Abstract
The hydrolysis of urea as a source of ammonia has been proposed as a mechanism for the nitrification of ammonia-oxidizing bacteria (AOB) in acidic soil. The growth of Nitrososphaera viennensis on urea suggests that the ureolysis of ammonia-oxidizing archaea (AOA) might occur in natural environments. In this study, 15N isotope tracing indicates that ammonia oxidation occurred upon the addition of urea at a concentration similar to the in situ ammonium content of tea orchard soil (pH 3.75) and forest soil (pH 5.4) and was inhibited by acetylene. Nitrification activity was significantly stimulated by urea fertilization and coupled well with abundance changes in archaeal amoA genes in acidic soils. Pyrosequencing of 16S rRNA genes at whole microbial community level demonstrates the active growth of AOA in urea-amended soils. Molecular fingerprinting further shows that changes in denaturing gradient gel electrophoresis fingerprint patterns of archaeal amoA genes are paralleled by nitrification activity changes. However, bacterial amoA and 16S rRNA genes of AOB were not detected. The results strongly suggest that archaeal ammonia oxidation is supported by hydrolysis of urea and that AOA, from the marine Group 1.1a-associated lineage, dominate nitrification in two acidic soils tested.
Keywords: acid soil, archaeal ammonia oxidation, urea hydrolysis, 15N pool enrichment, pyrosequencing
Ammonia, rather than ammonium ion, has been thought to be the actual substrate for ammonia-oxidizing bacteria (AOB) because the Michaelis constant calculated from ammonia plus ammonium exponentially declines with decreasing pH value but remains almost unchanged when calculated as a function of ammonia concentration (Suzuki et al., 1974; Frijlink et al., 1992). The ammonia concentration in acidic soil is generally too low to support the growth of all known AOB isolates because of the ionization of ammonia to ammonium (Allison and Prosser, 1991; Burton and Prosser, 2001; Gubry-Rangin et al., 2011). Until recently, the microorganisms responsible for the unsuspected nitrification activity in acidic soil have remained unidentified. The discovery of Nitrosotalea devanaterra provides conclusive evidence for the existence of acid-tolerant ammonia-oxidizing archaea (AOA) and demonstrates that at least some AOA possess extraordinary affinity to the substrate and grow at extremely low ammonia concentrations of 0.18 nℳ (Lehtovirta-Morley et al., 2011). The isolation of obligate acidophilic N. devanaterra revealed the important role of archaeal ammonia oxidation in acidic soils, as previously suggested in acidic forest (Boyle-Yarwood et al., 2008; Stopnisek et al., 2010) and agricultural soils (Nicol et al., 2008; Offre et al., 2009). However, there is also evidence for bacterial ammonia oxidation in acidic soil (Onodera et al., 2010; Ying et al., 2010). The existence of the mechanisms in AOB, which allow them to grow under highly acidic conditions, in AOA remains unclear (De Boer and Kowalchuk, 2001). For example, AOB isolated from acidic soil are generally urease positive, and ureolysis can afford AOB a greater advantage to nitrify in acidic soil (Burton and Prosser, 2001; Lehtovirta-Morley et al., 2011).
The first isolation of AOA from soil at pH 8.0 indicates that Nitrososphaera viennensis can grow on urea as an alternative energy source (Tourna et al., 2011). Both urea transporter and urease gene were identified in the genomes of N. viennensis (Tourna et al., 2011) and Cenarchaeum symbiosum (Hallam et al., 2006). Environmental genomics has also revealed the genes that encode urease and urea transporters in marine habitats (Konstantinidis et al., 2009; Yakimov et al., 2011; Tully et al., 2012). These results imply a mechanism in which ammonia is released through the intracellular hydrolysis of urea and subsequently oxidized by AOA, as previously shown for cultured AOB under acidic conditions (Burton and Prosser, 2001). However, Nitrosopumilus maritimus and Nitrosoarchaeum limnia showed no genetic potential to grow on urea because no urease genes were identified in their genomes (Walker et al., 2010; Blainey et al., 2011). Moreover, N. devanaterra does not grow at neutral pH (Lehtovirta-Morley et al., 2011) but urease-positive N. viennensis optimally grows at pH 7.5 (Tourna et al., 2011). These findings imply that urease activity may be a feature of some AOA and is not strictly restricted to acidic environments, as previously shown for AOB isolates (Allison and Prosser, 1991). Therefore, we used microcosm incubations to assess whether nitrification activity is affected by urea fertilization in tea orchard (pH 3.75) and forest (pH 5.40) soils using 15N pool enrichment and cultivation-independent techniques.
