable to specific phylotypes remains to be clarified.
Second, a HFD-disrupted gut microbiota has also been suggested as the primary mediator between obesity-related disorders and a primary pathological condition underlying the development of these diseases—low-grade, systemic and chronic inflammation (Cani et al., 2007; Cani et al., 2008; Cani et al., 2009). HFD feeding modulates the gut microbiota composition by decreasing the prevalence of specific gut barrier-protecting bacteria and increasing the prevalence of opportunistic pathogens that can release free antigens such as lipopolysaccharides. This imbalance may be associated with a higher gut permeability, leading to higher plasma levels of endotoxin, higher levels of inflammation and eventually the development of metabolic disorders, a phenomenon termed metabolic endotoxemia (Cani et al., 2007; Zhang et al., 2010).
Similar to other ecosystems, the complex microbial community in the mammalian gut is a dynamic system with a stable steady state (Sonnenburg et al., 2004), but this system can also be disrupted by many ‘environmental' factors, such as diets, drugs and changing host physiology (Ley et al., 2006a; Jia et al., 2008; De Filippo et al., 2010). Structural resilience is the capacity of a complex system to recover to its normal state after the perturbation has been removed. The gut microbiota may not completely recover to the baseline state after certain perturbations such as antibiotic treatment (Dethlefsen and Relman, 2010). Long-term high fat intake can shift the composition of the microbial community in the distal gut of mammals, which may be responsible for the development of obesity and related metabolic disorders (Cani et al., 2007; De Filippo et al., 2010; Zhang et al., 2010), but to what extent the HFD-disrupted structure of the gut microbiota can be reverted after returning to a balanced diet is unclear.
The dynamics of the complex community over time can reveal more about interactions between community members undergoing disturbance of external factors. Unfortunately, most of the existing reports only collected one-time snapshot samples at the end of interventions to detect the effect of dietary intervention on the gut microbiota (Turnbaugh et al., 2008; Murphy et al., 2010). Detailed trajectory analysis for finding out the actual route of structural recovery of the gut microbiota responding to dietary perturbations may help identify specific phylotypes in gut bacterial community, which are principally responding to dietary intervention in a way relevant to development of obesity and insulin resistance.
Therefore, in this study, we monitored the dynamic changes of the gut microbiota in a mouse model fed with a HFD for 12 weeks and then reverted to normal chow (NC) for 10 weeks. A microbiome-wide association study strategy was employed to understand structural responses of the gut microbiota to dietary perturbations relevant to development and alleviation of metabolic syndromes.
Materials and methods
Animal intervention
Male C57BL/6J mice at 10 weeks of age were purchased from National Rodent Laboratory Animal Resources, Shanghai Branch. Mice were maintained under a 12-h dark/light cycle (lights on at 0630 hours) at a temperature of 22±3 °C in accredited animal facilities of SLAC (Shanghai Laboratory Animal Center). All experimental procedures and protocols were approved by the Institutional Animal Care and Use Committee of the Shanghai Laboratory Animal Center, CAS. Before the experiments, mice were maintained on a NC diet (containing 5.28% fat, 61.3% carbohydrates, 3.25 kcal g−1, from SLAC Inc., Shanghai, China) (see Supplementary Table S3) for 2 weeks. At the beginning of the experiment (0 week), the mice were randomly assigned to two groups (n=10 for each group): a control group of NC-fed mice (CHO) group and a diet-induced obesity (DIO) group of mice fed with HFD (containing 34.9% fat, 26.3% carbohydrates, 5.21 kcal g−1, from Research Diets, Inc., New Brunswick, NJ, USA) (see Supplementary Table S4). After 12 weeks, the DIO group mice were switched to a NC diet, and then both groups were fed the NC diet for 10 weeks. The food intake and body weight of each animal was measured every 2 weeks. At 12 (n=1 for each group) and 22 weeks (n=5 for each group), mice were randomly selected from each group and killed to calculate the weight of their epididymal fat pads.
Glucose tolerance test
After overnight fasting, mice were injected intraperitoneally with 2 g kg−1 glucose. Blood glucose concentrations were measured before glucose injection and 15, 30, 60 and 120 min after glucose injection by using a glucometer (FreeStyle, Alameda, CA, USA). Mice underwent glucose tolerance tests at weeks 0, 4, 8, 12, 16, 20 and 22 of the experiment.
PCR amplification of the V3 region of 16S rRNA genes and pyrosequencing
Fresh fecal matter was collected from each mouse at weeks 0, 2, 4, 8, 12, 16, 20 and 22 of the experiment and immediately stored at −80 °C for subsequent analysis. The fecal samples used for analysis are shown as Supplementary Table S1. Fecal DNA was extracted using the PSPSpin Stool DNA Plus Kit (Invitek GmbH, Berlin, Germany) and used as the template for PCR amplification of the V3 region of 16S rRNA genes. The forward primer was 5′-NNNNNNCCTACGGGAGGCAGCAG-3′, and the reverse primer was 5′-NNNNNNATTACCGCGGCTGCT-3′, in which the underlined sequences are universal bacterial primers P1 and P2, respectively (Muyzer et al., 1993). NNNNNN is the unique 6-nucleotide barcode used to distinguish the PCR products from different samples. PCR was performed with a thermocycler PCR system (PCR Sprint, Thermo electron, Corp., Marietta, OH, USA) using the program described previously (Zhang et al., 2010). The products from different samples were mixed at equal ratios for pyrosequencing with the GS FLX platform (Roche, Branford, CT, USA).
Bioinformatics and statistical analysis
Based on several previous reports describing sources of errors in 454 sequencing runs (Margulies et al., 2005; Sogin et al., 2006; McKenna et al., 2008), we used standards for quality control as previously described (Zhang et al., 2010).