15N isotope tracing
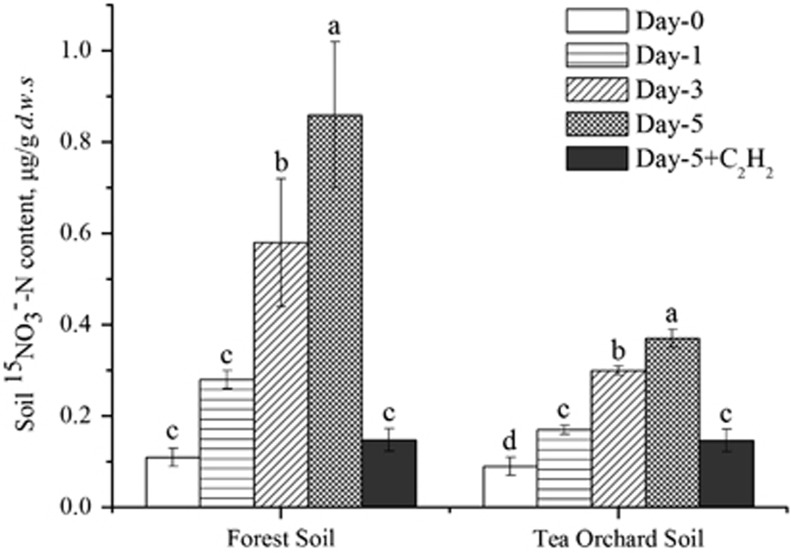
Soils at depths of 0–15 cm were collected on August 26, 2010 from tea orchards at the Tea Research Institute of the Chinese Academy of Agricultural Sciences and an adjacent forest site from which the tea orchard was converted in 1974. The tea orchard received approximately 600 kg of urea-N ha−1 a−1 with 3–4 split dressing applications over the last decades. Long-term field fertilization might have led to significant accumulation of nitrate in tea orchard field (Table 1). The site has been described previously in great detail (Han et al., 2007). The soil was sieved through a 2-mm mesh, and microcosms were established in triplicate by adding 10 g of fresh soil in 250 ml Erlenmeyer flasks. The labeled urea-N (15N atom, 98% excess) was added to the microcosms at a final concentration of 5 μg of N per gram of dry weight soil (d.w.s.), which is similar to the ammonium content in soil under in situ conditions (Table 1). The soil microcosms were incubated at 60% of the maximum water-holding capacity of the soil for 5 days with or without acetylene at 100 Pa and 25 °C under dark conditions (Jia and Conrad, 2009). The production of soil 15N-labeled nitrate and nitrite was measured as previously described (Zhang et al., 2011). Acetylene completely abolished nitrification activity in the forest soil, whereas a slight increase in 15NO3−-N was observed in the tea orchard soils after incubation for 5 days. In the absence of acetylene, the 15NO3−-N concentration showed linear increases over the course of incubation (Figure 1). The nitrification activities in the forest and tea orchard soils were estimated to be 0.150 and 0.056 μg of 15NO3−-N per gram d.w.s. per day, respectively. These activities fell well within the range from 0.051 to 0.459 μg of N per gram d.w.s. per day in acidic soils with pH <5.5 across China, as shown by the results of both 15N pool dilution and enrichment techniques (Zhang et al., 2011). The addition of considerably low concentrations of urea suggests that nitrification could likely occur under in situ conditions. The nitrification activity had a three-fold decrease after the forest soil was converted to tea orchard soil. The tea orchard cultivation decreased the soil pH by 1.75 units over the last 36 years. Thus, it seems plausible that the stress induced by highly acidic conditions increased the energy demand for the growth of ammonia oxidizers and reduced the nitrification activity in tea orchard soils (Baker-Austin and Dopson, 2007).
Table 1. Soil characteristics.
| Soila | pHb (1:2.5 H2O) | SOMc g kg−1 | TNc g kg−1 | Soil water content (%) | NO3−-N μg g−1 | NH4+-Nμg g−1 | Predicted soil NH3 concentration (nℳ)d | Nitrification activity μg NO3−-N·g−1 d−1e |
|---|---|---|---|---|---|---|---|---|
| TS | 3.75 | 53.2 | 2.95 | 22.9 | 92.9 | 7.02 | 7.00 | 0.056 |
| FS | 5.40 | 25.2 | 1.13 | 22.0 | 8.53 | 4.77 | 221 | 0.150 |
TS and FS represent tea orchard soil and forest soil, respectively.
pH was measured with water:soil ratio of 2.5.
SOM and TN denote soil organic matter and total nitrogen, respectively.
Soil NH3 concentration was estimated on the basis of soil water content, ammonium concentration, pH and adjusted for temperature (pKa of 9.245 at 25 °C).