Cluster Database at High Identity with Tolerance was used to cluster the unique V3 sequences at high identity (99–100%, step=0.1%) with tolerance (Turnbaugh et al., 2009). The most abundant sequence of each cluster was aligned using the Nearest Alignment Space Termination multi-aligner with a minimum template length of 90 bases and a minimum percent identity of 75% (DeSantis et al., 2006). Then, the resulting alignments were imported into the ARB (Ludwig et al., 2004). A distance matrix of these sequences from ARB was imported into Distance-based Operational Taxonomic Unit and Richness for phylotype binning (Schloss and Handelsman, 2005). An operational taxonomic unit (OTU) was defined using a threshold of 96% identity and measured for coverage (rarefaction analysis with Past software; free download from http://folk.uio.no/ohammer/past/) and diversity (Shannon index with R statistical software (2.11.1; http://cran.r-project.org)).
We used the most abundant sequence in each OTU as the representative sequence and inserted these representative sequences into a pre-established phylogenetic tree of the full-length 16S rRNA gene sequences with ARB. The phylogenetic tree was then used for UniFrac PCoA (principal coordinate analysis) and cluster environments with abundance weighting. The UniFrac software was obtained from http://bmf2.colorado.edu/unifrac/download/.
Taxonomic classifications of each OTU were obtained using Sapelo Island Microbial Observatory (SIMO) Taxnomic assignments (http://simo.marsci.uga.edu/public_db/taxonomy.htm), which are based on the similarity to vetted type species sequences in the Ribosomal Database Project database.
Redundancy analysis (RDA) was performed using the rda command of the vegan package (1.17–4) of R statistical software (2.11.1).
Results
Phenotype changes of mice during dietary perturbations
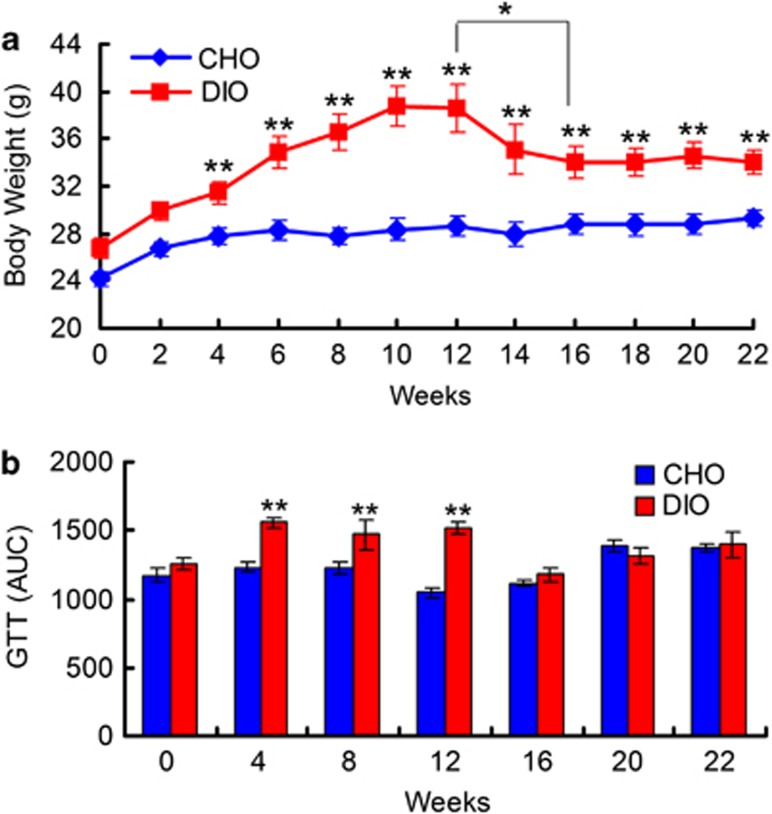
In the first 12 weeks of our experiment, the mice in the DIO group were fed a HFD, and these mice had higher daily fat and lower carbohydrate intake than their CHO group counterparts that were fed the NC diet (Supplementary Figure S1). The DIO group gained weight quickly during HFD feeding, and these mice were significantly heavier than their CHO group counterparts after only 2 weeks (29.9±0.8 g vs 26.7±0.6 g, P<0.01). The body weight of animals in the DIO group plateaued after 10 weeks (38.7±1.7 g vs 28.4±0.9 g, P<0.001) and remained unchanged over the following 2 weeks (Figure 1a). The DIO group mice also exhibited impaired glucose tolerance after 4 weeks of HFD feeding and had more epididymal fat after 12 weeks than their CHO group counterparts (Figure 1b and Supplementary Figure S2).
Figure 1.
Phenotype changes of mice during dietary alternations. (a) Body weight curve of each group. (b) Glucose tolerance tests (GTTs) were performed for each group. Shown are the calculated areas under the curve (AUC) of blood glucose from the GTT measurements. (a) and (b) for 0–12 weeks, CHO group and DIO group: n=10; for 14–22 weeks, CHO group: n=9, and DIO group: n=8. The data in (a) and (b) are shown as means±s.e.m. *P<0.05, **P<0.01 by one-way ANOVA.
After reverting to NC feeding, the DIO group exhibited similar fat and carbohydrate intake as the CHO group (Supplementary Figure S1). The body weight of DIO group mice was decreased after 2 weeks of NC feeding and was significantly decreased after 4 weeks of NC feeding compared with the values observed for HFD feeding (38.6±2.0 g vs 34.1±1.3 g, P<0.05). After 4 weeks of NC feeding, body weight began to stabilize in the DIO group mice. After 10 weeks of NC feeding, the body weight of DIO group mice became significantly lower than that observed during HFD feeding, although these animals remained significantly heavier than their CHO group counterparts (34.1±1.1 g vs 29.3±0.7 g, P<0.01; Figure 1a). The difference in epididymal fat levels between the DIO and CHO groups was insignificant at the end of the trial (end of the twenty-second week; Supplementary Figure S1). Additionally, glucose tolerance in the DIO group returned to normal levels after 2 weeks of NC feeding.
Pyrosequencing analysis of the gut microbiota
During the dietary intervention, we collected fecal samples from both groups intermittently for dynamic monitoring of the gut microbiota structural changes via bar-coded pyrosequencing of the V3 hypervariable region of 16S rRNA genes. After discarding 2079 reads that had no neighbors with higher than 75% homology in the Greengenes database, 256 278 reads (average of 1767 sequences per community tested, only two samples had less than 1000 reads; the reads of these two samples were 638 and 996) were clustered into 2744 OTUs at a cutoff of 96% homology. Good's coverage of all samples averaged 93.4±1.6% (mean±s.d., range=89.1–97.6%). Of the 2744 OTUs delineated in this study, 1425 (51.9%), 1027 (37.4%), 156 (5.6%), 61 (2.2%) and 75 (2.7%) OTUs were affiliated with Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria and with other phyla, respectively.
Changes in the richness and diversity of the gut microbiota in response to dietary perturbations
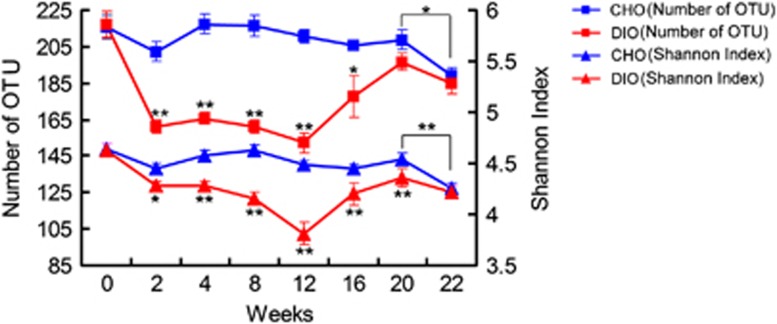
HFD feeding significantly influenced the richness and diversity of the bacterial community, which was reversible upon reverting to NC feeding. Disturbances associated with the dietary perturbations were indicated by plots of OTU richness and Shannon entropy over time (Figure 2). Compared with the CHO group values, richness and diversity (evaluated after rarefaction to 1000, reads/sample to normalize sampling intensity) declined significantly in the DIO group after only 2 weeks of HFD feeding (OTU number: 161.3±3.5 vs 202.4±5.3, P<0.001; Shannon index: 4.28±0.03 vs 4.45±0.06, P<0.05). The differences between DIO and CHO groups augmented during this stage and the lowest values of these parameters occurred after 12 weeks of HFD feeding (OTU number: 152.3±5.3 vs 210.8±3.8, P<0.001; Shannon index: 3.81±0.11 vs 4.49±0.03, P<0.001). The richness and diversity indices of the gut microbiota in the DIO group increased gradually after beginning NC feeding and became similar to those of the CHO group after 22 weeks (OTU number: 184.3±5.7 vs 189.0±4.6; Shannon index: 4.22±0.06 vs 4.26±0.05). For the CHO group, these parameters were stable in the first 20 weeks, but decreased notably in the twenty-second week (OTU number: 208.8±5.6 vs 189.0±4.6, P<0.05; Shannon index: 4.54±0.06 vs 4.26±0.05, P<0.01), indicating an age-related effect on the gut microbiota diversity.
Figure 2.
Changes in the richness and diversity of the gut microbiota during dietary alternations. OTU and Shannon diversity index numbers are shown for each group at eight time points (OTU cutoff, 96%). Calculations were performed after rarefying to an equal number of reads (1000) for all samples to control for unequal sampling effort. Data are shown as means±s.e.m., *P<0.05, **P<0.01 by one-way ANOVA.
Trajectory analysis of structural shifts of the gut microbiota during the dietary perturbations
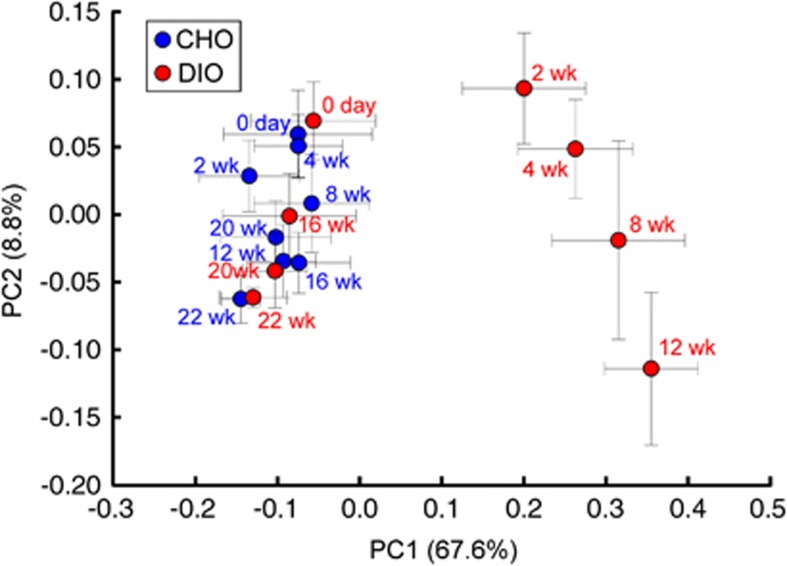
Time-course changes in the gut microbial community structures of the experimental mice in response to dietary perturbations were assessed by PCoA of weighted UniFrac distances, a measure of community dissimilarity based on OTU abundance and evolutionary relatedness (Figure 3). The structure of the gut microbiota at each time point exhibited no significant intragroup differences among our experimental animals. The PCoA score plot showed that HFD feeding induced significant alterations of the gut microbiota structure. The gut microbiota structure of DIO group animals was significantly different from that of CHO group animals after only 2 weeks of HFD feeding, and switched back to CHO space 4 weeks after changing to NC. After 10 weeks of NC feeding, the gut microbiota of the DIO group mice was virtually identical to that of the CHO group mice. The first principal coordinate (PC1), which is driven primarily by the response to HFD feeding, explained 67.6% of the inter-sample variance, with maximally perturbed samples on the right part clearly segregating from clusters of all samples on NC, indicating that diet is the major force shaping the gut microbiota. PC2, which reflects the effects of age on the gut microbiota structure of both DIO and CHO group mice, explained only 8.8% of the total variance. After 10 weeks of NC feeding, the gut microbiota of the DIO group mice matched that of age-matched CHO group mice, indicating that the structural recovery of the gut microbiota in the DIO group mice was also affected by the same age effect observed among CHO group mice.