Nitrification activity is determined by monitoring the 15NO3−-N production upon addition of 5 μg urea-N (99% 15N excess) to 1 g soil.
Figure 1.
15NO3−-N content in soil microcosms at days 0, 1, 3 and 5. Microcosm incubation was performed by adding 5 μg of urea-N (99% 15N excess) per gram d.w.s. at day 0. Day-5+C2H2 indicates incubation in the presence of 100 Pa C2H2. The error bars represent s.e.m. of the microcosm triplicates. The different letters above the columns indicate a significant difference (P<0.05) using analysis of variance.
Nitrification activity upon urea fertilization
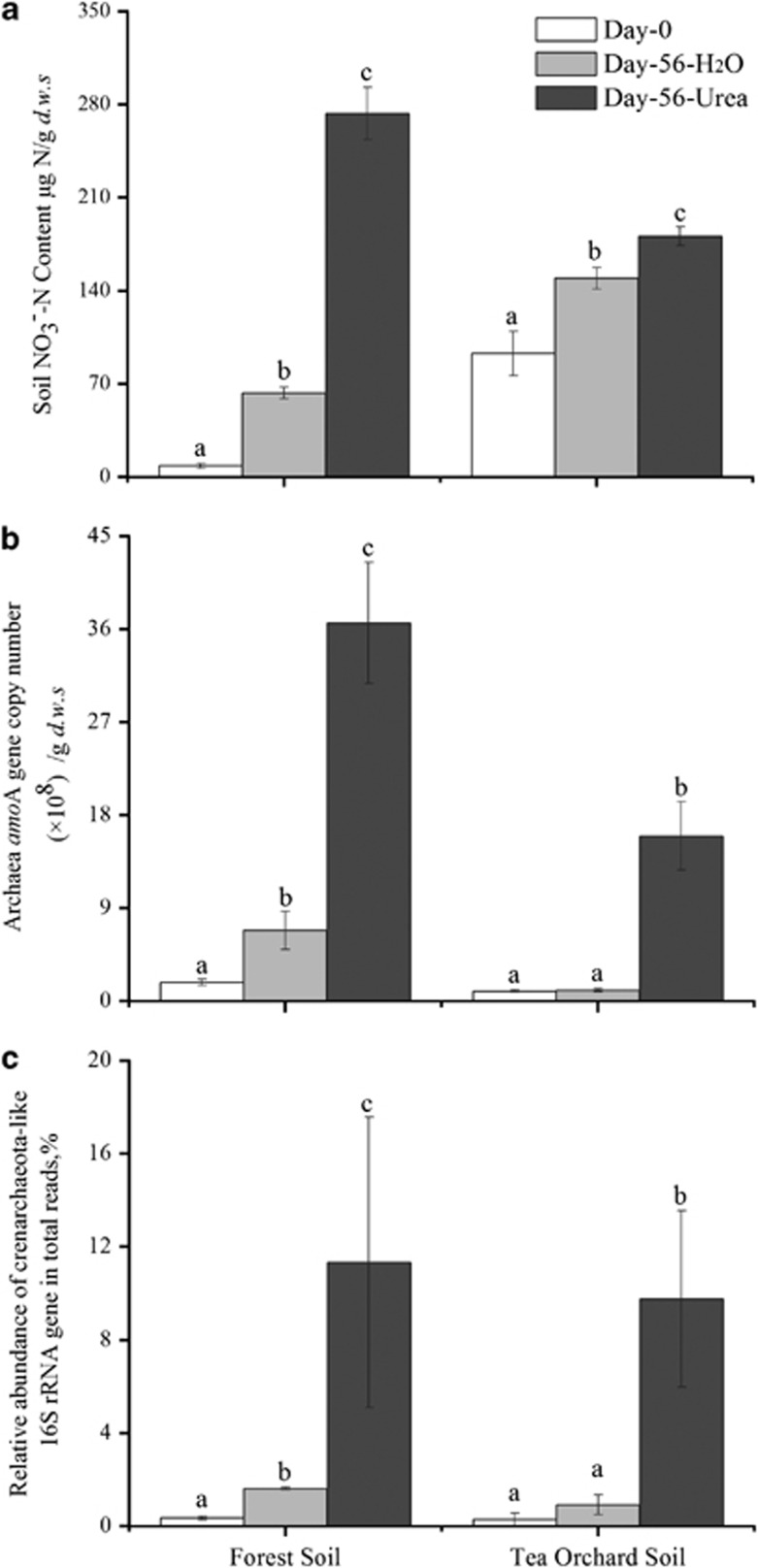
The microcosms were further established in triplicate by fertilizing the soil with 100 μg of urea-N per gram d.w.s. once a week as described previously with slight modifications (Xia et al., 2011). Thus, the soil microcosms received a total of 800 μg of urea-N per gram d.w.s. over an incubation period of 8 weeks. The control microcosm was established by adding equal amounts of sterilized distilled water to the soil. As a result of ammonia released during mineralization of organic matters, significant amounts of nitrate were produced in the control microcosms after incubation for 8 weeks (Figure 2a). The net nitrification activities in tea orchard and forest soils were estimated to be 1.01 and 0.97 μg of NO3−-N per gram d.w.s. per day, respectively. These results are similar to previous findings for acidic agricultural soils, from which acidophilic N. devanaterra was isolated (Nicol et al., 2008; Offre et al., 2009; Gubry-Rangin et al., 2010). Urea fertilization at a considerably high rate led to an almost four-fold increase in net nitrification activity in forest soil (4.73 μg of NO3−-N per gram d.w.s. per day) and 50% increase in tea orchard soil (1.58 μg of NO3−-N per gram d.w.s. per day). The activity changes were well paralleled by the abundance changes in archaeal amoA genes, as shown by the real-time quantitative PCR as described previously (Wu et al., 2011). The copy number of archaeal amoA genes significantly increased from 1.82 × 108 at day 0 to 6.86 × 108 and 3.66 × 109 at day 56, showing 4- and 20-fold increases, after incubation for 8 weeks in forest soil microcosms receiving water or urea-N, respectively (Figure 2b). For the tea orchard soil, urea fertilization resulted in a 16-fold increase in the copy number of amoA genes from 9.90 × 107 to 3.31 × 108 after incubation for 8 weeks. However, no statistically significant increase was observed in soil microcosms added with water, suggesting that the growth of AOA was so slow that the subtle changes in amoA genes could not be detected by the techniques used. Bacterial amoA genes were not amplified despite the use of a range of degenerate primers and combinations (Stephen et al., 1999; Nicolaisen and Ramsing, 2002). The absence of AOB in this study was further confirmed by pyrosequencing analysis of 16S rRNA genes at whole microbial community level in two acidic soils tested.