Figure 3.
Trajectory analysis of the gut microbiota during dietary alternations. Weighted UniFrac PCoA of the gut microbiota between different treatment groups at weeks 0, 2, 4, 8, 12, 16, 20 and 22 based on pyrosequencing OTU (96% identity) data. Each point represents the mean principal coordinate (PC) score from all of the mice in a group at one time point, and the error bar represents the s.e.m.
Key phylotypes in the gut microbiota in response to dietary perturbations and age
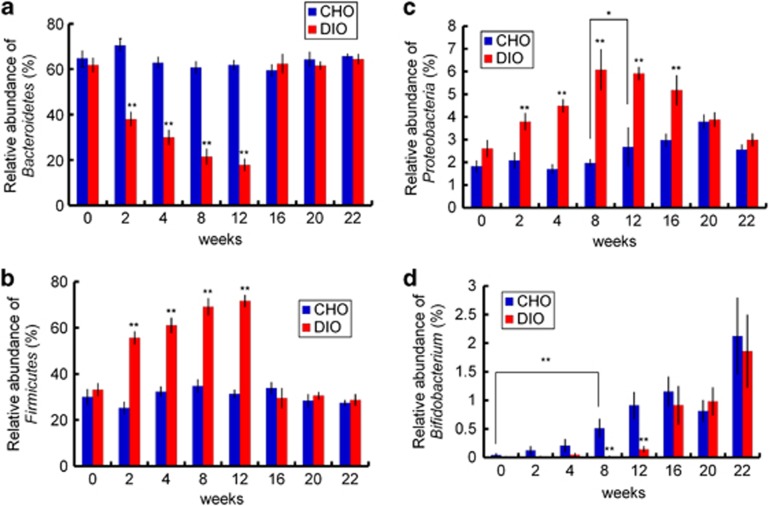
We observed a gradual and significant increase of the relative abundance of Firmicutes and a decrease in that of Bacteroidetes in HFD-fed DIO mice (Figures 4a and b). In addition to these two phyla, the relative abundance of Proteobacteria was also associated with dietary perturbation, as its abundance notably increased in response to HFD feeding in the DIO group and declined after these mice were switched to NC feeding (Figure 4c). In the CHO group, the abundance of Firmicutes and Bacteroidetes did not vary significantly during the entire experiment, but that of Proteobacteria increased beginning in the twelfth week. There were no differences in the relative abundances of Firmicutes, Bacteroidetes and Proteobacteria between DIO and CHO group after animals in the DIO group were switched to NC feeding.
Figure 4.
Changes of relative abundance of several important taxa during the trial. (a) Bacteroidetes, (b) Firmicutes, (c) Proteobacteria and (d) Bifidobacterium spp. in the DIO and CHO group at 0, 2, 4, 8, 12, 16, 20 and 22 weeks. DIO group: n=9 at weeks 0, 2, 4, 8 and 12; n=8 at weeks 16 and 22; n=7 at week 20. Control group: n=10 at weeks 0, 2, 4, 8 and 12; n=9 at weeks 16, 20 and 22. Data are shown as means±s.e.m., *P<0.05, **P<0.01 by one-way ANOVA.
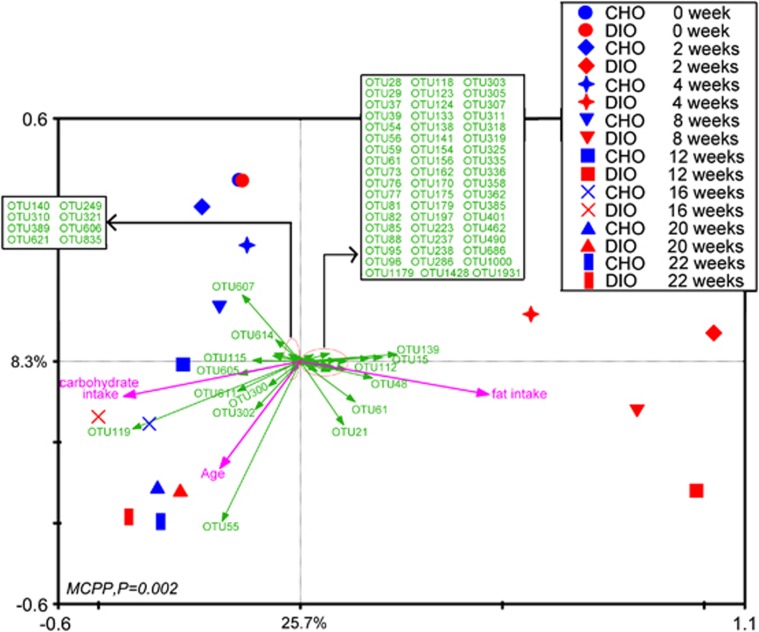
To determine whether any specific bacterial phylotypes are associated with dietary perturbation and age, RDA was conducted using fat intake, carbohydrate intake, and age as ‘environmental variables' and the relative abundances of the OTUs as ‘species variables' (Figure 5). The differences observed in RDA were significant (P=0.002) as assessed by Monte Carlo Permutation Procedure. In total, 34% of the total variation in the data set (microbiota composition at OTU level) was related to environmental factors (diet composition and age). In the resulting ordination plot, most of this variation (25.7%) was plotted on the first axis that separates the samples mainly based on the ratio of fat intake to carbohydrate intake. The second axis, which explains only 8.3% of the variability, was plotted with the alteration of the gut microbiota in response to age. Based on RDA, we found 77 OTUs that were closely associated with fat intake, carbohydrate intake, or age (explaining more than 10% of the variability of the samples; Figures 5 and 6 and Supplementary Table S2).
Figure 5.
Triplot of the RDA of the microbiota composition relative to fat intake, carbohydrate intake and age. Responding OTUs that explained more than 10% of the variability of the samples are indicated by green arrows. First and second ordination axes are plotted, representing 25.7% and 8.3% of the variability in the data set, respectively. Bottom-left, P-value obtained by Monte Carlo Permutation Procedure is reported.
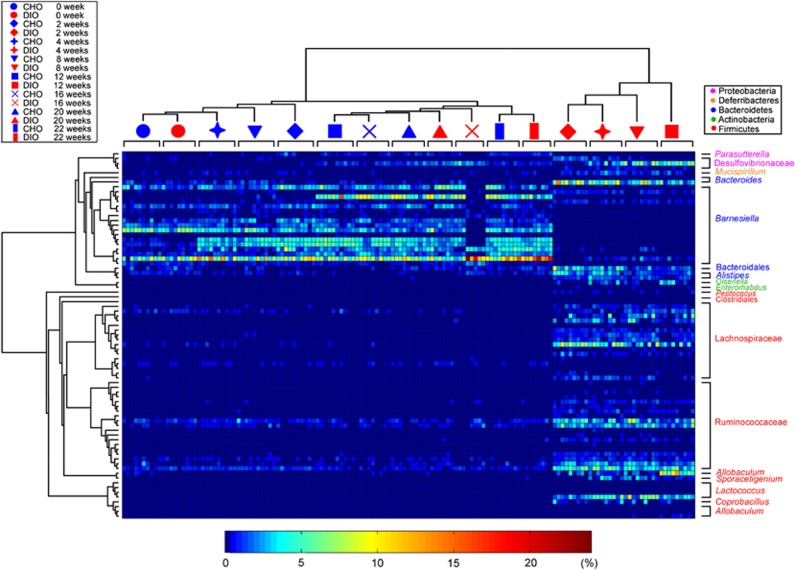
Figure 6.
RDA-derived abundance distribution pattern of the 77 key phylotypes responding to fat intake, carbohydrate intake and age. Heat map of the spot corresponds to the bacterial abundance in the sample. Cluster environments from UniFrac were used to group the mice. The OTUs were organized according to their phylogenetic positions. The taxa of the OTUs are shown on the right.
All of the OTUs identified here were associated with diet composition. Higher fat intake and lower carbohydrate intake enriched phylotypes belonging to Firmicutes in DIO group mice, such as Lachnospiraceae (15 OTUs), Ruminococcaceae (18 OTUs) and Lactococcus (4 OTUs). Interestingly, the abundance of OTUs in Allobaculum (a genus in Erysipelotrichaceae, previously identified as Mollicutes) only significantly increased in the twelfth week in DIO group mice. However, the OTUs associated with Bacteroidetes responded differentially to diet. The abundance of most of the OTUs in Bacteroidetes plunged rapidly when DIO group mice were fed an HFD, but the abundance of two OTUs in Barnesiella, two OTUs in Bacteroides and two OTUs in Alistipes increased significantly. The HFD-induced increase in the abundance of Proteobacteria was primarily due to changes in the abundance of three OTUs in Desulfovibrionaceae. However, the relative abundance of one OTU (Parasutterella) in this phylum was higher in the NC-fed mice than in the HFD-fed mice.
Some of the phylotypes were additionally associated with age. In the CHO group, the abundance of three predominant OTUs (OTU119, OTU55 and OTU115) in Barnesiella increased gradually with age. Although the abundance of these OTUs declined rapidly to a low level in DIO group mice during HFD feeding, the abundances of these OTUs exhibited age-related increases. This age-related response continued in DIO group animals after reverting to NC feeding. Similarly, irrespective of diet, the abundance of OTU21 in Lawsonia (in family Desulfovibrionaceae) also significantly increased with age in both the DIO and CHO groups.
None of these phylotypes was different between the two groups at the beginning of our experiment. Because of higher fat intake and lower carbohydrate intake for 12 weeks, the responsive OTUs were shifted significantly in DIO group mice. However, all of the bacterial phylotypes (OTUs) that had shifted in response to HFD feeding in DIO group mice had completely recovered to the same levels in CHO group mice at the twenty-second week, even if some of these OTUs exhibited large differences between the two groups at the end of HFD feeding. For example, the average relative abundance (6.9%) of one OTU in Allobaculum in the DIO group was 100-fold higher than that in the CHO group at the twelfth week, but its abundance decreased to less than 0.07% in DIO group mice at the twenty-second week, which was not significantly different from that in the CHO group (0.1%, P>0.05).