Figure 2.
Change in the concentrations of (a) NO3−-N, (b) number of archaeal amoA genes, and (c) relative abundance of crenarchaeota-like 16S rRNA gene reads in soil microcosms over an incubation period of 8 weeks. Day-56-H2O and day-56-Urea indicate that the soil microcosms received sterilized water or urea once a week, respectively. The error bars represent the s.e.m. of the microcosm triplicates. The relative abundance is expressed as the ratio of crenarchaeota-like 16S rRNA gene reads to total 16S rRNA gene reads in each microcosm. The pyrosequencing reads have been deposited at the DNA Data Bank of Japan with accession number DRA000546. The different letters above the columns indicate a significant difference (P<0.05) using analysis of variance.
Pyrosequencing at the whole microbial community level
The high-throughput sequencing of 16S rRNA genes at the whole community level provides an almost unbiased profiling strategy for measuring characteristic changes in relative proportions of the microorganisms involved in certain functional processes, such as nitrification, in a complex system. The results obtained from the improved bidirectional pyrosequencing reveal strong correlations between nitrification activity and crenarchaeota-like 16S rRNA genes. The total microbial communities in the triplicate microcosms were analyzed using a 454 FLX Titanium sequencer for the whole incubation period (Supplementary Table S1). Slight modification was made by PCR amplifying the 16S rRNA genes with the universal primers 515F and 907R extended as amplicon fusion primers using the respective primer A or B adapters, key sequence and taq sequence, as previously described (Xia et al., 2011). The 16S rRNA gene sequence reads were processed by the RDP pyrosequencing pipeline (Cole et al., 2009). We obtained 103 552 high-quality sequences from the 18 samples (Supplementary Table S1). The changes in the proportion of crenarchaeota-like 16S rRNA genes to the total number of 16S rRNA gene reads (Figure 2c) followed perfectly the changes in net nitrification activity (Figure 2a) and archaeal amoA gene abundances (Figure 2b) in the soil microcosms. The relative proportion of crenarchaeota-like 16S rRNA genes increased 32-fold and 7-fold over the whole incubation period in forest soils amended with urea or water, respectively. Similarly, the relative proportions of crenarchaeota-like 16S rRNA gene reads to the total 16S rRNA in the tea orchard soils significantly increased from 0.28 to 7.86% after incubation with urea for 8 weeks, representing a 28-fold increase. The positive correlation between nitrification activities and archaeal communities was further confirmed by denaturing gradient gel electrophoresis (DGGE) fingerprinting analysis of archaeal amoA genes in triplicate microcosms.