Some studies indicated that the abundance of Bifidobacterium in the mouse gut declined significantly in response to HFD feeding (Cani et al., 2007; Murphy et al., 2010). Our RDA did not identify OTUs in this genus that changed in response to dietary perturbation. There were 16 OTUs (containing 1468 reads, 0.57% of the total reads) that belonged to Bifidobacterium, and 96.7% of sequences were classified into one OTU (OTU92). At the beginning of our experiment, the abundance of Bifidobacterium was very low in all mice (less than 0.03% in the gut of each animal) but increased in the CHO group with age, being significantly enriched from the eighth week onward (Figure 4d). For the DIO group, HFD feeding retarded this increase in the abundance of Bifidobacterium, which remained at very low levels until reverting to NC feeding. The levels of phylotypes in Bifidobacterium increased rapidly in the DIO group and exhibited no difference to those in the CHO group 4 weeks after reverting to NC feeding.
Discussion
Although the mammalian distal gut could be considered as an efficient and stable natural bioreactor, many environmental factors, particularly diet, could affect the structure of the gut microbiota (Sonnenburg et al., 2004; Mueller et al., 2006; De Filippo et al., 2010; Zhang et al., 2010; Walker et al., 2011; Wu et al., 2011). By employing initially healthy animals with an established adult gut microbiota, we observed both a significant reduction of richness/diversity and overall structural shifts of bacterial communities in response to HFD feeding. Additionally, the relative abundances of 77 key phylotypes responded most strongly to HFD intake, and some of these phylotypes exhibited as much as two orders of magnitude differences between the DIO and CHO groups at the end of 12 weeks of HFD feeding. However, after 10 weeks of NC feeding in the DIO group, there were no differences in the diversity, overall structure and composition of the gut microbiota between the two groups, suggesting that HFD feeding could disrupt the gut microbiota structure as a potent external factor, but this alteration can be ‘corrected' by homeostatic mechanisms of the ecosystem when the disturbing factor is removed. The long-term stability of the distal gut community is thus not maintained by resistance to change (that is, robustness), but rather by the action of homeostatic forces that maintain the stability of this dynamic system within a certain range (that is, resilience) (Dethlefsen and Relman, 2010).
In the current study, diet was not the only factor that affected the gut microbiota of mice, as age also had a vital role. Interestingly, the abundance of some of the age-related phylotypes in DIO group mice were diminished significantly by HFD intake, but these bacteria still showed an age-related increase. Their relative abundances gradually returned to the levels observed in age-matched CHO group mice after reverting to NC feeding. Thus, diet does not seem to affect the age-related relationship between the host and its gut microbiota. Previous studies suggest that because of a global impact on the physiology of the intestinal tract, aging can seriously affect the composition of the gut microbiota (Kleessen et al., 1997; Flint et al., 2007; Guigoz et al., 2008; Ostan et al., 2008). The longitudinal age-related changes we observed here were over a relatively short period of time compared with other reports (Mariat et al., 2009; Biagi et al., 2010). The robustness of age-related change in our data indicates a possible close interaction between these bacteria and physiology of the host. The mechanisms behind this phenomenon warrant further studies.
Although the intestinal ecosystem has restoring forces, the microbial community is not completely recovered in all cases after the disturbance was removed. After disruption by antibiotics, the structure of the gut microbiota began to return to its initial state by 1 week after the end of each antibiotic course, but the return was not complete even after approximately 5 months (Dethlefsen and Relman, 2010). Antibiotics may induce a shift of the gut microbiota composition to an alternative stable state, making it impossible to revert to the original structure because some members may have been irreversibly removed from the ecosystem, whereas diet only provides growth advantages to certain individuals and disadvantages to others, permitting complete recovery to the normal state after the disturbance is removed. It may be possible to reverse HFD-induced damage to the gut microbiota structure by switching to a more balanced diet, providing that the host initially had a healthy gut microbiota.
The results from the current work echoed a previous discovery that significant compositional alterations in the gut microbiota linked with obesity are the result of HFD intake (Ley et al., 2005; Turnbaugh et al., 2008). However, the diet-induced changes in the composition of the gut microbiota did not occur at the division level, and the microbial lineages in the same phylum were actually shifted in different directions upon dietary intervention and aging. Our data indicate that some of the diet-induced changes at the phylotype level are more relevant to changes in the health of the host. Thus, when attempting to identify patterns of the gut microbiota relevant to obesity or insulin resistance, a microbiome-wide association study strategy should be employed to focus on phylotype-level changes rather than shifts at broad taxonomic levels.
In the past years, it has been a commonly accepted view that both abundance and species diversity of bifidobacteria in human gut microbiota decrease during aging (Mitsuoka, 1992; Hopkins and Macfarlane, 2002; Woodmansey et al., 2004; Mueller et al., 2006). However, several recent studies based on molecular techniques suggested that there was no difference between the bifidobacteria abundance in healthy elderly and young adults (Biagi et al., 2010). Our current work revealed an age-related increase of the relative abundance of Bifidobacterium spp. during the 22 weeks of the trial in control mice gut, which was retarded by HFD. This indicates that the behavior of Bifidobacterium spp. may be far more complicated than previously shown and further analysis with more robust methods such as qPCR is thus needed to resolve such discrepancies.
Genome sequencing and metabolic reconstructions of a related human gut-associated Mollicutes species (Eubacterium dolichum) revealed functions, which may metabolize the imported sugars to short-chain fatty acids that are readily absorbed by host. Thus, the Western diet-induced increase of Mollicutes in mouse gut may be responsible for the development of obesity (Turnbaugh et al., 2008). By using a dynamic monitoring strategy, we showed that the bacteria in Erysipelotrichaceae (the same as Mollicutes) remained low in abundance during the first 8 weeks on HFD feeding and significantly increased only in the twelfth week when the animals had developed obesity and insulin resistance. The changes of the composition and diversity of gut bacterial communities induced by HFD that are relevant to obesity could not thus be attributed to the variation of Mollicutes in our case. This finding suggests that the relationship between the microbial composition and energy-harvesting capacity is more complex than previously considered (Murphy et al., 2010). Therefore, dynamic monitoring of the complex community over time may help to reveal important details about the effects of external factors on the gut microbiota composition more relevant to changes in the health of the host.