DGGE fingerprinting of amoA genes
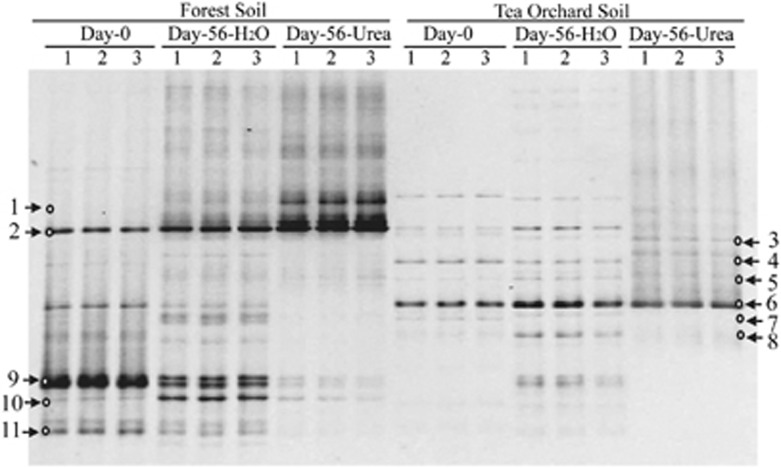
The DGGE analysis of archaeal amoA genes was performed by separating the PCR amplicons on 6% acrylamide gels with a 20 to 50% denaturing gradient, as previously described (Wu et al., 2011) but with primers of CrenamoA23f and CrenamoA616r (Nicol et al., 2008; Lehtovirta-Morley et al., 2011). The DGGE fingerprints of archaeal amoA genes showed highly similar patterns among the triplicate microcosms of each treatment (Figure 3). For the forest soil, the pairwise comparison of DGGE patterns between days 0 and 56 revealed that the relative intensities of DGGE bands 1 and 2 were enhanced after incubation for 56 days, particularly in the soil amended with urea. The opposite trend was observed for DGGE bands 6, 9 and 11. For the tea orchard soil, DGGE band 6 dominated the AOA communities and appeared to be enriched after incubation for 56 days. The conversion of forest soil to tea orchard soil significantly altered the DGGE fingerprints of archaeal amoA genes. The intensities of DGGE bands 2 and 9 significantly decreased, whereas only DGGE band 6 prevailed in the tea orchard soil. The dominant DGGE bands were excised for sequencing, and phylogenetic analysis of archaeal amoA genes was conducted (Figure 4) as previously described (Xia et al., 2011). DGGE bands 1 and 2 were phylogenetically most closely affiliated with the acidophilic N. devanaterra (Lehtovirta-Morley et al., 2011). DGGE band 6 was also related to N. devanaterra, but formed a unique cluster within the marine Group 1.1a-associated lineage. These results were further confirmed by the phylogenetic analysis of the 16S rRNA sequence reads. AOA members within the marine Group 1.1a-associated lineage dominated the archaeal communities, accounting for 97% and 95% of archaeal sequence reads in forest and tea orchard soils amended with urea, respectively (Figure 5 and Supplementary Table S2).
Figure 3.
DGGE fingerprints of archaeal amoA genes in soils. All designations are the same as those used in Figure 2. 1, 2 and 3 represent the three samples from triplicate microcosms for each treatment. The arrows indicate the DGGE bands excised for sequencing.
Figure 4.
Phylogenetic tree showing the relationship of archaeal amoA genes retrieved from the forest and tea orchard soils to those obtained from the GenBank. The excised DGGE bands are shown in bold. The nucleotide sequences of archaeal amoA genes have been deposited in the DDBJ with accession numbers from JQ972028 to JQ972041. Bootstrap values higher than 70% are indicated at branch nodes.
Figure 5.
Phylogenetic tree showing the relationship of archaeal 16S rRNA gene sequence reads in two acidic soils to those obtained from the GenBank. FS and TS indicate forest and tea orchard soils with symbols ▲ and ●, respectively. The designations are similar to those used in Figure 2. For instance, FS-Day-56-Urea-OTU-1-185 (56%) indicates that OTU-1 of archaeal 16S rRNA genes contains 185 sequence reads with >97% identity and accounts for 56% of the total archaeal 16S rRNA reads in forest soil after incubation with urea for 56 days. One representative sequence from each OTU was extracted using the mothur software package (Schloss et al., 2009) for tree construction. The representative sequences of archaeal 16S rRNA genes have been deposited in the DDBJ with accession numbers from JQ972042 to JQ972059. Bootstrap values higher than 70% are indicated at branch nodes.
The results of this study indicate that the nitrification of ammonia oxidizers is supported by the hydrolysis of urea in two acidic soils tested and most likely related to AOA rather than AOB. Since the first isolation of urease-positive AOB (De Boer et al., 1989), ureolysis has been proposed to promote nitrification in acidic soils (Allison and Prosser, 1991). The growth of AOB in low-pH cultures has strongly supported the mechanism of intracellular hydrolysis of urea as a source of ammonia for bacterial ammonia oxidation (Burton and Prosser, 2001). Our results demonstrate that nitrification occurred in both acidic soils when urea was added at considerably low rates of 5 μg of urea-N per gram d.w.s. The nitrification activities were also significantly enhanced when 800 μg of urea-N per gram d.w.s. was amended. The stimulated activities occurred as the abundance of archaeal amoA genes and the relative proportions of crenarchaeota-like 16S rRNA genes increased (Figure 2). Molecular fingerprinting of amoA genes further provided strong evidence that nitrification of archaeal ammonia oxidation was supported by urea hydrolysis. However, no conclusive evidence exists for intracellular urea hydrolysis by AOA in both acidic soils. We speculate that only a small proportion of the added urea entered the cells of AOA, and a portion of the ammonia produced due to this process was subsequently oxidized. Majority of the urea was most likely hydrolyzed by other organisms because extracellular ureolytic activity is a common feature in soil microbes (Mobley et al., 1995; Koper et al., 2004). The ammonia produced during these processes could have been ionized at low pH levels, making them effectively unavailable for oxidation by AOA in a manner similar to that in AOB as previously demonstrated (Burton and Prosser, 2001) This was further supported by the produced nitrate that accounted for only 11% and 33% of the urea added to tea orchard and forest soils, respectively. The key control of ammonia addition parallel to urea fertilization was not included in this study to assess whether the ammonia oxidized by AOA resulted solely from extracellular ureolytic activity in both acidic soils. Ammonia, rather than urea, is thought to be the substrate for ammonia monooxygenase. Thus, the stimulated activities of archaeal nitrification by urea hydrolysis are expected to be similar to a certain extent upon ammonia addition. However, recent studies have shown that nitrification activity was not affected by the addition of (NH4)2SO4 (Stopnisek et al., 2010) or NH4Cl (Levičnik-Höfferle et al., 2012) to acidic soil. We speculate that this might be attributed to the side effects due to the addition of inorganic ammonium, such as the introduction of anion to soil.
Phylogenetic analysis of both amoA (Figure 4) and 16S rRNA genes (Figure 5) suggests that N. devanaterra-like AOA dominated the nitrification activities in forest soil, whereas N. devanaterra-related AOA were active ammonia oxidizers in tea orchard soil. The relative abundance of N. devanaterra-related 16S rRNA sequences significantly increased from 56% to 95% in tea orchard soils after incubation with urea for 56 days (Supplementary Table S2), whereas N. devanaterra-like 16S rRNA genes showed a similar increase from 71% to 97% in forest soil amended with urea. N. devanaterra was isolated from acidic agricultural soils, in which the net nitrification activities (Tourna et al., 2008; Offre et al., 2009; Gubry-Rangin et al., 2010) were similar to those observed in the present study (Figure 2). The predominance of archaeal ammonia oxidation was also confirmed by the greatest substrate affinity of N. devanaterra determined by far (Lehtovirta-Morley et al., 2011). Soil NH3 concentration is estimated to be 7.0 nℳ in tea orchard soil and 221 nℳ in forest soil (Table 1). These values are sufficiently high for the growth of N. devanaterra but not of AOB in culture. Pyrosequencing analysis indeed reveals that none of the 16S rRNA genes are affiliated with AOB among the 103 552 sequence reads across all soil samples. This suggests that the abundance of AOB might be too low to be detected by bacterial amoA genes in both acidic soils. Soil nitrite concentration was below the detection limit, implying that nitrite toxicity against AOA and/or AOB may be excluded in this study (De Boer and Kowalchuk, 2001; Lehtovirta-Morley et al., 2011). Furthermore, 15N isotope tracing showed that nitrification activity was inhibited by acetylene, suggesting ammonia-monooxygenase-dependent activities as previously reported for N. devanaterra (Lehtovirta-Morley et al., 2011). These results strongly suggest that the active AOA observed in this study were very likely related to N. devanaterra and capable of ammonia oxidation through urea hydrolysis.
Taken together, our results indicate that nitrification of archaeal ammonia oxidizers was supported by urea hydrolysis in two acid soils tested, and was most likely attributed to AOA members within the marine Group 1.1a-associated lineage. The ability to hydrolyze urea may afford AOA a greater ecological advantage in ammonia-poor environments, but the metabolic mechanism remains unclear.