In the current work, we showed that the HFD-induced increases in the abundance of populations closely related to opportunistic pathogens and decreases in the abundance of probiotics could be reverted by NC feeding. Bifidobacteria are well-known probiotics that have been demonstrated to reduce intestinal endotoxin levels and improve mucosal barrier function (Griffiths et al., 2004, Wang et al., 2004; Wang et al., 2006). Our findings supported previous results that the high ratio of fat to carbohydrates in diets retarded the increase in the abundance of Bifidobacterium in the gut, which may be correlated with increased adiposity and insulin resistance. We also found that the abundance of some OTUs in Lawsonia, Desulfovibrio and Lactococcus increased in HFD-fed DIO group mice and declined to low levels after reverting to NC feeding. Many strains in these three genera are opportunistic pathogens that have been linked to some inflammatory diseases (Mosca et al., 1995; Loubinoux et al., 2000; Akhaddar et al., 2002; Weglarz et al., 2003; Michalski et al., 2006). On the other hand, members of Lawsonia and Desulfovibrio are endotoxins producers (Loubinoux et al., 2000; Weglarz et al., 2003) and are also capable of reducing sulphate to H2S, damaging the gut barrier (Beerens and Romond, 1977). Our analysis of serum levels of lipopolysaccharide-binding protein and adiponectin indicate that HFD-induced increase of antigen load from the gut microbiota and associated inflammatory condition in DIO mice can be reverted by switching back to NC (Supplementary Figure S3). The variations of those phylotypes modulated by diet might influence the chronic inflammatory condition of the host in a way that might be relevant to increases in the body weight and glucose tolerance of mice. Further work is needed to provide detailed molecular linkage between the patterns identified in this work and host metabolic responses.
In conclusion, HFD consumption can induce rather rapid changes of keystone species in the gut microbiota, and the disrupted structure of the gut microbiota can be completely reverted by reverting to the normal diet, indicating the high resilience of the gut microbiota in adult mice. The specific phylotypes responding to dietary perturbations may have significant roles in host health. These findings provide novel insights for designing clinical interventions for diet-related metabolic diseases in humans.
Acknowledgments
This work was supported by Project 30730005, 30800155 and 20875061 of the National Nature Science Foundation of China (NSFC), Key Project 2007DFC30450 and 075407001 of International Cooperation Program Grants.
Sequence information
All sequence data has been deposited in the GenBank Short Read Archive (SRP008754).
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Akhaddar A, El Mostarchid B, Gazzaz M, Boucetta M. Cerebellar abscess due to lactococcus lactis. A new pathogen. Acta Neurochir. 2002;144:305–306. doi: 10.1007/s007010200041. [DOI] [PubMed] [Google Scholar]
- Beerens H, Romond C. Sulfate-reducing anaerobic bacteria in human feces. Am J Clin Nutr. 1977;30:1770–1776. doi: 10.1093/ajcn/30.11.1770. [DOI] [PubMed] [Google Scholar]
- Biagi E, Nylund L, Candela M, Ostan R, Bucci L, Pini E, et al. Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS One. 2010;5:e10667. doi: 10.1371/journal.pone.0010667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cani PD, Bibiloni R, Knauf C, Waget A, Neyrinck AM, Delzenne NM, et al. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. 2008;57:1470–1481. doi: 10.2337/db07-1403. [DOI] [PubMed] [Google Scholar]
- Cani PD, Neyrinck AM, Fava F, Knauf C, Burcelin RG, Tuohy KM, et al. Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia. 2007;50:2374–2383. doi: 10.1007/s00125-007-0791-0. [DOI] [PubMed] [Google Scholar]
- Cani PD, Possemiers S, Van de Wiele T, Guiot Y, Everard A, Rottier O, et al. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut. 2009;58:1091–1103. doi: 10.1136/gut.2008.165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Keller K, Brodie EL, Larsen N, Piceno YM, et al. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 2006;34:W394–W399. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, Relman DA. Microbes and Health Sackler Colloquium: Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci USA. 2010;108:4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan SH, Lobley GE, Holtrop G, Ince J, Johnstone AM, Louis P, et al. Human colonic microbiota associated with diet, obesity and weight loss. Int J Obes. 2008;32:1720–1724. doi: 10.1038/ijo.2008.155. [DOI] [PubMed] [Google Scholar]
- Flint HJ, Duncan SH, Scott KP, Louis P. Interactions and competition within the microbial community of the human colon: links between diet and health. Environ Microbiol. 2007;9:1101–1111. doi: 10.1111/j.1462-2920.2007.01281.x. [DOI] [PubMed] [Google Scholar]
- Griffiths EA, Duffy LC, Schanbacher FL, Qiao H, Dryja D, Leavens A, et al. In vivo effects of bifidobacteria and lactoferrin on gut endotoxin concentration and mucosal immunity in Balb/c mice. Dig Dis Sci. 2004;49:579–589. doi: 10.1023/b:ddas.0000026302.92898.ae. [DOI] [PubMed] [Google Scholar]
- Guigoz Y, Dore J, Schiffrin EJ. The inflammatory status of old age can be nurtured from the intestinal environment. Curr Opin Clin Nutr Metab Care. 2008;11:13–20. doi: 10.1097/MCO.0b013e3282f2bfdf. [DOI] [PubMed] [Google Scholar]