Acknowledgments
This work was financially supported by the National Science Foundation of China (40971153 and 41090281), the Knowledge Innovation Programs of the Chinese Academy of Sciences (KSCX2-EW-G-16 and KZCX2-YW-BR-06). We are very grateful for the invaluable comments of reviewers, one of whom is solely responsible for the title. Dr Benli Chai at Michigan State University is gratefully acknowledged for bioinformatics assistance.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Allison SM, Prosser JI. Urease activity in neutrophilic autotrophic ammonia-oxidizing bacteria isolated from acid soils. Soil Biol Biochem. 1991;23:45–51. [Google Scholar]
- Baker-Austin C, Dopson M. Life in acid: pH homeostasis in acidophiles. Trends Microbiol. 2007;15:165–171. doi: 10.1016/j.tim.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Blainey PC, Mosier AC, Potanina A, Francis CA, Quake SR. Genome of a low-salinity ammonia-oxidizing archaeon determined by single-cell and metagenomic analysis. PLoS One. 2011;6:e16626. doi: 10.1371/journal.pone.0016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle-Yarwood SA, Bottomley PJ, Myrold DD. Community composition of ammonia-oxidizing bacteria and archaea in soils under stands of red alder and Douglas fir in Oregon. Environ Microbiol. 2008;10:2956–2965. doi: 10.1111/j.1462-2920.2008.01600.x. [DOI] [PubMed] [Google Scholar]
- Burton SAQ, Prosser JI. Autotrophic ammonia oxidation at low pH through urea hydrolysis. Appl Environ Microbiol. 2001;67:2952–2957. doi: 10.1128/AEM.67.7.2952-2957.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boer W, Duyts H, Laanbroek HJ. Urea stimulated autotrophic nitrification in suspensions of fertilized, acid heath soil. Soil Biol Biochem. 1989;21:349–354. [Google Scholar]
- De Boer W, Kowalchuk GA. Nitrification in acid soils: micro-organisms and mechanisms. Soil Biol Biochem. 2001;33:853–866. [Google Scholar]
- Frijlink MJ, Abee T, Laanbroek HJ, Boer W, Konings WN. The bioenergetics of ammonia and hydroxylamine oxidation in Nitrosomonas europaea at acid and alkaline pH. Arch Microbiol. 1992;157:194–199. [Google Scholar]
- Gubry-Rangin C, Hai B, Quince C, Engel M, Thomson BC, James P, et al. Niche specialization of terrestrial archaeal ammonia oxidizers. Proc Natl Acad Sci USA. 2011;108:21206–21211. doi: 10.1073/pnas.1109000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubry-Rangin C, Nicol GW, Prosser JI. Archaea rather than bacteria control nitrification in two agricultural acidic soils. FEMS Microbiol Ecol. 2010;74:566–574. doi: 10.1111/j.1574-6941.2010.00971.x. [DOI] [PubMed] [Google Scholar]
- Hallam SJ, Mincer TJ, Schleper C, Preston CM, Roberts K, Richardson PM, et al. Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine crenarchaeota. PLoS Biol. 2006;4:e95. doi: 10.1371/journal.pbio.0040095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han W, Kemmitt SJ, Brookes PC. Soil microbial biomass and activity in Chinese tea gardens of varying stand age and productivity. Soil Biol Biochem. 2007;39:1468–1478. [Google Scholar]
- Jia ZJ, Conrad R. Bacteria rather than Archaea dominate microbial ammonia oxidation in an agricultural soil. Environ Microbiol. 2009;11:1658–1671. doi: 10.1111/j.1462-2920.2009.01891.x. [DOI] [PubMed] [Google Scholar]
- Konstantinidis KT, Braff J, Karl DM, DeLong EF. Comparative metagenomic analysis of a microbial community residing at a depth of 4000 meters at station ALOHA in the north Pacific Subtropical Gyre. Appl Environ Microbiol. 2009;75:5345–5355. doi: 10.1128/AEM.00473-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koper TE, El-Sheikh AF, Norton JM, Klotz MG. Urease-encoding genes in ammonia-oxidizing bacteria. Appl Environ Microbiol. 2004;70:2342–2348. doi: 10.1128/AEM.70.4.2342-2348.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtovirta-Morley LE, Stoecker K, Vilcinskas A, Prosser JI, Nicol GW. Cultivation of an obligate acidophilic ammonia oxidizer from a nitrifying acid soil. Proc Natl Acad Sci USA. 2011;108:15892–15897. doi: 10.1073/pnas.1107196108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levičnik-Höfferle Š, Nicol GW, Ausec L, Mandić-Mulec I, Prosser JI. Stimulation of thaumarchaeal ammonia oxidation by ammonia derived from organic nitrogen but not added inorganic nitrogen. FEMS Microbiol Ecol. 2012;80:114–123. doi: 10.1111/j.1574-6941.2011.01275.x. [DOI] [PubMed] [Google Scholar]
- Mobley HL, Island MD, Hausinger RP. Molecular biology of microbial ureases. Microbiol Rev. 1995;59:451–480. doi: 10.1128/mr.59.3.451-480.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol GW, Leininger S, Schleper C, Prosser JI. The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ Microbiol. 2008;10:2966–2978. doi: 10.1111/j.1462-2920.2008.01701.x. [DOI] [PubMed] [Google Scholar]
- Nicolaisen MH, Ramsing NB. Denaturing gradient gel electrophoresis (DGGE) approaches to study the diversity of ammonia-oxidizing bacteria. J Microbiol Meth. 2002;50:189–203. doi: 10.1016/s0167-7012(02)00026-x. [DOI] [PubMed] [Google Scholar]
- Offre P, Prosser JI, Nicol GW. Growth of ammonia-oxidizing archaea in soil microcosms is inhibited by acetylene. FEMS Microbiol Ecol. 2009;70:99–108. doi: 10.1111/j.1574-6941.2009.00725.x. [DOI] [PubMed] [Google Scholar]
- Onodera Y, Nakagawa T, Takahashi R, Tokuyama T. Seasonal change in vertical distribution of ammonia-oxidizing archaea and bacteria and their nitrification in temperate forest soil. Microb Environ. 2010;25:28–35. doi: 10.1264/jsme2.me09179. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen JR, Chang Y-J, Macnaughton SJ, Kowalchuk GA, Leung KT, Flemming CA, et al. Effect of toxic netals on dndigenous soil β-subgroup proteobacterium ammonia oxidizer community structure and protection against toxicity by inoculated metal-resistant bacteria. Appl Environ Microbiol. 1999;65:95–101. doi: 10.1128/aem.65.1.95-101.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopnisek N, Gubry-Rangin C, Hofferle S, Nicol GW, Mandic-Mulec I, Prosser JI. Thaumarchaeal ammonia oxidation in an acidic forest peat soil is not influenced by ammonium amendment. Appl Environ Microbiol. 2010;76:7626–7634. doi: 10.1128/AEM.00595-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki I, Dular U, Kwok SC. Ammonia or ammonium ion as substrate for oxidation by nitrosomonas europaea cells and extracts. J Bacteriol. 1974;120:556–558. doi: 10.1128/jb.120.1.556-558.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourna M, Freitag TE, Nicol GW, Prosser JI. Growth, activity and temperature responses of ammonia-oxidizing archaea and bacteria in soil microcosms. Environ Microbiol. 2008;10:1357–1364. doi: 10.1111/j.1462-2920.2007.01563.x. [DOI] [PubMed] [Google Scholar]
- Tourna M, Stieglmeier M, Spang A, Könneke M, Schintlmeister A, Urich T, et al. Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc Natl Acad Sci USA. 2011;108:8420–8425. doi: 10.1073/pnas.1013488108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tully BJ, Nelson WC, Heidelberg JF. Metagenomic analysis of a complex marine planktonic thaumarchaeal community from the Gulf of Maine. Environ Microbiol. 2012;14:254–267. doi: 10.1111/j.1462-2920.2011.02628.x. [DOI] [PubMed] [Google Scholar]
- Walker CB, de la Torre JR, Klotz MG, Urakawa H, Pinel N, Arp DJ, et al. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc Natl Acad Sci USA. 2010;107:8818–8823. doi: 10.1073/pnas.0913533107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Lu L, Wang B, Lin X, Zhu J, Cai Z, et al. Long-term field fertilization significantly alters community structure of ammonia-oxidizing bacteria rather than archaea in a paddy soil. Soil Sci Soc Am J. 2011;75:1431–1439. [Google Scholar]
- Xia W, Zhang C, Zeng X, Feng Y, Weng J, Lin X, et al. Autotrophic growth of nitrifying community in an agricultural soil. ISME J. 2011;5:1226–1236. doi: 10.1038/ismej.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakimov MM, Cono VL, Smedile F, DeLuca TH, Juarez S, Ciordia S, et al. Contribution of crenarchaeal autotrophic ammonia oxidizers to the dark primary production in Tyrrhenian deep waters (Central Mediterranean Sea) ISME J. 2011;5:945–961. doi: 10.1038/ismej.2010.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying JY, Zhang LM, He JZ. Putative ammonia-oxidizing bacteria and archaea in an acidic red soil with different land utilization patterns. Environ Microbiol Rep. 2010;2:304–312. doi: 10.1111/j.1758-2229.2009.00130.x. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhu T, Cai Z, Müller C. Nitrogen cycling in forest soils across climate gradients in Eastern China. Plant Soil. 2011;342:419–432. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.