- Hopkins MJ, Macfarlane GT. Changes in predominant bacterial populations in human faeces with age and with Clostridium difficile infection. J Med Microbiol. 2002;51:448–454. doi: 10.1099/0022-1317-51-5-448. [DOI] [PubMed] [Google Scholar]
- Jia W, Li H, Zhao L, Nicholson JK. Gut microbiota: a potential new territory for drug targeting. Nat Rev. 2008;7:123–129. doi: 10.1038/nrd2505. [DOI] [PubMed] [Google Scholar]
- Kleessen B, Sykura B, Zunft HJ, Blaut M. Effects of inulin and lactose on fecal microflora, microbial activity, and bowel habit in elderly constipated persons. Am J Clin Nutr. 1997;65:1397–1402. doi: 10.1093/ajcn/65.5.1397. [DOI] [PubMed] [Google Scholar]
- Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006a;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006b;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- Loubinoux J, Mory F, Pereira IA, Le Faou AE. Bacteremia caused by a strain of desulfovibrio related to the provisionally named desulfovibrio fairfieldensis. J Clin Microbiol. 2000;38:931–934. doi: 10.1128/jcm.38.2.931-934.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariat D, Firmesse O, Levenez F, Guimaraes V, Sokol H, Dore J, et al. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 2009;9:123. doi: 10.1186/1471-2180-9-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna P, Hoffmann C, Minkah N, Aye PP, Lackner A, Liu Z, et al. The macaque gut microbiome in health, lentiviral infection, and chronic enterocolitis. PLoS Pathog. 2008;4:e20. doi: 10.1371/journal.ppat.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalski CW, Di Mola FF, Kummel K, Wendt M, Koninger JS, Giese T, et al. Human inflammatory bowel disease does not associate with Lawsonia intracellularis infection. BMC Microbiol. 2006;6:81. doi: 10.1186/1471-2180-6-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuoka T. Intestinal flora and aging. Nutr Rev. 1992;50:438–446. doi: 10.1111/j.1753-4887.1992.tb02499.x. [DOI] [PubMed] [Google Scholar]
- Mosca A, D'Alagni M, Del Prete R, De Michele GP, Summanen PH, Finegold SM, et al. Preliminary evidence of endotoxic activity of Bilophila wadsworthia. Anaerobe. 1995;1:21–24. doi: 10.1016/s1075-9964(95)80379-3. [DOI] [PubMed] [Google Scholar]
- Mueller S, Saunier K, Hanisch C, Norin E, Alm L, Midtvedt T, et al. Differences in fecal microbiota in different European study populations in relation to age, gender, and country: a cross-sectional study. Appl Environ Microbiol. 2006;72:1027–1033. doi: 10.1128/AEM.72.2.1027-1033.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy EF, Cotter PD, Healy S, Marques TM, O'Sullivan O, Fouhy F, et al. Composition and energy harvesting capacity of the gut microbiota: relationship to diet, obesity and time in mouse models. Gut. 2010;59:1635–1642. doi: 10.1136/gut.2010.215665. [DOI] [PubMed] [Google Scholar]
- Muyzer G, de Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostan R, Bucci L, Capri M, Salvioli S, Scurti M, Pini E, et al. Immunosenescence and immunogenetics of human longevity. Neuroimmunomodulation. 2008;15:224–240. doi: 10.1159/000156466. [DOI] [PubMed] [Google Scholar]
- Popkin B. The World is Fat: The Fads, Trends, Policies, and Products That Are Fattening the Human Race. Avery, New York, USA; 2007. [Google Scholar]
- Schloss PD, Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol. 2005;71:1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwiertz A, Taras D, Schafer K, Beijer S, Bos NA, Donus C, et al. Microbiota and SCFA in lean and overweight healthy subjects. Obesity. 2010;18:190–195. doi: 10.1038/oby.2009.167. [DOI] [PubMed] [Google Scholar]
- Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, Neal PR, et al. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl Acad Sci USA. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg JL, Angenent LT, Gordon JI. Getting a grip on things: how do communities of bacterial symbionts become established in our intestine. Nat Immunol. 2004;5:569–573. doi: 10.1038/ni1079. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Walker AW, Ince J, Duncan SH, Webster LM, Holtrop G, Ze X, et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISMEJ. 2011;5:220–230. doi: 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Xiao G, Yao Y, Guo S, Lu K, Sheng Z. The role of bifidobacteria in gut barrier function after thermal injury in rats. J Trauma. 2006;61:650–657. doi: 10.1097/01.ta.0000196574.70614.27. [DOI] [PubMed] [Google Scholar]
- Wang ZT, Yao YM, Xiao GX, Sheng ZY. Risk factors of development of gut-derived bacterial translocation in thermally injured rats. World J Gastroenterol. 2004;10:1619–1624. doi: 10.3748/wjg.v10.i11.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weglarz L, Dzierzewicz Z, Skop B, Orchel A, Parfiniewicz B, Wisniowska B, et al. Desulfovibrio desulfuricans lipopolysaccharides induce endothelial cell IL-6 and IL-8 secretion and E-selectin and VCAM-1 expression. Cell Mol Biol Lett. 2003;8:991–1003. [PubMed] [Google Scholar]
- Woodmansey EJ, McMurdo ME, Macfarlane GT, Macfarlane S. Comparison of compositions and metabolic activities of fecal microbiotas in young adults and in antibiotic-treated and non-antibiotic-treated elderly subjects. Appl Environ Microbiol. 2004;70:6113–6122. doi: 10.1128/AEM.70.10.6113-6122.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Zhang M, Wang S, Han R, Cao Y, Hua W, et al. Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISMEJ. 2010;4:232–241. doi: 10.1038/ismej.2009.112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.